Abstract
In this dataset we integrated figures comparing leaf number and rosette diameter in three Arabidopsis FT overexpressor lines (AtFTOE) driven by KNAT1 promoter, “A member of the KNOTTED class of homeodomain proteins encoded by the STM gene of Arabidopsis” [5], vs Wild Type (WT) Arabidopsis plats. Also, presented in the tables are some transcriptomic data obtained by RNA-seq Illumina HiSeq from rosette leaves of Arabidopsis plants of AtFTOE 2.1 line vs WT with accession numbers SRR2094583 and SRR2094587 for AtFTOE replicates 1–3 and AtWT for control replicates 1–2 respectively. Raw data of paired-end sequences are located in the public repository of the National Center for Biotechnology Information of the National Library of Medicine, National Institutes of Health, United States of America, Bethesda, MD, USA as Sequence Read Archive (SRA). Performed analyses of differential expression genes are visualized by Mapman and presented in figures. “Transcriptomic analysis of Arabidopsis overexpressing flowering locus T driven by a meristem-specific promoter that induces early flowering” [2], described the interpretation and discussion of the obtained data.
Keywords: Differential expression, Bioinformatics, Flowering
Specifications Table
| Subject area | Biology |
| More specific subject area | Plant Sciences |
| Type of data | Figure; tables |
| How data was acquired | PCR final point and ddPCR (The QX100 Droplet Digital PCR (ddPCR™ System), direct count of leaves and measurement of rosette diameter with image analysis software (Imagej [http://rsb.info.nih.gov]), RNA-seq by Illumina HiSeq Sequencing, |
| Data format | Analyzed |
| Experimental factors | Three lines overexpressing FT (AtFTOE) and WT Arabidopsis plants |
| Experimental features | Three lines of AtFTOE and WT Arabidopsis plants were grown on hydroponic under controlled conditions at 22 °C in short day (SD) photoperiod (8 h light /16 h dark) up to day 21 and then transferred to inductive conditions of long days (LD) photoperiod (16 h light /8 h dark) |
| Data source location | Mexico City, Mexico and at the National Center for Biotechnology Information (NCBI) |
| Data accessibility | Data is available with this article and at NCBI accession numbers SRR2094583 and SRR2094587 |
Value of the data
-
•
The amplification of transgene by PCR and copy variation number by ddPCR is fundamental to compare independent transgenic events.
-
•
ANOVA and T-Student test were employed to assess statistical significance of data (α=0.05) for leaf count and rosette diameter in order to compare three AtFTOE lines and WT.
-
•
Differentially expressed genes in the context of cellular functions are graphically presented by Mapman to understand the integrative changes in the metabolism.
-
•
Raw data from the Illumina HiSeq sequencing are available for further analyses.
1. Data
In this article are presented the data analyses (figures) from leaf count and rosette diameter for three lines AtFTOE (2.1, 3.1 an 4.3) compared with WT Arabidopsis plants (Fig. 2A and B respectively). Data corresponding to differential expression (log2 fold change) from AtFTOE 2.1 line vs WT Arabidopsis are visualized by Mapman (Fig. 3). Some data corresponding to down-regulated genes are presented in Table 3.
Fig. 2.
Interval plot from AtFTOE lines and WT for (A) Leaf count and (B) Rosette diameter at 21 days in SD conditions.
Fig. 3.
Mapman visualization showing the observed differential expression patterns, based on the Log2FCs of mRNA levels, in rosette leaves of three AtFTOE versus WT. Red color indicates up-regulated genes and blue color indicates down-regulated genes (A) Cell functions overview (B) Metabolism overview each gene is symbolized by a box.
Table 3.
List of Arabidopsis floral repressors downregulated in AtFTOE 2.1 line.
| Genes | Locus ID | Localization | Description | FC |
|---|---|---|---|---|
| At3g27200 | Plasma membrane | Plastocyanin-like domain-containing protein. Cupredoxin superfamily protein. | 0.11 | |
| SPL2, SQUAMOSA PROMOTER BINDING PROTEIN-LIKE 2 | At5g43270 | Nucleus | Member of the SPL (squamosa-promoter binding protein-like) gene family, a novel gene family encoding DNA binding proteins and putative transcription factors. | 0.57 |
| AP2, APETALA 2, ATAP2, FL1, FLO2, FLORAL MUTANT 2, FLOWER | AT4G36920 | Nucleus | Encodes a floral homeotic gene, a member of the AP2/EREBP (ethylene responsive element binding protein) class of transcription factors and is involved in the specification of floral organ identity, establishment of floral meristem identity, suppression of floral meristem indeterminacy, and development of the ovule and seed coat. AP2 also has a role in controlling seed mass. | 0.46 |
| At3g45160 | Putative membrane lipoprotein. | 0.47 | ||
| ATMYB29 | At5g07690 | Nucleus | Encodes a putative transcription factor. | 0.56 |
| CA1 | At3g01500 | Apoplast, chloroplast stroma, thylakoid membrane, plasma membrane | Encodes a putative beta-carbonic anhydrase betaCA1. | 0.44 |
| At3g05730 | Extracellular region | Encodes a defensin-like (DEFL) family protein. | 0.27 | |
| At5g05960 | Extracellular region | Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein. | 0.23 | |
| BETA CA2 | At5g14740 | Apoplast, chloroplast, chloroplast stroma, thylakoid membrane, cytoplasm | Encodes a beta carbonic anhydrase likely to be localized in the cytoplasm. | 0.41 |
| At3g26960 | Extracellular region | Pollen Ole e 1 allergen and extensin family protein. | 0.37 | |
| BETA GLUCOSIDASE 33, BGLU33 | At2g32860 | Chloroplast | Beta glucosidase | 0.27 |
| XCP2, XYLEM CYSTEINE PEPTIDASE 2 | At1g20850 | Chloroplast, extracellular space, lysosome | Cysteine-type peptidase activity, peptidase activity | 0.15 |
| At4g30650 | Cell wall, chloroplast, extracellular space, lysosome | Xylem cysteine peptidase 2 (XCP2) | 0.50 | |
| At1g29660 | GDSL-motif esterase/acyltransferase/lipase.Enzyme group with broad substrate specificity that may catalyze acyltransfer or hydrolase reactions with lipid and non-lipid substrates. | 0.24 | ||
| TON1 RECRUITING MOTIF 14, TRM14 | At3g61380 | Nucleus, plasma membrane | Phosphatidylinositol N-acetyglucosaminlytransferase subunit P-related. | 0.39 |
| ATIMD3, IMD3, IPMDH1 | At1g31180 | Chloroplast stroma, mitochondrion, plastid, thylakoid | The AtIMD3 is one out of 3 genes encoding the enzyme 3-isopropylmalate dehydrogenase involved in leucine biosynthesis in Arabidopsis. Its subcellular location has been targeted to plastids. | 0.61 |
| OEP6, OUTER ENVELOPE PROTEIN 6 | At3g63160 | Chloroplast, chloroplast envelope, chloroplast outer membrane, chloroplast thylakoid membrane, thylakoid | Molecular_function unknown | 0.48 |
| At1g35190 | Cytoplasm | 2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein | 0.55 | |
| AGAMOUS-LIKE 19, AGL19, GL19 | At4g22950 | Nucleus | MADS-box protein AGL19. | 0.23 |
| At3g27200 | Cupredoxin superfamily protein,electron carrier activity, copper ion binding. | 0.11 | ||
| At2g39850 | Extracellular region | Subtilisin-like serine endopeptidase family protein; metabolic process, proteolysis. | 0.56 | |
| At3g53100 | Extracellular region | GDSL-motif esterase/acyltransferase/lipase,hydrolase activity, acting on ester bonds. | 0.33 | |
| At5g44400 | Cell wall, cytoplasm, plasmodesma | FAD-binding Berberine family protein. | 0.11 | |
| MORPHOGENESIS OF ROOT HAIR 1, MRH1 | At4g18640 | Chloroplast | Protein serine/threonine kinase activity. | 0.11 |
| ATPAP3, PAP3, PURPLE ACID PHOSPHATASE 3 | At1g14700 | Extracellular region, vacuole | acid phosphatase activity, metal ion binding, protein serine/threonine phosphatase activity. | 0.25 |
| At3g29030 | Cell wall, extracellular region, membrane | Encodes an expansin. Naming convention from the Expansin Working Group. | 0.17 | |
| At1g04040 | Cell wall, plant-type cell wall, plasmodesma, vacuolar membrane, vacuole | HAD superfamily, subfamily IIIB acid phosphatase | 0.27 | |
| TPPH, TREHALOSE-6-PHOSPHATE PHOSPHATASE H | At4g39770 | Cytoplasm, cytosol, nucleus | Trehalose biosynthetic process. | 0.61 |
| At5g63180 | Pectin lyase fold/virulence factor | Pectin lyase-like superfamily protein | 0.35 |
2. Experimental design, materials and methods
2.1. Determination of transgene (FT) and copy variation number in three AtFTOE lines
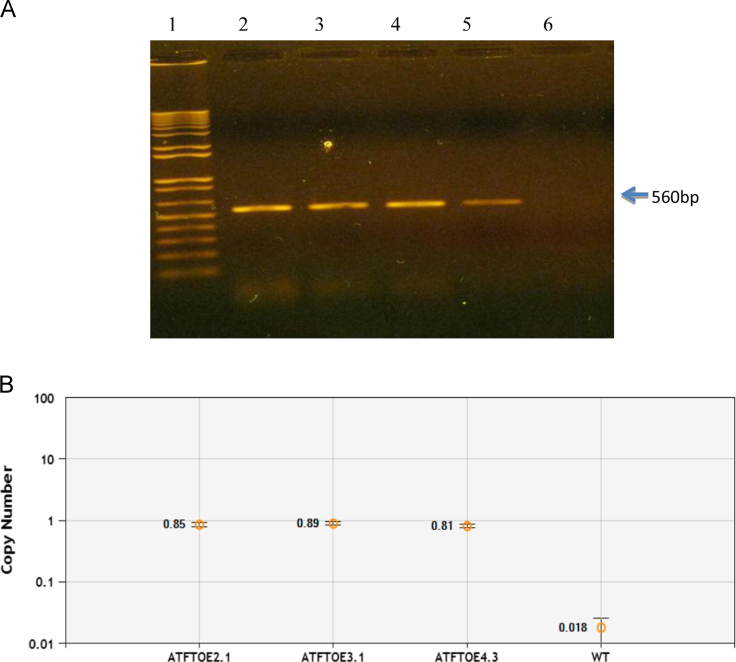
Transgene FT (560 bp) was amplified by PCR from three AtFTOE (2.1,3.1 and 4.3) lines (Fig. 1A). Droplet digital PCR (ddPCR) was employed to determine transgene (FT) copy variation number (CVN) (Fig. 1B). As template were used 2.5 ng of genomic DNA previously digested with HindIII. Droplets were generated for PCR reaction with the specific primers AtFT-qPCR (F) (5′-TCCGTTTAATAGATCAATCAC-3′), FT-qPCR (R) (5′-CCACCATAACCAAAGTATAG-3) and probe TaqMan ddPCRFT [5′FAM] TCCTGAGGTCTTCTCCACCA [3′BHQ1]. The 152 bp of PCR-amplified product Arabidopsis HMGB1 (AT3G51880) was used as an internal, reference gene. The primers used as reference were HMGB1 probe [5′HEX]AGGCACCGGCTGAGAAGCCT[3′BHQ1], HMGB1F (5′-CAGAAAGGTGGGAAAGAGGA-3′) and HMGB1-R (5′-AAGGACCCAAACAAACCAAA-3′). The HMGB1 PCR-amplified product was 96 bp. After cycling, the PCR nano droplets were counted using the droplet reader Bio-Rad QX100 system (Bio-Rad 2012).
Fig. 1.
Amplification of transgene FT and copy number Variation in three AtFTOE lines (A) 560 bp amplified fragment correspondent to the size of the FT transgene visualized in 0.8% agarose gel stained with ethidium bromide. Carril 1: 1 kb plus DNA ladder (Thermo Fisher Scientific®); Carril 2: AtFTOE 2.1; Carril 3: AtFTOE 3.1; Carril 4: AtFTOE 4.3; Carril 5: WT Copy variation number of three AtFTOE lines determined by dPCR™ System (The QX100 Droplet Digital PCR) (B) Copy variation number of three AtFTOE lines determined by dPCR™ System (The QX100 Droplet Digital PCR).
2.2. Determination of leaf number and rosette diameter in three AtFTOE and WT plants
Three AtFTOE (2.1,3.1 and 4.3) lines and WT Arabidopsis Columbia-0 ecotype were employed. Seeds were stratified, kept at 4 °C for 3 days in the dark and then germinated and grown in hydroponic system [1] at 22 °C under controlled conditions, initially in short days (8 h light, 16 h dark) and after 21 days the seedlings were transferred to long-day (16 h light, 8 h dark) photoperiod under 100–120 mmol m−2 s−1. Plants grown under these conditions were used for determining rosette leaf number (Fig. 2A) by individual counting and rosette diameter (Fig. 2B) quantified by image analyses using the software Imagej (http://rsb.info.nih.gov). Cuantitative data were statistically analyzed with ANOVA and T-Student test at (α=0.05).
2.3. Massive mRNA-sequencing by Illumina, bioinformatics analysis and data processing
For RNA-Sequencing by illumina HiSeq was used total RNA from rosette leaves of 35-day old of AtFTOE 2.1 line and Wild Type (WT) plants. RNA was extracted with the RNeasy Plant Kit (Qiagen). The RNA-seq experiments were conducted with RNA isolated from three biological replicates per AtFTOE 2.1 line and two biological replicates for WT with accession numbers SRR2094583 and SRR2094587 respectively. Illumina sequencing was performed at Otogenetics (Georgia-USA). Illumina HiSeq Sequencing with PE50 yielded 20 million reads by triplicate (Table 2). The raw data files are available at the National Center for Biotechnology Information (NCBI) Sequence Read Archive (SRA) accession numbers SRR2094583 and SRR2094587 for AtFTOE replicates 1–3 and AtWT for control replicates 1–2 respectively. The paired-end reads were aligned to the reference Arabidopsis genome using Tophat (version 2.0.10) [7] and Bowtie2 (version 2.1.0) [4]. The reference Arabidopsis genome and gene model annotation files (TAIR10<ftp://igenome:G3nom3s4u@ussd-ftp.illumina.com/Arabidopsis_thaliana/NCBI/TAIR10/Arabidopsis_thaliana_NCBI_TAIR10.tar.gz>) were downloaded from the Illumina iGenomes (http://support.illumina.com/sequencing/sequencing_software/igenome.html). Differential expression was determined by Cufflinks (version 2.1.1) as described by Trapnell et al. [8] and then was visualized by CummeRbound, an edgeR package [3]. To analyze the variation in expression between two replicates from WT and three replicates from AtFTOE it was calculated the absolute difference of the log2 fold change and adjusted to P-value ≤0.05.
Table 2.
RNA sequences obtained by Illumina HighSeq2000/2500 PE100 sequencer.
| Sample | Total bases* | Total reads |
|---|---|---|
| WT1 | 2,612,645,388 | 24,647,598 |
| WT2 | 2,770,468,788 | 26,136,498 |
| FT1 | 2,474,087,064 | 23,340,444 |
| FT2 | 2,455,442,088 | 23,164,548 |
| FT3 | 2,257,321,728 | 21,295,488 |
2.4. Visualization of differential expression by Mapman
In order to visualize differential expression we used Mapman tool, mapping the Mapman databases (http://mapman.gabipd.org/web/guest/mapman; Ath_AGI_LOCUS_TAIR10_Aug2012-3.m02; [6]) using raw data of differential expression log2 fold change and adjusted to P-value ≤0.05 (Fig. 3).
2.5. Primer design
Primers design (Table 1) was performed by OligoArchitect™ Primer and Probe Design Solutions (http://www.sigmaaldrich.com/technical-documents/articles/biology/probe-design-services.html). Gene sequences were obtained from Tair database (https://www.arabidopsis.org).
Table 1.
Primers to detect differentially expressed genes.
| Gene | Locus ID | Primer Forward 5′−3′ | Primer Reverse 5′−3′ |
|---|---|---|---|
| AGAMOUS-LIKE 68,/MAF5 | AT5g65080 | MAF5- RTSYBG (F) GGAAGAAGAAGAGTAGAGAT | MAF5-RTSYBG (R) ACAGAGAATTGAGAGTTGA |
| Squamosa promoter-binding-like protein 4 SPL4 | AT5g65080 | SPL4-RTSYBG (F)GTCGGAGAGGAATCAATG | SPL4-RTSYBG (R) GGCATAGGAAGTGTCATC |
| MADS – BOX PROTEIN SOC1 | AT2g45660 | SOC1- RTSYBG (F) GAAGAGAATAGAGAATGC | SOC1-RTSYBG (R) AGAGAAGATGATAAGAGAA |
| ATSTP13, MSS1, STP13, SUGAR TRANSPORT PROTEIN 13 | AT5G26340 | AtSTP13-qPCR(F) ACTCCGTTGACAAAGTCGGT | AtSTP13-qPCR(R) TGGCGATTACGACTTGAGAG |
Acknowledgments
This work was supported by Departmental funds from CINVESTAV-IPN, by CONACyT grants nos. 156162 to RR-M and 105985 to BX-C, and a SENASICA-SAGARPA grant to BX-C and RR-M. LD-B was supported by a doctoral fellowship from CONACyT no 388937. This research was partially supported by the Intramural Research Program of the NIH, NLM, NCBI.
Footnotes
Transparency data associated with this article can be found in the online version at 10.1016/j.dib.2016.06.002.
Supplementary data associated with this article can be found in the online version at 10.1016/j.dib.2016.06.002.
Transparency document. Supplementary material
Supplementary material
Appendix A. Supplementary material
Supplementary material
Supplementary material
Supplementary material
References
- 1.Conn S.J., Hocking B., Dayod M., Xu B., Athman A., Henderson S., Gilliham M. Protocol: optimising hydroponic growth systems for nutritional and physiological analysis of Arabidopsis thaliana and other plants. Plant Methods. 2013;9(1):4. doi: 10.1186/1746-4811-9-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Duplat-Bermúdez L., Ruiz-Medrano R., Landsman D., Mariño-Ramírez L., Xoconostle-Cázares Transcriptomic analysis of Arabidopsis overexpressing flowering locus T driven by a meristem-specific promoter that induces early flowering. Gene. 2016 doi: 10.1016/j.gene.2016.04.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.L. Goff, C. Trapnell, D. Kelley, CummeRbund: analysis, exploration, manipulation, and visualization of Cufflinks high-throughput sequencing data. R package version 2.8.2., 2013.
- 4.Langmead B., Salzberg S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods. 2012;9(4):357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Long J., Moan E.E., Medford J., Barton K. A member of the KNOTTED class of homeodomain proteins encoded by the STM gene of Arabidopsis. Lett. Nat. 1996;379:66–69. doi: 10.1038/379066a0. [DOI] [PubMed] [Google Scholar]
- 6.Thimm O., Bläsing O., Gibon Y., Nagel A., Meyer S., Krüger P., Selbig J., Müller L.A., Rhee S.Y., Stitt M. mapman: a user-driven tool to display genomics data sets onto diagrams of metabolic pathways and other biological processes. Plant J. 2004;37:914–939. doi: 10.1111/j.1365-313x.2004.02016.x. [DOI] [PubMed] [Google Scholar]
- 7.Trapnell C., Pachter L., Salzberg S.L. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25(9):1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Trapnell C., Roberts A., Goff L., Pertea G., Kim D., Kelley D.R., Pachter L. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protoc. 2012;7(3):562–578. doi: 10.1038/nprot.2012.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material
Supplementary material
Supplementary material
Supplementary material