Abstract
Although both C-C chemokine receptor 5 (CCR5)- and CXC chemokine receptor 4 (CXCR4)-using HIV-1 strains cause AIDS, the emergence of CXCR4-utilizing variants is associated with an accelerated decline in CD4 + T cells. It remains uncertain if CXCR4-using viruses hasten disease or if these variants only emerge after profound immunological damage. We show that exclusively CXCR4- as compared to cocirculating CCR5-utilizing variants are less sensitive to neutralization by both contemporaneous autologous plasma and plasma pools from individuals that harbor only CCR5-using HIV-1. The CXCR4-utilizing variants, however, do not have a global antigenic change because they remain equivalently susceptible to antibodies that do not target coreceptor binding domains. Studies with envelope V3 loop directed antibodies and chimeric envelopes suggest that the neutralization susceptibility differences are potentially influenced by the V3 loop. In vitro passage of a neutralization sensitive CCR5-using virus in the presence of autologous plasma and activated CD4 + T cells led to the emergence of a CXCR4-utilizing virus in 1 of 3 cases. These results suggest that in some but not necessarily all HIV-1 infected individuals humoral immune pressure against the autologous virus selects for CXCR4-using variants, which potentially accelerates disease progression. Our observations have implications for using antibodies for HIV-1 immune therapy.
Keywords: HIV, Envelope, Neutralization, Co-receptor, Evolution, Resistance, Tropism
Graphical abstract
Highlights
-
•
CXCR4- as compared to coexisting CCR5-using HIV are less neutralization sensitive.
-
•
CXCR4-using virus can emerge by passaging a CCR5 HIV in the presence of plasma.
-
•
Humoral immune response may lead to the emergence of CXCR4-utilizing variants.
The emergence of HIV-1 variants that use the CXCR4 as opposed to strictly utilizing the CCR5 receptor (termed R5) has been associated with an accelerated progression to AIDS. Potentially, the emergence of CXCR4-using viruses hastens disease or individuals with profound immunosuppression have a greater likelihood of producing CXCR4-utilizing variants. We show that variants that exclusively use the CXCR4 (X4) are neutralization escape variants. Furthermore, CXCR4-using virus can emerge by passaging a R5 variant in the presence of autologous contemporaneous plasma. This suggests that in some individuals, humoral immune pressure against HIV-1 selects for CXCR4-utilizing variants, which accelerates disease progression.
1. Introduction
In nearly all cases HIV-1 infection leads to a progressive loss of CD4 + T cells, and both host immunologic factors and viral characteristics influence the rate of progression towards AIDS. Although HIV-1 can enter target cells by either using the C-C chemokine receptor 5 (CCR5) or CXC-chemokine receptor 4 (CXCR4) co-receptor after CD4 receptor engagement (Deng et al., 1996, Feng et al., 1996), presence of syncytium-inducing (SI) CXCR4-using viruses is associated with accelerated disease progression (Koot et al., 1993, Richman and Bozzette, 1994, Connor et al., 1997). In general, viruses that exclusively utilize the CCR5 receptor (termed R5 or non-syncytium-inducing (NSI)) establish initial infection, and these viruses persist throughout the course of disease (Connor et al., 1997, van't Wout et al., 1994, Schuitemaker et al., 1992). During the chronic phase of infection, some individuals have a dual-mixed (DM) population consisting of R5 viruses and CXCR4-using variants, which can either only use CXCR4 (termed X4) or be dual-tropic that can engage both co-receptors (termed dual or R5X4) (Schuitemaker et al., 1992, Connor et al., 1997). Previous studies have not clarified if CXCR4-using viruses accelerate disease progression or if faster CD4 + T cell decline facilitates the emergence of DM virus populations. Deciphering mechanisms for the emergence of CXCR4-utilizing variants will provide insights into this viral factor important for progression towards AIDS.
It is generally agreed that R5 variants give rise to R5X4 viruses, which can further evolve to X4 HIV-1 (Pastore et al., 2004). Specific modifications in the envelope glycoprotein (env) variable loop 3 (V3), which binds the co-receptor, are required for the transition from R5 to X4 virus (Huang et al., 2007, Chesebro et al., 1996, Cocchi et al., 1996). The selective pressures which induce the transition from CCR5 to CXCR4 usage during the natural course of infection remain poorly understood. It has been suggested that CXCR4-using viruses potentially emerge because of a reduced number of susceptible CCR5 + target cells or replication differences in various immune cells among the variants that use different co-receptors (Regoes and Bonhoeffer, 2005, Ribeiro et al., 2006). Indeed, individuals heterozygous for a 32 base pair deletion in CCR5 gene (CCRΔ32), who have lower levels of the CCR5 receptor on their cells, have an increased likelihood of harboring CXCR4-using viruses compared to people with both wild-type alleles (Brumme et al., 2005, Henrich et al., 2014). Other non-mutually exclusive hypotheses argue that CXCR4-utilizing variants arise because of random mutations, and these new viruses persist because declining adaptive and innate immune responses fail to clear the less fit dual-tropic and/or X4 HIV-1. Previous studies have also suggested that neutralizing antibody (nAb) pressure do not drive co-receptor switching (Trkola et al., 1998, Montefiori et al., 1998, Cecilia et al., 1998, Lacasse et al., 1998). Other investigations have argued that CXCR4-using viruses are highly neutralization susceptible, and the variants that can use the CXCR4 receptor only emerge after collapse of the humoral immune response (Bunnik et al., 2007, Ho et al., 2007). Within weeks to months after HIV-1 acquisition, infected individuals develop nAbs against their autologous viruses, but the virus changes its env to escape this humoral response (Richman et al., 2003, Wei et al., 2003). Neutralization resistance is frequently associated with modifications in and around the env variable portions, such as the env V1–V2 and V3 loop (Sagar et al., 2006, Rong et al., 2009, Moore et al., 2009). In a cohort of HIV-1 subtype C (HIV-1C) infected individuals, we have previously shown that the env quasispecies diversity was significantly higher among individuals with DM as compared to strictly R5 viruses (Lin et al., 2012). In addition, we observed that all HIV-1C X4 as compared to R5 variants had either a 2 amino acid insertion prior to or basic amino acid substitutions in the generally invariant V3 loop crown. Together, these observations led us to hypothesize that difference in host antibody responses drive the greater env diversity observed in individuals with DM as compared to R5 virus populations, and the V3 loop genotypic signatures associated with X4 variants potentially modify neutralization susceptibility. We also postulated that the emergence of specific host antibodies in some but not all individuals force env sequence and structural changes. These env modifications influence co-receptor switching, and the continual presence of the specific host antibodies allows for the persistence of CXCR4-using viruses. In this study, we examined the role of nAbs on HIV-1C co-receptor switching.
2. Materials and Methods
2.1. Subjects
HIV-1 envs were isolated from plasma collected from women in the Mashi study previously shown to harbor DM virus (Thior et al., 2006).
2.2. Ethics Statement
The use of samples was approved by the institutional review boards at the Harvard School of Public Health, Partners Healthcare Systems, Boston University, and by the Botswana Ministry of Health. Investigators who analyzed the data had no contact with the women enrolled in the Mashi study, and thus all analysis was done in an anonymous fashion.
2.3. Viruses
The env expression fragments were generated as previously described using bulk nested RT-PCRs (Lin et al., 2010). Briefly, RNA was isolated from 140 μl of individual plasma samples using QIAamp Viral RNA mini kit (Qiagen). For each sample, 1st round RT-PCR was performed in triplicate to limit re-sampling (Liu et al., 1996). In order to minimize possible recombination associated with bulk PCR cloning (Salazar-Gonzalez et al., 2008, Keele et al., 2008), the RT-PCR reactions were performed with long extension times, limited cycle numbers, and use of proofreading polymerase (Lahr and Katz, 2009, Judo et al., 1998, Meyerhans et al., 1990, Fang et al., 1998). Each reaction was done with 4 μl of the extracted viral RNA, SuperScript III RT/Platinum Taq HiFi mix (ThermoFisher Scientific), and an extension time of 7 min for each of the 20 cycles. These 1st round reactions were combined and cloned using TOPO TA cloning (Invitrogen). A CMV promoter was attached to each individually isolated env clone using overlap PCR. All primers and amplification conditions have been described previously (Lin et al., 2010). All env sequences have been submitted to Genbank previously (accession numbers KF770248–KF770455, KX060565–KX060578) (Lin et al., 2012). Pseudotyped viruses were generated by co-transfecting HEK293T cells with the CMV promoter PCR product attached to the amplified env and an env deficient HIV-1 backbone (pNL4-3R-E-) plasmid expressing luciferase using the Fugene protocol (Roche Molecular Biochemicals).
2.4. Generation of Replication Competent Viruses
Replication competent recombinant viruses were generated with minor modifications from a previously described protocol (Chatziandreou et al., 2012). In the majority of cases, we found that full-length HIV-1C envs did not yield replication competent recombinant viruses when incorporated into a NL4-3 (HIV-1B) or Q23-17 (HIV-1A) background (Etemad et al., 2014, Pena-Cruz et al., 2013). Because the same envs yielded infectious pseudotypes, we assumed that incompatible cis interactions between the env tail and matrix protein accounted for the lack of infectious virus (Freed and Martin, 1995). To overcome this issue, we generated a plasmid pCMV-Q23-PBS → LTRΔEnvEcto in which the env ecto-domain was replaced with a uracil biosynthesis (URA3) gene. Briefly, the env ecto-domain in pCMV-Q23-17-PBS → LTR was replaced with URA3 PCR product, which was amplified with the primers AGAAAGAGCAGAAGACAGTGGCAATGATTAATTAAACCACCTTTTCAATTCATC and AAGCCTCCTACTATCATTATCTGATATAATTAAATTGAAGC from pRS316. The underlined portions correspond to primer sequences used to amplify env ectodomains (HXB2 env amino acid numbering 1–692) from subject samples, and the sequences in italics contain a PacI restriction site. Different env ecto-domains were shuttled into Q23 plasmid by transforming yeast with pCMV-Q23-PBS → LTRΔEnvEcto linearized with PacI and amplified env ecto-domain PCR products from subject samples. Replication competent viruses were generated by co-transfecting HEK293T cells with a plasmid containing the subject env ecto-domain within pCMV-Q23-17-PBS → LTR and another plasmid with Q23–17 sequences from 5′ LTR to early portion of gag, pCMV-Q23-17-LTR → Gag4 (Chatziandreou et al., 2012). All virus stocks were collected 48 h post transfection and titered on TZM-bl cells.
2.5. Reagents
The pNL4-3R-E- plasmid, U87 and TZM-bl cell lines, and VRC01 and 10E8 monoclonal antibody (mAb) were obtained through the NIH AIDS Reagent Program. The 293T cells were obtained from American Type Culture Collection. The PGT128 and 447-52D mAbs were provided by International AIDS Vaccine Initiative and Dr. Susan Zolla-Pazner respectively. The CD4-Ig was produced by gene synthesis and cloning into pcDNA3.1 (Invitrogen).
2.6. Co-receptor Phenotype
Co-receptor usage was determined by assessing entry capacity in the presence and absence of a co-receptor antagonist on U87 cells expressing CD4 and either CXCR4 or CCR5 as previously described (Lin et al., 2010). To examine replication, U87 cells were incubated with virus and polybrene (8 μg/ml) for approximately 16 h. Post-incubation, cells were washed to remove virus. Culture supernatants were examined for p24gag antigen 5 days post-infection. In all cases NL4-3 (X4) and YU-2 (R5) were used as controls.
2.7. HIV-1 gp120 Expression and Purification
The codon-optimized gene encoding the R5 DM268M full-length gp120 was synthesized with a C-terminal Avi-6xHis tag and cloned into the pcDNA3.1(−) expression vector (Life Technologies). The X4 V3 (DM268Y), differed from the R5 V3 (DM268M) by 5 amino acids; these modifications and the CD4 knock-out mutation D368R were introduced by sequential site-directed mutagenesis using the TagMaster Site-Directed Mutagenesis Kit (GM Biosciences) (Lin et al., 2012). The gp120 proteins were expressed by transient transfection using the expi293 system (Life Technologies). Six days post transfection, culture supernatants were harvested, filtered, and buffer-exchanged into PBS, and then purified using HisTALON Gravity Column Purification Kit (Clontech Laboratories).
2.8. Neutralization Assay
Infection of TZM-bl cells in the absence and presence of 2-fold serial dilutions was used to estimate the 50% inhibitory concentration (IC50) as previously described (Etemad et al., 2009). All reported IC50s are mean estimates from a minimum of 3 independent assays. For virus/plasma combinations that failed to achieve more than or equal to 50% inhibition at the highest tested dilution, they were assigned values of 25, which is the midpoint between 0 and highest tested dilution 50. For virus/mAb combinations, a value equal to the highest tested concentration was assigned when 50% inhibition could not be achieved. For the neutralization competition assays, plasma at various dilutions was incubated with media alone or 500 μg/ml of gp120 for 1 h at 37 °C. Virus was added, and wells were incubated for an additional 30 min. Then TZM-bls were added and infections were allowed to proceed for 48 h. Relative light units generated in the absence (positive wells) as compared to the presence of plasma were used to estimate % inhibition at specific plasma dilutions. For estimating inhibition in the presence DM268M-R5V3 or DM268M-X4V3 gp120, the positive wells also contained the respective gp120. In all cases, plasmas showed no neutralization capacity against NL4-3 pseudovirions with a vesicular stomatitis virus (VSV) envelope G protein (data not shown). Thus, observed neutralization was specific against the HIV-1 envelope.
2.9. ELISA
Plates were coated with gp120 antigen in PBS (pH 7.4) at 2 μg/ml overnight at 4 °C, blocked with 1% BSA in PBS for 1 h at 37 °C, and then incubated with serially diluted plasma or antibodies in blocking buffer for 1 h at 37 °C. Horseradish peroxidase (HRP)-conjugated goat anti-human IgG Fc antibody (Jackson ImmunoResearch Laboratories Inc.) was added in blocking buffer at 1:10,000 for 1 h at 37 °C. Plates were washed between each step with 0.1% Tween 20 in PBS and developed with 3,3′,5,5′-tetramethylbenzidine (TMB) (Life Technologies) for 20 min at room temperature. 1 M sulfuric acid was added to terminate the reaction and the plates were read at 450 nm.
2.10. Virus Passage
Viruses were passaged on CD4 + T cells in the presence of increasing concentrations of heat inactivated autologous plasma. Peripheral blood mononuclear cells (PBMCs) were isolated from HIV-1 negative blood donors' buffy packs using Ficoll-Hypaque density centrifugation method. CD4 + T cells were isolated from the PBMCs using magnetic bead isolation (Stem Cell Technologies). Around 2 × 106 phytohemagglutinin (PHA) and interleukin-2 (IL-2) activated CD4 + T cells were exposed to infectious virus in the presence of 20 μg/ml diethylaminoethyl (DEAE) dextran. Plasma was added after infections were established, generally two days after infection. Cultures were monitored for viral replication twice a week using a p24gag antigen assay (Perkin Elmer). The cultures were fed twice weekly with activated CD4 + T cells, and plasma concentration was doubled if virus showed continued growth.
2.11. Statistical Analysis
In the majority of cases, individual env variant's IC50s was a mean of 3 independent assays. Comparisons within an individual were done using Wilcoxon rank sum test. For comparisons across multiple individuals, the median IC50 for strains with specific co-receptor usage was determined for each subject. Inter-subject comparison were done using Kruskal Wallis test, and pair-wise inter-subject comparisons were done using the Wilcoxon rank sum test. The matched pairs Wilcoxon rank sum test was used when all individuals had variants with the co-receptor usage being compared. All statistical analysis were done using Stata version 8.0 (Stata Corporation). All p-values are based on two sided tests.
3. Results
3.1. Subjects
Previously, we determined the co-receptor usage in 148 HIV-1C infected women in Botswana, and we found that 22 of them harbored DM populations while 126 had strictly R5 virus (Lin et al., 2011). Sequence characteristics and co-receptor use of individual envs were examined from 10 of the DM samples that showed relatively high CXCR4 usage by the phenotypic assay (Lin et al., 2012). Examination of individuals that harbored X4 variants were prioritized because X4 as compared to co-existing R5 and R5X4 envs were demonstrated to harbor unique genotypic features (Lin et al., 2012). In addition, individual envs were not examined from DM samples that demonstrated low amount of CXCR4 usage in the co-receptor assay because it would likely require more extensive cloning to isolate CXCR4-using variants. Only 5 from the original 148 samples fulfilled these criteria. We chose to examine an additional 2 samples with only co-circulating R5 and R5X4 variants. We assessed neutralization susceptibility of a median of 11 co-circulating envs (range: 9–18) from these 7 different women (Table 1). The co-existing env populations consisted of X4 and R5 variants in 3 (DM8, DM178, and DM268), R5X4 and R5 variants in 2 (DM146 and DM269), R5X4 and X4 variants in 1 (DM173), and all three co-receptor phenotypes in 1 (DM159) of the individuals. An additional 6 DM8 clones failed to yield other X4 envs. Intra-individual env variants with the different co-receptor usage were phylogenetically related, which ruled out contamination and super-infection (Fig. S1). Even though all 88 primary envs examined in this study were generated using bulk-cloning PCR as described previously (Lin et al., 2010, Lin et al., 2012), all variants had unique sequences. Thus, we avoided re-sampling the same variant multiple times, which is a bias often associated with the bulk PCR cloning technique used to isolate the envs in this study (Salazar-Gonzalez et al., 2008, Keele et al., 2008).
Table 1.
Subject demographics and patient characteristics.
| Subject | Plasma HIV (copies/ml) |
CD4 count (cells/μl) |
Previous ARTa | Env variants |
||
|---|---|---|---|---|---|---|
| R5 | Dual | X4 | ||||
| DM8 | 522,000 | 43 | N | 10 | 0 | 1 |
| DM146 | 653,000 | 172 | N | 10 | 8 | 0 |
| DM159 | 154,000 | 116 | N | 5 | 2 | 3 |
| DM173 | 1290 | 316 | N | 0 | 6 | 3 |
| DM178 | 230,000 | 21 | Y | 8 | 0 | 6 |
| DM268 | 333,000 | 375 | Y | 5 | 0 | 9 |
| DM269 | 10,600 | 319 | Y | 9 | 3 | 0 |
Women were sampled either prior to or a minimum of 6 months after the discontinuation of antiretroviral therapy (ART) because of virological failure.
3.2. X4 Are Less Neutralization Susceptible to Contemporaneous Plasma
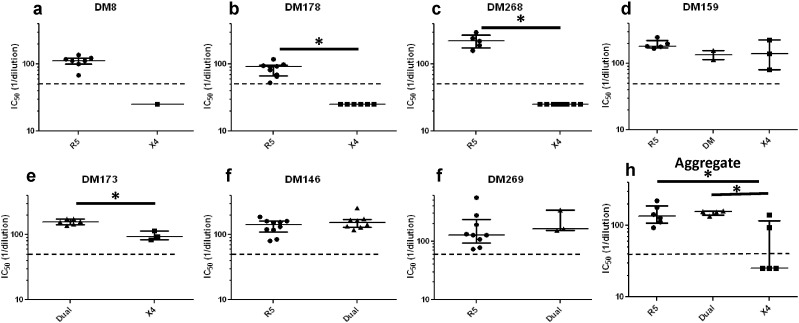
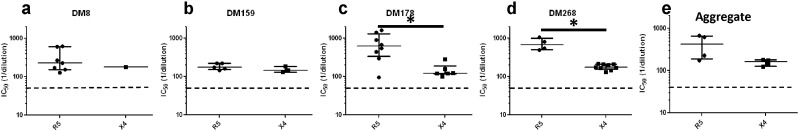
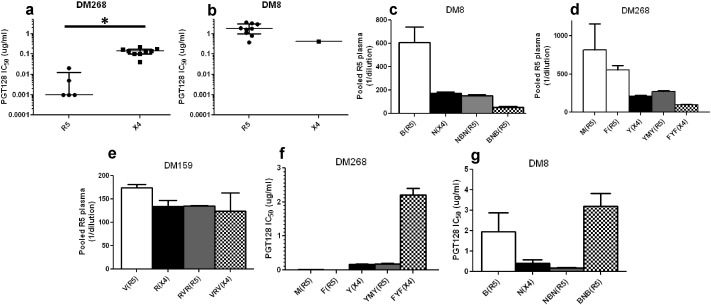
To assess neutralization sensitivity differences among 88 co-circulating variants with different observed co-receptor usage, we first examined susceptibility to contemporaneous autologous plasma. In 5 individuals, X4 strains were less sensitive to contemporaneous autologous plasma as compared to co-existing R5 (DM8, DM178, DM268, DM159) or R5X4 (DM173) viruses (Fig. 1a–e). Indeed, in 3 individuals (DM8, DM178, and DM268), the X4 envs were not neutralized above 50% at the highest tested plasma dilution (1:50) (Fig. 1a–c). These virus-plasma combinations were assigned a value of 25, which is the mid-point value between 0 and 50, for statistical comparisons. In 3 individuals (DM178, DM268, and DM173) differences were statistically significant (p < 0.05, Wilcoxon rank sum test), while in the other 2 subjects (DM8 and DM159), limited number of envs with different co-receptor usage potentially limited the power to detect significant differences. In 2 individuals (DM146 and DM269) co-circulating R5 and dual-tropic envs showed similar neutralization sensitivity (p > 0.05, Wilcoxon rank sum test) (Fig. 1f–g). Aggregate comparisons were done by estimating a median IC50 for each individual's envs that used different co-receptor. In aggregate, neutralization IC50s to autologous contemporaneous plasma was significantly different among R5, R5X4, and X4 envs (p = 0.03, Kruskal Wallis test). The median neutralization susceptibility to contemporaneous autologous plasma of the X4 viruses (median IC50: 25.0, range: 25.0–140.7) was around 5 fold lower compared to R5 (median IC50: 136.0, range: 94.2–222.7, p = 0.03, Wilcoxon rank sum test) and R5X4 viruses (median IC50: 156.5, range: 135.2–160.8, p = 0.03, Wilcoxon rank sum test) (Fig. 1h).
Fig. 1.
The X4 as compared to co-circulating R5 and R5X4 envs are more neutralization resistant to autologous contemporaneous plasma. (a–g) Figures show neutralization IC50s for R5 (circles), dual-tropic (triangles), and X4 (boxes) viruses against autologous contemporaneous plasma. Symbols represent one of the 88 independent envs from the 7 subjects labeled above each graph. Y-axis shows the IC50 against autologous contemporaneous plasma as reciprocal dilution. Figure h shows the aggregate comparisons in all subjects, and each symbol shows the median IC50s within a subject for envs with the defined co-receptor phenotype. In every case, the long line represents the median of the dot plots, and the whiskers show the interquartile range. Star indicates statistically significant difference (p < 0.05). The dashed line shows the highest tested plasma dilution (1:50). Symbols below the dotted lines are envs that were not neutralized by at least 50% at the highest tested plasma dilution. For statistical comparisons, these envs were assigned a midpoint value between 0 and 50.
In preliminary data, we also found an HIV-1B infected individual also harbored X4 as compared to CCR5-using variants that were significantly less sensitive to neutralization by autologous contemporaneous plasma (Fig. S2). This suggests our observations are not specific to HIV-1C infections only. Thus, in every individual with isolated X4 viruses (n = 6), the X4 as compared to co-existing R5 and/or R5X4 variants had greater neutralization resistance to autologous contemporaneous plasma (Figs. 1 and S2). Previous studies have noted similar IC50 fold changes among longitudinally collected viruses to sequential autologous plasma (Richman et al., 2003). In addition, the observed neutralization response against contemporaneous viruses in the individuals examined in this study is not necessarily unique because recent work shows that plasma and isolated antibodies neutralize some autologous contemporaneous viruses to a similar extent, which subsequently provides pressure for envelope evolution (Moody et al., 2015). In aggregate, this suggests that the × X4 as compared to co-existing CCR5-using viruses have both statistical and biological meaningful differences in neutralization susceptibility. Thus, X4 variants are potentially neutralization escape variants to the antibodies present in the contemporaneous autologous plasma.
3.3. X4 Are More Resistant to Heterologous Pooled Plasma
We next hypothesized that X4 envs are escape variants from antibodies often present in individuals that harbor only CCR5-using viruses. To examine this possibility, we compared sensitivity of X4 and R5 strains to heterologous pooled R5 plasma, which was generated by pooling plasma from 5 women documented to harbor R5 viruses only (Lin et al., 2012). We examined 47 envs from the 4 individuals (DM8, DM159, DM178, and DM268) in which the median IC50 against autologous contemporaneous plasma was lower for the X4 as compared to co-circulating R5 strains. We did not examine variants from DM146, DM173, and DM269 because in these individuals we did not isolate co-existing R5 and X4 strains. In all 4 individuals, the X4 viruses demonstrated decreased sensitivity to this pooled R5 plasma compared to the co-existing CCR5 utilizing viruses (Fig. 2a–d). Significant differences (p < 0.05, Wilcoxon rank sum test) were observed in 2 cases (DM178 and DM268) but not in the other 2 individuals (DM8 and DM159) with a limited number of envs with different co-receptor usage. In aggregate, X4 (median IC50: 161.7, range: 122.8–179.6) as compared to R5 (median IC50: 425.3, range: 174.5–684.8) viruses showed a statistical trend of around 3 fold greater neutralization resistance to the pooled R5 plasma (p = 0.1, paired Wilcoxon rank sum test) (Fig. 2e). This suggests that X4 as compared to co-existing R5 viruses are less neutralization susceptible to antibodies commonly circulating in individuals that harbor only R5 variants.
Fig. 2.
The X4 as compared to co-circulating R5 are more neutralization resistant to heterologous pooled R5 plasma. (a–d) Graphs show neutralization IC50s (reciprocal plasma dilution) for R5 (circles) and X4 (boxes) viruses against heterologous pooled R5 plasma for 4 subjects. Each symbol represents one of the 47 independent envs from the 4 individuals shown above the graph. Figure E shows the aggregate comparisons in the 4 subjects, and each symbol shows the median IC50s for the R5 or X4 envs within an individual. In every case, the long line represents the median of the dot plots with the whiskers showing the interquartile range. Star indicates statistically significant difference (p < 0.05). The dashed line shows the highest tested plasma dilution (1:50).
3.4. X4 Variants Remain Sensitive to Antibodies Targeting env Non-co-receptor Binding Sites
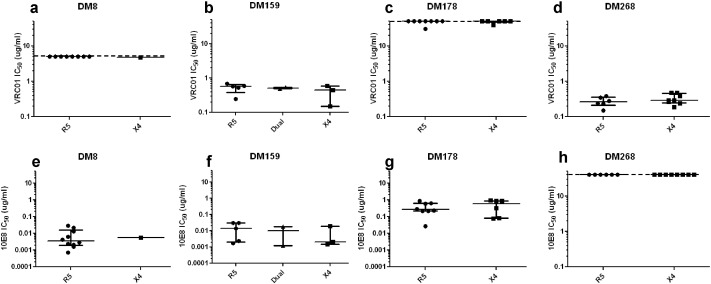
It is possible that mutations in the X4 viruses compared to the R5 variants leads to a “global” antigenic change, similar to some previously described strains (del Prete et al., 2009, Roederer et al., 2013). If the X4 as compared to the R5 variants have an overall antigenic difference, they may have different neutralization susceptibility to antibodies targeting env regions not implicated in co-receptor binding. To explore this possibility, we assessed sensitivity to two broadly neutralizing antibodies (bnAbs), VRC01, which targets the CD4 binding site (CD4bs) (Wu et al., 2010, Zhou et al., 2010), and 10E8, which binds the membrane proximal external region (MPER) (Huang et al., 2012). In individual subjects, the X4 as compared to R5 viruses demonstrated similar sensitivity to neutralization (p > 0.05, Wilcoxon rank sum test) to VRC01 (Fig. 3a–d) and 10E8 (Fig. 3e–h). Although in some individuals the highest tested bnAb concentration failed to yield 50% inhibition, this lack of neutralization was observed in both the X4 and co-existing R5 variants. In these cases, virus-antibody combination was assigned a value equivalent to the highest tested concentration for statistical comparisons. In aggregate, the median VRC01 and 10E8 IC50 of an individual's X4 and co-existing R5 variants were not significantly different (p > 0.05, paired Wilcoxon rank sum test). Thus, the co-existing X4 and R5 variants demonstrated equivalent sensitivity to antibodies that target env regions not involved in co-receptor binding, which suggests that changes that confer co-receptor switching do not lead to overall structural change in CD4 binding site or domains close to the membrane. Comparing sensitivity to other bnAbs that target unique sites, such as the V1–V2 loop or the interface between the trans-membrane domain and surface unit, will need to be examined to assess structural changes in other env regions.
Fig. 3.
The X4 variants do not have a “global” antigenic change. Neutralization IC50s for R5 (circles), dual-tropic (triangles), and X4 (boxes) viruses to VRC01 (a–d) and 10E8 (e–h). Y-axis shows the bnAb concentration in μg/ml. Dotted line shows the highest tested antibody concentration. For statistical comparisons, virus antibody combination that failed to yield > 50% neutralization at the highest tested antibody concentration were assigned an IC50 value equivalent to the highest tested antibody concentration. In every case, the long line represents the median of the dot plots with the whiskers showing the interquartile range.
3.5. Infected Hosts Develop Antibodies Against X4 Variants
A study using a simian-human immunodeficiency virus with a subtype B env (SHIV-1B) has suggested that CXCR4-using variants are exquisitely neutralization sensitive, and these viruses potentially arise in setting where there are limited nAb responses in the infected host (Ho et al., 2007). If CXCR4 viruses emerge after immunological collapse then infected hosts will potentially not be able to develop antibody responses against X4 variants over time. We assessed sensitivity to an autologous plasma sample collected around 6 months after the time point at which the DM8 X4 and R5 variants were isolated to assess this notion. The DM8 X4 compared to the co-circulating R5 variants demonstrated similar sensitivity to plasma collected 6 month after env isolation (Fig. S3a). Importantly, all R5 and the one X4 env were more susceptible to the later compared to the contemporaneous plasma sample (p = 0.008, paired Wilcoxon rank sum test) (Fig. S3b). Autologous plasma samples from later time-points were not available from other individuals to further examine if infected hosts could generate nAbs against previously circulating envs after the emergence of DM virus populations. These observations suggest that subject DM8 with a DM virus population continues to develop humoral responses over time against previously co-circulating R5 and X4 variants.
3.6. Neutralization Susceptibility Differences Are Influenced by env V3 Loop Changes
Our results suggest that X4 variants are relatively insensitive to specific antibodies present in autologous plasma and heterologous antibodies in individuals with only R5 circulating variants. We hypothesized that these antibodies potentially target the env V3 loop because the V3 loop is the primary determinant for co-receptor usage (Cann et al., 1992). We compared sensitivity of co-existing X4 and R5 variants to the bnAb, PGT128, because V3 loop stem sequence partially influences susceptibility to this bnAb (Walker et al., 2011, Pejchal et al., 2011). In DM268, X4 viruses (median IC50: 0.14 μg/ml, range: 0.04–0.22) were > 100 fold less neutralization susceptible to PGT128 compared to co-circulating R5 (median IC50: 0.001 μg/ml, range: 0.001–0.02) variants (p = 0.003, Wilcoxon rank sum test) (Fig. 4a). In contrast, neutralization sensitivity to PGT128 for the one DM8 X4 variant (IC50: 0.41 μg/ml) was within the range for the co-circulating R5 variants (median IC50: 1.8 μg/ml, range: 0.37–3.67) (Fig. 4b). None of the DM159 and DM178 X4 and R5 variants were inhibited > 50% at the highest tested PGT128 concentration (5 μg/ml), and thus an IC50s could not be calculated for comparisons.
Fig. 4.
Neutralization susceptibility differences among X4 and R5 viruses are influenced by env V3 loop. Figures a–b show mean IC50s (μg/ml) to PGT128 among co-circulating variants with different co-receptor usage in DM268 (a) and DM8 (b). Figures c–g show IC50 against pooled R5 plasma (reciprocal dilution) (c–e) and PGT128 (μg/ml) (f–g) for the original R5 (white), original X4 (black), env with a R5 V3 loop in a X4 background (gray) or X4 V3 loop in a R5 background (checkers). In each graph, the subject is denoted above. Label below the x-axis denotes the env clone followed by the observed co-receptor usage in parenthesis. Star denotes significant p-value for the comparison (p < 0.05, Wilcoxon rank sum test). DM268 chimeric env MYM was not infectious (Lin et al., 2012).
To further examine the role of V3 loop in the observed neutralization differences among co-circulating R5 and X4 variants, we generated V3 loop chimeric envs. In 3 subjects (DM8, DM159, and DM268) with documented differences in neutralization sensitivity among co-existing X4 and R5 variants, we exchanged V3 loops between a R5 and X4 variant. The chimeric DM178 envs with V3 loop exchanges did not yield infectious virus. Swapping the V3 loops resulted in co-receptor changes in every case other than in DM8 (Fig. 4c–e) (Lin et al., 2012). The chimeric envs with X4 V3s were less sensitive to pooled R5 heterologous plasma compared to the envs containing the R5 V3 loops in every case. The differences, however, were relatively small especially compared to the susceptibility differences among the original envs. Furthermore, in both DM268 and DM8, introduction of an X4 V3 loop within the R5 background yielded a virus with greater resistance to PGT128 compared to the original env or the recombinant with a R5 V3 loop (Fig. 4f–g). In aggregate, these results suggest that the V3 loop potentially influences the observed neutralization differences among co-circulating R5 and X4 variants.
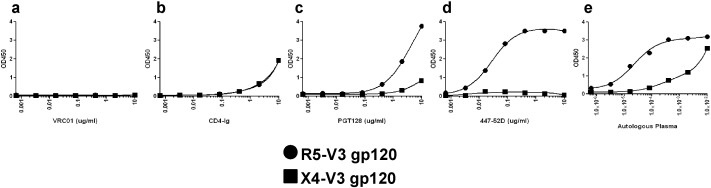
ELISAs were used to further examine V3 directed antibody binding variation to co-existing DM268 X4 and R5 variants. We only assessed DM268 envs because of the observed sensitivity difference to bnAb, PGT128. We generated gp120 proteins based on the predicted amino acids in a DM268 R5 variant (clone M). One gp120 contained an R5 V3, DM268-M-R5V3, and the other harbored an X4 V3 from clone Y, DM268-M-X4V3. Both contained a D368R mutation to avoid CD4 interference (Olshevsky et al., 1990). As expected, the CD4 binding site bnAb, VRC01, showed no binding to either gp120 because of the D368R mutation (Fig. 5a). On the other hand, both gp120s showed relatively equivalent weak binding to CD4-Ig (Fig. 5b), which is similar to what has been shown with other gp120s containing the D368R mutation (Jia et al., 2016). The DM268-M-R5V3 gp120 as compared to DM268-M-X4V3, however, showed markedly greater binding to PGT128 (Fig. 5c), V3 loop crown antibody, 447-52D (Fig. 5d), and contemporaneous autologous serum (Fig. 5e) (Gorny et al., 1992). Although, 447-52D demonstrated ELISA binding, this mAb was unable to neutralize any DM268M variant at the highest tested concentration (5 μg/ml) (data not shown). These ELISA results suggest that the DM268 plasma harbors anti V3 loop antibodies that preferentially bind R5 but not X4 envs.
Fig. 5.
Co-circulating R5 and X4 viruses have different reactivity to anti-env V3 loop antibodies and autologous plasma. Figure a thru e show ELISA binding profiles of the DM268_D368R gp120 with either a R5 V3 loop (circle) or X4 V3 loop (square) to VRC01 (a), CD4-Ig (b), PGT128 (c), 447-52D (d), and autologous contemporary plasma (e). The y-axis shows the OD and the x-axis shows the antibody concentration or plasma dilution.
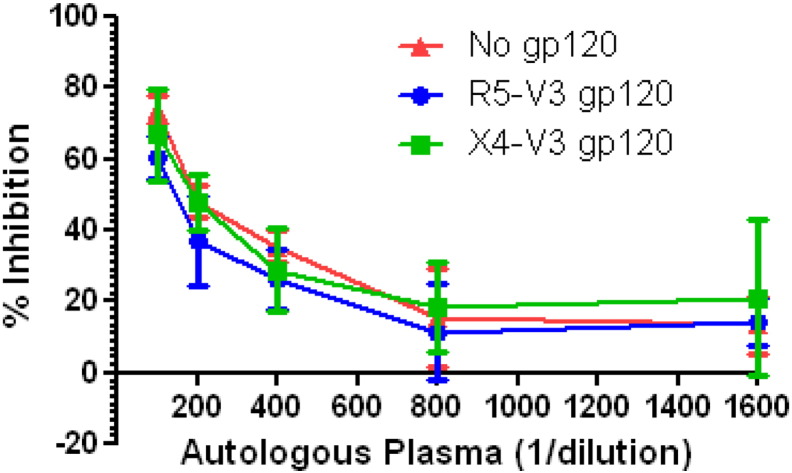
Neutralization competition experiments were carried out to assess if the presence anti V3 loop antibodies in the autologous plasma is responsible for the differential neutralization of DM268 R5 and X4 variants. If anti V3 loop antibodies that preferentially bind R5 but not X4 envs were primarily responsible for neutralization susceptibility differences among DM268 X4 and R5 variants, it would be expected that gp120s with R5 but not X4 V3 would prevent autologous contemporaneous plasma from neutralizing the virus. In the presence of DM268-M-R5V3 gp120 (reciprocal plasma dilution IC50: 132.1) neutralization of the DM268M virus required greater amount of plasma compared to in the presence of DM268-M-X4V3 gp120 (reciprocal plasma dilution IC50: 168.3) or the absence of gp120 (reciprocal plasma dilution IC50: 200.0) (Fig. 6). These estimated IC50 differences, however, were not statistically significantly different. This suggests that even though the plasma contains anti V3 loop antibodies that preferentially bind R5 but not X4 gp120s, these antibodies had a small impact in neutralizing a DM268 R5 strain.
Fig. 6.
gp120s with different env V3 loops fails to prevent neutralization. Percent inhibition observed in the absence or presence of DM268_D368R gp120 with R5 V3 or X4 V3. The y-axis shows the percent inhibition and the x-axis shows the reciprocal plasma dilution.
3.7. Emergence of CXCR4-using Virus With Serial Passage in Plasma
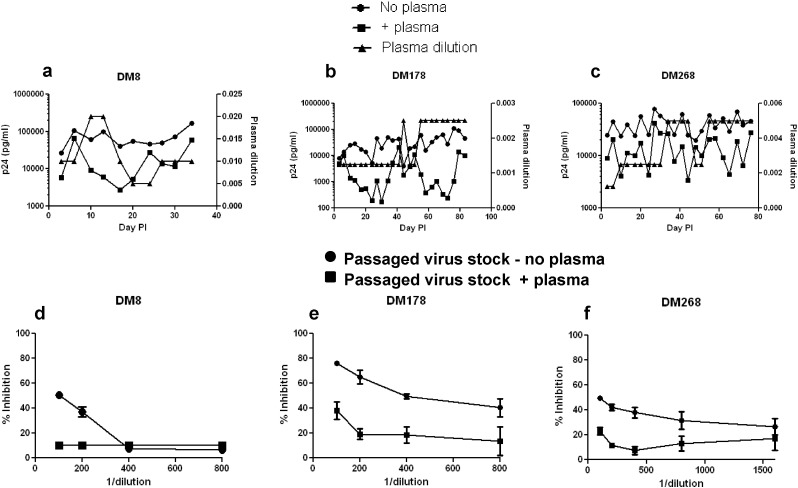
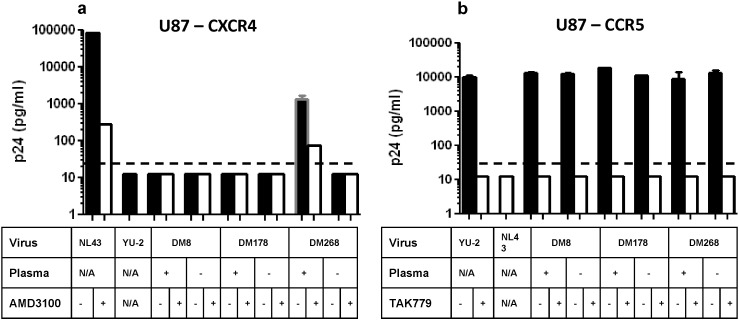
To understand if CXCR4-using variants emerge from R5 viruses as a consequence of nAb pressure, we independently passaged a neutralization sensitive R5 variant from 3 different subjects (DM8, DM178, and DM268) in activated CD4 + T cells in the absence or presence of heat inactivated contemporaneous autologous plasma (Fig. 7a–c). While all the previous experiments used pseudovirions with full-length envs, replication competent recombinant viruses were used in these passages. These replication competent recombinant viruses incorporated an individual's neutralization sensitive R5 env ecto-domain rather than the full-length env. Incorporation of full-length envs within a heterologous HIV-1 backbone did not yield infectious virus. In all 3 cases, virus passaged in the presence as compared to the absence of autologous plasma yielded virus supernatants that were neutralization resistant (Fig. 7d–f). Passage of DM268M R5 variant in the presence but not the absence of serially increasing amounts of contemporaneous autologous plasma led to the emergence of a CXCR4-using virus (Fig. 8a–b). The day 75 DM268 virus supernatant passaged in the presence as opposed to the absence of autologous plasma replicated in U87–CXCR4 cells that lacked the CCR5 receptor (Fig. 8a). Replication decreased in the presence of CXCR4 inhibitor, AMD3100, suggesting that cell entry was mediated by engaging the CXCR4 co-receptor. CXCR4 usage was highly inefficient because day 75 plasma passaged DM268 virus supernatant yielded nearly 10 fold less HIV-1 antigen (p24gag) in the U87–CXCR4 as compared to U87–CCR5 cells (Fig. 8b). Similar results were observed with day 68 DM268 virus supernatant passaged in the presence of autologous plasma (data not shown). In contrast, all other virus supernatants passaged either in the absence or presence of plasma infected U87–CCR5 cells but not U87–CXCR4 cells.
Fig. 7.
Passage in the presence of autologous contemporaneous plasma leads to neutralization resistant virus. Figure a thru c show passage of DM8 clone S (a), DM178 clone A (b), and DM268 clone M (c) in the absence (circle) or presence (square) of autologous contemporaneous plasma. In each graph, the x-axis shows the days post-infection. The left y-axis shows the estimated p24 concentration in the culture and the right y-axis shows contemporaneous autologous plasma dilution in the culture (triangles). Figures d thru f show neutralization susceptibility of passaged virus stock to contemporaneous autologous plasma. The y-axis shows the percent inhibition and the x-axis shows the reciprocal of the autologous plasma dilution. Any virus/plasma combination that did not show a positive inhibition value was assigned a % inhibition of 10% to improve visualization on the graph. Virus stocks passaged in the presence (square) or absence of (circle) of autologous plasma were from day 34 (DM8) (d), day 92 (DM178) (e), and day 75 (DM268) (f) post-infection.
Fig. 8.
A CXCR4-using virus emerged with passage in the presence as opposed to the absence of autologous plasma in DM268. Replication in U87-CD4-CXCR4 (a) and U87-CD4-CCR5 (b) cells for day 34 (DM8), day 92 (DM178), and day 75 (DM268) virus stocks. The table below indicates whether the virus stock was from passage in the presence or absence of autologous plasma. The table also shows replication observed in the presence (white bars) or absence (black bars) of specific CXCR4 (AMD3100) and CCR5 (TAK779) inhibitors. Graphs also show replication of control viruses NL43 (X4) and YU-2(R5). Each bar shows standard error from 2 independent measurements. Dotted line shows the limit of detection (25 pg/ml). Cultures in which the p24 antigen level was below the limit of detection were assigned a value midpoint between 0 and 25 to improve visualization on the graph.
We hypothesized that the day 75 DM268 virus stock passaged in the presence of plasma (DM268 + plasma-d75) contained a possible mixture of R5, R5X4, and X4 viruses. To efficiently isolate the CCR5- versus CXCR4-using viruses in DM268 + plasma-d75, we examined viruses in the U87–CXCR4 and U87–CCR5 cell culture supernatants. We reasoned that the U87–CXCR4 and the U87–CCR5 cell culture supernatants should only contain CXCR4- and CCR5-utilizing viruses respectively. Interestingly, the culture supernatants from the U87–CCR5 and U87–CXCR4 cells exposed to DM268 + plasma-d75 showed identical env bulk sequences. This suggests that passage in the presence of autologous plasma led to the emergence of a neutralization resistant R5X4 variant with relatively inefficient CXCR4 usage. Surprisingly, this R5X4 variant had an amino acid change at position 88 (HXB2 numbering), which introduced a predicted asparagine (N) linked glycosylation site (PNGS) (Fig. S4). In contrast, passage in the absence of plasma generated virus stocks that contained a mixture of amino acids at 2 positions but was otherwise invariant from the starting input virus. Interestingly, all other R5 and X4 DM268 variants (n = 17) previously isolated from this individual harbored the PNGS at position 88. Site directed mutagenesis could not be used to assess if this modification would change co-receptor usage in other strains because all other isolated R5 envs contained the position 88 PNGS. The effect of the position 88 change on co-receptor usage, however, is context dependent because none of the other previously isolated DM268 env variants with a position 88 PNGS were able to use both the CCR5 and CXCR4 co-receptor for cell entry. These observations imply that passage in the presence of autologous plasma led to the emergence of a PNGS at position 88, which subsequently conferred CXCR4 usage.
4. Discussion
This study demonstrates that X4 variants are more neutralization resistant compared to R5 and R5X4 viruses and provides a potential biological mechanism for HIV-1 co-receptor switching. In multiple subjects, we show that co-existing X4 as compared to R5 variants are more resistant to antibodies present in autologous contemporaneous and heterologous pooled plasma from individuals with only R5 strains. This decreased neutralization sensitivity did not result from changes that led to a global antigenic change. Thus, this argues that X4 viruses are more resistant to certain antibodies, likely against the env V3 loop, generated by the infected individual. Furthermore, we show that passage of neutralization sensitive CCR5-using variants in the presence of increasing concentrations of autologous contemporaneous plasma was sufficient to force the evolution of a CXCR4-using virus from an R5 strain in 1 of 3 subjects. In aggregate, our studies suggest that humoral immune pressure can drive the emergence of CXCR4-utilizing viruses in some individuals.
Studies have suggested that dual-tropic strains evolve from R5 variants, and X4 viruses arise from the R5X4 strains (Pastore et al., 2004). It is unlikely that the emergence of a dual-tropic virus during the passage experiments occurred by chance because CXCR4-using variants were not detected in the absence of plasma and changes required for co-receptor switching are generally associated with diminished replication fitness (Pastore et al., 2004). The DM268 passage results were surprising because we observed no significant neutralization susceptibility difference to contemporaneous autologous plasma among co-existing R5 and dual-tropic strains (Fig. 1). Although previous studies have demonstrated that diverse glycosylation changes can render viruses neutralization resistant (Sagar et al., 2006, Wei et al., 2003), it remains unclear how the addition of PNGS at position 88 leads to low level CXCR4 usage. It has been shown that co-circulating R5 and R5X4 variants often have the same predicted V3 loop sequence (Huang et al., 2007, Lin et al., 2012), and thus it possible that modifications in other env segments lead to a structural change that can influence co-receptor usage. Interestingly, DM viruses did not emerge in the other 2 cases (DM8 and DM178) even though their X4 as compared to R5 envs were more neutralization resistant to autologous contemporaneous plasma. Our observations suggest that the passaged DM8 and DM178 env variants accommodated changes required for neutralization resistance to autologous plasma without co-receptor switching. Other R5 variants from these individuals, not examined in the passaging experiments, may acquire the ability to use the CXCR4 receptor as they become resistant to autologous antibodies.
The earliest studies implied that lab-adapted SI, presumably CXCR4-using, strains were more neutralization sensitive compared to NSI–CCR5-utilizing variants (Mascola et al., 1996, Montefiori et al., 1998, D'Souza et al., 1997, Sawyer et al., 1994). Subsequent studies showed that compared to CCR5-utilizing strains primary as opposed to lab-adapted CXCR4-using envs displayed a relatively similar range of neutralization susceptibilities to some mAbs and unrelated heterologous plasma (Trkola et al., 1998, Montefiori et al., 1998, Cecilia et al., 1998, Lacasse et al., 1998). This led to the notion that nAbs are unlikely to be the major selection pressure that drives co-receptor switching. Our results also show that X4 and co-existing R5 variants have equivalent sensitivity to antibodies targeting env domains not involved in co-receptor binding. More recent investigations with SHIV-1B and HIV-1B infected subjects argue that relatively neutralization sensitive CXCR4-utilizing variants emerge later in disease after HIV-1 induced immunological damage has led to a compromised humoral immune response (Bunnik et al., 2007, Ho et al., 2007). We, however, observed no case in which X4 or R5X4 as compared to co-circulating R5 envs were significantly more neutralization susceptible to autologous contemporaneous plasma, even though we sampled more envs from each subject and examined a greater number of individuals. Furthermore, the previous human study examined PBMC passaged variants (Bunnik et al., 2007), which may have led to the different results because passaging virus in PBMCs significantly impacts its neutralization properties in a unique manner for each virus antibody combination (Etemad et al., 2015, Provine et al., 2012).
Our results also differ from the previous human study (Bunnik et al., 2007) potentially because we examined HIV-1C versus HIV-1B infected individuals. Our preliminary results, however, suggest this is not the case (Fig. S2). While HIV-1C constitutes the majority of worldwide infections, emergence of CXCR4 usage occurs at a lower frequency compared to HIV-1B (Lin et al., 2011, Ataher et al., 2012). Reasons for the difference in incidence remain uncertain. Although it has been reported that CCR5 and CXCR4-using viruses preferentially infect memory and naïve T cells respectively (Davenport et al., 2002, Blaak et al., 2000), there is no evidence that this preference differs among HIV-1B versus HIV-1C variants (Cashin et al., 2014, Ribeiro et al., 2006). While, it is known that the CCR5Δ32 allele frequency is higher among Europeans than Africans (Galvani and Novembre, 2005), the density of the CCR5 receptor on CD4 + T cells is not significantly different among people with European ancestry who constitute the majority of HIV-1B infections versus those with African heritage who are predominantly infected with HIV-1C (Picton et al., 2012). Thus, the higher incidence of DM virus population in HIV-1B as compared to HIV-1C infected individuals is unlikely because of lower availability of appropriate CCR5 expressing target cells. It has been suggested that HIV-1B and HIV-1C envs have structural differences, especially in the V3 loop (Patel et al., 2008). Potentially in response to these env structural differences, HIV-1C infected individuals develop antibodies targeting the C3-V4 region while those with HIV-1B generate responses against the V1-V2 domain and the base of the V3 loop (Rong et al., 2009, Moore et al., 2008, Tang et al., 2011). Interestingly, the main determinants for co-receptor switching are located in the V1 to V3 domains (O'Brien et al., 1990, Chesebro et al., 1991, Shioda et al., 1992, Groenink et al., 1993, Fouchier et al., 1995). Greater prevalence of anti-V1-V2 or V3 loop antibodies may correlate with the greater frequency of DM virus populations in HIV-1B as compared to HIV-1C infected individuals.
While most HIV-1 infected individuals develop nAbs against their circulating variants and viruses escape this humoral pressure, there are potentially two important factors that explain why CXCR4-using variants do not evolve in everyone. First, CXCR4-utilizing viruses will not emerge if the circulating env variants cannot accommodate changes required for CXCR4 usage regardless of the selection pressure. Both laboratory and human studies have documented that some HIV-1 env variants do not evolve to use the CXCR4 receptor even in setting where they are exposed to CCR5 inhibitors (Westby et al., 2007, Marozsan et al., 2005, Tsibris et al., 2008). Second, similar to the observation that some but not all HIV-1 infected individuals develop cross-neutralizing antibodies (Doria-Rose et al., 2009), co-receptor switching due to humoral selection may only occur in people that develop certain types of antibodies over the course of infection. Thus, in people that do not generate antibodies with the appropriate specificity neutralization escape may occur without changes in co-receptor usage. Unfortunately, we did not have access to live PBMCs to generate mAbs from these individuals, which would have allowed us to map the epitope for the antibodies that neutralize R5 but not co-existing X4 variants.
Interestingly, treatment of HIV-1 infected humanized mice with anti-V3 loop bnAb, PGT128, but not anti-CD4bs bnAb, 45-46GW, has been predicted to select for a significantly higher proportion of CXCR4-using variants (Pfeifer et al., 2014). Furthermore, CXCR4 as compared to CCR5-using variants are overrepresented among variants resistant to env V1–V2 directed bnAbs, PG9 and PG16. Thus similar to our suggestions, this investigation implies that antibodies directed against the env variable loops potentially select for neutralization resistant CXCR4-utilizing variants (Pfeifer et al., 2014). One study has argued that this may also be the case during longitudinal HIV-2 infection (Marcelino et al., 2012). Even though our conclusions are based on observations from a small number of individuals, and in some cases only a limited number of envs with different co-receptor phenotype, our studies strongly support the idea that X4 variants are neutralization escape variants.
In contrast to the previously proposed mechanisms that lead to co-receptor switching, such as random mutations, waning immunity, and/or decreasing number of CCR5 + T cells, our observations suggest that antibody pressure selects for CXCR4-utilizing variants (Regoes and Bonhoeffer, 2005). It is likely that in some but not necessarily all individuals that eventually develop CXCR4-using viruses antibodies force env structural changes that directly influence co-receptor switching. Continual presence of these humoral responses eventually leads to the emergence of X4 variants resistant to these and other host generated anti V3 antibodies. This new biological mechanism for co-receptor switching has a number of important implications. It is generally agreed that presence of CXCR4-using variants leads to faster CD4 decline and earlier onset of AIDS (Koot et al., 1993, Richman and Bozzette, 1994, Connor et al., 1997), although the causal link is unclear. Our studies would argue that an infected individual's humoral immune response leads to the emergence of CXCR4-utilizing variants, which results in an adverse outcome. Thus, our observations imply that CXCR4-using variants cause and are not a result of decreasing immunity. Importantly, this suggests that in some individuals, the humoral immune response against their autologous virus worsens infection outcome. Although antibodies have been suggested to adversely affect disease progression in other viral infections, notably dengue and influenza, no previous study has implied this for HIV-1 (Halstead and O'Rourke, 1977, Khurana et al., 2013). In addition, in contrast to dengue or influenza where antibodies enhance entry of the infecting virus into host cells, our studies suggest a mechanism where antibodies force phenotypic changes in replicating variants to expand the cellular tropism. Finally, our results have implications for proposed strategies of using bnAbs as prophylaxis, treatment, or cure (Caskey et al., 2015, Lynch et al., 2015). Our results and previous studies suggest that specific antibodies may be less potent against CXCR4-utilizing variants, which may select for their emergence in settings where antibodies are unable to fully inhibit virus replication (Pfeifer et al., 2014). Thus, the bnAbs selected for immunotherapy will need to be evaluated carefully to ensure that they do not promote the emergence of CXCR4-using strains, which can accelerate disease progression among infected individuals.
Funding Sources
This project was supported by NIH grants AI081545 (NL) and AI122209 (MS) and BU Pilot Grant (MS). No author received compensation from a pharmaceutical company or other agency for completion of this manuscript.
Conflict of Interest Statement
There are no conflicts of interest associated with this study. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Author Contributions
NL, XW, DRK, and MS designed the study. NL, OG, CB, LR, BE, and HL performed all the experiments. SL, ME, and SM oversaw the original trial and sample collection. MS wrote the paper with input from co-authors.
Acknowledgements
We would like to thank all Mashi trial participants, without whom this study would not have been possible.
Footnotes
Supplementary data to this article can be found online at http://dx.doi.org/10.1016/j.ebiom.2016.04.040.
Appendix A. Supplementary Data
Model for antibody induced emergence of CXCR4-using virus
References
- Ataher Q., Portsmouth S., Napolitano L.A., ENG S., Greenacre A., Kambugu A., Wood R., Badal-Faesen S., Tressler R. The epidemiology and clinical correlates of HIV-1 co-receptor tropism in non-subtype B infections from India, Uganda and South Africa. J. Int. AIDS Soc. 2012;15:2. doi: 10.1186/1758-2652-15-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blaak H., Van't Wout A.B., Brouwer M., Hooibrink B., Hovenkamp E., Schuitemaker H. In vivo HIV-1 infection of CD45RA(+)CD4(+) T cells is established primarily by syncytium-inducing variants and correlates with the rate of CD4(+) T cell decline. Proc. Natl. Acad. Sci. U. S. A. 2000;97:1269–1274. doi: 10.1073/pnas.97.3.1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brumme Z.L., Goodrich J., Mayer H.B., Brumme C.J., Henrick B.M., Wynhoven B., Asselin J.J., Cheung P.K., Hogg R.S., Montaner J.S., Harrigan P.R. Molecular and clinical epidemiology of CXCR4-using HIV-1 in a large population of antiretroviral-naive individuals. J. Infect. Dis. 2005;192:466–474. doi: 10.1086/431519. [DOI] [PubMed] [Google Scholar]
- Bunnik E.M., Quakkelaar E.D., van Nuenen A.C., Boeser-Nunnink B., Schuitemaker H. Increased neutralization sensitivity of recently emerged CXCR4-using human immunodeficiency virus type 1 strains compared to coexisting CCR5-using variants from the same patient. J. Virol. 2007;81:525–531. doi: 10.1128/JVI.01983-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cann A.J., Churcher M.J., Boyd M., O'Brien W., Zhao J.Q., Zack J., Chen I.S. The region of the envelope gene of human immunodeficiency virus type 1 responsible for determination of cell tropism. J. Virol. 1992;66:305–309. doi: 10.1128/jvi.66.1.305-309.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cashin K., Paukovics G., Jakobsen M.R., Ostergaard L., Churchill M.J., Gorry P.R., Flynn J.K. Differences in coreceptor specificity contribute to alternative tropism of HIV-1 subtype C for CD4(+) T-cell subsets, including stem cell memory T-cells. Retrovirology. 2014;11:97. doi: 10.1186/s12977-014-0097-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caskey M., Klein F., Lorenzi J.C., Seaman M.S., West A.P., JR., Buckley N., Kremer G., Nogueira L., Braunschweig M., Scheid J.F., Horwitz J.A., Shimeliovich I., Ben-Avraham S., Witmer-Pack M., Platten M., Lehmann C., Burke L.A., Hawthorne T., Gorelick R.J., Walker B.D., Keler T., Gulick R.M., Fatkenheuer G., Schlesinger S.J., Nussenzweig M.C. Viraemia suppressed in HIV-1-infected humans by broadly neutralizing antibody 3BNC117. Nature. 2015;522:487–491. doi: 10.1038/nature14411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cecilia D., Kewalramani V.N., O'Leary J., Volsky B., Nyambi P., Burda S., Xu S., Littman D.R., Zolla-Pazner S. Neutralization profiles of primary human immunodeficiency virus type 1 isolates in the context of coreceptor usage. J. Virol. 1998;72:6988–6996. doi: 10.1128/jvi.72.9.6988-6996.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatziandreou N., Arauz A.B., Freitas I., Nyein P.H., Fenton G., Mehta S.H., Kirk G.D., Sagar M. Sensitivity changes over the course of infection increases the likelihood of resistance against fusion but not CCR5 receptor blockers. AIDS Res. Hum. Retrovir. 2012;28:1584–1593. doi: 10.1089/aid.2011.0319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chesebro B., Nishio J., Perryman S., Cann A., O'Brien W., Chen I.S., Wehrly K. Identification of human immunodeficiency virus envelope gene sequences influencing viral entry into CD4-positive HeLa cells, T-leukemia cells, and macrophages. J. Virol. 1991;65:5782–5789. doi: 10.1128/jvi.65.11.5782-5789.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chesebro B., Wehrly K., Nishio J., Perryman S. Mapping of independent V3 envelope determinants of human immunodeficiency virus type 1 macrophage tropism and syncytium formation in lymphocytes. J. Virol. 1996;70:9055–9059. doi: 10.1128/jvi.70.12.9055-9059.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cocchi F., Devico A.L., Garzino-Demo A., Cara A., Gallo R.C., Lusso P. The V3 domain of the HIV-1 gp120 envelope glycoprotein is critical for chemokine-mediated blockade of infection. Nat. Med. 1996;2:1244–1247. doi: 10.1038/nm1196-1244. [DOI] [PubMed] [Google Scholar]
- Connor R.I., Sheridan K.E., Ceradini D., Choe S., Landau N.R. Change in coreceptor use correlates with disease progression in HIV-1-infected individuals. J. Exp. Med. 1997;185:621–628. doi: 10.1084/jem.185.4.621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davenport M.P., Zaunders J.J., Hazenberg M.D., Schuitemaker H., Van Rij R.P. Cell turnover and cell tropism in HIV-1 infection. Trends Microbiol. 2002;10:275–278. doi: 10.1016/s0966-842x(02)02370-3. [DOI] [PubMed] [Google Scholar]
- del Prete G.Q., Haggarty B., Leslie G.J., Jordan A.P., Romano J., Wang N., Wang J., Holmes M.C., Montefiori D.C., Hoxie J.A. Derivation and characterization of a simian immunodeficiency virus SIVmac239 variant with tropism for CXCR4. J. Virol. 2009;83:9911–9922. doi: 10.1128/JVI.00533-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng H., Liu R., Ellmeier W., Choe S., Unutmaz D., Burkhart M., di Marzio P., Marmon S., Sutton R.E., Hill C.M., Davis C.B., Peiper S.C., Schall T.J., Littman D.R., Landau N.R. Identification of a major co-receptor for primary isolates of HIV-1. Nature. 1996;381:661–666. doi: 10.1038/381661a0. [DOI] [PubMed] [Google Scholar]
- Doria-Rose N.A., Klein R.M., Manion M.M., O'Dell S., Phogat A., Chakrabarti B., Hallahan C.W., Migueles S.A., Wrammert J., Ahmed R., Nason M., Wyatt R.T., Mascola J.R., Connors M. Frequency and phenotype of human immunodeficiency virus envelope-specific B cells from patients with broadly cross-neutralizing antibodies. J. Virol. 2009;83:188–199. doi: 10.1128/JVI.01583-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Souza M.P., Livnat D., Bradac J.A., Bridges S.H. Evaluation of monoclonal antibodies to human immunodeficiency virus type 1 primary isolates by neutralization assays: performance criteria for selecting candidate antibodies for clinical trials. AIDS Clinical Trials Group Antibody Selection Working Group. J. Infect. Dis. 1997;175:1056–1062. doi: 10.1086/516443. [DOI] [PubMed] [Google Scholar]
- Etemad B., Fellows A., Kwambana B., Kamat A., Feng Y., Lee S., Sagar M. Human immunodeficiency virus type 1 V1-to-V5 envelope variants from the chronic phase of infection use CCR5 and fuse more efficiently than those from early after infection. J. Virol. 2009;83:9694–9708. doi: 10.1128/JVI.00925-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Etemad B., Gonzalez O.A., White L., Laeyendecker O., Kirk G.D., Mehta S., Sagar M. Characterization of HIV-1 envelopes in acutely and chronically infected injection drug users. Retrovirology. 2014;11:106. doi: 10.1186/s12977-014-0106-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Etemad B., Ghulam-Smith M., Gonzalez O., White L.F., Sagar M. Single genome amplification and standard bulk PCR yield HIV-1 envelope products with similar genotypic and phenotypic characteristics. J. Virol. Methods. 2015;214:46–53. doi: 10.1016/j.jviromet.2015.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang G., Zhu G., Burger H., Keithly J.S., Weiser B. Minimizing DNA recombination during long RT-PCR. J. Virol. Methods. 1998;76:139–148. doi: 10.1016/s0166-0934(98)00133-5. [DOI] [PubMed] [Google Scholar]
- Feng Y., Broder C.C., Kennedy P.E., Berger E.A. HIV-1 entry cofactor: functional cDNA cloning of a seven-transmembrane, G protein-coupled receptor. Science. 1996;272:872–877. doi: 10.1126/science.272.5263.872. [DOI] [PubMed] [Google Scholar]
- Fouchier R.A., Broersen S.M., Brouwer M., Tersmette M., van't Wout A.B., Groenink M., Schuitemaker H. Temporal relationship between elongation of the HIV type 1 glycoprotein 120 V2 domain and the conversion toward a syncytium-inducing phenotype. AIDS Res. Hum. Retrovir. 1995;11:1473–1478. doi: 10.1089/aid.1995.11.1473. [DOI] [PubMed] [Google Scholar]
- Freed E.O., Martin M.A. Virion incorporation of envelope glycoproteins with long but not short cytoplasmic tails is blocked by specific, single amino acid substitutions in the human immunodeficiency virus type 1 matrix. J. Virol. 1995;69:1984–1989. doi: 10.1128/jvi.69.3.1984-1989.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galvani A.P., Novembre J. The evolutionary history of the CCR5-Delta32 HIV-resistance mutation. Microbes Infect. 2005;7:302–309. doi: 10.1016/j.micinf.2004.12.006. [DOI] [PubMed] [Google Scholar]
- Gorny M.K., Conley A.J., Karwowska S., Buchbinder A., Xu J.Y., Emini E.A., Koenig S., Zolla-Pazner S. Neutralization of diverse human immunodeficiency virus type 1 variants by an anti-V3 human monoclonal antibody. J. Virol. 1992;66:7538–7542. doi: 10.1128/jvi.66.12.7538-7542.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groenink M., Fouchier R.A., Broersen S., Baker C.H., Koot M., van't Wout A.B., Huisman H.G., Miedema F., Tersmette M., Schuitemaker H. Relation of phenotype evolution of HIV-1 to envelope V2 configuration. Science. 1993;260:1513–1516. doi: 10.1126/science.8502996. [DOI] [PubMed] [Google Scholar]
- Halstead S.B., O'Rourke E.J. Dengue viruses and mononuclear phagocytes. I. Infection enhancement by non-neutralizing antibody. J. Exp. Med. 1977;146:201–217. doi: 10.1084/jem.146.1.201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henrich T.J., McLaren P.J., Rao S.S., Lin N.H., Hanhauser E., Giguel F., Gulick R.M., Ribaudo H., de Bakker P.I., Kuritzkes D.R. Genome-wide association study of human immunodeficiency virus (HIV)-1 coreceptor usage in treatment-naive patients from an AIDS clinical trials group study. Open Forum Infect. Dis. 2014;1:ofu018. doi: 10.1093/ofid/ofu018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho S.H., Tasca S., Shek L., Li A., Gettie A., Blanchard J., Boden D., Cheng-Mayer C. Coreceptor switch in R5-tropic simian/human immunodeficiency virus-infected macaques. J. Virol. 2007;81:8621–8633. doi: 10.1128/JVI.00759-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang W., Eshleman S.H., Toma J., Fransen S., Stawiski E., Paxinos E.E., Whitcomb J.M., Young A.M., Donnell D., Mmiro F., Musoke P., Guay L.A., Jackson J.B., Parkin N.T., Petropoulos C.J. Coreceptor tropism in human immunodeficiency virus type 1 subtype D: high prevalence of CXCR4 tropism and heterogeneous composition of viral populations. J. Virol. 2007;81:7885–7893. doi: 10.1128/JVI.00218-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J., Ofek G., Laub L., Louder M.K., Doria-Rose N.A., Longo N.S., Imamichi H., Bailer R.T., Chakrabarti B., Sharma S.K., Alam S.M., Wang T., Yang Y., Zhang B., Migueles S.A., Wyatt R., Haynes B.F., Kwong P.D., Mascola J.R., Connors M. Broad and potent neutralization of HIV-1 by a gp41-specific human antibody. Nature. 2012;491:406–412. doi: 10.1038/nature11544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia M., Lu H., Markowitz M., Cheng-Mayer C., Wu X. Development of broadly neutralizing antibodies and their mapping by monomeric gp120 in HIV-1 infected humans and SHIVSF162P3N infected macaques. J. Virol. 2016;90:4017–4031. doi: 10.1128/JVI.02898-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Judo M.S., Wedel A.B., Wilson C. Stimulation and suppression of PCR-mediated recombination. Nucleic Acids Res. 1998;26:1819–1825. doi: 10.1093/nar/26.7.1819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keele B.F., Giorgi E.E., Salazar-Gonzalez J.F., Decker J.M., Pham K.T., Salazar M.G., Sun C., Grayson T., Wang S., Li H., Wei X., Jiang C., Kirchherr J.L., Gao F., Anderson J.A., Ping L.H., Swanstrom R., Tomaras G.D., Blattner W.A., Goepfert P.A., Kilby J.M., Saag M.S., Delwart E.L., Busch M.P., Cohen M.S., Montefiori D.C., Haynes B.F., Gaschen B., Athreya G.S., Lee H.Y., Wood N., Seoighe C., Perelson A.S., Bhattacharya T., Korber B.T., Hahn B.H., Shaw G.M. Identification and characterization of transmitted and early founder virus envelopes in primary HIV-1 infection. Proc. Natl. Acad. Sci. U. S. A. 2008;105:7552–7557. doi: 10.1073/pnas.0802203105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khurana S., Loving C.L., Manischewitz J., King L.R., Gauger P.C., Henningson J., Vincent A.L., Golding H. Vaccine-induced anti-HA2 antibodies promote virus fusion and enhance influenza virus respiratory disease. Sci. Transl. Med. 2013;5:200ra114. doi: 10.1126/scitranslmed.3006366. [DOI] [PubMed] [Google Scholar]
- Koot M., Keet I.P., Vos A.H., de Goede R.E., Roos M.T., Coutinho R.A., Miedema F., Schellekens P.T., Tersmette M. Prognostic value of HIV-1 syncytium-inducing phenotype for rate of CD4 + cell depletion and progression to AIDS. Ann. Intern. Med. 1993;118:681–688. doi: 10.7326/0003-4819-118-9-199305010-00004. [DOI] [PubMed] [Google Scholar]
- Lacasse R.A., Follis K.E., Moudgil T., Trahey M., Binley J.M., Planelles V., Zolla-Pazner S., Nunberg J.H. Coreceptor utilization by human immunodeficiency virus type 1 is not a primary determinant of neutralization sensitivity. J. Virol. 1998;72:2491–2495. doi: 10.1128/jvi.72.3.2491-2495.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lahr D.J., Katz L.A. Reducing the impact of PCR-mediated recombination in molecular evolution and environmental studies using a new-generation high-fidelity DNA polymerase. Biotechniques. 2009;47:857–866. doi: 10.2144/000113219. [DOI] [PubMed] [Google Scholar]
- Lin N.H., Negusse D.M., Beroukhim R., Giguel F., Lockman S., Essex M., Kuritzkes D.R. The design and validation of a novel phenotypic assay to determine HIV-1 coreceptor usage of clinical isolates. J. Virol. Methods. 2010;169:39–46. doi: 10.1016/j.jviromet.2010.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin N.H., Smeaton L.M., Giguel F., Novitsky V., Moyo S., Mitchell R.M., Makhema J., Essex M., Lockman S., Kuritzkes D.R. Prevalence and clinical associations of CXCR4-using HIV-1 among treatment-naive subtype C-infected women in Botswana. J. Acquir. Immune Defic. Syndr. 2011;57:46–50. doi: 10.1097/QAI.0b013e318214fe27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin N.H., Becerril C., Giguel F., Novitsky V., Moyo S., Makhema J., Essex M., Lockman S., Kuritzkes D.R., Sagar M. Env sequence determinants in CXCR4-using human immunodeficiency virus type-1 subtype C. Virology. 2012;433:296–307. doi: 10.1016/j.virol.2012.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S.L., Rodrigo A.G., Shankarappa R., Learn G.H., Hsu L., Davidov O., Zhao L.P., Mullins J.I. HIV quasispecies and resampling. Science. 1996;273:415–416. doi: 10.1126/science.273.5274.415. [DOI] [PubMed] [Google Scholar]
- Lynch R.M., Boritz E., Coates E.E., Dezure A., Madden P., Costner P., Enama M.E., Plummer S., Holman L., Hendel C.S., Gordon I., Casazza J., Conan-Cibotti M., Migueles S.A., Tressler R., Bailer R.T., Mcdermott A., Narpala S., O'Dell S., Wolf G., Lifson J.D., Freemire B.A., Gorelick R.J., Pandey J.P., Mohan S., Chomont N., Fromentin R., Chun T.W., Fauci A.S., Schwartz R.M., Koup R.A., Douek D.C., Hu Z., Capparelli E., Graham B.S., Mascola J.R., Ledgerwood J.E. Virologic effects of broadly neutralizing antibody VRC01 administration during chronic HIV-1 infection. Sci. Transl. Med. 2015;7:319ra206. doi: 10.1126/scitranslmed.aad5752. [DOI] [PubMed] [Google Scholar]
- Marcelino J.M., Borrego P., Nilsson C., Familia C., Barroso H., Maltez F., Doroana M., Antunes F., Quintas A., Taveira N. Resistance to antibody neutralization in HIV-2 infection occurs in late stage disease and is associated with X4 tropism. AIDS. 2012;26:2275–2284. doi: 10.1097/QAD.0b013e328359a89d. [DOI] [PubMed] [Google Scholar]
- Marozsan A.J., Kuhmann S.E., Morgan T., Herrera C., Rivera-Troche E., Xu S., Baroudy B.M., Strizki J., Moore J.P. Generation and properties of a human immunodeficiency virus type 1 isolate resistant to the small molecule CCR5 inhibitor, SCH-417690 (SCH-D) Virology. 2005;338:182–199. doi: 10.1016/j.virol.2005.04.035. [DOI] [PubMed] [Google Scholar]
- Mascola J.R., Snyder S., Weislow O., Belay S., Belshe R., Schwartz D., Clements M., Dolin R., Graham B., Gorse G., Keefer M., Mcelrath M., Walker M., Wagner K., Mcneil J., Mccutchan F., Burke D. Immunization with envelope subunit vaccine products elicits neutralizing antibodies against laboratory-adapted but not primary isolates of human immunodeficiency virus type 1. The National Institute of Allergy and Infectious Diseases AIDS Vaccine Evaluation Group. J. Infect. Dis. 1996;173:340–348. doi: 10.1093/infdis/173.2.340. [DOI] [PubMed] [Google Scholar]
- Meyerhans A., Vartanian J.P., Wain-Hobson S. DNA recombination during PCR. Nucleic Acids Res. 1990;18:1687–1691. doi: 10.1093/nar/18.7.1687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montefiori D.C., Collman R.G., Fouts T.R., Zhou J.Y., Bilska M., Hoxie J.A., Moore J.P., Bolognesi D.P. Evidence that antibody-mediated neutralization of human immunodeficiency virus type 1 by sera from infected individuals is independent of coreceptor usage. J. Virol. 1998;72:1886–1893. doi: 10.1128/jvi.72.3.1886-1893.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moody M.A., Gao F., Gurley T.C., Amos J.D., Kumar A., Hora B., Marshall D.J., Whitesides J.F., Xia S.M., Parks R., Lloyd K.E., Hwang K.K., Lu X., Bonsignori M., Finzi A., Vandergrift N.A., Alam S.M., Ferrari G., Shen X., Tomaras G.D., Kamanga G., Cohen M.S., Sam N.E., Kapiga S., Gray E.S., Tumba N.L., Morris L., Zolla-Pazner S., Gorny M.K., Mascola J.R., Hahn B.H., Shaw G.M., Sodroski J.G., Liao H.X., Montefiori D.C., Hraber P.T., Korber B.T., Haynes B.F. Strain-specific V3 and CD4 binding site autologous HIV-1 neutralizing antibodies select neutralization-resistant viruses. Cell Host Microbe. 2015;18:354–362. doi: 10.1016/j.chom.2015.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore P.L., Gray E.S., Choge I.A., Ranchobe N., Mlisana K., Abdool Karim S.S., Williamson C., Morris L. The c3-v4 region is a major target of autologous neutralizing antibodies in human immunodeficiency virus type 1 subtype C infection. J. Virol. 2008;82:1860–1869. doi: 10.1128/JVI.02187-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore P.L., Ranchobe N., Lambson B.E., Gray E.S., Cave E., Abrahams M.R., Bandawe G., Mlisana K., Abdool Karim S.S., Williamson C., Morris L. Limited neutralizing antibody specificities drive neutralization escape in early HIV-1 subtype C infection. PLoS Pathog. 2009;5:e1000598. doi: 10.1371/journal.ppat.1000598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Brien W.A., Koyanagi Y., Namazie A., Zhao J.Q., Diagne A., Idler K., Zack J.A., Chen I.S. HIV-1 tropism for mononuclear phagocytes can be determined by regions of gp120 outside the CD4-binding domain. Nature. 1990;348:69–73. doi: 10.1038/348069a0. [DOI] [PubMed] [Google Scholar]
- Olshevsky U., Helseth E., Furman C., Li J., Haseltine W., Sodroski J. Identification of individual human immunodeficiency virus type 1 gp120 amino acids important for CD4 receptor binding. J. Virol. 1990;64:5701–5707. doi: 10.1128/jvi.64.12.5701-5707.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pastore C., Ramos A., Mosier D.E. Intrinsic obstacles to human immunodeficiency virus type 1 coreceptor switching. J. Virol. 2004;78:7565–7574. doi: 10.1128/JVI.78.14.7565-7574.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel M.B., Hoffman N.G., Swanstrom R. Subtype-specific conformational differences within the V3 region of subtype B and subtype C human immunodeficiency virus type 1 Env proteins. J. Virol. 2008;82:903–916. doi: 10.1128/JVI.01444-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pejchal R., Doores K.J., Walker L.M., Khayat R., Huang P.S., Wang S.K., Stanfield R.L., Julien J.P., Ramos A., Crispin M., Depetris R., Katpally U., Marozsan A., Cupo A., Maloveste S., Liu Y., Mcbride R., Ito Y., Sanders R.W., Ogohara C., Paulson J.C., Feizi T., Scanlan C.N., Wong C.H., Moore J.P., Olson W.C., Ward A.B., Poignard P., Schief W.R., Burton D.R., Wilson I.A. A potent and broad neutralizing antibody recognizes and penetrates the HIV glycan shield. Science. 2011;334:1097–1103. doi: 10.1126/science.1213256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pena-Cruz V., Etemad B., Chatziandreou N., Nyein P.H., Stock S., reynolds S.J., Laeyendecker O., Serwadda D., Gray R.H., Lee S., Quinn T.C., Sagar M. HIV-1 envelope replication and α4β7 utilization among newly infected subjects and their corresponding heterosexual partners. Retrovirology. 2013;10:162. doi: 10.1186/1742-4690-10-162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeifer N., Walter H., Lengauer T. Association between HIV-1 coreceptor usage and resistance to broadly neutralizing antibodies. J. Acquir. Immune Defic. Syndr. 2014;67:107–112. doi: 10.1097/QAI.0000000000000283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picton A.C., Shalekoff S., Paximadis M., Tiemessen C.T. Marked differences in CCR5 expression and activation levels in two South African populations. Immunology. 2012;136:397–407. doi: 10.1111/j.1365-2567.2012.03592.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Provine N.M., Cortez V., Chohan V., Overbaugh J. The neutralization sensitivity of viruses representing human immunodeficiency virus type 1 variants of diverse subtypes from early in infection is dependent on producer cell, as well as characteristics of the specific antibody and envelope variant. Virology. 2012;427:25–33. doi: 10.1016/j.virol.2012.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Regoes R.R., Bonhoeffer S. The HIV coreceptor switch: a population dynamical perspective. Trends Microbiol. 2005;13:269–277. doi: 10.1016/j.tim.2005.04.005. [DOI] [PubMed] [Google Scholar]
- Ribeiro R.M., Hazenberg M.D., Perelson A.S., Davenport M.P. Naive and memory cell turnover as drivers of CCR5-to-CXCR4 tropism switch in human immunodeficiency virus type 1: implications for therapy. J. Virol. 2006;80:802–809. doi: 10.1128/JVI.80.2.802-809.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richman D.D., Bozzette S.A. The impact of the syncytium-inducing phenotype of human immunodeficiency virus on disease progression. J. Infect. Dis. 1994;169:968–974. doi: 10.1093/infdis/169.5.968. [DOI] [PubMed] [Google Scholar]
- Richman D.D., Wrin T., Little S.J., Petropoulos C.J. Rapid evolution of the neutralizing antibody response to HIV type 1 infection. Proc. Natl. Acad. Sci. U. S. A. 2003;100:4144–4149. doi: 10.1073/pnas.0630530100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roederer M., Keele B.F., Schmidt S.D., Mason R.D., Welles H.C., Fischer W., LABRANCHE C., Foulds K.E., Louder M.K., Yang Z.Y., Todd J.P., Buzby A.P., Mach L.V., Shen L., Seaton K.E., Ward B.M., Bailer R.T., Gottardo R., Gu W., Ferrari G., Alam S.M., Denny T.N., Montefiori D.C., Tomaras G.D., Korber B.T., Nason M.C., Seder R.A., Koup R.A., Letvin N.L., Rao S.S., Nabel G.J., Mascola J.R. Immunological and virological mechanisms of vaccine-mediated protection against SIV and HIV. Nature. 2013;505:502–508. doi: 10.1038/nature12893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rong R., Li B., Lynch R.M., Haaland R.E., Murphy M.K., Mulenga J., Allen S.A., Pinter A., Shaw G.M., Hunter E., Robinson J.E., Gnanakaran S., Derdeyn C.A. Escape from autologous neutralizing antibodies in acute/early subtype C HIV-1 infection requires multiple pathways. PLoS Pathog. 2009;5:e1000594. doi: 10.1371/journal.ppat.1000594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sagar M., Wu X., Lee S., Overbaugh J. HIV-1 V1-V2 envelope loop sequences expand and add glycosylation sites over the course of infection and these modifications affect antibody neutralization sensitivity. J. Virol. 2006;80:9586–9598. doi: 10.1128/JVI.00141-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salazar-Gonzalez J.F., Bailes E., Pham K.T., Salazar M.G., Guffey M.B., Keele B.F., Derdeyn C.A., Farmer P., Hunter E., Allen S., Manigart O., Mulenga J., Anderson J.A., Swanstrom R., Haynes B.F., Athreya G.S., Korber B.T., Sharp P.M., Shaw G.M., Hahn B.H. Deciphering human immunodeficiency virus type 1 transmission and early envelope diversification by single-genome amplification and sequencing. J. Virol. 2008;82:3952–3970. doi: 10.1128/JVI.02660-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawyer L.S., Wrin M.T., Crawford-Miksza L., Potts B., Wu Y., Weber P.A., Alfonso R.D., Hanson C.V. Neutralization sensitivity of human immunodeficiency virus type 1 is determined in part by the cell in which the virus is propagated. J. Virol. 1994;68:1342–1349. doi: 10.1128/jvi.68.3.1342-1349.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuitemaker H., Koot M., Kootstra N.A., Dercksen M.W., de Goede R.E., van Steenwijk R.P., Lange J.M., Schattenkerk J.K., Miedema F., Tersmette M. Biological phenotype of human immunodeficiency virus type 1 clones at different stages of infection: progression of disease is associated with a shift from monocytotropic to T-cell-tropic virus population. J. Virol. 1992;66:1354–1360. doi: 10.1128/jvi.66.3.1354-1360.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shioda T., Levy J.A., Cheng-Mayer C. Small amino acid changes in the V3 hypervariable region of gp120 can affect the T-cell-line and macrophage tropism of human immunodeficiency virus type 1. Proc. Natl. Acad. Sci. U. S. A. 1992;89:9434–9438. doi: 10.1073/pnas.89.20.9434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang H., Robinson J.E., Gnanakaran S., Li M., Rosenberg E.S., Perez L.G., Haynes B.F., Liao H.X., Labranche C.C., Korber B.T., Montefiori D.C. Epitopes immediately below the base of the V3 loop of gp120 as targets for the initial autologous neutralizing antibody response in two HIV-1 subtype B-infected individuals. J. Virol. 2011;85:9286–9299. doi: 10.1128/JVI.02286-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thior I., Lockman S., Smeaton L.M., Shapiro R.L., Wester C., Heymann S.J., Gilbert P.B., Stevens L., Peter T., Kim S., van Widenfelt E., Moffat C., Ndase P., Arimi P., Kebaabetswe P., Mazonde P., Makhema J., Mcintosh K., Novitsky V., Lee T.H., Marlink R., Lagakos S., Essex M. Breastfeeding plus infant zidovudine prophylaxis for 6 months vs formula feeding plus infant zidovudine for 1 month to reduce mother-to-child HIV transmission in Botswana: a randomized trial: the Mashi Study. JAMA. 2006;296:794–805. doi: 10.1001/jama.296.7.794. [DOI] [PubMed] [Google Scholar]
- Trkola A., Ketas T., Kewalramani V.N., Endorf F., Binley J.M., Katinger H., Robinson J., Littman D.R., Moore J.P. Neutralization sensitivity of human immunodeficiency virus type 1 primary isolates to antibodies and CD4-based reagents is independent of coreceptor usage. J. Virol. 1998;72:1876–1885. doi: 10.1128/jvi.72.3.1876-1885.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsibris A.M., Sagar M., Gulick R.M., Su Z., Hughes M., Greaves W., Subramanian M., Flexner C., Giguel F., Leopold K.E., Coakley E., Kuritzkes D.R. In vivo emergence of vicriviroc resistance in a human immunodeficiency virus type 1 subtype C-infected subject. J. Virol. 2008;82:8210–8214. doi: 10.1128/JVI.00444-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van't Wout A.B., Kootstra N.A., Mulder-Kampinga G.A., Albrecht-van Lent N., Scherpbier H.J., Veenstra J., Boer K., Coutinho R.A., Miedema F., Schuitemaker H. Macrophage-tropic variants initiate human immunodeficiency virus type 1 infection after sexual, parenteral, and vertical transmission. J. Clin. Invest. 1994;94:2060–2067. doi: 10.1172/JCI117560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker L.M., Huber M., Doores K.J., Falkowska E., Pejchal R., Julien J.P., Wang S.K., Ramos A., Chan-Hui P.Y., Moyle M., Mitcham J.L., Hammond P.W., Olsen O.A., Phung P., Fling S., Wong C.H., Phogat S., Wrin T., Simek M.D., Koff W.C., Wilson I.A., Burton D.R., Poignard P. Broad neutralization coverage of HIV by multiple highly potent antibodies. Nature. 2011;477:466–470. doi: 10.1038/nature10373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei X., Decker J.M., Wang S., Hui H., Kappes J.C., Wu X., Salazar-Gonzalez J.F., Salazar M.G., Kilby J.M., Saag M.S., Komarova N.L., Nowak M.A., Hahn B.H., Kwong P.D., Shaw G.M. Antibody neutralization and escape by HIV-1. Nature. 2003;422:307–312. doi: 10.1038/nature01470. [DOI] [PubMed] [Google Scholar]
- Westby M., Smith-Burchnell C., Mori J., Lewis M., Mosley M., Stockdale M., Dorr P., Ciaramella G., Perros M. Reduced maximal inhibition in phenotypic susceptibility assays indicates that viral strains resistant to the CCR5 antagonist maraviroc utilize inhibitor-bound receptor for entry. J. Virol. 2007;81:2359–2371. doi: 10.1128/JVI.02006-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X., Yang Z.Y., Li Y., Hogerkorp C.M., Schief W.R., Seaman M.S., Zhou T., Schmidt S.D., Wu L., Xu L., Longo N.S., McKee K., O'Dell S., Louder M.K., Wycuff D.L., Feng Y., Nason M., Doria-Rose N., Connors M., Kwong P.D., Roederer M., Wyatt R.T., Nabel G.J., Mascola J.R. Rational design of envelope identifies broadly neutralizing human monoclonal antibodies to HIV-1. Science. 2010;329:856–861. doi: 10.1126/science.1187659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou T., Georgiev I., Wu X., Yang Z.Y., Dai K., Finzi A., Kwon Y.D., Scheid J.F., Shi W., Xu L., Yang Y., Zhu J., Nussenzweig M.C., Sodroski J., Shapiro L., Nabel G.J., Mascola J.R., Kwong P.D. Structural basis for broad and potent neutralization of HIV-1 by antibody VRC01. Science. 2010;329:811–817. doi: 10.1126/science.1192819. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Model for antibody induced emergence of CXCR4-using virus