Abstract
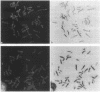
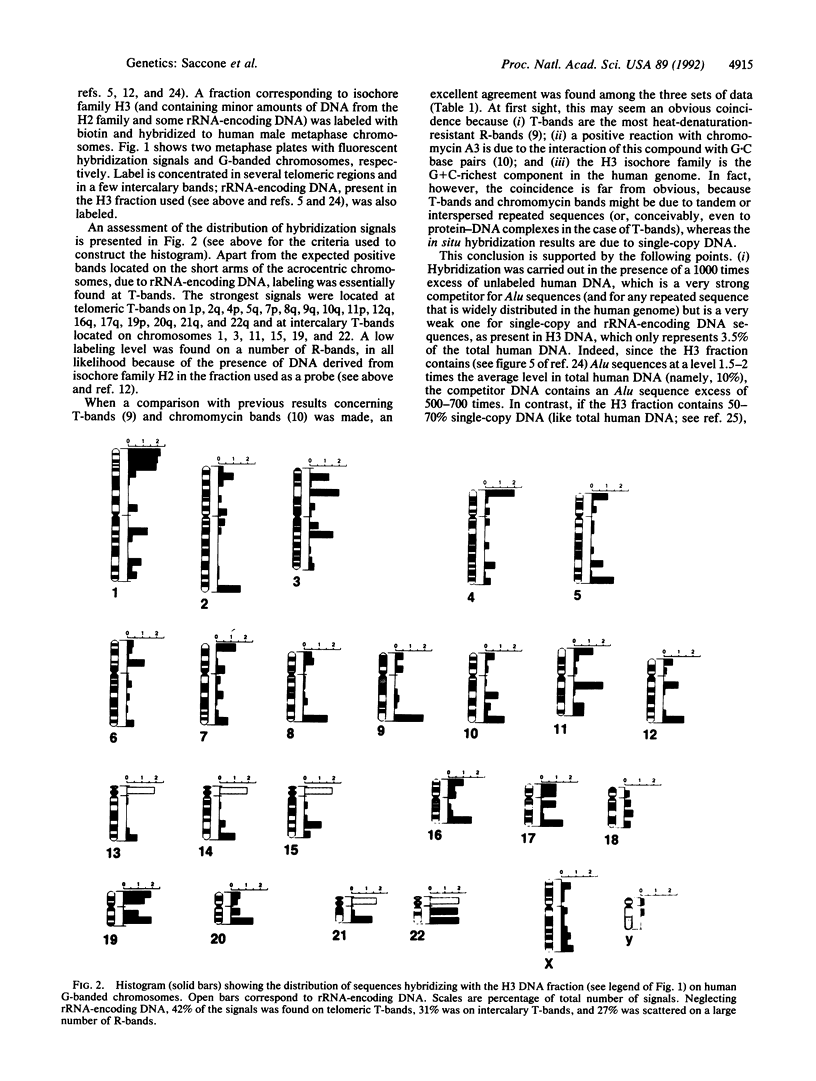
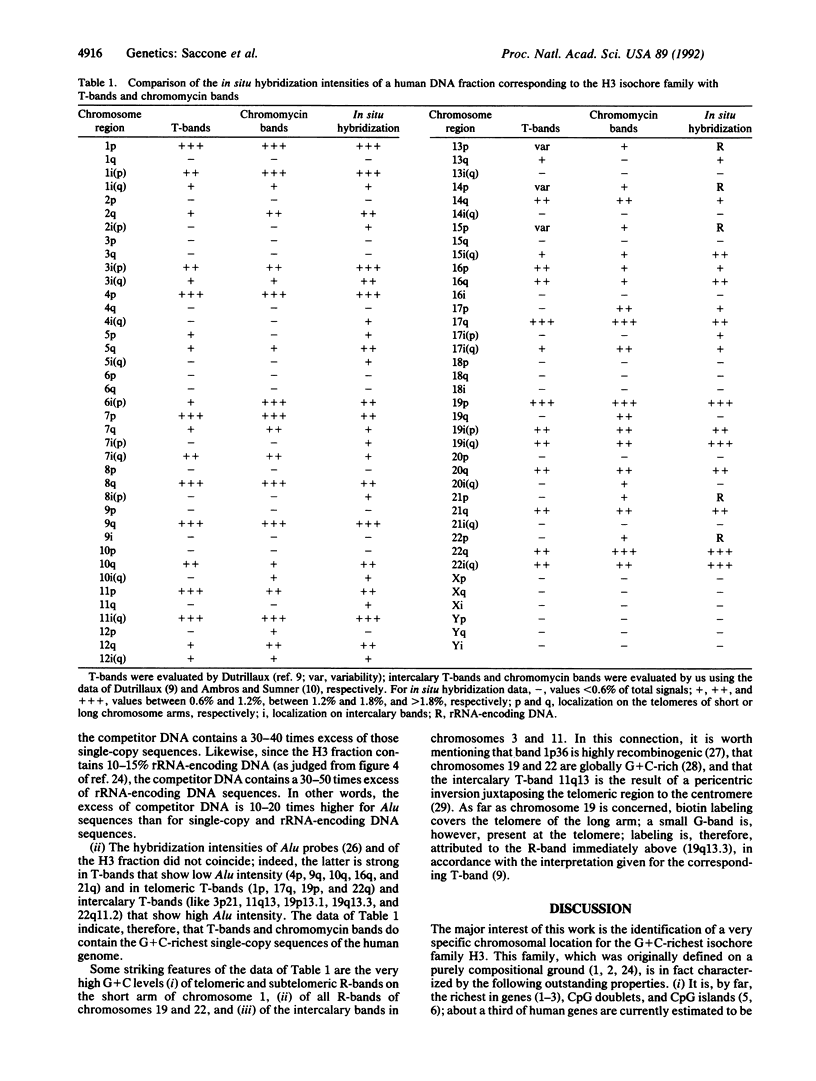
Chromosome in situ suppression hybridization has been carried out on human metaphase chromosomes to localize the G+C-richest human DNA fraction (which only represents 3.5% of the genome), as isolated by preparative equilibrium centrifugation in Cs2SO4/3,6-bis(acetatomercurimethyl)-1,4-dioxane density gradient. This fraction essentially corresponds to isochore family H3. The rationale for carrying out this experiment is that this isochore family has, by far, the highest gene concentration, the highest concentration in CpG islands, the highest transcriptional and recombinational activity, and a distinct chromatin structure. The in situ hybridization results obtained show that the H3 isochore family is localized in two coincident sets of bands of human metaphase chromosomes: telomeric bands and chromomycin A3-positive 4',6-diamidino-2-phenylindole-negative bands. This result is the first step toward a complete compositional map of the human karyotype. Because the G+C gradient across isochore families is paralleled by a gene concentration gradient, such a map has structural, functional, and evolutionary relevance.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Ambros P. F., Sumner A. T. Correlation of pachytene chromomeres and metaphase bands of human chromosomes, and distinctive properties of telomeric regions. Cytogenet Cell Genet. 1987;44(4):223–228. doi: 10.1159/000132375. [DOI] [PubMed] [Google Scholar]
- Aïssani B., Bernardi G. CpG islands, genes and isochores in the genomes of vertebrates. Gene. 1991 Oct 15;106(2):185–195. doi: 10.1016/0378-1119(91)90198-k. [DOI] [PubMed] [Google Scholar]
- Aïssani B., Bernardi G. CpG islands: features and distribution in the genomes of vertebrates. Gene. 1991 Oct 15;106(2):173–183. doi: 10.1016/0378-1119(91)90197-j. [DOI] [PubMed] [Google Scholar]
- Bernardi G., Mouchiroud D., Gautier C., Bernardi G. Compositional patterns in vertebrate genomes: conservation and change in evolution. J Mol Evol. 1988 Dec;28(1-2):7–18. doi: 10.1007/BF02143493. [DOI] [PubMed] [Google Scholar]
- Bernardi G., Olofsson B., Filipski J., Zerial M., Salinas J., Cuny G., Meunier-Rotival M., Rodier F. The mosaic genome of warm-blooded vertebrates. Science. 1985 May 24;228(4702):953–958. doi: 10.1126/science.4001930. [DOI] [PubMed] [Google Scholar]
- Bernardi G. The isochore organization of the human genome. Annu Rev Genet. 1989;23:637–661. doi: 10.1146/annurev.ge.23.120189.003225. [DOI] [PubMed] [Google Scholar]
- Cortadas J., Macaya G., Bernardi G. An analysis of the bovine genome by density gradient centrifugation: fractionation in Cs2SO4/3,6-bis(acetatomercurimethyl)dioxane density gradient. Eur J Biochem. 1977 Jun 1;76(1):13–19. doi: 10.1111/j.1432-1033.1977.tb11565.x. [DOI] [PubMed] [Google Scholar]
- Cuny G., Soriano P., Macaya G., Bernardi G. The major components of the mouse and human genomes. 1. Preparation, basic properties and compositional heterogeneity. Eur J Biochem. 1981 Apr;115(2):227–233. doi: 10.1111/j.1432-1033.1981.tb05227.x. [DOI] [PubMed] [Google Scholar]
- De Sario A., Aïssani B., Bernardi G. Compositional properties of telomeric regions from human chromosomes. FEBS Lett. 1991 Dec 16;295(1-3):22–26. doi: 10.1016/0014-5793(91)81375-i. [DOI] [PubMed] [Google Scholar]
- Dutrillaux B. Chromosomal evolution in primates: tentative phylogeny from Microcebus murinus (Prosimian) to man. Hum Genet. 1979 May 10;48(3):251–314. doi: 10.1007/BF00272830. [DOI] [PubMed] [Google Scholar]
- Dutrillaux B. Nouveau système de marquage chromosomique: Les bandes. Chromosoma. 1973 Apr 27;41(4):395–402. doi: 10.1007/BF00396497. [DOI] [PubMed] [Google Scholar]
- Gardiner K., Aissani B., Bernardi G. A compositional map of human chromosome 21. EMBO J. 1990 Jun;9(6):1853–1858. doi: 10.1002/j.1460-2075.1990.tb08310.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikemura T., Wada K. Evident diversity of codon usage patterns of human genes with respect to chromosome banding patterns and chromosome numbers; relation between nucleotide sequence data and cytogenetic data. Nucleic Acids Res. 1991 Aug 25;19(16):4333–4339. doi: 10.1093/nar/19.16.4333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korenberg J. R., Engels W. R. Base ratio, DNA content, and quinacrine-brightness of human chromosomes. Proc Natl Acad Sci U S A. 1978 Jul;75(7):3382–3386. doi: 10.1073/pnas.75.7.3382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korenberg J. R., Rykowski M. C. Human genome organization: Alu, lines, and the molecular structure of metaphase chromosome bands. Cell. 1988 May 6;53(3):391–400. doi: 10.1016/0092-8674(88)90159-6. [DOI] [PubMed] [Google Scholar]
- Landegent J. E., Jansen in de Wal N., Dirks R. W., Baao F., van der Ploeg M. Use of whole cosmid cloned genomic sequences for chromosomal localization by non-radioactive in situ hybridization. Hum Genet. 1987 Dec;77(4):366–370. doi: 10.1007/BF00291428. [DOI] [PubMed] [Google Scholar]
- Lichter P., Cremer T., Borden J., Manuelidis L., Ward D. C. Delineation of individual human chromosomes in metaphase and interphase cells by in situ suppression hybridization using recombinant DNA libraries. Hum Genet. 1988 Nov;80(3):224–234. doi: 10.1007/BF01790090. [DOI] [PubMed] [Google Scholar]
- Lichter P., Tang C. J., Call K., Hermanson G., Evans G. A., Housman D., Ward D. C. High-resolution mapping of human chromosome 11 by in situ hybridization with cosmid clones. Science. 1990 Jan 5;247(4938):64–69. doi: 10.1126/science.2294592. [DOI] [PubMed] [Google Scholar]
- Montanaro V., Casamassimi A., D'Urso M., Yoon J. Y., Freije W., Schlessinger D., Muenke M., Nussbaum R. L., Saccone S., Maugeri S. In situ hybridization to cytogenetic bands of yeast artificial chromosomes covering 50% of human Xq24-Xq28 DNA. Am J Hum Genet. 1991 Feb;48(2):183–194. [PMC free article] [PubMed] [Google Scholar]
- Mouchiroud D., D'Onofrio G., Aïssani B., Macaya G., Gautier C., Bernardi G. The distribution of genes in the human genome. Gene. 1991 Apr;100:181–187. doi: 10.1016/0378-1119(91)90364-h. [DOI] [PubMed] [Google Scholar]
- Moyzis R. K., Buckingham J. M., Cram L. S., Dani M., Deaven L. L., Jones M. D., Meyne J., Ratliff R. L., Wu J. R. A highly conserved repetitive DNA sequence, (TTAGGG)n, present at the telomeres of human chromosomes. Proc Natl Acad Sci U S A. 1988 Sep;85(18):6622–6626. doi: 10.1073/pnas.85.18.6622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinkel D., Landegent J., Collins C., Fuscoe J., Segraves R., Lucas J., Gray J. Fluorescence in situ hybridization with human chromosome-specific libraries: detection of trisomy 21 and translocations of chromosome 4. Proc Natl Acad Sci U S A. 1988 Dec;85(23):9138–9142. doi: 10.1073/pnas.85.23.9138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinkel D., Straume T., Gray J. W. Cytogenetic analysis using quantitative, high-sensitivity, fluorescence hybridization. Proc Natl Acad Sci U S A. 1986 May;83(9):2934–2938. doi: 10.1073/pnas.83.9.2934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romani M., De Ambrosis A., Alhadeff B., Purrello M., Gluzman Y., Siniscalco M. Preferential integration of the Ad5/SV40 hybrid virus at the highly recombinogenic human chromosomal site 1p36. Gene. 1990 Nov 15;95(2):231–241. doi: 10.1016/0378-1119(90)90366-y. [DOI] [PubMed] [Google Scholar]
- Rynditch A., Kadi F., Geryk J., Zoubak S., Svoboda J., Bernardi G. The isopycnic, compartmentalized integration of Rous sarcoma virus sequences. Gene. 1991 Oct 15;106(2):165–172. doi: 10.1016/0378-1119(91)90196-i. [DOI] [PubMed] [Google Scholar]
- Schmid C. W., Jelinek W. R. The Alu family of dispersed repetitive sequences. Science. 1982 Jun 4;216(4550):1065–1070. doi: 10.1126/science.6281889. [DOI] [PubMed] [Google Scholar]
- Soriano P., Macaya G., Bernardi G. The major components of the mouse and human genomes. 2. Reassociation kinetics. Eur J Biochem. 1981 Apr;115(2):235–239. doi: 10.1111/j.1432-1033.1981.tb05228.x. [DOI] [PubMed] [Google Scholar]
- Tazi J., Bird A. Alternative chromatin structure at CpG islands. Cell. 1990 Mar 23;60(6):909–920. doi: 10.1016/0092-8674(90)90339-g. [DOI] [PubMed] [Google Scholar]
- Wada M., Little R. D., Abidi F., Porta G., Labella T., Cooper T., Della Valle G., D'Urso M., Schlessinger D. Human Xq24-Xq28: approaches to mapping with yeast artificial chromosomes. Am J Hum Genet. 1990 Jan;46(1):95–106. [PMC free article] [PubMed] [Google Scholar]
- Zerial M., Salinas J., Filipski J., Bernardi G. Gene distribution and nucleotide sequence organization in the human genome. Eur J Biochem. 1986 Nov 3;160(3):479–485. doi: 10.1111/j.1432-1033.1986.tb10064.x. [DOI] [PubMed] [Google Scholar]