Main Text
Members of the two families of endo/sarcoplasmic reticulum calcium release channels, ryanodine receptors (RyRs) and IP3 receptors, share the same general architecture. They consist of an ionic channel domain inserted into an internal cell membrane, coupled to a very large cytoplasmic domain through which the channels are controlled and then interact with each other. RyRs were originally identified and purified based on the high affinity for the plant alkaloid ryanodine, defined by the work of I. N. Pessah (1). Although present in a variety of tissues, RyRs have been the special subjects of long-term inquiries in cross-striated muscles where they are relatively abundant and are located at calcium release units (CRUs). RyRs, within CRUs, are the sites of the rapid calcium release that initiates muscle activation in excitation-contraction coupling. In vertebrates, RyR1, RyR2, and RyR3 are the products of three separate genes (2) with various splice variants. In insects, a single gene, first identified by Takeshima et al. (3) in 1994, codes for invertebrate RyR.
The three vertebrate isoforms differ significantly in properties and their role is defined by their muscle- and fiber-type-specific expression and further by their location and/or interaction with other proteins of CRUs. RyR1 is an essential component of all skeletal muscle fibers: the channel is strictly located in the junctional gap between T tubules and sarcoplasmic reticulum (SR) junctional position and is involved in a direct, reciprocal talk with the T tubules’ voltage-sensing channel. RyR3 is an accessory channel that is present in some but not all skeletal muscle fibers, and is located in a well-defined parajunctional position proximal to, but separated from, the junctional gap. It has a peripheral enhancing effect on Ca release events, which, however, is not essential to E-C coupling. RyR2 is limited to cardiac muscle and, interestingly it has a more varied disposition than the other two varieties: it may be located either in the junctional gap between SR and T tubules/plasmalemma, or on the free surface of corbular or extended junctional SR elements within the depth of the cell. As RyR1, RyR2 has an essential role in E-C coupling, but differently for RyR1 an indirect calcium-activated calcium release mechanism is invoked as its mode of activation, and this may even involve a spread from one RyR2 cluster to another in the absence of T tubules.
In a muscle like myocardium, which requires continuous repetitive activity, calcium homeostasis has to be continuously balanced, because even a minor imbalance has dire effects; hence the necessity for a full understanding of RyR2-RyR2 relationships in cardiac muscle, which is the subject of a recent publication in this issue of the Biophysical Journal (4). The article explores the interactions between cytoplasmic domains of RyR2 revealed by their self-association in vitro. The data are most timely because, 44 years after their first detection and 27 years after their initial purification and characterization, RyRs have come of age with the publication of three reports of near-atomic resolution structure for the entire rabbit RyR1 (e.g., Yan et al. (5)) based on recent advances in single-particle cryo-electron microscopy. The high-resolution electron tomography images were preceded and more recently supplemented by MRI and x-ray diffraction images of several key domains that could not be sufficiently well resolved in the images of the whole molecule (6). The structure of the permeation channel, and its connections to the cytoplasmic vestibule that harbors numerous disease-related hot spots, are well defined and questions of molecular interactions can be addressed directly.
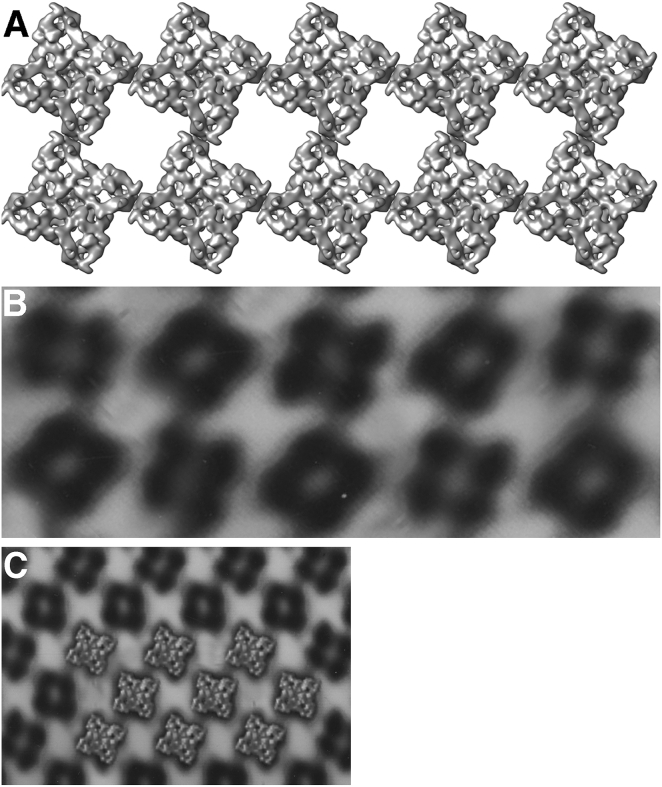
This brings us to the question of why definition of the spatial relationship between clustered RyR2s in cardiac cells is still a matter for debate. By contrast, skeletal muscle RyR1s are well known to form a stable checkerboard pattern in vivo that extends along the junctional SR-facing T tubule and which is sufficiently stable to survive at least part of the time in isolated SR vesicles, despite the drastic change in shape (Fig. 1 A; Paolini et al. (7)). The same pattern is formed in vitro, demonstrating that it is based exclusively on an interaction between the channels’ cytoplasmic domains that come to contact with each other. RyR3s, although parajunctional and thus facing the cytoplasm rather than T tubules, also form a specific arrangement separate and distinct from that of RyR1. RyR2s are grouped within clusters in both junctional and corbular SR, although the grouping is often incomplete, with some uneven gaps (8, 9). However, no images of fully ordered cardiac RyR2 arrays, comparable to those seen in skeletal muscle, have been provided to-date. One possible key to the mystery is provided by en face views of junctional SR surfaces obtained by dual-tilt electron tomography suggesting that the arrangement is quite variable, consisting of at least two predominant channel-to channel positions that are also labile and capable of relatively rapid changes (10).
Figure 1.
Illustration of possible RyR arrays. (A) The checkerboard array of RyR1 in skeletal muscle involving domain 6 is well established, quite stable, and is achieved in vivo, within CRUs as well as in vitro, in the absence of other proteins. Diagram is courtesy of Monserrat Samsó. (B) Body muscles of various arthropods have CRUs with extensive arrays of RyRs. A filtered and two-dimensional reconstructed image from the RyR array of a scorpion tail muscle shows two alternate orientations of RyRs. (C) Profiles of RyR2, oriented as predicted by the “oblique interaction” (Cabra et al. (4), their Fig. 4 C) fit quite well over the scorpion array, suggesting that this may be a possible disposition of RyR2 in cardiac muscle (Cabra et al. (4), their Fig. 5 C). Courtesy of Monserrat Samsó.
Cabra et al. (4) offer a somewhat similar solution, based on an independent approach. They simply observe pairwise interactions formed by channels in a dilute sample and convincingly argue for selecting one configuration in which the two participating channels show the most stable positioning as representing the most likely mode of RyR2-RyR2 interaction. The proposed model in Fig. 4 C in Cabra et al. (4) is quite consistent with the latest high-resolution modeling of the spry1 and tandem repeat domains in association with FKBP12 at the relevant corner of the cytoplasmic domain (11). The stochastic model of Fig. 5 in Cabra et al. (4) would again suggest a less rigid configuration than seen in skeletal muscle, but one that may still allow the various types of functional interactions of which the channels are capable. On the other hand, muscles of invertebrates offer an alternative, highly ordered and apparently quite stable disposition of feet with RyR-RyR contacts that are more similar to those of RyR2 than RyR1 and a dimeric unit cell (Fig. 1 B, Loesser et al. (12)). This disposition is actually consistent with the oblique interaction modeled in Cabra et al. (4), as illustrated in Fig. 1 C here and more precisely in Fig. 5 C in Cabra et al. (4), suggesting that a stable configuration may be achieved by RyR2. This author hopes that the new data represent a beginning of a fruitful discussion, and she looks forward to the next installments of the RyR array saga.
Acknowledgments
I thank M. Samsó for discussion.
Supported by grant No. 2PO1-AR-052354-06A1 to P.D. Allen (UC Davis, CA).
Editor: Eric Sobie.
References
- 1.Pessah I.N., Waterhouse A.L., Casida J.E. The calcium-ryanodine receptor complex of skeletal and cardiac muscle. Biochem. Biophys. Res. Comm. 1985;128:449–456. doi: 10.1016/0006-291x(85)91699-7. [DOI] [PubMed] [Google Scholar]
- 2.Sutko J.L., Airey J.A. Ryanodine receptor Ca2+ release channels: does diversity in form equal diversity in function? Physiol. Rev. 1996;76:1027–1071. doi: 10.1152/physrev.1996.76.4.1027. [DOI] [PubMed] [Google Scholar]
- 3.Takeshima H., Nishi M., Hotta Y. Isolation and characterization of a gene for a ryanodine receptor/calcium release channel in Drosphila melanogaster. FEBS Lett. 1994;337:81–87. doi: 10.1016/0014-5793(94)80634-9. [DOI] [PubMed] [Google Scholar]
- 4.Cabra V., Murayama T., Samsó M. Ultrastructural analysis of self-associated RyR2s. Biophys. J. 2016;110:2651–2662. doi: 10.1016/j.bpj.2016.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yan Z., Bai X.C., Yan N. Structure of the rabbit ryanodine receptor RyR1 at near-atomic resolution. Nature. 2015;517:50–55. doi: 10.1038/nature14063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tung C.C., Lobo P.A., van Petegem F. The amino-terminal disease hotspot of ryanodine receptors forms a cytoplasmic vestibule. Nature. 2010;468:585–588. doi: 10.1038/nature09471. [DOI] [PubMed] [Google Scholar]
- 7.Paolini C., Protasi F., Franzini-Armstrong C. The relative position of RyR feet and DHPR tetrads in skeletal muscle. J. Mol. Biol. 2004;342:145–153. doi: 10.1016/j.jmb.2004.07.035. [DOI] [PubMed] [Google Scholar]
- 8.Hayashi T., Martone M.E., Hoshijima M. Three-dimensional electron microscopy reveals new details of membrane systems for Ca2+ signaling in the heart. J. Cell Sci. 2009;122:1005–1013. doi: 10.1242/jcs.028175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Baddeley D., Jayasinghe I.D., Soeller C. Optical single-channel resolution imaging of the ryanodine receptor distribution in rat cardiac myocytes. Proc. Natl. Acad. Sci. USA. 2009;106:22275–22280. doi: 10.1073/pnas.0908971106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Asghari P., Scriven D.R., Moore E.D. Nonuniform and variable arrangements of ryanodine receptors within mammalian ventricular couplons. Circ. Res. 2014;115:252–262. doi: 10.1161/CIRCRESAHA.115.303897. [DOI] [PubMed] [Google Scholar]
- 11.Yuchi Z., Yuen S.M., van Petegem F. Crystal structures of ryanodine receptor SPRY1 and tandem-repeat domains reveal a critical FKBP12 binding determinant. Nat. Commun. 2015;6:7947. doi: 10.1038/ncomms8947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Loesser K.E., Castellani L., Franzini-Armstrong C. Dispositions of junctional feet in muscles of invertebrates. J. Muscle Res. Cell Motil. 1992;13:161–173. doi: 10.1007/BF01874153. [DOI] [PubMed] [Google Scholar]