Abstract
Genetic factors affect host susceptibility to pathogens. In this population-based case control study, we explored the genetic polymorphisms of IL-17, TLR4 and miR-146a in association with pulmonary tuberculosis in a Chinese Han population. We recruited 1601 pulmonary tuberculosis patients matched with 1526 healthy controls and genotyped twelve functional single nucleotide polymorphisms (SNPs). After the correction for multiple comparisons, two SNPs (rs10759932 and rs2737190) in the TLR4 gene remained significant. Individuals carrying the rs2737190-AG genotype (vs. AA) had a significantly increased risk of either clinical tuberculosis (OR: 1.31, 95% CI: 1.11–1.53) or sputum smear-positive tuberculosis (OR: 1.35, 95% CI: 1.13–1.61). Stratification analysis revealed that the effects of genetic variations on tuberculosis were more evident among non-smokers. People with haplotype TLR4 rs10983755G–rs10759932C had a significantly increased risk of tuberculosis (OR: 3.43, 95% CI: 2.34–5.05). Moreover, we found that SNPs of rs3819024 in IL-17A and rs763780 in IL-17F were weakly related to a prognosis of tuberculosis. Our results suggest that genetic polymorphisms of IL-17 and TLR4 may play a role in host susceptibility to tuberculosis in the Chinese Han population. More work is necessary to identify specific causative variants of tuberculosis underlying the observed associations.
Tuberculosis is a chronic infectious disease caused by the pathogen of Mycobacterium tuberculosis (MTB), and has been a major public health problem worldwide1. An estimated 9 million people developed active tuberculosis, and 1.5 million died from it in 2013, mostly in developing countries2. The outcome of MTB infection ranges from complete pathogen clearance to asymptomatic latent infection to active tuberculosis disease. Most infected individuals are in the latent period, and only 5–10% will progress to the active phase during their lifetimes3,4,5. Researchers have shown that the innate and adaptive immune responses play an important role in the control of MTB infection6.
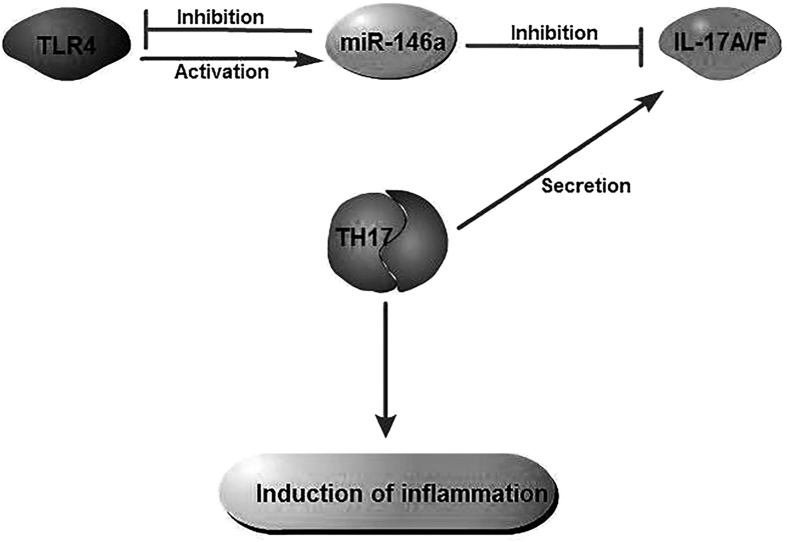
CD4(+) T cells play a critical role during MTB infection by regulating the immune response and mediating host protection. Th1 and Th17 cells are the main effector CD4(+) T cells7. Th1 cells contribute to tuberculosis protection by secreting IFN-γ and activating the antimycobacterial reaction in macrophages7. Th17 cells are interleukin (IL)-17-producing CD4+ T cells with implications in inducing neutrophilic inflammation and mediate tissue damage7,8. Antimicrobial inflammatory response primarily begins through the initial sensing of different pathogen-associated molecular patterns by the pattern recognition receptors of the host9. Amongst the innate immune receptors, Toll-like receptors (TLRs) have the unique capacity to sense the initial infection and are the most potent inducers of the immune responses9. Toll-like receptor 4 (TLR4) is the main receptor mediating the signals responsible for the production of IL-17A induced by MTB10. The deficiency of TLR4 inhibits Th17 cell differentiation by suppressing the Signal Transducer and Activator of Transcription 3 (STAT3) pathway and promoting Th1 cell differentiation by enhancing the STAT1 pathway11. As shown in Fig. 1, microRNA-146a (miR-146a) is also involved in the host immune response to MTB infection by acting as a negative feedback regulator of the TLR/NF-kB pathway and potentially participating in regulating IL-17 expression by targeting the 3′-untranslated region (UTR) of the TRAF6 and the IRAK-1 genes12,13. The activation of innate immunity receptors via a pathogen induces the up-regulation of miR-146a expression and will in turn exert a negative feedback on TLR4, leading to an inhibition of Th17 pathway molecules and pro-inflammatory cytokines (IL-17A, IL-17F, IL-6 and TNF-α) and an attenuation of the inflammatory effect of Th17 cells12.
Figure 1.
Both IL-17A and IL-17F are members of the IL-17 cytokine family. They are located adjacent to one another on the same human chromosome, 6p12, and have similar expression profiles14. The TLR4 gene is located on the long arm of chromosome 9 at position 33.115. Although genetic polymorphisms of IL-17 and TLR4 have gained much more interest in the risk of tuberculosis16,17,18,19,20, few studies have examined their synergistic effect, and a small number of these studies were performed in China. Considering the roles of TLR4, IL-17 and miR-146a in the pro-inflammatory response12, we conducted a population-based case control study in a Chinese Han population, with the goals of exploring whether genetic polymorphisms in IL-17, TLR4, and miR-146a are associated with susceptibility to and the prognosis of pulmonary tuberculosis.
Materials and Methods
Study design and study population
This study has a mixed case control and prospective follow-up design. We recruited 1601 pulmonary tuberculosis patients from Jiangsu province, China since 2011. They were genetically-unrelated Chinese Han individuals. Patients were aged 18 years or older, without HIV infection, cancer or autoimmune diseases. Tuberculosis cases were group-matched (by sex and age) with 1526 controls from a pool of individuals who participated in the community-based health examination programs. Individuals with a history of tuberculosis, diabetes, malignancy, HIV and immunosuppressive conditions were excluded. This study was approved by the ethics committee of Nanjing Medical University (No: 2012-0105, Date: Jan 5, 2012). The methods were carried out in accordance with the approved guidelines. Written informed consent was obtained from all participants. The manuscript was drafted according to the STROBE statement (http://www.strobe-statement.org/).
Diagnosis of tuberculosis
Tuberculosis cases were diagnosed by specialized doctors following the guidelines recommended by the China Ministry of Health, which were based on clinical symptoms and signs, chest x-ray examination, sputum smear tests or sputum culture (http://www.chinatb.org). Three sputum samples were collected from each subject with labelled plastic bottles. The Ziehl-Neelsen hot staining method was used for sputum smear microscopy. If the equipment and technology allowed, the culture was carried out. In brief, sputum samples were decontaminated with 4% sodium hydroxide (NaOH), centrifuged and then cultured on Lowenstein-Jensen (LJ) culture media21. The LJ culture media were incubated at 37 °C. Identification of MTB was done using the p-nitrobenzoic acid (PNB) and thiophene carboxylic acid hydrazine (TCH) resistance test. Growth in LJ medium containing PNB indicates that the bacilli do not belong to the MTB complex. Species other than MTB were excluded from the current analysis.
Data collection
Trained local health facility staff interviewers administered a risk factor questionnaire to all participants. The collected data included demographic characteristics, tobacco smoking, alcohol drinking, medical history and laboratory tests. Patients were followed to obtain information on their therapeutic regimens, treatment adherence and outcomes. After informed consent was obtained, a blood sample was collected from each participant for molecular analyses.
SNP selection and genotyping
We selected SNPs in the IL-17 and TLR4 genes based on the following criteria: (1) minor allele frequency (MAF) ≥ 0.05 in the Chinese Han population; (2) Hardy-Weinberg equilibrium test: P ≥ 0.05; and (3) SNPs located in the functional areas such as 5′-UTR, 5′ near the gene, exon or 3′-UTR. In addition, a functional polymorphism in the miR-146a gene was also selected for genotyping (http://www.bioguo.org/miRNASNP2/). As a result, twelve SNPs were genotyped, including four SNPs in IL-17A (rs2275913, rs3819024, rs8193036 and rs3748067), one SNP in IL-17F (rs763780), six SNPs in TLR4 (rs1927914, rs10759932, rs2737190, rs10983755, rs7873784, rs11536889) and one SNP in miR-146a (rs2910164). Genomic DNA was extracted from leukocytes in the peripheral blood sample by proteinase K digestion and phenol/chloroform extraction. The primer and probe sequences for each SNP were showed in Table 1. According to the manufacturer’s instructions, we genotyped SNPs using the TaqMan allelic discrimination technology on the 384-well ABI 7900HT Real-Time PCR System (Applied Biosystems, Foster City, CA, USA), ascertained using SDS software (version 2.3)22. Amplification was performed under the following conditions: 50 °C for 2 min, 95 °C for 10 min followed by 45 cycles of 95 °C for 15 s and 60 °C for 1 min. The success rate for each SNP was over 96%. To avoid batch bias, we allocated DNA samples of both cases and controls in each plate with no discrepancies between the reaction conditions. Approximately 10% of the samples were randomly selected for repeat genotyping for confirmation, and the results were 100% concordant.
Table 1. Primers and probes designed for genotyping.
| Gene | SNPs | Primer (5′-3′) | Probe |
|---|---|---|---|
| IL-17A | rs2275913 | F-TGAATTTCTGCCCTTCCCATT | A: FAM-CTTCAGAAGAAGAGATT-MGB |
| G > A | R-GGTTCAGGGGTGACACCATTT | G: HEX-TTCAGAAGGAGAGATT-MGB | |
| rs3819024 | F-CCGGAATTGTCTCCACAACAC | G: FAM-AATCTGTGAGGGAAAG-MGB | |
| A > G | R-TGTACCTTGATTTTCCATTTGATCTT | A: HEX-AGGAATCTGTGAGGAAA-MGB | |
| rs8193036 | F-CTCCTTTCTAGTTCTCATCACTCTCTACTC | G: FAM-CTTTTCTCCATCTTCA-MGB | |
| C > T | R-TGTTTTGAGGAAGGAATTGAAAATG | A: HEX-CTTTTCTCCATCTCCA-MGB | |
| rs3748067 | F-TGAGTTTTTATTTTACTTGGGCTGAA | G: FAM-TTCTCATACTTAAAGTTC-MGB | |
| G > A | R-CAACCCAGAAAGGAGCTGATG | A: HEX-TTCTCATACTTAAAATTC-MGB | |
| IL-17F | rs763780 | F-GAGAAGGTGCTGGTGACTGTTG | G: FAM-CCTGTCATCCACCGTG-MGB |
| T > C | R-CTTCTTCAGCTGAGTGGATATGCA | A: HEX-CCTGTCATCCACCATG-MGB | |
| TLR4 | rs1927914 | F-GAAGTGCTTGGAGGATATTACAGTAGAA | G: FAM-CTAGGACTTAGCATGCATA-MGB |
| T > C | R-GAACTGGCATTTGTAAAGCTTTTAGG | A: HEX-ACTTAGCATACATAATATT-MGB | |
| rs10759932 | F-CCCACAAATGGTGTACAGGAGTT | G: FAM-ATCTTCACCAACGCT-MGB | |
| T > C | R-TGCAAGCTTCTGCTATGATTAAAAG | A: HEX-CATCTTCACCAACACT-MGB | |
| rs10983755 | F-ACCACAAAATGGTCCCTCACA | G: FAM-CTTGGTTTTTGACACGTT-MGB | |
| G > A | R-TTCTACTGTAATATCCTCCAAGCACTTC | A: HEX-TTGGTTTTTGACACATTG-MGB | |
| rs2737190 | F-GGAGCATGCCTTATGCACACT | T: FAM-ACCCAAGTAGACACTGT-MGB | |
| A > G | R-GACCTGTGATGATTAGGGCTGAA | C: HEX-ACCCAAGTAGACACCGT-MGB | |
| rs7873784 | F-AGAACACTTAACATGAGAGGTACCC | C: FAM-TTCATTATACGAACTCTGC-MGB | |
| C > G | R-GATGAATTAGCTCTAAAGATCAGCTGT | G: HEX-TTCATTATAGGAACTCTGC-MGB | |
| rs11536889 | F-GTTGGGCAATGCTCCTTGA | G: FAM-ATTTTGGGAAGAGTGGAT-MGB | |
| G > C | R-GAACCCCATTAATTCCAGACACA | C: HEX-CACATTTTGGGAACAGT-MGB | |
| miR-146a | rs2910164 | F-GAACTGAATTCCATGGGTTGTGT | G: FAM-TCAGACCTGTGAAATT-MGB |
| C > G | R-GCCCACGATGACAGAGATATCC | C: HEX-TCAGACCTCTGAAATT-MGB |
Statistical analysis
Data were entered with EpiData 3.1 software (Denmark) and analyzed using STATA 10.0 (StataCorp, College Station, TX, USA). Student’s t-test (for continuous variables) and the χ2 test (for categorical variables) were used to analyze the differences in demographic variables and potential risk factors between cases and controls. Hardy-Weinberg equilibrium (HWE) was tested using a goodness-of-fit χ2 test by comparing the observed genotype frequencies with the expected frequencies among the controls to make sure that the alleles were independently segregated. An unconditional logistic regression model was carried out to analyze the associations between genotypes and the risk of tuberculosis by calculating the odds ratio (OR) and 95% confidence interval (CI). The relative risk (RR) and 95% CI were calculated to evaluate the effect of genetic polymorphisms on the patient prognoses. To control for potential confounding, we adjusted for age, sex, tobacco smoking and alcohol drinking. To comprehensively analyze the effect of SNPs, we applied three different genetic models: additive model, dominant model and recessive model. IL-17A and TLR4 haplotypes were performed using phase 2.1 software. Bonferroni corrections were applied for multiple comparisons.
Results
General characteristics of the study subjects
Demographic characteristics of the cases and controls are shown in Table 2. In total, 1601 tuberculosis cases (73.8% males and 26.2% females) and 1526 controls (72.9% males and 27.1% females) were recruited. The average (±standard deviation, SD) age was 52.1(±17.7) years in cases and 52.4(±17.0) years in controls. Due to the frequency matching, there was no significant difference in the distribution of age and sex between the two groups. The proportion of ever smokers was 52.4% among cases, which was significantly higher than that in controls (36.0%) (χ2 = 84.73, P < 0.001). Alcohol drinking was inversely related to tuberculosis, and 22.2% of the cases vs. 26.8% of the controls had a history of alcohol consumption (χ2 = 9.06, P = 0.003).
Table 2. General characteristics of the cases and controls.
| Variables | Case (n = 1601) n(%) | Control (n = 1526) n(%) | t/χ2 | P |
|---|---|---|---|---|
| Age(years) | ||||
| Mean ± SD | 52.1 ± 17.7 | 52.4 ± 17.0 | 0.564 | 0.573 |
| Sex | 0.321 | 0.571 | ||
| Male | 1181(73.8) | 1112(72.9) | ||
| Female | 420(26.2) | 414(27.1) | ||
| Smoking | 84.730 | <0.001 | ||
| Never | 762(47.6) | 976(64.0) | ||
| Ever | 839(52.4) | 550(36.0) | ||
| Drink | 9.065 | 0.003 | ||
| Never | 1246(77.8) | 1117(73.2) | ||
| Ever | 355(22.2) | 409(26.8) | ||
| Sputum smear test | – | – | ||
| Positive | 1080(67.5) | – | ||
| Negative | 521(32.5) | – | ||
Risk analysis
Except for rs1927914 (P = 0.012), the genotype distributions of the eleven SNPs were all in HWE in the controls (P = 0.43 for rs2275913, P = 0.41 for rs3819024, P = 0.84 for rs8193036, P = 0.12 for rs3748067, P = 0.06 for rs763780, P = 0.10 for rs10759932, P = 0.07 for rs2737190, P = 0.34 for rs10983755, P = 0.60 for rs7873784, P = 0.98 for rs11536889 and P = 0.33 for rs2910164). As shown in Table 3, if we set the test level at 0.002 (0.05/11*2) to consider both the multiple comparisons of 11 SNPs and genotypes of each SNP, two SNPs (rs10759932 and rs2737190) were found to be significantly associated with the risk of tuberculosis. For SNP rs2737190, individuals carrying the AG genotype had a significantly increased risk of either clinical tuberculosis (OR: 1.31, 95% CI: 1.11–1.53) or sputum smear-positive tuberculosis (OR: 1.35, 95% CI: 1.13–1.61). For SNP rs10759932, the association was only significant for clinical tuberculosis, where the TC/CC carrier had a 27% increased risk (OR: 1.27, 95% CI: 1.09–1.46).
Table 3. Genotype distributions of the eleven SNPs among the cases and controls.
| Gene | SNPs | Control (n = 1526) n(%) | Total cases(n = 1601) | Smear-positive cases (n = 1080) | ||||
|---|---|---|---|---|---|---|---|---|
| n(%) | OR(95%Cl)a | P | n(%) | OR(95%Cl)a | P | |||
| IL-17A | rs2275913 | |||||||
| GG | 450(29.6) | 477(31.7) | 1 | 309(30.7) | 1 | |||
| GA | 741(48.7) | 729(48.4) | 0.89(0.75–1.06) | 0.192 | 494(49.2) | 0.94(0.77–1.13) | 0.500 | |
| AA | 331(21.7) | 301(20.0) | 0.79(0.64–0.97) | 0.028 | 202(20.1) | 0.82(0.65–1.04) | 0.098 | |
| Add | 0.89(0.80–0.99) | 0.027 | 0.91(0.81–1.02) | 0.106 | ||||
| Dom | 0.86(0.73–1.01) | 0.066 | 0.90(0.75–1.08) | 0.249 | ||||
| Rec | 0.85(0.71–1.02) | 0.074 | 0.85(0.70–1.05) | 0.129 | ||||
| G | 1641(53.9) | 1683(55.8) | 1 | 1112(55.3) | 1 | |||
| A | 1403(46.1) | 1331(44.2) | 0.93(0.84–1.02) | 0.131 | 898(44.7) | 0.95(0.84–1.06) | 0.323 | |
| rs3819024 | ||||||||
| AA | 422(27.7) | 442(28.2) | 1 | 284(26.8) | 1 | |||
| AG | 745(48.9) | 784(50.0) | 0.98(0.83–1.16) | 0.816 | 544(51.4) | 1.06(0.88–1.29) | 0.547 | |
| GG | 358(23.5) | 341(21.8) | 0.85(0.69–1.04) | 0.110 | 230(21.7) | 0.90(0.71–1.13) | 0.355 | |
| Add | 0.92(0.83–1.02) | 0.124 | 0.95(0.85–1.07) | 0.401 | ||||
| Dom | 0.94(0.80–1.10) | 0.421 | 1.01(0.84–1.21) | 0.942 | ||||
| Rec | 0.86(0.72–1.02) | 0.081 | 0.86(0.71–1.05) | 0.137 | ||||
| A | 1589(52.1) | 1668(53.2) | 1 | 1112(52.6) | 1 | |||
| G | 1461(47.9) | 1466(46.8) | 0.96(0.87–1.06) | 0.376 | 1004(47.4) | 0.98(0.88–1.10) | 0.748 | |
| rs8193036 | ||||||||
| CC | 783(51.4) | 789(51.1) | 1 | 515(49.7) | 1 | |||
| CT | 621(40.7) | 618(40.0) | 1.00(0.86–1.17) | 0.966 | 429(41.4) | 1.07(0.90–1.27) | 0.433 | |
| TT | 120(7.9) | 137(8.9) | 1.22(0.93–1.60) | 0.156 | 92(8.9) | 1.26(0.93–1.71) | 0.128 | |
| Add | 1.06(0.95–1.19) | 0.312 | 1.10(0.97–1.25) | 0.135 | ||||
| Dom | 1.04(0.90–1.20) | 0.623 | 1.10(0.94–1.30) | 0.246 | ||||
| Rec | 1.22(0.93–1.58) | 0.146 | 1.23(0.92–1.64) | 0.172 | ||||
| C | 2187(71.8) | 2196(71.1) | 1 | 1459(70.4) | 1 | |||
| T | 861(28.2) | 892(28.9) | 1.03(0.92–1.15) | 0.580 | 613(29.6) | 1.07(0.94–1.21) | 0.300 | |
| rs3748067 | ||||||||
| GG | 1094(71.8) | 1135(71.2) | 1 | 757(70.5) | 1 | |||
| GA | 385(25.3) | 415(26.1) | 1.06(0.89–1.25) | 0.528 | 286(26.6) | 1.09(0.90–1.31) | 0.369 | |
| AA | 45(3.0) | 43(2.7) | 1.02(0.66–1.58) | 0.935 | 31(2.9) | 1.13(0.70–1.83) | 0.607 | |
| Add | 1.04(0.90–1.19) | 0.592 | 1.08(0.93–1.26) | 0.325 | ||||
| Dom | 1.05(0.90–1.23) | 0.540 | 1.09(0.92–1.31) | 0.327 | ||||
| Rec | 1.00(0.65–1.55) | 0.984 | 1.11(0.69–1.79) | 0.670 | ||||
| G | 2573(84.4) | 2685(84.3) | 1 | 1800(83.8) | 1 | |||
| A | 475(15.6) | 501(15.7) | 1.01(0.88–1.16) | 0.878 | 348(16.2) | 1.05(0.90–1.22) | 0.549 | |
| IL-17F | rs763780 | |||||||
| TT | 1175(77.0) | 1225(77.6) | 1 | 840(79.0) | 1 | |||
| TC | 318(20.9) | 323(20.5) | 1.00(0.84–1.20) | 0.974 | 207(19.5) | 0.95(0.77–1.16) | 0.586 | |
| CC | 32(2.1) | 31(2.0) | 0.91(0.54–1.52) | 0.713 | 16(1.5) | 0.67(0.36–1.25) | 0.207 | |
| Add | 0.99(0.85–1.15) | 0.866 | 0.91(0.76–1.08) | 0.269 | ||||
| Dom | 0.99(0.84–1.18) | 0.947 | 0.92(0.76–1.12) | 0.397 | ||||
| Rec | 0.91(0.54–1.51) | 0.710 | 0.68(0.36–1.26) | 0.219 | ||||
| T | 2668(87.5) | 2773(87.8) | 1 | 1887(88.8) | 1 | |||
| C | 382(12.5) | 385(12.2) | 0.97(0.83–1.13) | 0.690 | 239(11.2) | 0.89(0.75–1.05) | 0.162 | |
| TLR4 | rs10759932 | |||||||
| TT | 779(51.4) | 722(45.7) | 1 | 505(47.2) | 1 | |||
| TC | 597(39.4) | 697(44.1) | 1.27(1.09–1.48) | 0.002b | 458(42.8) | 1.18(1.00–1.40) | 0.054 | |
| CC | 140(9.2) | 161(10.2) | 1.25(0.97–1.61) | 0.090 | 107(10.0) | 1.20(0.91–1.60) | 0.199 | |
| Add | 1.17(1.05–1.31) | 0.005 | 1.13(1.00–1.28) | 0.053 | ||||
| Dom | 1.27(1.09–1.46) | 0.001b | 1.19(1.01–1.39) | 0.037 | ||||
| Rec | 1.12(0.87–1.43) | 0.378 | 1.12(0.85–1.47) | 0.430 | ||||
| T | 2155(71.1) | 2141(67.8) | 1 | 1468(68.6) | 1 | |||
| C | 877(28.9) | 1019(32.2) | 1.17(1.05–1.30) | 0.005 | 672(31.4) | 1.13(1.00–1.27) | 0.055 | |
| rs2737190 | ||||||||
| AA | 557(37.0) | 518(32.6) | 1 | 343(32.0) | 1 | |||
| AG | 690(45.8) | 840(52.9) | 1.31(1.11–1.53) | 0.001b | 580(54.1) | 1.35(1.13–1.61) | 0.001b | |
| GG | 259(17.2) | 231(14.5) | 0.95(0.76–1.18) | 0.628 | 150(14.0) | 0.93(0.73–1.19) | 0.566 | |
| Add | 1.03(0.93–1.15) | 0.559 | 1.03(0.92–1.16) | 0.601 | ||||
| Dom | 1.21(1.04–1.41) | 0.015 | 1.23(1.04–1.46) | 0.015 | ||||
| Rec | 0.81(0.66–0.99) | 0.038 | 0.78(0.62–0.98) | 0.030 | ||||
| A | 1804(59.9) | 1876(59.0) | 1 | 1266(59.0) | 1 | |||
| G | 1208(40.1) | 1302(41.0) | 1.04(0.94–1.15) | 0.489 | 880(41.0) | 1.04(0.93–1.16) | 0.516 | |
| rs10983755 | ||||||||
| GG | 793(52.1) | 806(50.7) | 1 | 551(51.5) | 1 | |||
| GA | 600(39.4) | 644(40.5) | 1.06(0.91–1.23) | 0.450 | 424(39.6) | 1.02(0.86–1.21) | 0.829 | |
| AA | 128(8.4) | 139(8.7) | 1.06(0.81–1.38) | 0.672 | 95(8.9) | 1.07(0.80–1.44) | 0.636 | |
| Add | 1.04(0.93–1.17) | 0.472 | 1.03(0.91–1.17) | 0.650 | ||||
| Dom | 1.06(0.92–1.22) | 0.428 | 1.03(0.88–1.21) | 0.731 | ||||
| Rec | 1.03(0.80–1.34) | 0.808 | 1.07(0.80–1.42) | 0.665 | ||||
| G | 2186(71.9) | 2256(71.0) | 1 | 1526(71.3) | 1 | |||
| A | 856(28.1) | 922(29.0) | 1.04(0.94–1.17) | 0.446 | 614(28.7) | 1.03(0.91–1.16) | 0.664 | |
| rs7873784 | ||||||||
| GG | 1271(83.9) | 1310(83.1) | 1 | 876(82.2) | 1 | |||
| GC | 235(15.5) | 256(16.2) | 1.07(0.88–1.31) | 0.476 | 181(17.0) | 1.13(0.91–1.41) | 0.265 | |
| CC | 9(0.6) | 11(0.7) | 1.38(0.56–3.41) | 0.489 | 9(0.8) | 1.76(0.68–4.54) | 0.244 | |
| Add | 1.09(0.91–1.31) | 0.362 | 1.16(0.95–1.42) | 0.144 | ||||
| Dom | 1.09(0.89–1.32) | 0.411 | 1.15(0.93–1.43) | 0.192 | ||||
| Rec | 1.36(0.55–3.37) | 0.505 | 1.72(0.67–4.44) | 0.262 | ||||
| G | 2777(91.7) | 2876(91.2) | 1 | 1933(90.7) | 1 | |||
| C | 253(8.3) | 278(8.8) | 1.06(0.89–1.27) | 0.515 | 199(9.3) | 1.13(0.93–1.37) | 0.218 | |
| rs11536889 | ||||||||
| GG | 891(58.7) | 953(60.2) | 1 | 637(59.5) | 1 | |||
| GC | 545(35.9) | 535(33.8) | 0.95(0.81–1.11) | 0.506 | 372(34.7) | 0.99(0.83–1.18) | 0.911 | |
| CC | 83(5.5) | 94(5.9) | 1.04(0.76–1.43) | 0.799 | 62(5.8) | 1.03(0.72–1.47) | 0.859 | |
| Add | 0.98(0.87–1.11) | 0.780 | 1.00(0.88–1.15) | 0.970 | ||||
| Dom | 0.96(0.83–1.11) | 0.602 | 1.00(0.85–1.17) | 0.962 | ||||
| Rec | 1.06(0.78–1.45) | 0.704 | 1.04(0.73–1.47) | 0.840 | ||||
| G | 2327(76.6) | 2441(77.1) | 1 | 1646(76.8) | 1 | |||
| C | 711(23.4) | 723(22.9) | 0.97(0.86–1.09) | 0.606 | 496(23.2) | 0.99(0.87–1.12) | 0.836 | |
| miR-146a | rs2910164 | |||||||
| CC | 537(35.4) | 550(34.7) | 1 | 387(36.1) | 1 | |||
| CG | 715(47.2) | 775(48.9) | 1.08(0.92–1.27) | 0.360 | 505(47.2) | 1.01(0.84–1.21) | 0.927 | |
| GG | 264(17.4) | 259(16.4) | 0.96(0.77–1.19) | 0.691 | 179(16.7) | 0.94(0.74–1.19) | 0.620 | |
| Add | 1.00(0.90–1.10) | 0.934 | 0.98(0.87–1.10) | 0.693 | ||||
| Dom | 1.05(0.90–1.22) | 0.567 | 0.99(0.84–1.17) | 0.909 | ||||
| Rec | 0.92(0.76–1.11) | 0.378 | 0.94(0.76–1.16) | 0.554 | ||||
| C | 1789(59.0) | 1875(59.2) | 1 | 1279(59.7) | 1 | |||
| G | 1243(41.0) | 1293(40.8) | 0.99(0.90–1.10) | 0.884 | 863(40.3) | 0.97(0.87–1.09) | 0.610 | |
aOR: odds ratio; CI: confidence interval, adjusted for age, sex, smoking and drinking.
bSignificant after the Bonferroni correction for multiple comparisons. Add: additive model; Dom: dominant model; Rec: recessive model.
Stratification analysis revealed that the effects of genetic variations on tuberculosis were more evident among non-smokers. Two SNPs of rs10759932 and rs2737190 remained significant after correcting for multiple comparisons among non-smokers (Table 4).
Table 4. The association between eleven SNPs and the risk of tuberculosis stratified by smoking.
| Gene | SNPs | Never(n = 1738) | Ever (n = 1389) | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Control(%) | Case(%) | OR(95%Cl)a | P | Control(%) | Case(%) | OR(95%Cl)a | P | ||
| IL-17A | rs2275913 | ||||||||
| GG | 290(29.7) | 257(36.3) | 1 | 160(29.3) | 220(27.5) | 1 | |||
| GA | 487(49.9) | 318(44.9) | 0.74(0.59–0.92) | 0.007 | 254(46.4) | 411(51.4) | 1.12(0.87–1.46) | 0.380 | |
| AA | 198(20.3) | 133(18.8) | 0.73(0.55–0.97) | 0.028 | 133(24.3) | 168(21.0) | 0.88(0.65–1.21) | 0.439 | |
| rs3819024 | |||||||||
| AA | 268(27.5) | 234(31.7) | 1 | 154(28.0) | 208(25.1) | 1 | |||
| AG | 492(50.5) | 354(47.9) | 0.80(0.64–1.01) | 0.057 | 253(46.0) | 430(51.9) | 1.25(0.96–1.63) | 0.100 | |
| GG | 215(22.1) | 151(20.4) | 0.77(0.58–1.01) | 0.063 | 143(26.0) | 190(22.9) | 0.96(0.71–1.31) | 0.796 | |
| rs8193036 | |||||||||
| CC | 502(51.5) | 347(47.9) | 1 | 281(51.1) | 442(54.0) | 1 | |||
| CT | 401(41.2) | 300(41.4) | 1.08(0.88–1.33) | 0.458 | 220(40.0) | 318(38.8) | 0.91(0.72–1.15) | 0.437 | |
| TT | 71(7.3) | 78(10.8) | 1.64(1.15–2.33) | 0.006 | 49(8.9) | 59(7.2) | 0.83(0.54–1.26) | 0.372 | |
| rs3748067 | |||||||||
| GG | 696(71.4) | 529(70.0) | 1 | 398(72.5) | 606(72.4) | 1 | |||
| GA | 256(26.3) | 202(26.7) | 1.08(0.87–1.35) | 0.484 | 129(23.5) | 213(25.4) | 1.02(0.79–1.32) | 0.882 | |
| AA | 23(2.4) | 25(3.3) | 1.49(0.83–2.67) | 0.181 | 22(4.0) | 18(2.2) | 0.66(0.34–1.26) | 0.206 | |
| IL-17F | rs763780 | ||||||||
| TT | 747(76.6) | 570(76.2) | 1 | 428(77.8) | 655(78.8) | 1 | |||
| TC | 205(21.0) | 165(22.1) | 1.06(0.84–1.34) | 0.625 | 113(20.5) | 158(19.0) | 0.94(0.71–1.24) | 0.651 | |
| CC | 23(2.4) | 13(1.7) | 0.70(0.35–1.41) | 0.318 | 9(1.6) | 18(2.2) | 1.29(0.57–2.95) | 0.545 | |
| TLR4 | rs10759932 | ||||||||
| TT | 507(52.3) | 323(43.4) | 1 | 272(49.8) | 399(47.8) | 1 | |||
| TC | 372(38.4) | 348(46.7) | 1.47(1.20–1.81) | <0.001b | 225(41.2) | 349(41.8) | 1.04(0.82–1.31) | 0.751 | |
| CC | 91(9.4) | 74(9.9) | 1.24(0.88–1.75) | 0.214 | 49(9.0) | 87(10.4) | 1.23(0.83–1.82) | 0.299 | |
| rs2737190 | |||||||||
| AA | 363(37.8) | 238(31.6) | 1 | 194(35.6) | 280(33.5) | 1 | |||
| AG | 437(45.5) | 402(53.3) | 1.43(1.15–1.78) | 0.001b | 253(46.4) | 438(52.5) | 1.17(0.91–1.49) | 0.215 | |
| GG | 161(16.8) | 114(15.1) | 1.05(0.78–1.41) | 0.757 | 98(18.0) | 117(14.0) | 0.80(0.58–1.12) | 0.200 | |
| rs10983755 | |||||||||
| GG | 514(52.8) | 369(48.9) | 1 | 279(50.9) | 437(52.4) | 1 | |||
| GA | 375(38.5) | 322(42.6) | 1.20(0.98–1.47) | 0.074 | 225(41.1) | 322(38.6) | 0.89(0.71–1.13) | 0.338 | |
| AA | 84(8.6) | 64(8.5) | 1.03(0.72–1.47) | 0.867 | 44(8.0) | 75(9.0) | 1.09(0.72–1.64) | 0.685 | |
| rs7873784 | |||||||||
| GG | 812(83.9) | 606(81.7) | 1 | 459(83.9) | 704(84.3) | 1 | |||
| GC | 151(15.6) | 129(17.4) | 1.14(0.88–1.48) | 0.332 | 84(15.4) | 127(15.2) | 0.99(0.73–1.34) | 0.939 | |
| CC | 5(0.5) | 7(0.9) | 1.92(0.60–6.15) | 0.270 | 4(0.7) | 4(0.5) | 0.89(0.22–3.67) | 0.873 | |
| rs11536889 | |||||||||
| GG | 561(57.8) | 456(61.0) | 1 | 330(60.2) | 497(59.6) | 1 | |||
| GC | 355(36.6) | 254(34.0) | 0.89(0.73–1.10) | 0.276 | 190(34.7) | 281(33.7) | 1.04(0.82–1.32) | 0.720 | |
| CC | 55(5.7) | 38(5.1) | 0.87(0.56–1.35) | 0.538 | 28(5.1) | 56(6.7) | 1.32(0.81–2.14) | 0.267 | |
| miR-146a | rs2910164 | ||||||||
| CC | 327(33.8) | 266(35.3) | 1 | 210(38.3) | 284(34.2) | 1 | |||
| CG | 471(48.7) | 364(48.3) | 0.93(0.75–1.16) | 0.530 | 244(44.4) | 411(49.5) | 1.27(0.99–1.62) | 0.057 | |
| GG | 169(17.5) | 123(16.3) | 0.87(0.66–1.16) | 0.352 | 95(17.3) | 136(16.4) | 1.07(0.77–1.48) | 0.693 | |
aOR: odds ratio; CI: confidence interval, adjusted for age, sex and drinking.
bSignificant after the Bonferroni correction for multiple comparisons.
Prognosis analysis
We followed the treatment outcomes of all tuberculosis cases. Among the cases, 874(54.6%) were cured, 480(30.0%) completed treatment, 57(3.6%) failed to be treated, and 190(11.9%) defaulted. We categorized the outcomes as successful (cured or completed treatment) and unsuccessful. Single SNP analysis showed that rs3819024 in IL-17A and rs763780 in IL-17F were significantly associated with the treatment outcomes of tuberculosis. For the SNP rs3819024, individuals carrying the AG genotype were likely to have a decreased risk of treatment failure when compared with the AA genotype, with an adjusted RR of 0.56 (95% CI: 0.31–1.00) (Table 5). The dominant model showed a 41% decreased risk among individuals carrying variant genotypes (AG/GG), with the adjusted RR of 0.59 (95% CI: 0.34–0.99, P = 0.045). For the SNP rs763780, individuals carrying the TC genotype were likely to have a significantly increased risk of treatment failure when compared with the TT genotype (adjusted RR: 1.84, 95% CI: 1.05–3.14) (Table 5). The dominant model showed a 77% increased risk among individuals carrying variant genotypes (TC/CC), with an adjusted RR of 1.77(95% CI: 1.02–2.99). However, these differences were not significant after Bonferroni correction.
Table 5. The association analysis of genetic polymorphisms and treatment outcomes.
| Gene | SNP | Success (n%) | Failure (n%) | RR(95%Cl)a | P |
|---|---|---|---|---|---|
| IL-17A | rs2275913 | ||||
| GG | 397(30.9) | 23(43.4) | 1 | ||
| GA | 635(49.4) | 21(39.6) | 0.60(0.33–1.08) | 0.089 | |
| AA | 253(19.7) | 9(17.0) | 0.62(0.29–1.32) | 0.219 | |
| Add | 0.75(0.50–1.10) | 0.140 | |||
| Dom | 0.61(0.35–1.04) | 0.069 | |||
| Rec | 0.82(0.40–1.64) | 0.576 | |||
| rs3819024 | |||||
| AA | 376(28.1) | 23(41.1) | 1 | ||
| AG | 680(50.8) | 22(39.3) | 0.56(0.31–1.00) | 0.049 | |
| GG | 283(21.1) | 11(19.6) | 0.64(0.31–1.30) | 0.219 | |
| Add | 0.75(0.52–1.10) | 0.143 | |||
| Dom | 0.59(0.34–0.99) | 0.045 | |||
| Rec | 0.89(0.46–1.68) | 0.719 | |||
| rs8193036 | |||||
| CC | 675(51.3) | 23(42.6) | 1 | ||
| CT | 531(40.3) | 23(42.6) | 1.21(0.69–2.12) | 0.503 | |
| TT | 111(8.4) | 8(14.8) | 2.09(0.95–4.36) | 0.067 | |
| Add | 1.38(0.93–2.05) | 0.107 | |||
| Dom | 1.36(0.80–2.30) | 0.251 | |||
| Rec | 1.91(0.91–3.86) | 0.085 | |||
| rs3748067 | |||||
| GG | 971(71.9) | 38(66.7) | |||
| AG | 343(25.4) | 18(31.6) | 1.29(0.74–2.21) | 0.373 | |
| AA | 37(2.7) | 1(1.8) | 0.75(0.10–4.75) | 0.769 | |
| Add | 1.15(0.71–1.85) | 0.581 | |||
| Dom | 1.24(0.72–2.11) | 0.440 | |||
| Rec | 0.69(0.10–4.38) | 0.712 | |||
| IL-17F | rs763780 | ||||
| TT | 1056(78.6) | 37(66.1) | 1 | ||
| TC | 260(19.3) | 18(32.1) | 1.84(1.05–3.14) | 0.032 | |
| CC | 28(2.1) | 1(1.8) | 1.06(0.14–6.50) | 0.955 | |
| Add | 1.52(0.95–2.43) | 0.082 | |||
| Dom | 1.77(1.02–2.99) | 0.041 | |||
| Rec | 0.90(0.12–5.50) | 0.918 | |||
| TLR4 | rs10759932 | ||||
| TT | 622(46.2) | 23(41.1) | 1 | ||
| TC | 594(44.1) | 27(48.2) | 1.16(0.67–1.98) | 0.601 | |
| CC | 130(9.7) | 6(10.7) | 1.29(0.53–3.02) | 0.573 | |
| Add | 1.14(0.77–1.70) | 0.515 | |||
| Dom | 1.18(0.70–1.98) | 0.537 | |||
| Rec | 1.20(0.51–2.68) | 0.675 | |||
| rs2737190 | |||||
| AA | 452(33.6) | 18(32.1) | 1 | ||
| AG | 703(52.2) | 28(50.0) | 0.98(0.55–1.75) | 0.958 | |
| GG | 191(14.2) | 10(17.9) | 1.34(0.62–2.78) | 0.449 | |
| Add | 1.13(0.76–1.67) | 0.552 | |||
| Dom | 1.06(0.61–1.82) | 0.840 | |||
| Rec | 1.35(0.69–2.60) | 0.378 | |||
| rs10983755 | |||||
| GG | 682(50.7) | 26(45.6) | 1 | ||
| GA | 547(40.6) | 27(47.4) | 1.24(0.73–2.08) | 0.431 | |
| AA | 117(8.7) | 4(7.0) | 0.96(0.34–2.63) | 0.937 | |
| Add | 1.09(0.73–1.62) | 0.690 | |||
| Dom | 1.19(0.71–1.97) | 0.501 | |||
| Rec | 0.87(0.31–2.30) | 0.778 | |||
| rs7873784 | |||||
| GG | 1120(83.3) | 46(82.1) | 1 | ||
| GC | 215(16.0) | 9(16.1) | 1.03(0.51–2.06) | 0.924 | |
| CC | 9(0.7) | 1(1.8) | 2.61(0.35–12.18) | 0.337 | |
| Add | 1.16(0.62–2.17) | 0.643 | |||
| Dom | 1.10(0.56–2.13) | 0.780 | |||
| Rec | 2.60(0.35–12.17) | 0.339 | |||
| rs11536889 | |||||
| GG | 811(60.3) | 31(55.4) | 1 | ||
| GC | 453(33.7) | 23(41.1) | 1.31(0.77–2.21) | 0.312 | |
| CC | 81(6.0) | 2(3.6) | 0.66(0.16–2.63) | 0.568 | |
| Add | 1.07(0.70–1.64) | 0.753 | |||
| Dom | 1.22(0.72–2.03) | 0.453 | |||
| Rec | 0.60(0.14–2.32) | 0.464 | |||
| miR-146a | rs2910164 | ||||
| CC | 468(34.9) | 20(35.7) | 1 | ||
| GC | 660(49.3) | 25(44.6) | 0.91(0.51–1.60) | 0.737 | |
| GG | 212(15.8) | 11(19.6) | 1.18(0.57–2.38) | 0.645 | |
| Add | 1.06(0.73–1.54) | 0.766 | |||
| Dom | 0.98(0.57–1.66) | 0.930 | |||
| Rec | 1.25(0.65–2.36) | 0.495 |
aRR: rate ratio; CI: confidence interval, adjusted for age and sex.
Linkage analysis and haplotype construction
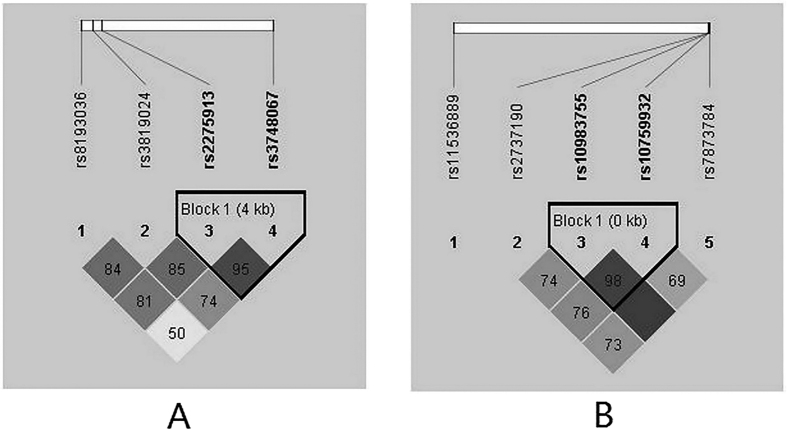
To better understand the genetic associations, the linkage disequilibrium (LD) and haplotype blocks were further assessed. LD analysis was carried out on four SNPs of IL-17A and five SNPs of TLR4. Figure 2 displays the LD plot of SNPs on the same chromosome. With a D’ ≥0.95, two SNPs (rs2275913 and rs3748067) of IL-17A on chromosome 6, as well as two SNPs (rs10983755 and rs10759932) of TLR4 on chromosome 9, were in relatively strong linkage disequilibrium with one another. Thus, we performed a haplotype analysis based on these four SNPs. As shown in Table 6, compared with the common haplotype rs10983755G–rs10759932T, rs10983755G–rs10759932C had a significantly increased risk of tuberculosis (OR: 3.43, 95% CI: 2.34–5.05). This increased risk remained significant after Bonferroni correction for multiple comparisons. No significant haplotypes were found be related to the treatment outcome (data not shown).
Figure 2. Graphical representation of the SNP locations and LD structure.
The SNP distribution and haplotype block structure across IL-17A and TLR4 genes are shown. The measure of LD (D’) among all possible pairs of SNPs is shown graphically according to the shade of color (A/B), where white represents very low D’, and dark represents very high D’. The numbers in squares are D’ values (D’ × 100).
Table 6. The haplotype analysis on the risk of tuberculosis.
| Haplotype | Control, n(%) | Case, n(%) | OR(95%Cl)a | P |
|---|---|---|---|---|
| rs2275913-rs3748067 | ||||
| AG | 1401(45.90) | 1416(44.22) | 1 | |
| GG | 1176(38.53) | 1285(40.13) | 1.12(1.00–1..25) | 0.046 |
| GA | 469(15.37) | 493(15.4) | 1.09(0.94–1.27) | 0.265 |
| AA | 6(0.20) | 8(0.25) | 1.28(0.42–3.87) | 0.667 |
| rs10983755-rs10759932 | ||||
| GT | 2159(70.74) | 2156(67.33) | 1 | |
| AC | 846(27.72) | 909(28.39) | 1.08(0.96–1.21) | 0.198 |
| GC | 36(1.18) | 120(3.75) | 3.43(2.34–5.05) | <0.001b |
| AT | 11(0.36) | 17(0.53) | 1.42(0.65–3.09) | 0.375 |
aOR: odds ratio; CI: confidence interval, adjusted for age, sex, smoking and drinking.
bSignificant after the Bonferroni correction for multiple comparisons.
Discussion
The magnitude and complexity of the human immune response to mycobacteria have historically been underestimated23. It is vital to determine whether those who remain healthy have a genetically endowed high level of resistance to tuberculosis or whether the resistance is affected by environmental or other exogenous factors24. The genome-wide association study (GWAS) identified several susceptibility loci for tuberculosis in sub-Saharan African, Russian and Moroccan populations25,26,27. However, the follow-up studies reported conflicting results28.
In the present study, we explored the genetic polymorphisms of IL-17, TLR4 and miR-146a in association with pulmonary tuberculosis in a Chinese Han population. To our knowledge, this is the first study revealing the effect of genetic variations of rs10759932 and rs2737190 of TLR4 on the risk of tuberculosis. Haplotype analysis found an increased risk for tuberculosis among individuals carrying TLR4 rs10983755G–rs10759932C. Moreover, we found that SNPs of rs3819024 in IL-17A and rs763780 in IL-17F might be weakly related to the tuberculosis prognosis.
Cytokine secretion is initiated by different immune cells interacting with bacteria29. IL-17 acts as a pro-inflammatory cytokine by recruiting granulocytes to the sites of infection17. Previous studies have suggested the association between genetic polymorphisms of IL-17A/IL-17F and susceptibility to tuberculosis but with inconsistent results18,30,31,32. Du et al. observed that the rs763780-CC polymorphisms of the IL-17F gene were more likely to have an increased risk30. Ocejo-Vinyals et al. investigated the IL-17A rs2275913 polymorphisms and suggested that the GG genotype was related to an increased risk of tuberculosis18. Shi et al. genotyped rs2275913 and rs3748067 in IL-17A and rs763780 in IL-17F and found that the CC genotype of rs763780 was associated with an increased risk of tuberculosis32. Peng et al. conducted a study in a Chinese population and found that those carrying the CT/TT genotype of rs763780 were more susceptible to tuberculosis, but no significant association was found for rs227591331. The discrepancies between these results may be due to the different ethnicities, study design and sample sizes32.
TLR4 is expressed on the plasma membrane and bind lipoprotein or lipid components of bacteria, and it may sense and simultaneously recognize various MTB-encoded factors. TLR4 signaling may have a critical function in fine tuning inflammation during chronic mycobacterial infection33. The SNP rs10759932 is located in the 5′ flanking region of the TLR4 gene34. It has been reported to be associated with the risk of precancerous lesions in the stomach35, gastric carcinogenesis34 or prostate cancer36. In contrast to the findings of a study in a Sudanese population37, we found that variations of this SNP were related to an increased risk of tuberculosis. The SNP rs2737190 is located in the 5′-UTR of TLR4 gene. As 5′-UTR influences the translation of regulatory proteins, modulation of 5′-UTR activity plays a role in the development or progress of specific forms of disease38. Zhou et al. have observed that the G allele was more frequent among preterm gram-negative bacterial infection neonates with a 32% increased risk39. We first explored the effect of the polymorphism at this locus on susceptibility to pulmonary tuberculosis. Our findings support the hypothesis that genetic polymorphisms of the TLR4 gene affect the host’s susceptibility to infectious diseases.
MiR-146a has been previously described as a negative regulator of the immune response and its systemic down-regulation may be associated with the exacerbated inflammatory response in tuberculosis patients40. Pre-miR-146a C/G polymorphism, designated rs2910164, is encoded on chromosome 5q33 and located in the precursor stem region, +60 relative to the first nucleotide of pre-miR-146a, opposite the mature miR-146a sequence41. The change from the G:U pair to the C:U mismatch in the stem structure of the miR-146a precursor might reduce the stability of the pri-miR, the efficiency of processing pri-miR into pre-miR, or processing pre-miR into mature miR42. Previous studies indicated that miR-146a rs2910164 was related to an altered risk of colorectal cancer43, breast cancer or ovarian cancer44. To date, two studies have described the association between this SNP and tuberculosis45,46. One was performed in a Kazak population45, and another was conducted in a Tibetan/Han population46. However, our study did not replicate the previous significant findings in the Chinese Han population. This difference might be attributed to the variations in allelic frequencies of genetic polymorphisms, and therefore, it is not surprising that the genetic association analyses yielded conflicting results in different populations47.
Haplotype-based methods offer a powerful approach to disease gene mapping, based on the association between causal mutations and the ancestral haplotypes from which they arose48. In this study, we constructed an LD analysis and identified SNPs of IL-17A and TLR4 in a Chinese Han population. Our data showed a combined effect of rs2275913 together with rs3748067 on the risk of tuberculosis. Additionally, a LD was found between rs10983755 and rs10759932, contributing to the susceptibility of tuberculosis. LD is a concept of statistical correlation between alleles segregated at two or more loci. Population genetic factors can produce LD through a variety of processes such as natural selection, strong genetic drift, admixture and new mutations49. The association between each mutant allele and its ancestral haplotype is disrupted only by mutation and recombination in subsequent generations48. Further approaches should be carried out to identify the responsible functional SNPs in the LD areas where we identified risk haplotype alleles.
There are several limitations in this study. First, we purposely selected functional SNPs in the IL-17A, IL-17F and TLR4 gene. Although the analysis of the Encyclopedia of DNA Elements (ENCODE) as implemented in Regulome DB indicated that some SNPs might influence the binding of specified transcription factors, their real functions were not proven with experimental evidence. Further work with both knockout and overexpression models is likely to be the most fruitful approach for understanding the mechanisms through which these variants influence the risk of tuberculosis. Second, due to the weak effect of a single genetic polymorphism, other genes in the immunity pathway, together with environmental factors, should also be considered.
Conclusions
Taken together, our results suggest that genetic polymorphisms of rs10759932 and rs2737190 in TLR4 gene may play a role in susceptibility to tuberculosis in the Chinese population.
Additional Information
How to cite this article: Wang, M. et al. Genetic polymorphisms of IL-17A, IL-17F, TLR4 and miR-146a in association with the risk of pulmonary tuberculosis. Sci. Rep. 6, 28586; doi: 10.1038/srep28586 (2016).
Acknowledgments
The National Natural Science Foundation of China (81473027), Jiangsu Science Supported Planning/Social Development Foundation (BE2011841), Qing Lan Project (2014), Six Talent Peaks Project in Jiangsu Province (2014-YY-023), Zhenjiang Key Lab for Drug Resistant Tuberculosis (SS2013018), Jiangsu Provincial Clinical Medical Science and Technology Project (BL2014067) and Priority Academic Program Development of Jiangsu Higher Education Institutions (PAPD) supported this study. The funders had no role in the study design, data collection and analysis, decision to publish, or in the preparation of the manuscript.
Footnotes
Author Contributions M.W., G.X. and J.W. conceived the study. Y.C., H.P. and J.W. collected data. M.W., G.X., L.L. and K.X. performed the experiment. M.W. and G.X. performed the analysis. M.W., G.X. and J.W. drafted the manuscript. B.B. and K.B. refined the manuscript. All authors reviewed the manuscript.
References
- Sulis G., Roggi A., Matteelli A. & Raviglione M. C. Tuberculosis: epidemiology and control. Mediterr J Hematol Infect Dis 6, e2014070, doi: 10.4084/mjhid.2014.070 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zumla A. et al. The WHO 2014 global tuberculosis report–further to go. Lancet Glob Health 3, e10–12, doi: 10.1016/s2214-109x(14)70361-4 (2015). [DOI] [PubMed] [Google Scholar]
- Flynn J. L. Immunology of tuberculosis and implications in vaccine development. Tuberculosis (Edinb) 84, 93–101 (2004). [DOI] [PubMed] [Google Scholar]
- Lawn S. D. & Zumla A. I. Tuberculosis. Lancet 378, 57–72, doi: 10.1016/s0140-6736(10)62173-3 (2011). [DOI] [PubMed] [Google Scholar]
- Young D. B., Perkins M. D., Duncan K. & Barry C. E. 3rd Confronting the scientific obstacles to global control of tuberculosis. J Clin Invest 118, 1255–1265, doi: 10.1172/jci34614 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leandro A. C., Rocha M. A., Cardoso C. S. & Bonecini-Almeida M. G. Genetic polymorphisms in vitamin D receptor, vitamin D-binding protein, Toll-like receptor 2, nitric oxide synthase 2, and interferon-gamma genes and its association with susceptibility to tuberculosis. Braz J Med Biol Res 42, 312–322 (2009). [DOI] [PubMed] [Google Scholar]
- Lyadova I. V. & Panteleev A. V. Th1 and Th17 Cells in Tuberculosis: Protection, Pathology, and Biomarkers. Mediators Inflamm 2015, 854507, doi: 10.1155/2015/854507 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karakas-Celik S. et al. May TLR4 Asp299Gly and IL17 His161Arg polymorphism be associated with progression of primary measles infection to subacute sclerosing panencephalitis? Gene 547, 186–190, doi: 10.1016/j.gene.2014.03.056 (2014). [DOI] [PubMed] [Google Scholar]
- Mukherjee S., Karmakar S. & Babu S. P. TLR2 and TLR4 mediated host immune responses in major infectious diseases: a review. Braz J Infect Dis 20, 193–204, doi: 10.1016/j.bjid.2015.10.011 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- van de Veerdonk F. L. et al. Mycobacterium tuberculosis induces IL-17A responses through TLR4 and dectin-1 and is critically dependent on endogenous IL-1. J Leukoc Biol 88, 227–232, doi: 10.1189/jlb.0809550 (2010). [DOI] [PubMed] [Google Scholar]
- Xu Q. Q. et al. Toll-like receptor 4 signaling inhibits malignant pleural effusion by altering Th1/Th17 responses. Cell Biol Int 39, 1120–1130, doi: 10.1002/cbin.10485 (2015). [DOI] [PubMed] [Google Scholar]
- Omrane I. & Benammar-Elgaaied A. The immune microenvironment of the colorectal tumor: Involvement of immunity genes and microRNAs belonging to the TH17 pathway. Biochim Biophys Acta 1856, 28–38, doi: 10.1016/j.bbcan.2015.04.001 (2015). [DOI] [PubMed] [Google Scholar]
- Aalaei-andabili S. H. & Rezaei N. Toll like receptor (TLR)-induced differential expression of microRNAs (MiRs) promotes proper immune response against infections: a systematic review. J Infect 67, 251–264, doi: 10.1016/j.jinf.2013.07.016 (2013). [DOI] [PubMed] [Google Scholar]
- Rutitzky L. I., Lopes da Rosa J. R. & Stadecker M. J. Severe CD4 T cell-mediated immunopathology in murine schistosomiasis is dependent on IL-12p40 and correlates with high levels of IL-17. J Immunol 175, 3920–3926 (2005). [DOI] [PubMed] [Google Scholar]
- Branger J. et al. Toll-like receptor 4 plays a protective role in pulmonary tuberculosis in mice. Int Immunol 16, 509–516 (2004). [DOI] [PubMed] [Google Scholar]
- Abhimanyu, Bose M., Komal & Varma-Basil M. Lack of association between IL17A and IL17F polymorphisms and related serum levels in north Indians with tuberculosis. Gene 529, 195–198, doi: 10.1016/j.gene.2013.06.090 (2013). [DOI] [PubMed] [Google Scholar]
- Bulat-Kardum L. J., Etokebe G. E., Lederer P., Balen S. & Dembic Z. Genetic Polymorphisms in the Toll-like Receptor 10, Interleukin (IL)17A and IL17F Genes Differently Affect the Risk for Tuberculosis in Croatian Population. Scand J Immunol 82, 63–69, doi: 10.1111/sji.12300 (2015). [DOI] [PubMed] [Google Scholar]
- Ocejo-Vinyals J. G. et al. The IL-17 G-152A single nucleotide polymorphism is associated with pulmonary tuberculosis in northern Spain. Cytokine 64, 58–61, doi: 10.1016/j.cyto.2013.05.022 (2013). [DOI] [PubMed] [Google Scholar]
- Velez D. R. et al. NOS2A, TLR4, and IFNGR1 interactions influence pulmonary tuberculosis susceptibility in African-Americans. Hum Genet 126, 643–653, doi: 10.1007/s00439-009-0713-y (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arji N. et al. Genetic diversity of TLR2, TLR4, and VDR loci and pulmonary tuberculosis in Moroccan patients. J Infect Dev Ctries 8, 430–440, doi: 10.3855/jidc.3820 (2014). [DOI] [PubMed] [Google Scholar]
- Shao Y. et al. Epidemiology of anti-tuberculosis drug resistance in a Chinese population: current situation and challenges ahead. BMC Public Health 11, 110, doi: 10.1186/1471-2458-11-110 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teuber M., Wenz M. H., Schreiber S. & Franke A. GMFilter and SXTestPlate: software tools for improving the SNPlex genotyping system. BMC Bioinformatics 10, 81, doi: 10.1186/1471-2105-10-81 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scriba T. J. et al. Distinct, specific IL-17- and IL-22-producing CD4 + T cell subsets contribute to the human anti-mycobacterial immune response. J Immunol 180, 1962–1970 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies P. D. & Grange J. M. Factors affecting susceptibility and resistance to tuberculosis. Thorax 56 Suppl 2, ii23–29 (2001). [PMC free article] [PubMed] [Google Scholar]
- Thye T. et al. Genome-wide association analyses identifies a susceptibility locus for tuberculosis on chromosome 18q11.2. Nat Genet 42, 739–741, doi: 10.1038/ng.639 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtis J. et al. Susceptibility to tuberculosis is associated with variants in the ASAP1 gene encoding a regulator of dendritic cell migration. Nat Genet 47, 523–527, doi: 10.1038/ng.3248 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant A. V. et al. A genome-wide association study of pulmonary tuberculosis in Morocco. Hum Genet 135, 299–307, doi: 10.1007/s00439-016-1633-2 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji L. D. et al. Lack of association between polymorphisms from genome-wide association studies and tuberculosis in the Chinese population. Scand J Infect Dis 45, 310–314, doi: 10.3109/00365548.2012.726739 (2013). [DOI] [PubMed] [Google Scholar]
- Etna M. P., Giacomini E., Severa M. & Coccia E. M. Pro- and anti-inflammatory cytokines in tuberculosis: a two-edged sword in TB pathogenesis. Semin Immunol 26, 543–551, doi: 10.1016/j.smim.2014.09.011 (2014). [DOI] [PubMed] [Google Scholar]
- Du J. et al. StIL-17 gene polymorphisms in the development of pulmonary tuberculosis. Int J Clin Exp Pathol 8, 3225–3229 (2015). [PMC free article] [PubMed] [Google Scholar]
- Peng R. et al. The IL-17F sequence variant is associated with susceptibility to tuberculosis. Gene 515, 229–232, doi: 10.1016/j.gene.2012.11.017 (2013). [DOI] [PubMed] [Google Scholar]
- Shi G. C. & Zhang L. G. Influence of interleukin-17 gene polymorphisms on the development of pulmonary tuberculosis. Genet Mol Res 14, 8526–8531, doi: 10.4238/2015.July.28.22 (2015). [DOI] [PubMed] [Google Scholar]
- Fremond C. M., Nicolle D. M., Torres D. S. & Quesniaux V. F. Control of Mycobacterium bovis BCG infection with increased inflammation in TLR4-deficient mice. Microbes Infect 5, 1070–1081 (2003). [DOI] [PubMed] [Google Scholar]
- Huang H. et al. A 5′-flanking region polymorphism in toll-like receptor 4 is associated with gastric cancer in a Chinese population. J Biomed Res 24, 100–106, doi: 10.1016/s1674-8301(10)60017-6 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan Y. F. et al. TLR4 polymorphisms associated with developing gastric pre-cancer lesions in a Chinese Han population. Hum Immunol 75, 176–181, doi: 10.1016/j.humimm.2013.11.002 (2014). [DOI] [PubMed] [Google Scholar]
- Cheng I., Plummer S. J., Casey G. & Witte J. S. Toll-like receptor 4 genetic variation and advanced prostate cancer risk. Cancer Epidemiol Biomarkers Prev 16, 352–355, doi: 10.1158/1055-9965.epi-06-0429 (2007). [DOI] [PubMed] [Google Scholar]
- Zaki H. Y. et al. Common polymorphisms in TLR4 gene associated with susceptibility to pulmonary tuberculosis in the Sudanese. Int J Tuberc Lung Dis 16, 934–940, doi: 10.5588/ijtld.11.0517 (2012). [DOI] [PubMed] [Google Scholar]
- Minmin S. et al. Single nucleotide polymorphisms of Toll-like receptor 4 decrease the risk of development of hepatocellular carcinoma. PLoS One 6, e19466, doi: 10.1371/journal.pone.0019466 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J. G. et al. Toll-like receptor 4 polymorphisms in gram-negative bacterial infections of Han Chinese neonates. Am J Perinatol 32, 363–370, doi: 10.1055/s-0034-1387929 (2015). [DOI] [PubMed] [Google Scholar]
- Spinelli S. V. et al. Altered microRNA expression levels in mononuclear cells of patients with pulmonary and pleural tuberculosis and their relation with components of the immune response. Mol Immunol 53, 265–269, doi: 10.1016/j.molimm.2012.08.008 (2013). [DOI] [PubMed] [Google Scholar]
- Zhang X. et al. Association of Pre-miR-146a rs2910164 Polymorphism with Papillary Thyroid Cancer. Int J Endocrinol 2015, 802562, doi: 10.1155/2015/802562 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jazdzewski K. et al. Common SNP in pre-miR-146a decreases mature miR expression and predisposes to papillary thyroid carcinoma. Proc Natl Acad Sci USA 105, 7269–7274, doi: 10.1073/pnas.0802682105 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chae Y. S. et al. A miR-146a polymorphism (rs2910164) predicts risk of and survival from colorectal cancer. Anticancer Res 33, 3233–3239 (2013). [PubMed] [Google Scholar]
- Shen J. et al. A functional polymorphism in the miR-146a gene and age of familial breast/ovarian cancer diagnosis. Carcinogenesis 29, 1963–1966, doi: 10.1093/carcin/bgn172 (2008). [DOI] [PubMed] [Google Scholar]
- Zhang X. et al. Association of the miR-146a, miR-149, miR-196a2 and miR-499 polymorphisms with susceptibility to pulmonary tuberculosis in the Chinese Uygur, Kazak and Southern Han populations. BMC Infect Dis 15, 41, doi: 10.1186/s12879-015-0771-9 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li D. et al. Genetic study of two single nucleotide polymorphisms within corresponding microRNAs and susceptibility to tuberculosis in a Chinese Tibetan and Han population. Hum Immunol 72, 598–602, doi: 10.1016/j.humimm.2011.03.004 (2011). [DOI] [PubMed] [Google Scholar]
- Ansari A. et al. Cytokine gene polymorphisms across tuberculosis clinical spectrum in Pakistani patients. PLoS One 4, e4778, doi: 10.1371/journal.pone.0004778 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabriel S. B. et al. The structure of haplotype blocks in the human genome. Science (New York, N.Y.) 296, 2225–2229, doi: 10.1126/science.1069424 (2002). [DOI] [PubMed] [Google Scholar]
- Schrodi S. J., Garcia V. E., Rowland C. & Jones H. B. Pairwise linkage disequilibrium under disease models. Eur J Hum Genet 15, 212–220, doi: 10.1038/sj.ejhg.5201731 (2007). [DOI] [PubMed] [Google Scholar]