Abstract
The outer membrane of Gram-negative bacteria is an asymmetric membrane with lipopolysaccharides on the external leaflet and phospholipids on the periplasmic leaflet. This outer membrane contains mainly β-barrel transmembrane proteins and lipidated periplasmic proteins (lipoproteins). The multisubunit protein β-barrel assembly machine (BAM) catalyzes the insertion and folding of the β-barrel proteins into this membrane. In Escherichia coli, the BAM complex consists of five subunits, a core transmembrane β-barrel with a long periplasmic domain (BamA) and four lipoproteins (BamB/C/D/E). The BamA periplasmic domain is composed of five globular subdomains in tandem called POTRA motifs that are key to BAM complex formation and interaction with the substrate β-barrel proteins. The BAM complex is believed to undergo conformational cycling while facilitating insertion of client proteins into the outer membrane. Reports describing variable conformations and dynamics of the periplasmic POTRA domain have been published. Therefore, elucidation of the conformational dynamics of the POTRA domain in full-length BamA is important to understand the function of this molecular complex. Using molecular dynamics simulations, we present evidence that the conformational flexibility of the POTRA domain is modulated by binding to the periplasmic surface of a native lipid membrane. Furthermore, membrane binding of the POTRA domain is compatible with both BamB and BamD binding, suggesting that conformational selection of different POTRA domain conformations may be involved in the mechanism of BAM-facilitated insertion of outer membrane β-barrel proteins.
Introduction
Gram-negative bacteria have an asymmetric outer membrane with lipopolysaccharides (LPS) in the external leaflet and phospholipids in the internal leaflet (1). The transmembrane proteins in these outer membranes are almost exclusively β-barrel structures and are important for transmembrane transport, cell recognition, protein domain anchoring, and membrane biogenesis (2, 3). Although β-barrel outer membrane proteins (OMPs) are able to spontaneously insert and fold into phospholipid membranes in vitro (4, 5, 6), in the cell the OMPs diffuse from the Sec translocon in the inner membrane to the outer membrane surface across the periplasmic space while interacting with various chaperones (7, 8, 9, 10). At the outer membrane, they are recognized by a multiprotein complex called the β-barrel assembly machine (BAM) that catalyzes insertion and folding of OMPs into the outer membrane (11, 12, 13, 14, 15, 16).
The BAM complex consists of one copy each of five subunits (BamA/B/C/D/E) (17). The BamA subunit contains a transmembrane β-barrel and a large periplasmic domain composed of five globular polypeptide transport-associated (POTRA) motifs linked in tandem and numbered 1–5 from the N-terminal end (18, 19, 20, 21, 22). The POTRA motifs display an evolutionarily conserved fold but have low sequence conservation (19, 22, 23, 24, 25, 26). The BamB/C/D/E subunits are lipoproteins bound to the periplasmic surface of the outer membrane through a covalent lipid modification (27, 28). Although only BamA and BamD are essential for cell viability, individual deletion of any of the other subunits causes defects in OMP biogenesis and decreased cell viability under certain conditions (8, 12, 13, 16, 28, 29, 30, 31, 32).
A structural understanding of the subunit arrangement in the BAM complex has emerged from several independent x-ray crystallographic, NMR, and electron microscopy studies of either individual subunits or complexes of fragments of BamA/B, BamA/D, and BamC/D (18, 19, 21, 23, 24, 25, 26, 33, 34, 35, 36, 37) and recent x-ray structures of the BamA/C/D/E (38, 39) and BamA/B/C/D/E complexes (39, 40). In addition, the POTRA domains have independently been shown to bind to BamB and BamD (29, 32), and it has been suggested that they may template β-strand formation in client OMP polypeptides (23, 24, 39). BamA has been cross-linked to the periplasmic chaperone SurA (8, 11), potentially indicating a docking site where unfolded OMPs are delivered to the BAM complex for membrane integration.
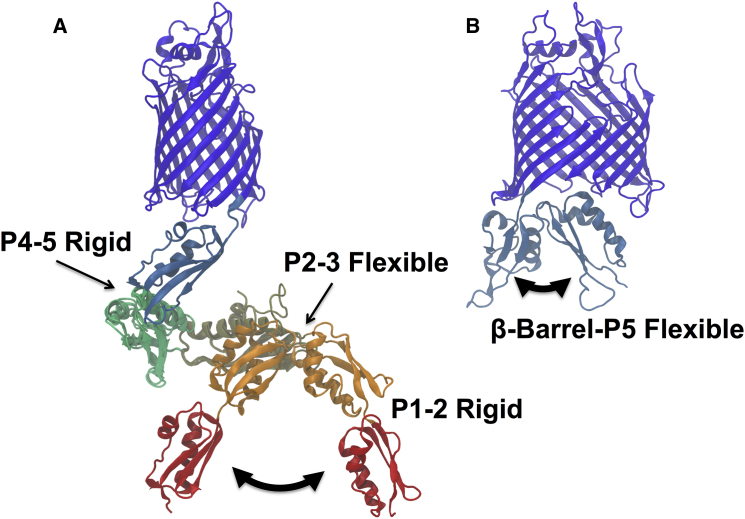
The multistep process of client OMP folding into the bacterial outer membrane together with the cyclic nature of the BAM catalysis (41, 42, 43) suggest that distinct conformations of the BAM complex may be required during different steps of OMP folding and membrane insertion. In addition, recent models of the involvement of BAM in the function of bacterial surface autotransporters include a hybrid two-β-barrel complex that necessarily invokes large conformational rearrangements of BamA during secretion of the autotransporter passenger domains (44). Results of some crystallographic studies support the idea of multiple conformations as revealed in several distinct structures of BamA alone (19, 23, 24) and of BAM complexes (38, 39, 40). Two of these conformations are shown (Fig. 1 A) where the POTRA2–3 hinge exhibits the most flexibility in crystalline BamA alone. Similarly, NMR studies on isolated POTRA motif fragments in solution have shown various degrees of mobility at POTRA intermotif hinges, indicating either significant conformational flexibility or limited mobility depending on the domains in question and on experimental conditions (21, 25). Recent molecular dynamics (MD) simulations of various assemblies of BAM embedded in a membrane have also shown variable flexibilities of the POTRA domain depending on the presence or absence of BAM lipoproteins (39). Because the POTRA motifs are connected in a linear fashion, the extent of local flexibilities at POTRA intermotif joints will have large effects on the set of global conformations available to the BamA periplasmic domain. Similarly, binding of the BamA POTRA domain to either the membrane or other BAM subunits could selectively stabilize certain POTRA conformations. It is not known whether or not the complex undergoes subunit dissociation during its functional cycle.
Figure 1.
X-ray crystal structures suggest variable flexibility at POTRA interdomain linkers and β-barrel-POTRA5 joint. (A) Full-length BamA overlaid with two extreme POTRA domain conformations found in x-ray crystallographic structures of BamA and POTRA domain fragments. Backbone Cα of POTRA4 were used for alignment. The POTRA2–3 hinge appears to have the most flexibility. Red, POTRA1; orange, POTRA2; khaki, POTRA3; light green, POTRA4; light blue, POTRA5; dark blue, β-barrel. Structures used were PDB: 4K3C (POTRA4–5-β-barrel from H. ducreyi), PDB: 2QCZ and PDB: 3EFC (POTRA1–4 from E. coli). The same color scheme for individual POTRA motifs is used throughout this report. (B) BamA POTRA5-β-barrel fragments with two different POTRA5 orientations. Structures used were PDB: 4K3B (N. gonorrheae) and PDB: 4K3C (H. ducreyi). To see this figure in color, go online.
Because conformational cycling may be essential to the BAM mechanism, we investigated BamA flexibility using MD simulations of BamA embedded in a model Escherichia coli outer membrane. Here, we report that POTRA-membrane interactions influence the set of observed conformations. The molecular basis of POTRA-membrane binding is encoded by specific, evolutionarily conserved hydrophobic interactions and by a large number of nonspecific interactions. Two POTRA3 tryptophan side chains insert into the lipid interface region. An important finding is the observation that the membrane-associated POTRA domain samples many orientations that are compatible with the known BamB-POTRA3 and BamD-POTRA5 interactions. Taken together, our simulations suggest that the POTRA domain’s binding to lipoproteins and the membrane may act in concert to populate the functional conformations of the POTRA domain during the OMP insertion and folding cycle.
Materials and Methods
MD simulations
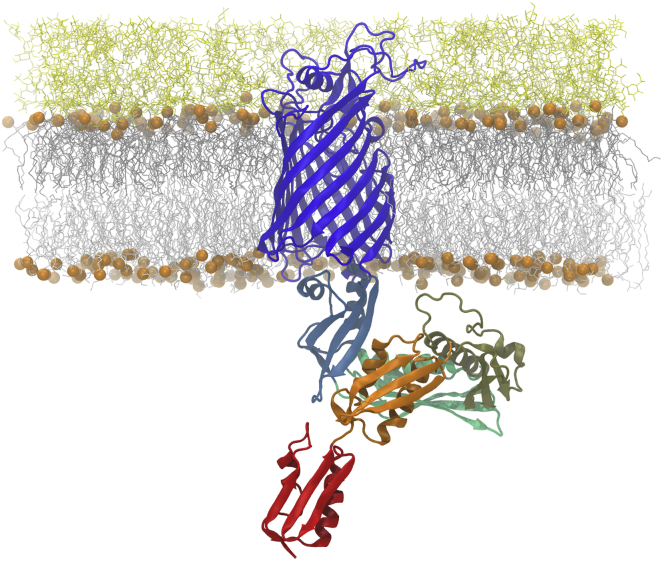
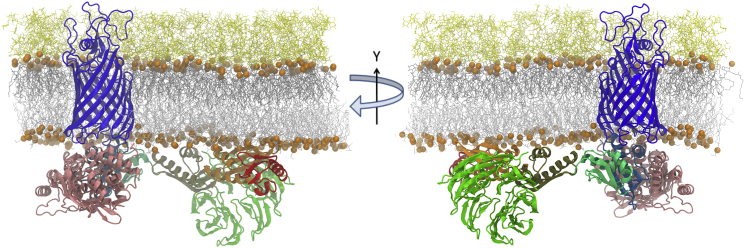
An E. coli full-length BamA structural model was assembled using the N-terminal POTRA domain (residues 23–426) from the E. coli homology model reported by Noinaj et al. (19): most of the β-barrel domain from crystal structure Protein Data Bank (PDB): 4N75 (20) and residues 715–740 from crystal structure PDB: 4C4V (18). The resultant full-length BamA model was embedded into an asymmetric bilayer whose acyl chain and headgroup composition was chosen to mimic a typical E. coli outer membrane (45) using the protocol implemented in CHARMM-GUI Membrane Builder (46, 47) as previously described in (48) and also recently applied for OmpF simulations (49). The outer leaflet of the membrane was composed entirely of rough LPS with lipid A and R1 core (50), and the inner leaflet consisted of a mixture of PPPE, PVPG, and PVCL with a ratio of 15:4:1 (51), where PPPE is 16:0 (palmitoyl) - 16:1 cis-9 (palmitoleoyl) phosphatidylethanolamine, PVPG is 16:0 (palmitoyl) - 18:1 cis-11 (vacenoyl) phosphatidylglycerol, and PVCL is 1,1′-palmitoyl-2,2′-vacenoyl cardiolipin with a net charge of −2e. The MD systems were built with approximate dimensions of 150 × 150 × 200 Å to allow space for full conformational freedom of the POTRA domain. This size resulted in a membrane with 225 PPPE, 60 PVPG, 15 PVCL, and 113 LPS molecules per BamA. In addition to 565 calcium ions (to neutralize LPS molecules), ∼300 potassium and ∼180 chloride ions (0.15 M KCl) were included to act as counter ions. The starting structure of full-length BamA embedded in the asymmetric LPS/PL membrane is shown in Fig. 2. The different domains of BamA are colored as in Fig. 1. Three independent systems with 391,584, 391,709, and 391,636 atoms (including water molecules) were separately built with different LPS orientations and phospholipid conformations and simulated for comparison of simulation results.
Figure 2.
Starting structure of system for simulation. BamA is depicted as a ribbon drawing with coloring of motifs as in Fig. 1. Membrane lipid phosphates delimiting the hydrophobic core are shown as orange spheres, other membrane components as white sticks with LPS on top and phospholipid on the bottom. Note the extended loop in POTRA3 (khaki) that includes residues 200–213. To see this figure in color, go online.
The POTRA4–5 and POTRA2–3 fragment structural models were separately constructed using crystal structures PDB: 3Q6B (26) and PDB: 3EFC (24), respectively. The MD systems of these fragments were generated using CHARMM-GUI Quick MD Simulator. The system dimensions were ∼90 × 90 × 90 Å, and the systems were neutralized with 0.15 M KCl.
All MD systems were initially equilibrated with CHARMM (52) and then NAMD (53) was used for continued NPT (constant particle number, pressure, and temperature) simulations for 540 ns (BamA systems) and 400–500 ns (POTRA fragment systems) with the inputs provided by CHARMM-GUI (54) using the CHARMM C36 force field for protein (55), LPS (56), lipids (55), carbohydrates (57, 58, 59), and the TIP3P water model (60). During equilibration various planar and dihedral restraints were applied to the LPS molecules, phospholipids, and water molecules, and the restraint forces were gradually reduced during this process (see (48, 50, 56) for details). Additional dihedral angle restraints were applied to restrain all sugar rings to the pertinent chair conformation, which were maintained during the production simulations. Langevin dynamics was used to maintain constant temperature with a Langevin coupling coefficient set to 1 ps−1, and a Nosé-Hoover Langevin-piston (61, 62) was used to maintain constant pressure with a piston period of 50 fs and a piston decay time of 25 fs. A 2-fs timestep was used with the SHAKE algorithm (63). The van der Waals interactions were smoothly switched off at 10–12 Å by a force-switching function (64), and the long-range electrostatic interactions were calculated using the particle-mesh Ewald (65) method. The temperature and pressure were held at 310.15 K and 1 bar, respectively. Both CHARMM (52) and VMD (66) were used for trajectory analysis.
Evolutionary structural alignment
ConSurf (67) was used to create a multiple sequence alignment of BamA from sequences corresponding to 149 species of Gram-negative bacteria. A WebLogo graphical sequence alignment was created from the aligned sequences of POTRA1, 2, and 3 (68).
Motif definitions and intermotif angle analysis
The POTRA domain mobility was analyzed using structural motifs of BamA defined as follows. POTRA1 corresponds to residues 23–90; POTRA2, residues 91–174; POTRA3, residues 175–264; POTRA4, residues 265–346; POTRA5, residues 347–422; β-barrel, residues describing the barrel strands only. The intermotif angles are defined for each linked pair of motifs using the angle between the major principal axes. To minimize the effects of backbone fluctuations on the orientation of the major principal axis, a copy of the POTRA1 structure from the BamA starting structure (Fig. 2) was first aligned to each different POTRA motif using Cα atoms of selected secondary structural elements. The principal axes were then calculated for the individual aligned POTRA1 motifs using the ORIENT plugin in VMD, and the respective major principal axes angles were calculated.
Results and Discussion
Properties of the membrane systems containing full-length BamA
Three independent replicas of the BamA system (Fig. 2) were simulated for 540 ns each for a total of ∼1.5 μs. Fig. S1 in the Supporting Material shows a time series of the membrane (i.e., x-y) and system z dimensions for the systems, indicating that system equilibrium was achieved at ∼200 ns. This result is consistent with previous work in which the outer membrane showed slow movement of the LPS molecules (48). Fig. S2 shows a time series of the individual POTRA motif Cα root mean-square deviations (RMSD). Some backbone flexibility was observed in POTRA1 and POTRA3 with RMSD values transiently reaching up to ∼6 Å for replica 2 (POTRA1) and ∼4 Å for replica 3 (POTRA3). Significant deviations from the x-ray crystallographic structures in these POTRA motifs are localized in specific segments: the N-terminal β-strand (residues 23–34) in POTRA1 and the loop (residues 200–213) between helices in POTRA3. This loop is uniquely longer than other POTRA loops as can be seen in Fig. 2. POTRA2 and POTRA5 in all three replicas exhibited RMSD values of <2 Å for most simulation time. POTRA5 has the smallest backbone flexibility of the POTRA motifs.
The BamA POTRA domain partitions onto the membrane surface
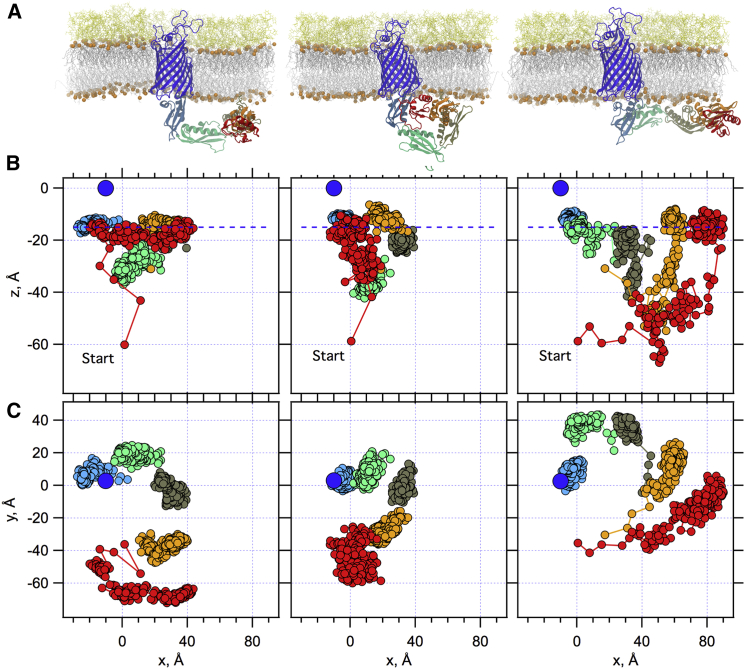
Crystal structures of the BAM complex have the POTRA domain projecting into the periplasm (38, 39, 40) with a conformation similar to the structures originally found for BamA alone (19) as shown in Fig. 1 A. Although these structures are relatively low resolution (2.7– 3.3 Å) the fact that similar extended structures are observed in multiple space groups suggests that this conformation represents a low energy form with limited conformations. In contrast to this view, the results shown in Fig. 3 illustrate a striking finding that the POTRA domain interacts extensively with the periplasmic phospholipid membrane surface in the absence of BamB/C/D/E lipoproteins. Animations of the BamA POTRA domain conformational dynamics are further illustrated in Movies S1, S2, S3, S4, S5, and S6. Replicas 1 and 2 partition onto the membrane surface within 100 ns and 200 ns from the start of simulation. The POTRA domain in replica 3 (Fig. 3, right column) is the last to make contact (at ∼230 ns) following a conformational diffusion in the aqueous space. Membrane binding is most clearly reflected in all three replicas by an increase in the z-coordinate (perpendicular to the membrane plane) of the center of mass (COM) of a particular POTRA motif to −15 Å below the phospholipid surface that is at z = 0 (Fig. 3 B). Once bound to the membrane surface, the POTRA domain remains there for the rest of each simulation trajectory.
Figure 3.
BamA POTRA domain mobility is modulated by membrane binding. (A) Snapshots at the end of each replica trajectory; (B) x, z plots of the positional time evolution of individual POTRA motif's COM for full-length BamA embedded in LPS/phospholipid membranes; (C) x, y plots of the same. The membrane periplasmic phospholipid phosphate atoms are centered at z = 0 Å. The dark blue circle represents the phospholipid membrane surface at the center of the BamA β-barrel. Red, POTRA1; orange, POTRA2; khaki, POTRA3; light green, POTRA4; light blue, POTRA5. Replicas 1, 2, and 3 from left to right, respectively. Each POTRA excursion represents 540 ns of simulation using 10 ns steps; the starting position of POTRA1 is labeled, “Start” in (B). Individual x, y plots are animated in Movies S4, S5, and S6. To see this figure in color, go online.
There are two major membrane-bound conformations observed in the simulations. Replicas 1 and 3 show that the POTRA domain undergoes a continued lateral diffusion in the x-y plane on the membrane surface, illustrated by plots in Fig. 3 C and Movies S4, S5, and S6. POTRA1 (red) in replicas 1 and 3 laterally diffuses up to 60 Å and 90 Å with angular differences of up to 56° and 78°, respectively, relative to the initial structure. In contrast, replica 2 displays a distinct, more compact conformation in which there is a relatively smaller drift (30 Å and 35°) on the x-y plane.
By comparing the space explored by each of the POTRA motifs within a replica, Fig. 3, B and C, shows that there is a gradient of positional accessibilities along the POTRA domain. In all three replicas, the volume explored by POTRA1 is the highest, and this volume decreases as the motif number increases. In part, this gradient is expected as a consequence of the serial connectivity of the POTRA motifs. POTRA5 only has a restricted space available to it at the base of the membrane because one end is fixed to the BamA β-barrel. In contrast, the diffusion volume available to POTRA1 is a result of a concatenation of the spatial volumes available to each POTRA motif with respect to each other.
Membrane partitioning by the POTRA domain involves both hydrophobic interactions and multiple hydrogen bonds with phospholipids
The molecular interactions of the POTRA domain with the E. coli model membrane surface are predominantly due to multivalent hydrogen bonding between phospholipid headgroups and both protein side chain and backbone atoms. These are illustrated by the time series of backbone and side-chain hydrogen bond formation with phospholipids (Figs. S3 and S4) and comprehensive contact maps (Fig. S5).
Fig. S3 shows that there are frequently >20 simultaneous hydrogen bonds between the POTRA domain and the membrane. Hydrogen bonds between POTRA side chains and phospholipids are not static because an individual side chain often hydrogen bonds with a succession of lipid partners (see Fig. S4, A–D, for four typical POTRA residue side-chain hydrogen bonding patterns from replica 1). Other residues have similar dynamic hydrogen bonding interactions. Tryptophan residues W205 and W206 in POTRA3 are also observed to dynamically insert into the membrane interfacial region as shown in the contact maps (Fig. S5). These multivalent interactions with different phospholipids and through hydrophobic partitioning allows the POTRA domain to remain on the membrane surface while simultaneously showing lateral drifting in a manner that is independent of the mobility of individual lipid components.
The contact maps (Fig. S5) illustrate the fraction of time that each residue is in contact with all other molecules for the last 140 ns (400–540 ns) of each trajectory. Each POTRA motif has different interaction patterns with the membrane, representing the dynamic nature of these interactions. Interestingly, POTRA2 has the most extensive contacts with the membrane in all three replicas. For replicas 1 and 2, the POTRA3 interhelical loop (residues 200–213) that contains W205 and W206 contacts the membrane early in the simulations and remains in contact throughout the simulations. In replica 3, this loop is rotated away from the membrane and the tryptophans do not partition into the membrane interface within the present simulation time.
The promiscuity and multiplicity of amino acid residue interactions between the POTRA domain and the membrane surface indicate that no particular residue interaction is a dominant energetic determinant of POTRA membrane binding. Fig. S6 shows the relative sequence conservation of POTRA motifs 1–3 based on an alignment of 149 homologous bacterial sequences (68). The membrane surface interacting with the POTRA domain has a net negative charge due to the presence of both phosphatidylglycerol (20%) and cardiolipin (5%). Khalid et al. (69) have reported the results of a MD simulation of OmpA in a dimyristoyl-phosphatidylcholine membrane where the periplasmic domain of OmpA shows conformational dynamics and interaction with the membrane similar to the POTRA domain observed here. Although the phosphatidylcholine membrane has a net neutral surface charge the interactions of the OmpA periplasmic domain with the membrane were different in zero bulk salt versus 1 M NaCl conditions, suggesting that soluble protein/membrane interactions were electrostatic. The simulations reported here were carried out at 0.15 M KCl. However, specific POTRA basic residues interacting with the negative membrane surface are neither completely conserved nor are the membrane interacting residues of a specific physical/chemical type (Fig. S6, gray arrows). Of the POTRA residue positions that interact with the membrane, highly conserved residues in terms of amino acid attributes are W205 and W206 in the POTRA3 loop. These two aromatic residues are completely exposed on the surface of the POTRA3 motif, which is unusual in that tryptophans typically have only 10% of their accessible surface area exposed to the surface (70). These two residues are among the first to interact with the membrane potentially because the starting structure positions these exposed residues close to the membrane surface (see the residue 200–213 loop in Fig. 2). This segment in POTRA3 is different from all other POTRAs as was noted when the structure was solved (24). Sequence alignment shows that the loop containing residues 205–206 is rich in aromatic residues with at least one being present in all bacterial species.
The POTRA1–2 and POTRA2–3 joints are highly flexible
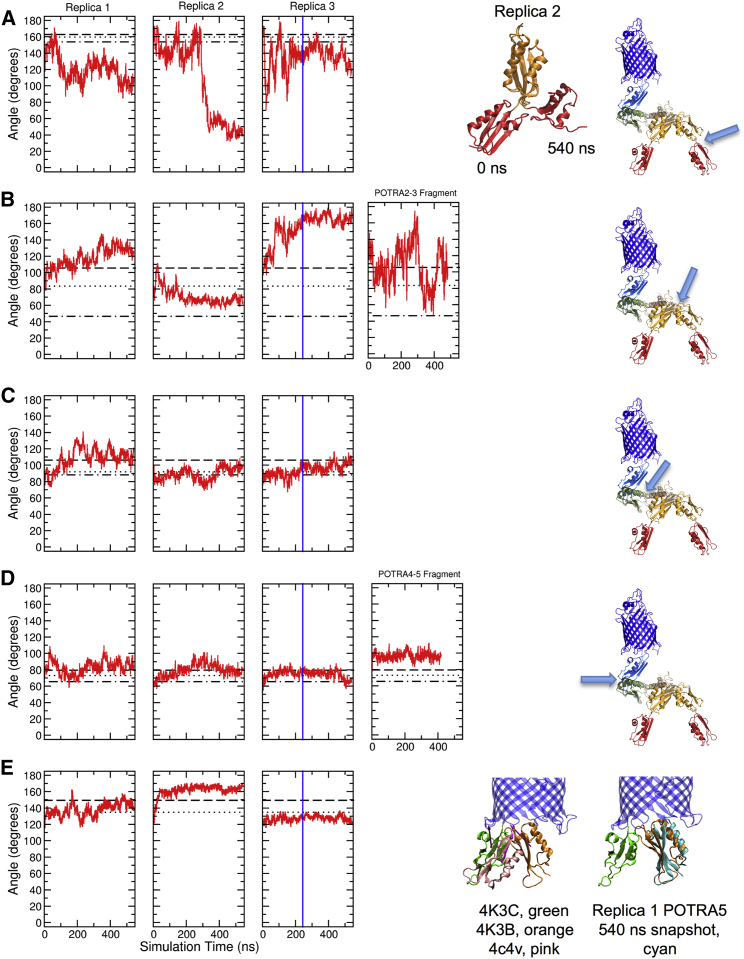
The POTRA1–2 intermotif linker regions have been observed to be relatively inflexible between crystal structures (cf. Fig. 1 A) (19, 24), and this rigidity is consistent with small-angle x-ray scattering (SAXS) results of a POTRA1–5 fragment (25). In contrast, some NMR and SAXS results suggested that this intermotif linker region is very flexible (21). The results from the current simulations of the full-length BamA in E. coli outer membranes show extreme flexibility of the POTRA1–2 joint as illustrated in Fig. 4 A. In this figure the time series of the intermotif angles (red curves) are overlaid upon the angles observed in several crystal structures (black horizontal lines). In replica 2, POTRA1 bends around to contact the side of POTRA2 as shown in the ribbons drawing in Fig. 4 A. This dramatic bend occurs immediately after POTRA1 contacts the membrane (see Movie S2). The flexibility of the POTRA1-2 joint in replica 2 can also be visualized in Movie S5 where the POTRA1 COM (red circles) moves to the side of the POTRA2 COM (orange circles) after 350 ns.
Figure 4.
POTRA intermotif angles in full-length BamA in E. coli outer membranes. (A–E, left three panels) POTRA1-2, POTRA2–3, POTRA3–4, POTRA4–5, and POTRA5-β-barrel angles, respectively. Red curves, time series of intermotif POTRA major principal axes angles during MD simulations of full-length BamA/membrane systems. The vertical blue line in the third panel indicates time of contact between the POTRA domain and membrane in replica 3. (B and D, fourth panels) The same angles of a POTRA2–3 or POTRA4–5 fragment in solution, respectively, are plotted. Black horizontal lines represent respective crystal structure angles: (A) PDB: 2QDF, 163°, dashed lines; PDB: 3EFC, 160°, dotted lines; PDB: 4K3B, 154°, dot-dashed lines, (B) PDB: 3EFC, 106°, dashed lines; PDB: 4K3B, 84°, dotted lines; PDB: 2QDF, 47°, dot-dashed lines, (C) PDB: 2QDF, 106°, dashed lines; PDB: 4K3B, 92°, dotted lines; PDB: 3EFC, 88°, dot-dashed lines, (D) PDB: 4K3C, 79°, dashed lines; PDB: 3OG5 and PDB: 3Q6B (73° together), dotted lines; PDB: 4K3B, 66°, dot-dashed lines, (E) PDB: 4C4V, 149°, dashed lines; PDB: 4K3B and PDB: 4K3C (135° together), dotted lines. The replica 2 POTRA1–2 fragments from simulation times zero and 540 ns are shown as ribbon drawings aligned on POTRA2 at the right of (A); the respective intermotif angles plotted are indicated in the ribbons drawings with arrows in (A)–(D); the different POTRA5-β-barrel orientations for three crystal structures are illustrated in the first ribbons drawings in (E). Although the intermotif principal axis angles are similar, precession of POTRA5 around the β-barrel axis (equivalent to the membrane normal) results in different POTRA orientations. The POTRA5-β-barrel orientation of simulated BamA for all three replicas (cyan ribbons) was similar to that of PDB: 4K3B (orange ribbons) as illustrated in the second ribbons drawing in (E). To see this figure in color, go online.
Several different POTRA2–3 intermotif angles have been observed in different crystal structures (cf. Fig. 1 A) (23, 24), and NMR and SAXS results also indicate significant flexibility at this link (21). Crystallographic intermotif joint angles for POTRA2–3 fragments observed in several crystal structures are 106° (PDB: 3EFC), 84° (PDB: 4K3B), and 47° (PDB: 2QDF). We simulated a POTRA2–3 fragment alone in solution and observed that this fragment had extreme intermotif flexibility as shown in Fig. 4 B (right panel). This fragment samples intermotif angles from ∼50° to ∼170°. The left three panels in Fig. 4 B show that during simulation of the full-length BamA in E. coli outer membranes, the POTRA2–3 joint samples the same range of angles as the POTRA2–3 fragment in solution, although each full-length replica has a different range of angles as modulated by different interactions with the membranes.
The POTRA3–4, POTRA4–5, and POTRA5–β-barrel joints capture the limited flexibilities implied by crystal structures
In simulations of full-length BamA, the POTRA3–4 intermotif joint samples similar angles compared to the crystal structures (88° to 106°) but replica 1 samples a slightly larger range of angles as shown in Fig. 4 C.
For the POTRA4–5 fragment, several independent crystal structures (PDB: 3OG5, 4K3B, 4K3C, 3Q6B) as well as solution SAXS and NMR studies show consistent intermotif angles (19, 25, 26). Taken together, these results suggest that the POTRA4–5 joint should be relatively fixed. As illustrated in Fig. 4 D the POTRA4–5 joint does show relatively low flexibility during simulation consistent with these structural studies. The intermotif angles are stabilized throughout the trajectories by interactions of R314 and R346 across the linker region as described for the crystal structure PDB: 3OG5 except that R314 at times exchanges D383 for D380. Fig. S7 illustrates these hydrogen bonds at a time when R314 is interacting with both D380 and D383.
The diffusion volumes illustrated in Fig. 3 and Movies S4, S5, and S6 are defined primarily by the level of flexibility of the intermotif joint regions that connect each of the POTRA motifs to its neighbor. However, because crystal structures of BamA from two different species show different POTRA5–β-barrel orientations from each other (Fig. 1 B) (19), we examined the possibility that the joint between the BamA β-barrel and POTRA5 might be a fulcrum point whose pivot capabilities could exert a large influence on the conformations available to the entire POTRA domain. A flexible joint at this position would allow a great deal of conformational space to be explored by the periplasmic domain of BamA, even if other POTRA interdomain joints were relatively rigid.
Fig. 4 E illustrates the time evolutions of the POTRA5–β-barrel intermotif angles overlaid upon the angles observed in several crystal structures. This result shows that the mobility of this region in the simulations captures the limited flexibility of the crystal structures in terms of principal axis angles. Movies S4 and S6 show that the lateral drift of the POTRA123 COMs in the x-y plane is not correlated with movement of the POTRA5 COM. Furthermore, Movie S7 shows that POTRA5 does not undergo a rotation around its major principal axis. The large drift of the POTRA domain observed in replicas 1 and 3 cannot therefore be completely explained by a fulcrum function of the POTRA5–β-barrel joint.
The lateral diffusion on the membrane surface is a concatenation of the POTRA intermotif joint flexibilities
Our conclusion from inspection of the individual intermotif flexibilities is that the lateral drift of the POTRA domain seen in Fig. 3 and Movies S1, S2, S3, S4, S5, and S6 is not due to a rigid arm rotating on a fulcrum at the base of the β-barrel but rather due to the concatenation of individual flexibilities at each intermotif joint. Additionally, the simulations suggest that the BamA POTRA domain generally has more flexibility than observed in crystal structures. It is possible that crystallization traps the POTRA domain in one of several lowest energy conformations but that in solution or while interacting with the membrane, the POTRA domain samples a wider range of conformations. The fact that we observed both a wide range of POTRA conformations and a frequent sampling of x-ray crystal structure intermotif angles suggest that the time of simulation was adequate to sample representative states of the BamA soluble domain.
The lateral diffusion of the POTRA domain observed in Fig. 3 is consistent with the 30° rotation of the POTRA domains revealed by two different crystal structures of the BAM complex (39). This rotation of the POTRA domains in crystal structures is coincident with tilting of β-barrel strands 1–6 (39). However, we did not observe any changes in the BamA β-barrel during simulations as discussed below.
Binding of BamB and BamD is compatible with selected conformations of the membrane bound POTRA domain
BamB is known to bind to BamA (37, 71), and a crystal structure of a BamB-POTRA3–5 fragment complex (71) in addition to recent crystal structures of the BamA/B/C/D/E complex (39, 40) show that the BamB β-propeller contacts the face of the POTRA3 β-sheet. For BamD, the only essential BAM lipoprotein (29), a crystal structure of a BamD-POTRA4–5 fragment identified the interface structural elements between subunits (33) and this interface has been confirmed in crystal structures of the BAM complex (38, 39, 40). To test whether the orientations of the POTRA domain in the membrane bound state are compatible with BamB and BamD binding, we docked both BamB and BamD onto the respective POTRA interfaces in individual frames of the simulations. A crystal structure BamB-POTRA3 fragment (PDB: 4PK1) and a separate BamD-POTRA5 fragment structure (PDB: 5EFR) were used for superposition; only the backbone atoms of the respective POTRA subunits were fit onto corresponding atoms of the simulated BamA structure. During the simulation trajectories of replicas 1 and 3 the POTRA domain is frequently in a compatible conformation for binding to BamB and BamD. A snapshot of such a conformation with both BamB and BamD docked in silico is shown in Fig. 5. In contrast, after superposition with replica 2, BamD is partially inserted into the membrane for most frames (not shown). These results suggest that lipoprotein binding to the BamA POTRA domain could select specific conformations from the range observed in this study.
Figure 5.
BamB and BamD binding is compatible with membrane bound POTRA domain. Trajectory snapshots of replica 3 showing BamB (green) docked to POTRA3 and BamD (pink) docked to POTRA5. The BamB-POTRA3 interface contacts are based on crystal structure PDB: 4PK1 and the BamD-POTRA5 interface contacts are based on crystal structure PDB: 5EFR. To see this figure in color, go online.
Gu et al. recently published results of MD simulations of BAM in a similar asymmetric membrane where the BAM contained various combinations of BAM assembles (39). They report that in the absence of lipoproteins the POTRA domain of BamA exhibits greater dynamics. However, they did not observe extensive interaction of the POTRA motifs with the membrane. Gu et al. simulated individual systems for 100 ns and this may not have been long enough for the POTRA domain to diffuse up to the membrane as seen here for replica 3 where full membrane contact did not occur until ∼230 ns of simulation (Fig. 3).
POTRA-membrane interaction does not affect POTRA5 internal conformational dynamics
POTRA5 has been reported to exhibit internal conformational plasticity that may be important for the binding of BamD to BamA and to the overall function of the BAM complex (29, 72). Because interactions of the POTRA domain with the membrane may influence the internal conformational dynamics of POTRA5, we investigated its intramotif interactions in more detail. Unlike POTRA1–4 motifs, which have a hydrophobic core, POTRA5 has a network of electrostatic interactions that extend to the domain surface. Sinnige et al. (72) used NMR to show conformational plasticity of this network on the micro- to millisecond timescale and employed MD simulations to demonstrate interchanging electrostatic interactions between K351, D362, R366, and E373. We compared the hydrogen bonding patterns of this region of full-length BamA in outer membranes to determine if the POTRA domain mobility influences the core of POTRA5. Fig. S8 shows three snapshots from a trajectory of this region revealing distinct hydrogen bonding patterns. In addition to the interactions described by Sinnige et al. we also observed hydrogen bonding between R366 and E521, which is in a β-barrel loop. Fig. S9, A–C, shows the time evolved interchange of hydrogen bonding partners of the central residue R366 in all three replicas. We observed similar dynamic exchange to that reported by Sinnige et al. Although the POTRA1–3 motifs are partially limited in their conformational dynamics by interaction with the membrane, this distal interaction does not seem to affect the internal conformational plasticity of the POTRA5 electrostatic network. Interestingly, despite this internal plasticity, POTRA5 exhibited the smallest backbone RMSD of all POTRA motifs during simulation (Fig. S2).
Protein crowding limits POTRA domain dynamics
In contrast to the results presented here, Sinnige et al. previously reported limited dynamics in the POTRA domain of membrane embedded BamA using solid-state NMR (73). In these experiments BamA was reconstituted into liposomes with lipid/protein ratios of 25:1 to 150:1. To approximate this condition in silico, we separately built and simulated two systems containing full-length BamA in smaller asymmetric membrane surface areas for ∼1.2 μs. These systems contained a membrane surface of 90 × 90 Å and inner leaflet phospholipid/protein molar ratio of 100:1 (equivalent to a total phospholipid/protein ratio of 200:1 in a symmetric phospholipid membrane). It was clear that these systems were too small to allow the POTRA domain to explore complete conformational freedom. As shown in Fig. S10, the BamA molecules in periodic boundary neighboring cells came into stable contact. Such neighbor contacts occurred in both systems within 250 ns simulation time and remained intact throughout the remainder of the simulations. The contacts were not the same in both systems: in one case POTRA1 contacted POTRA4 and in the other POTRA2 contacted a different segment of POTRA4. In proteoliposomes with a phospholipid ratio of 25:1 to 150:1, a similar neighbor-neighbor contact would occur and limit the mobility of the POTRA domains. Such inter-BamA interaction would not be expected in the bacterial cell membrane. The estimated cell copy number of BamA is ∼200 molecules (74); these BamA molecules appear to be dispersed in the bacterial outer membrane and surrounded by recently folded client OMPs (75).
The BamA β-barrel has a small effect on membrane thickness
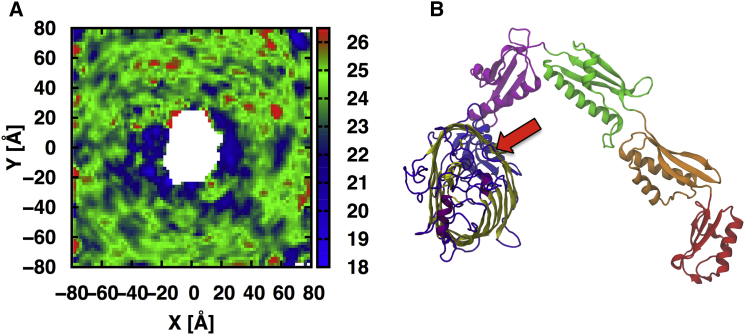
The BamA β-barrel exhibits an asymmetric transmembrane hydrophobic thickness; the side containing the strand1/16 seam is between 12 Å (L780 to Y432) and 16 Å (W810 to V438) thick compared to ∼24 Å on the opposite side (48). Because the hydrophobic thickness of the E. coli outer membrane is ∼24–25 Å (76), it has been proposed that the membrane in proximity to the strand1–16 seam is destabilized by hydrophobic mismatch. Previous simulation results of the Neisseria gonorrheae and Haemophilus ducreyi BamA β-barrel in gel-like dimyristoyl-phosphatidylethanolamine bilayers showed a dramatic thinning near the strand1–16 seam (19). However, as shown in Fig. 6 we observed membrane thinning of only a few Ångstroms (to ∼20 Å thickness) of small patches of the outer membrane in the vicinity of the BamA β-barrel. Furthermore, areas of this thinning were not consistently proximate to the strand1/16 seam among the three replicate systems (Fig. S11). Similar thinning was observed during simulations of the β-barrel protein OmpLA in the outer membrane of the same lipid composition (48).
Figure 6.
(A) Two-dimensional z-thickness of the LPS/phospholipid membrane with embedded full-length BamA. Average of replicas 1–3. Thickness is defined as the distance between the average of C2 and C4 atoms of lipid A (combined) and the average C2 positions of phospholipids within a 2 Å grid and represents the hydrophobic thickness. (B) The relative orientation of the embedded BamA; the red arrow points to the strand 1–16 seam position. The z-thickness of individual replicas is shown in Fig. S11. To see this figure in color, go online.
Lateral gate opening is not observed in these simulations using the E. coli outer membrane
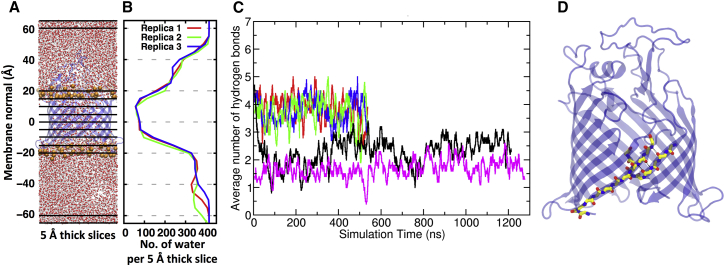
One of the most interesting discoveries from early simulations of BamA barrel structures from N. gonorrheae and H. ducreyi in symmetric dimyristoyl-phosphatidylethanolamine membranes was the lateral opening of the β-barrel between strand1 and stand16 (19). In addition, recent crystal structures of BamA/C/D/E complexes in detergent show twisting of strands 1–8 and opening of the barrel between strands 1 and 16 (38, 39). The cavity of the BamA β-barrel contains significant water as illustrated in Fig. 7, A and B. The average numbers of waters inside the cavity (z = ±10 Å) are 403, 377, and 427 for replicas 1, 2, and 3, respectively. As we observed only approximately a 4 Å thinning of the LPS/phospholipid membrane (to ∼20 Å hydrophobic core thickness proximal to the BamA barrel) during our simulations (Fig. 6), opening of the barrel would expose this cavity water to the hydrophobic core of the membrane. Similar water numbers in the cavity of the SecY translocon were recently found to have anomalous behavior and were partially displaced by lipid acyl chain incursions into the translocon cavity (77).
Figure 7.
The BamA β-barrel is water-filled and stable. (A) Water distribution within the BamA cavity. A snapshot of replica 1 trajectory cropped to a 50 × 50 × 130 Å prism highlighting the continuous water filled cavity. Water is shown in red (oxygen) and white (hydrogen) ball-stick representation. Lipid phosphates delimiting the hydrophobic core of the membrane are shown as orange spheres, BamA is shown as a ribbon representation. Black horizontal lines represent 5 Å thick slabs parallel to the membrane plane used to subdivide the prism for calculating water density as a function of position along the membrane normal (for clarity, 5-Å-thick slabs are not shown inside whole box). (B) Time-averaged number of water molecules within 5-Å thick slabs from 100 to 540 ns of the simulations in all three replicas. (C) BamA β-barrel strands 1–16 remain hydrogen bonded. A time series of the average number backbone hydrogen bonds between strand 1 (residues 427 to 434) and adjacent strand 16 (residues 803 to 810). Criteria were distance of 3.0 Å and minimum angle cutoff of 45°, averages were over 10 ns. Red, blue, and green curves represent replicas 1, 2, and 3 of the main systems in this study, respectively. Black and magenta curves represent average hydrogen bonds for the smaller membrane systems shown in Fig. S10. (D) In these smaller systems the C-terminus (residues 808–810) curled into the barrel cavity and therefore the number of interstrand hydrogen bonds decreased to an average of 2–3 but the strands 1–16 never opened (replica 2 is shown). An inward curl of the C-terminus was also seen the in the crystal structure of N. gonorrheae BamA (19). To see this figure in color, go online.
In ∼5-μs total simulation of full-length BamA at 310 K in membranes that mimic the E. coli outer membrane, we observed neither a separation of strands 1 and 16 nor a register shift of hydrogen bonding in this region. Fig. 7 C shows the time series of the average number of hydrogen bonds between strands 1 and 16 for several systems. The three replicate systems with large membrane surface areas (150 × 150 Å) maintained an average of ∼4 interstrand hydrogen bonds. Two systems with smaller membrane surface area (90 × 90 Å) started out with four interstrand hydrogen bonds but the C-terminus curled into the barrel cavity breaking ∼2 hydrogen bonds (Fig. 7 D). A similar inward curl of the C-terminus is seen in the crystal structure of N. gonorrheae BamA (19). At no time during our simulations did the strand1/16 seam open. In fact, the β-barrel strands showed small fluctuations. Fig. S12, A–C, shows images of the BamA β-barrel average structures from simulations of the full-length BamA in outer membranes color keyed to Cα root mean-square fluctuation (RMSF). The barrel strands were very stable with Cα RMSF values of <1 Å but the external loops showed higher dynamic flexibilities. This difference in relative movement can be observed in Fig. S13, where the calculated RMSF values of Cα atoms in the BamA β-barrel are plotted as a function of residue number. Both the C-terminal segment of loop 6 (residues 670–705) and loop 7 extend to the very top of the barrel and exhibit the largest RMSF values. Other loops have RMSF values between 1 and 2 Å, mobility slightly larger than loops of OmpLA in a similar E. coli membrane (48). No consistent difference in the mobility of loops and turns is observed.
Conclusion
The POTRA domain of BamA without BamB/C/D/E lipoproteins partitions onto the surface of the phospholipid leaflet of an E. coli outer membrane in orientations that are competent to interact with BamB and BamD. Both hydrophobic insertions of the conserved aromatic residues (W205 and W206) into the membrane and numerous POTRA side-chain hydrogen bonds with the membrane phospholipid raise the idea that membrane-bound orientations of the BamA POTRA domain may be long lived. Significant conformational flexibility of the intermotif linker regions is observed, which results in lateral drift of the POTRA domains along the membrane surface. BamA alone shows extreme conformational flexibility and long-lived interactions with the membrane. However, only a subset of these conformations is compatible with the binding of BamB and the BamC/D/E subcomplex. This suggests that one important role of BAM lipoproteins is to modulate the BamA conformations to select those that are important for the OMP folding and insertion cycle. Weak binding between the POTRA domain and lipoproteins also could facilitate conformational cycling of the BamA during its functional cycle. Finally, with ∼4 Å thinning of the outer membrane near BamA, a lateral gate opening between strand1 and strand16 is not observed in our simulations.
Author Contributions
P.J.F., D.S.P., M.C.S., K.G.F., and W.I. designed the research; P.J.F., D.S.P., E.L.W., M.C.S., Y.Q., and M.S.Y. performed the research; P.J.F. and D.S.P. analyzed the data; P.J.F., K.G.F., M.C.S., D.S.P., and W.I. wrote the article.
Acknowledgments
This work was supported by National Institutes of Health (NIH) grants U54GM087519 (to W.I.), R01AI080709 (to M.C.S.), and R01GM079440 (to K.G.F.) and National Science Foundation (NSF) grants MCB-1516154 (to W.I.), DBI-1145987 (to W.I.), and MCB1412108 (to K.G.F.). This work used the computational resources at the National Institute of Supercomputing and Networking/Korea Institute of Science and Technology Information including technical support (KSC-2015-C3-004 to M.S.Y.), the Maryland Advanced Research Computing Center (MARCC to K.G.F.), and the Extreme Science and Engineering Discovery Environment (XSEDE MCB070009 to W.I. and MCB120050 to K.G.F.), which is supported by National Science Foundation grant No. ACI-1053575.
Editor: Emad Tajkhorshid.
Footnotes
Thirteen figures and seven movies are available at http://www.biophysj.org/biophysj/supplemental/S0006-3495(16)30287-9.
Supporting Material
Full-length BamA embedded in an LPS/phospholipid membrane; simulation time shown is 1–540 ns. Red, POTRA1; orange, POTRA2; khaki, POTRA3; green, POTRA4; light blue, POTRA5; dark blue, β-barrel. Membrane phosphates delimiting the hydrophobic core are shown as orange spheres, polysaccharide as dark yellow lines, and other lipid as gray lines.
Full-length BamA embedded in an LPS/phospholipid membrane; simulation time shown is 1–540 ns. Red, POTRA1; orange, POTRA2; khaki, POTRA3; green, POTRA4; light blue, POTRA5; dark blue, β-barrel. Membrane phosphates delimiting the hydrophobic core are shown as orange spheres, polysaccharide as dark yellow lines, and other lipid as gray lines.
Full-length BamA embedded in an LPS/phospholipid membrane; simulation time shown is 1–540 ns. The POTRA domains contact the membrane at ∼230 ns of simulation. Red, POTRA1; orange, POTRA2; khaki, POTRA3; green, POTRA4; light blue, POTRA5; dark blue, β-barrel. Membrane phosphates delimiting the hydrophobic core are shown as orange spheres, polysaccharide as dark yellow lines, and other lipid as gray lines.
The view is looking down the membrane normal toward the phospholipid surface of the membrane. The dark blue circle represents the center of the BamA β-barrel. Red, POTRA1; orange, POTRA2; khaki, POTRA3; light green, POTRA4; light blue, POTRA5. Note that the drift of POTRAs 1, 2, and 3 in this x, y plane (parallel to the membrane surface) is occasionally correlated whole POTRA domain drift is not due correlated with POTRA5 movement.
The view is looking down the membrane normal toward the phospholipid surface of the membrane. The dark blue circle represents the center of the BamA β-barrel. Red, POTRA1; orange, POTRA2; khaki, POTRA3; light green, POTRA4; light blue, POTRA5. Note that the drift of POTRAs 1, 2, and 3 in this x, y plane (parallel to the membrane surface) is occasionally correlated whole POTRA domain drift is not due correlated with POTRA5 movement.
The view is looking down the membrane normal toward the phospholipid surface of the membrane. The dark blue circle represents the center of the BamA β-barrel. Red, POTRA1; orange, POTRA2; khaki, POTRA3; light green, POTRA4; light blue, POTRA5. Note that the drift of POTRAs 1, 2, and 3 in this x, y plane (parallel to the membrane surface) is occasionally correlated whole POTRA domain drift is not due correlated with POTRA5 movement.
The β-barrel and POTRA5 fragments from crystal structure 4K3B (translucent blue and orange, respectively) are aligned to the same from replica 1 simulation at time zero (translucent blue and light blue, respectively). Simulation time shown is 1–540 ns.
References
- 1.Nikaido H., Vaara M. Molecular basis of bacterial outer membrane permeability. Microbiol. Rev. 1985;49:1–32. doi: 10.1128/mr.49.1.1-32.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nikaido H. Molecular basis of bacterial outer membrane permeability revisited. Microbiol. Mol. Biol. Rev. 2003;67:593–656. doi: 10.1128/MMBR.67.4.593-656.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Koebnik R., Locher K.P., Van Gelder P. Structure and function of bacterial outer membrane proteins: barrels in a nutshell. Mol. Microbiol. 2000;37:239–253. doi: 10.1046/j.1365-2958.2000.01983.x. [DOI] [PubMed] [Google Scholar]
- 4.Surrey T., Jähnig F. Refolding and oriented insertion of a membrane protein into a lipid bilayer. Proc. Natl. Acad. Sci. USA. 1992;89:7457–7461. doi: 10.1073/pnas.89.16.7457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kleinschmidt J.H., Tamm L.K. Folding intermediates of a beta-barrel membrane protein. Kinetic evidence for a multi-step membrane insertion mechanism. Biochemistry. 1996;35:12993–13000. doi: 10.1021/bi961478b. [DOI] [PubMed] [Google Scholar]
- 6.Burgess N.K., Dao T.P., Fleming K.G. Beta-barrel proteins that reside in the Escherichia coli outer membrane in vivo demonstrate varied folding behavior in vitro. J. Biol. Chem. 2008;283:26748–26758. doi: 10.1074/jbc.M802754200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bos M.P., Robert V., Tommassen J. Biogenesis of the gram-negative bacterial outer membrane. Annu. Rev. Microbiol. 2007;61:191–214. doi: 10.1146/annurev.micro.61.080706.093245. [DOI] [PubMed] [Google Scholar]
- 8.Sklar J.G., Wu T., Silhavy T.J. Defining the roles of the periplasmic chaperones SurA, Skp, and DegP in Escherichia coli. Genes Dev. 2007;21:2473–2484. doi: 10.1101/gad.1581007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tommassen J. Assembly of outer-membrane proteins in bacteria and mitochondria. Microbiology. 2010;156:2587–2596. doi: 10.1099/mic.0.042689-0. [DOI] [PubMed] [Google Scholar]
- 10.Mogensen J.E., Otzen D.E. Interactions between folding factors and bacterial outer membrane proteins. Mol. Microbiol. 2005;57:326–346. doi: 10.1111/j.1365-2958.2005.04674.x. [DOI] [PubMed] [Google Scholar]
- 11.Bennion D., Charlson E.S., Misra R. Dissection of β-barrel outer membrane protein assembly pathways through characterizing BamA POTRA 1 mutants of Escherichia coli. Mol. Microbiol. 2010;77:1153–1171. doi: 10.1111/j.1365-2958.2010.07280.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bos M.P., Robert V., Tommassen J. Functioning of outer membrane protein assembly factor Omp85 requires a single POTRA domain. EMBO Rep. 2007;8:1149–1154. doi: 10.1038/sj.embor.7401092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gentle I., Gabriel K., Lithgow T. The Omp85 family of proteins is essential for outer membrane biogenesis in mitochondria and bacteria. J. Cell Biol. 2004;164:19–24. doi: 10.1083/jcb.200310092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hagan C.L., Kim S., Kahne D. Reconstitution of outer membrane protein assembly from purified components. Science. 2010;328:890–892. doi: 10.1126/science.1188919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Voulhoux R., Tommassen J. Omp85, an evolutionarily conserved bacterial protein involved in outer-membrane-protein assembly. Res. Microbiol. 2004;155:129–135. doi: 10.1016/j.resmic.2003.11.007. [DOI] [PubMed] [Google Scholar]
- 16.Werner J., Misra R. YaeT (Omp85) affects the assembly of lipid-dependent and lipid-independent outer membrane proteins of Escherichia coli. Mol. Microbiol. 2005;57:1450–1459. doi: 10.1111/j.1365-2958.2005.04775.x. [DOI] [PubMed] [Google Scholar]
- 17.Ricci D.P., Silhavy T.J. The Bam machine: a molecular cooper. Biochim. Biophys. Acta. 2012;1818:1067–1084. doi: 10.1016/j.bbamem.2011.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Albrecht R., Schütz M., Zeth K. Structure of BamA, an essential factor in outer membrane protein biogenesis. Acta Crystallogr. D Biol. Crystallogr. 2014;70:1779–1789. doi: 10.1107/S1399004714007482. [DOI] [PubMed] [Google Scholar]
- 19.Noinaj N., Kuszak A.J., Buchanan S.K. Structural insight into the biogenesis of β-barrel membrane proteins. Nature. 2013;501:385–390. doi: 10.1038/nature12521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ni D., Wang Y., Huang Y. Structural and functional analysis of the β-barrel domain of BamA from Escherichia coli. FASEB J. 2014;28:2677–2685. doi: 10.1096/fj.13-248450. [DOI] [PubMed] [Google Scholar]
- 21.Knowles T.J., Jeeves M., Henderson I.R. Fold and function of polypeptide transport-associated domains responsible for delivering unfolded proteins to membranes. Mol. Microbiol. 2008;68:1216–1227. doi: 10.1111/j.1365-2958.2008.06225.x. [DOI] [PubMed] [Google Scholar]
- 22.Sánchez-Pulido L., Devos D., Valencia A. POTRA: a conserved domain in the FtsQ family and a class of beta-barrel outer membrane proteins. Trends Biochem. Sci. 2003;28:523–526. doi: 10.1016/j.tibs.2003.08.003. [DOI] [PubMed] [Google Scholar]
- 23.Kim S., Malinverni J.C., Kahne D. Structure and function of an essential component of the outer membrane protein assembly machine. Science. 2007;317:961–964. doi: 10.1126/science.1143993. [DOI] [PubMed] [Google Scholar]
- 24.Gatzeva-Topalova P.Z., Walton T.A., Sousa M.C. Crystal structure of YaeT: conformational flexibility and substrate recognition. Structure. 2008;16:1873–1881. doi: 10.1016/j.str.2008.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gatzeva-Topalova P.Z., Warner L.R., Sousa M.C. Structure and flexibility of the complete periplasmic domain of BamA: the protein insertion machine of the outer membrane. Structure. 2010;18:1492–1501. doi: 10.1016/j.str.2010.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang H., Gao Z.Q., Dong Y.H. High-resolution structure of a new crystal form of BamA POTRA4-5 from Escherichia coli. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 2011;67:734–738. doi: 10.1107/S1744309111014254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wu T., Malinverni J., Kahne D. Identification of a multicomponent complex required for outer membrane biogenesis in Escherichia coli. Cell. 2005;121:235–245. doi: 10.1016/j.cell.2005.02.015. [DOI] [PubMed] [Google Scholar]
- 28.Sklar J.G., Wu T., Silhavy T.J. Lipoprotein SmpA is a component of the YaeT complex that assembles outer membrane proteins in Escherichia coli. Proc. Natl. Acad. Sci. USA. 2007;104:6400–6405. doi: 10.1073/pnas.0701579104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Malinverni J.C., Werner J., Silhavy T.J. YfiO stabilizes the YaeT complex and is essential for outer membrane protein assembly in Escherichia coli. Mol. Microbiol. 2006;61:151–164. doi: 10.1111/j.1365-2958.2006.05211.x. [DOI] [PubMed] [Google Scholar]
- 30.Ricci D.P., Hagan C.L., Silhavy T.J. Activation of the Escherichia coli β-barrel assembly machine (Bam) is required for essential components to interact properly with substrate. Proc. Natl. Acad. Sci. USA. 2012;109:3487–3491. doi: 10.1073/pnas.1201362109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rigel N.W., Schwalm J., Silhavy T.J. BamE modulates the Escherichia coli beta-barrel assembly machine component BamA. J. Bacteriol. 2012;194:1002–1008. doi: 10.1128/JB.06426-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vuong P., Bennion D., Misra R. Analysis of YfgL and YaeT interactions through bioinformatics, mutagenesis, and biochemistry. J. Bacteriol. 2008;190:1507–1517. doi: 10.1128/JB.01477-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bergal H.T., Hopkins A.H., Sousa M.C. The structure of a BamA-BamD fusion illuminates the architecture of the beta-barrel assembly machine core. Structure. 2016;24:243–251. doi: 10.1016/j.str.2015.10.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Albrecht R., Zeth K. Structural basis of outer membrane protein biogenesis in bacteria. J. Biol. Chem. 2011;286:27792–27803. doi: 10.1074/jbc.M111.238931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dong C., Hou H.F., Dong Y.H. Structure of Escherichia coli BamD and its functional implications in outer membrane protein assembly. Acta Crystallogr. D Biol. Crystallogr. 2012;68:95–101. doi: 10.1107/S0907444911051031. [DOI] [PubMed] [Google Scholar]
- 36.Kim K.H., Paetzel M. Crystal structure of Escherichia coli BamB, a lipoprotein component of the β-barrel assembly machinery complex. J. Mol. Biol. 2011;406:667–678. doi: 10.1016/j.jmb.2010.12.020. [DOI] [PubMed] [Google Scholar]
- 37.Noinaj N., Fairman J.W., Buchanan S.K. The crystal structure of BamB suggests interactions with BamA and its role within the BAM complex. J. Mol. Biol. 2011;407:248–260. doi: 10.1016/j.jmb.2011.01.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bakelar J., Buchanan S.K., Noinaj N. The structure of the β-barrel assembly machinery complex. Science. 2016;351:180–186. doi: 10.1126/science.aad3460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gu Y., Li H., Dong C. Structural basis of outer membrane protein insertion by the BAM complex. Nature. 2016;531:64–69. doi: 10.1038/nature17199. [DOI] [PubMed] [Google Scholar]
- 40.Han L., Zheng J., Huang Y. Structure of the BAM complex and its implications for biogenesis of outer-membrane proteins. Nat. Struct. Mol. Biol. 2016;23:192–196. doi: 10.1038/nsmb.3181. [DOI] [PubMed] [Google Scholar]
- 41.Hagan C.L., Silhavy T.J., Kahne D. β-Barrel membrane protein assembly by the Bam complex. Annu. Rev. Biochem. 2011;80:189–210. doi: 10.1146/annurev-biochem-061408-144611. [DOI] [PubMed] [Google Scholar]
- 42.Plummer A.M., Fleming K.G. BamA alone accelerates outer membrane protein folding in vitro through a catalytic mechanism. Biochemistry. 2015;54:6009–6011. doi: 10.1021/acs.biochem.5b00950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rigel N.W., Ricci D.P., Silhavy T.J. Conformation-specific labeling of BamA and suppressor analysis suggest a cyclic mechanism for β-barrel assembly in Escherichia coli. Proc. Natl. Acad. Sci. USA. 2013;110:5151–5156. doi: 10.1073/pnas.1302662110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bernstein H.D. Looks can be deceiving: recent insights into the mechanism of protein secretion by the autotransporter pathway. Mol. Microbiol. 2015;97:205–215. doi: 10.1111/mmi.13031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lugtenberg E.J., Peters R. Distribution of lipids in cytoplasmic and outer membranes of Escherichia coli K12. Biochim. Biophys. Acta. 1976;441:38–47. doi: 10.1016/0005-2760(76)90279-4. [DOI] [PubMed] [Google Scholar]
- 46.Jo S., Kim T., Im W. CHARMM-GUI: a web-based graphical user interface for CHARMM. J. Comput. Chem. 2008;29:1859–1865. doi: 10.1002/jcc.20945. [DOI] [PubMed] [Google Scholar]
- 47.Wu E.L., Cheng X., Im W. CHARMM-GUI Membrane Builder toward realistic biological membrane simulations. J. Comput. Chem. 2014;35:1997–2004. doi: 10.1002/jcc.23702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wu E.L., Fleming P.J., Im W. E. coli outer membrane and interactions with OmpLA. Biophys. J. 2014;106:2493–2502. doi: 10.1016/j.bpj.2014.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Patel D.S., Re S., Im W. Dynamics and interactions of OmpF and LPS: influence on pore accessibility and ion permeability. Biophys. J. 2016;110:930–938. doi: 10.1016/j.bpj.2016.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wu E.L., Engström O., Im W. Molecular dynamics and NMR spectroscopy studies of E. coli lipopolysaccharide structure and dynamics. Biophys. J. 2013;105:1444–1455. doi: 10.1016/j.bpj.2013.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Vance D.E., Vance J.E. Elsevier; Amsterdam; Boston: 2002. Biochemistry of Lipids, Lipoproteins, and Membranes. [Google Scholar]
- 52.Brooks B.R., Brooks C.L., 3rd, Karplus M. CHARMM: the biomolecular simulation program. J. Comput. Chem. 2009;30:1545–1614. doi: 10.1002/jcc.21287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Phillips J.C., Braun R., Schulten K. Scalable molecular dynamics with NAMD. J. Comput. Chem. 2005;26:1781–1802. doi: 10.1002/jcc.20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lee J., Cheng X., Im W. CHARMM-GUI input generator for NAMD, GROMACS, AMBER, OpenMM, and CHARMM/OpenMM simulations using the CHARMM36 additive force field. J. Chem. Theory Comput. 2016;12:405–413. doi: 10.1021/acs.jctc.5b00935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Klauda J.B., Venable R.M., Pastor R.W. Update of the CHARMM all-atom additive force field for lipids: validation on six lipid types. J. Phys. Chem. B. 2010;114:7830–7843. doi: 10.1021/jp101759q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Jo S., Wu E.L., Im W. Lipopolysaccharide membrane building and simulation. Methods Mol. Biol. 2015;1273:391–406. doi: 10.1007/978-1-4939-2343-4_24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Guvench O., Greene S.N., Mackerell A.D., Jr. Additive empirical force field for hexopyranose monosaccharides. J. Comput. Chem. 2008;29:2543–2564. doi: 10.1002/jcc.21004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hatcher E., Guvench O., Mackerell A.D. CHARMM additive all-atom force field for aldopentofuranoses, methyl-aldopentofuranosides, and fructofuranose. J. Phys. Chem. B. 2009;113:12466–12476. doi: 10.1021/jp905496e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Guvench O., Hatcher E.R., Mackerell A.D. CHARMM additive all-atom force field for glycosidic linkages between hexopyranoses. J. Chem. Theory Comput. 2009;5:2353–2370. doi: 10.1021/ct900242e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Jorgensen W.L., Chandrasekhar J., Klein M.L. Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 1983;79:926–935. [Google Scholar]
- 61.Feller S.E., Zhang Y., Brooks B.R. Constant pressure molecular dynamics simulation: the Langevin piston method. J. Chem. Phys. 1995;103:4613–4621. [Google Scholar]
- 62.Martyna G.J., Tobias D.J., Klein M.L. Constant-pressure molecular-dynamics algorithms. J. Chem. Phys. 1994;101:4177–4189. [Google Scholar]
- 63.Ryckaert J.P., Ciccotti G., Berendsen H.J.C. Numerical integration of the cartesian equations of motion of a system with constraints: molecular dynamics of n-alkanes. J. Comput. Phys. 1977;23:327–341. [Google Scholar]
- 64.Steinbach P.J., Brooks B.R. New spherical-cutoff methods for long-range forces in macromolecular simulation. J. Comput. Chem. 1994;15:667–683. [Google Scholar]
- 65.Essmann U., Perera L., Pedersen L.G. A smooth particle mesh Ewald method. J. Chem. Phys. 1995;103:8577–8593. [Google Scholar]
- 66.Humphrey W., Dalke A., Schulten K. VMD: visual molecular dynamics. J. Mol. Graph. 1996;14:33–38. doi: 10.1016/0263-7855(96)00018-5. 27–38. [DOI] [PubMed] [Google Scholar]
- 67.Ashkenazy H., Erez E., Ben-Tal N. ConSurf 2010: calculating evolutionary conservation in sequence and structure of proteins and nucleic acids. Nucleic Acids Res. 2010;38:W529–W533. doi: 10.1093/nar/gkq399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Crooks G.E., Hon G., Brenner S.E. WebLogo: a sequence logo generator. Genome Res. 2004;14:1188–1190. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Khalid S., Bond P.J., Sansom M.S. OmpA: gating and dynamics via molecular dynamics simulations. Biochim. Biophys. Acta. 2008;1778:1871–1880. doi: 10.1016/j.bbamem.2007.05.024. [DOI] [PubMed] [Google Scholar]
- 70.Lins L., Thomas A., Brasseur R. Analysis of accessible surface of residues in proteins. Protein Sci. 2003;12:1406–1417. doi: 10.1110/ps.0304803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Jansen K.B., Baker S.L., Sousa M.C. Crystal structure of BamB bound to a periplasmic domain fragment of BamA, the central component of the β-barrel assembly machine. J. Biol. Chem. 2015;290:2126–2136. doi: 10.1074/jbc.M114.584524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Sinnige T., Weingarth M., Baldus M. Conformational plasticity of the POTRA 5 domain in the outer membrane protein assembly factor BamA. Structure. 2015;23:1317–1324. doi: 10.1016/j.str.2015.04.014. [DOI] [PubMed] [Google Scholar]
- 73.Sinnige T., Weingarth M., Baldus M. Solid-state NMR studies of full-length BamA in lipid bilayers suggest limited overall POTRA mobility. J. Mol. Biol. 2014;426:2009–2021. doi: 10.1016/j.jmb.2014.02.007. [DOI] [PubMed] [Google Scholar]
- 74.Masuda T., Saito N., Ishihama Y. Unbiased quantitation of Escherichia coli membrane proteome using phase transfer surfactants. Mol. Cell. Proteomics. 2009;8:2770–2777. doi: 10.1074/mcp.M900240-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kleanthous C., Rassam P., Baumann C.G. Protein-protein interactions and the spatiotemporal dynamics of bacterial outer membrane proteins. Curr. Opin. Struct. Biol. 2015;35:109–115. doi: 10.1016/j.sbi.2015.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Lomize M.A., Lomize A.L., Mosberg H.I. OPM: orientations of proteins in membranes database. Bioinformatics. 2006;22:623–625. doi: 10.1093/bioinformatics/btk023. [DOI] [PubMed] [Google Scholar]
- 77.Capponi S., Heyden M., White S.H. Anomalous behavior of water inside the SecY translocon. Proc. Natl. Acad. Sci. USA. 2015;112:9016–9021. doi: 10.1073/pnas.1424483112. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Full-length BamA embedded in an LPS/phospholipid membrane; simulation time shown is 1–540 ns. Red, POTRA1; orange, POTRA2; khaki, POTRA3; green, POTRA4; light blue, POTRA5; dark blue, β-barrel. Membrane phosphates delimiting the hydrophobic core are shown as orange spheres, polysaccharide as dark yellow lines, and other lipid as gray lines.
Full-length BamA embedded in an LPS/phospholipid membrane; simulation time shown is 1–540 ns. Red, POTRA1; orange, POTRA2; khaki, POTRA3; green, POTRA4; light blue, POTRA5; dark blue, β-barrel. Membrane phosphates delimiting the hydrophobic core are shown as orange spheres, polysaccharide as dark yellow lines, and other lipid as gray lines.
Full-length BamA embedded in an LPS/phospholipid membrane; simulation time shown is 1–540 ns. The POTRA domains contact the membrane at ∼230 ns of simulation. Red, POTRA1; orange, POTRA2; khaki, POTRA3; green, POTRA4; light blue, POTRA5; dark blue, β-barrel. Membrane phosphates delimiting the hydrophobic core are shown as orange spheres, polysaccharide as dark yellow lines, and other lipid as gray lines.
The view is looking down the membrane normal toward the phospholipid surface of the membrane. The dark blue circle represents the center of the BamA β-barrel. Red, POTRA1; orange, POTRA2; khaki, POTRA3; light green, POTRA4; light blue, POTRA5. Note that the drift of POTRAs 1, 2, and 3 in this x, y plane (parallel to the membrane surface) is occasionally correlated whole POTRA domain drift is not due correlated with POTRA5 movement.
The view is looking down the membrane normal toward the phospholipid surface of the membrane. The dark blue circle represents the center of the BamA β-barrel. Red, POTRA1; orange, POTRA2; khaki, POTRA3; light green, POTRA4; light blue, POTRA5. Note that the drift of POTRAs 1, 2, and 3 in this x, y plane (parallel to the membrane surface) is occasionally correlated whole POTRA domain drift is not due correlated with POTRA5 movement.
The view is looking down the membrane normal toward the phospholipid surface of the membrane. The dark blue circle represents the center of the BamA β-barrel. Red, POTRA1; orange, POTRA2; khaki, POTRA3; light green, POTRA4; light blue, POTRA5. Note that the drift of POTRAs 1, 2, and 3 in this x, y plane (parallel to the membrane surface) is occasionally correlated whole POTRA domain drift is not due correlated with POTRA5 movement.
The β-barrel and POTRA5 fragments from crystal structure 4K3B (translucent blue and orange, respectively) are aligned to the same from replica 1 simulation at time zero (translucent blue and light blue, respectively). Simulation time shown is 1–540 ns.