Abstract
Background
Several clinical trials have shown that immune treatment focus on programmed death-1 and programmed death-ligand 1 (PD-L1) yields a good clinical efficacy in advanced non-small cell lung cancer (NSCLC). We investigated whether the PD-L1 expression was related to clinicopathologic and molecular characteristics in patients with surgically resected NSCLC.
Methods
Between December 2008 and 2013, formalin-fixed, paraffin-embedded samples were obtained from patients with lung adenocarcinoma at Zhejiang Cancer Hospital. RT-PCR was used to analyze EGFR, KRAS, NRAS, PIK3CA, BRAF, HER2 mutations and ALK, ROS1, RET fusion genes. The PD-L1 expression was evaluated by immunohistochemistry and staining of 5 % or more was scored as positive expression. Survival analysis was evaluated using the Kaplan–Meier method. Multivariate regression was performed using the Cox proportional hazards model.
Results
Mutations were detected in 76.6 % of the 385 patients tested: EGFR mutation (n = 205, 53.2 %), followed by EML4–ALK rearrangement (n = 18, 4.7 %), KRAS (n = 16, 4.2 %), HER2 (n = 9, 2.3 %), ROS1 rearrangement (n = 8, 2.1 %), PIK3CA (n = 6, 1.6 %), RET rearrangement (n = 6,1.6 %), BRAF (n = 2, 0.5 %), and NRAS mutations (n = 1, 0.2 %). Twenty-four (6.2 %) patients carried coexisting mutations. PD-L1 expression was detected in 48.3 % (186/385) of all the patients. PD-L1 positive patients more frequently carried coexisting mutations (18/24, 75 %), followed by single-gene (145/271, 53.5 %) and pan-negative mutations (23/90, 25.6 %). PD-L1 expression decreased disease-free survival (DFS) in univariate analysis (P = 0.014). Multivariate analysis revealed that PD-L1 expression was not an independent risk factor for poor DFS and overall survival (OS) (P = 0.22 and 0.37, respectively).
Conclusions
PD-L1 overexpression is more frequently observed in oncogene-mediated lung adenocarcinoma, especially with coexisting mutation subtypes. PD-L1 expression is not a prognostic factor in surgically resected lung adenocarcinoma patients.
Keywords: Non-small cell lung cancer, Programmed cell deathligand 1, Lung adenocarcinoma, Gene mutation, Coexisting mutations, Prognosis
Background
Lung cancer is the leading cause of cancer-related death in China [1]. The standard treatment of lung cancer, especially non-small cell lung cancer (NSCLC) comprises platinum-based chemotherapy and driver gene-based targeted therapy, which resulted in extended survival and increased the quality of life in NSCLC patients [2–7]. However, drug resistance is a major challenge in most patients [8]. The median survival time in advanced NSCLC is no more than 2 years because of limited treatments available excluding chemotherapy and targeted therapy [9, 10].
Blockade of immune checkpoints in cancer with monoclonal antibodies has recently emerged as a promising approach to the treatment of solid tumors. Programmed death 1 (PD1), which belongs to the CD28 family of proteins, is a T cell surface receptor that regulates T cell activation and proliferation. Its ligand, programmed death-ligand 1 (PD-L1), is frequently expressed in many types of carcinomas [11–14]. Recent clinical trials found that inhibition of the PD-L1-PD1 interaction using specific antibodies resulted in promising antitumor efficacy in patients with various carcinomas [15, 16]. PD-L1 overexpression in NSCLC was reported ranging from 19 to 100 % [17–19]. Although several studies elucidated the association between common driver genes and PD-L1 expression in NSCLC, the results remain controversial and the prognostic value of PD-L1 expression is unclear [20].
This study focused on patients with completely resected lung adenocarcinoma and evaluated the association of PD-L1 expression with clinicopathologic parameters and driver genes, as well as its prognosis value in Chinese patients.
Patients and methods
Patients
A total of 385 adenocarcinoma patients underwent resection between December 2008 and 2013 in Zhejiang Cancer Hospital. Histological typing was determined according to the 2004 World Health Organization classification [21]. Tumor-node-metastasis (TNM) staging was based on the 7th edition of the lung cancer staging system. The recurrence or metastases were confirmed using chest CT, brain MRI, and bone scan as well as ultrasound and/or CT of the abdomen. The exclusion criteria included: (1) preoperative chemotherapy or radiation therapy, (2) death from other diseases unrelated to NSCLC. The Ethics Committee of Zhejiang Cancer Hospital approved this study and written informed consent was obtained from each participant.
Immunohistochemical analysis of PD-L1 expression
Immunohistochemical (IHC) staining of PD-L1 expression was performed on 4-6 μm thick formalin-fiated, paraffi-embedded tissue. The concentration of rabbit primary antibody that reacts to PD-L1 The concentration of rabbit primary antibody that reacts to PD-L1 (Proteintech Group Inc., Chicago, IL, USA, Catalog number: 66248-1-Ig) was 1:100 in Dako antibody diluent; slides were incubated with this antibody overnight at 4 °C. Then, the slides were incubated with Ventana Omni Mapanti-rabbit secondary antibody for 60 min. AVentana Chromo MapKit was used for antibody detection, and then the slides were counterstained with hematoxylin. Next, the slides were dehydrated and cover slipped as per normal laboratory protocol. Two independent pathologists (Wei Wu and Guoping Cheng) assessed the expressions.
PD-L1 immunostaining results were classified into two groups based on the degree and intensity of staining: (1) negative, when staining was absent or detected in <5 % of the cells; and (2) positive, when membranous staining was present in ≥5 % of the cells.
We used another antibody (5H1, Cell Signaling Technology, Beverly, MA, USA) to confirmed the PD-L1 expression in 102 patients. The PD-L1 immunostaining criterion is same with the former antibody.
Gene analysis
Genomic DNA or RNA was extracted from tumor tissues according to standard protocols (RNeasy Mini Kit, and QiAamp DNA Mini Kit, Qiagen, Hilden, Germany). Briefly, the isolated RNA samples were used for reverse transcription into cDNA using Revert Aid First Strand cDNA Synthesis Kit (Fermentas, St Leon-Rot, Germany). Either genomic DNA or cDNA was used for PCR amplification and sequencing. EGFR, HER2, KRAS, NRAS, BRAF, and PIK3CA were PCR amplified using genomic DNA. Cycle sequencing of the purified PCR products was carried out with PCR primers using the commercially available ADx Mutation Detection Kits (Amory, Xiamen, China).
The ALK, ROS1, and RET fusion mRNA was detected by PCR with fusion gene detection kit (Amory, Xiamen, China). In brief, total RNA was extracted with QiagenRNeasy FFPE Kit. The mRNA was reverse-transcribed to cDNA at 42 °C for 1 h. β-actin was used as the internal control. The RT-PCR conditions were as follows: an initial denaturation at 95 °C for 5 min, followed by 95 °C for 25 s, 64 °C for 20 s, and 72 °C for 20 s to ensure the specificity; and 31 cycles at 93 °C for 25 s, 60 °C for 35 s, 72 °C for 20 s were performed for data collection and sensitivity analysis. All of the positive genes including mutations or fusions were confirmed with Sanger sequencing. All the experiments were performed according to the user manual as described previously [22].
Statistical analysis
The Chi squared test was used to evaluate the relationships between different driver genes and PD-L1 expression. Survival curves of pathologically confirmed samples were calculated using the Kaplan–Meier method until death or last follow-up. Multivariate analysis was performed using the Cox regression model. Statistical analysis was performed with the SPSS 18 software (Chicago, IL, USA). P < 0.05 was considered statistically significant. The median follow-up was 54 months (6.5–65) and the last follow-up date was July 31, 2015.
Results
Patient characteristics
Patients’ clinical profile is presented in Table 1. One hundred and ninety-eight patients (51.4 %) were male with a median age of 59 years. One hundred and fifty (39.0 %) patients were former or current smokers. Pathologic stage I was seen in 121 patients, stage II in 79 patients, and stage III in 185 Patients.
Table 1.
Demographic characteristics of the study population (n = 385)
| Number | |
|---|---|
| Gender | |
| Male | 198 |
| Female | 187 |
| Age | |
| Range | 28–79 |
| Median | 59 |
| <60 | 207 |
| ≥60 | 178 |
| Smoking status | |
| Never | 235 |
| Former/current | 150 |
| Stage | |
| I | 121 |
| II | 79 |
| III | 185 |
| PD-L1 expression | |
| Yes | 186 |
| No | 199 |
| Gene alteration | |
| EGFR | 205 |
| ALK | 18 |
| KRAS | 16 |
| HER2 | 9 |
| ROS1 | 8 |
| PIK3CA | 6 |
| RET | 6 |
| BRAF | 2 |
| NRAS | 1 |
| Concurrent alteration | 24 |
| Pan-negative | 90 |
| Adjuvant treatment | |
| Yes | 269 |
| No | 116 |
Gene analysis results
All the patients were analyzed for EGFR, KRAS, NRAS, PIK3CA, BRAF and HER2 mutations and ALK, ROS1, RET fusion genes. This analysis included EGFR mutations (n = 205, 53.2 %), followed by EML4–ALK rearrangements (n = 18, 4.7 %), KRAS (n = 16, 4.2 %), HER2 (n = 9, 2.3 %), ROS1 (n = 8, 2.1 %), PIK3CA (n = 6, 1.6 %), RET (n = 6, 1.6 %), BRAF (n = 2, 0.5 %), and NRAS (n = 1, 0.2 %), and 24 coexisting mutations (6.2 %). All the nine genes were negative in 90 patients, defined as pan-negative. The details of coexisting mutations are listed in Table 2.
Table 2.
Clinical characteristics and PD-L1 expression in concurrent gene alteration patients
| Case | Gender | Age | Stage | Smoking | Gene type | PD-L1 expression | OS (month) |
|---|---|---|---|---|---|---|---|
| 1 | Male | 43 | IB | Yes | EGFR+PIK3CA | Yes | 67+ |
| 2 | Female | 51 | IIIA | No | RET+PIK3CA | Yes | 42 |
| 3 | Female | 58 | IIIA | No | EGFR+ALK | Yes | 34 |
| 4 | Male | 74 | IA | No | EGFR+PIK3CA | No | 66+ |
| 5 | Male | 60 | IIIA | Yes | KRAS+ALK | Yes | 35+ |
| 6 | Female | 60 | IB | No | EGFR+RET-M2 | Yes | 54 |
| 7 | Female | 60 | IA | No | EGFR+PIK3CA | Yes | 36+ |
| 8 | Male | 64 | IIA | No | RET+PIK3CA | No | 55 |
| 9 | Male | 69 | IB | No | KRAS6+HER2 | Yes | 43+ |
| 10 | Male | 45 | IIIA | Yes | KRAS+PIK3CA | Yes | 25 |
| 11 | Female | 64 | IIB | Yes | EGFR+HER2 | No | 46+ |
| 12 | Female | 75 | IIIA | No | EGFR+PIK3CA | No | 24+ |
| 13 | Female | 69 | IIA | No | KRAS+PIK3CA | Yes | 36 |
| 14 | Female | 49 | IB | No | EGFR+HER2 | Yes | 48+ |
| 15 | Female | 55 | IIB | Yes | ROS1+HER2 | Yes | 37+ |
| 16 | Male | 62 | IB | Yes | EGFR+ALK | No | 46 |
| 17 | Male | 55 | IIIA | No | EGFR+PIK3CA | Yes | 39 |
| 18 | Female | 68 | IB | No | EGFR+PIK3CA | Yes | 58+ |
| 19 | Female | 76 | IB | No | ALK+RET-M16 | No | 28 |
| 20 | Male | 43 | IB | No | EGFR+PIK3CA | Yes | 55+ |
| 21 | Male | 59 | IIIA | Yes | KRAS+PIK3CA | Yes | 18+ |
| 22 | Female | 61 | IB | No | EGFR+PIK3CA | Yes | 66+ |
| 23 | Female | 68 | IIA | No | EGFR+HER2 | Yes | 45 |
| 24 | Male | 62 | IIIA | Yes | KRAS+HER2 | Yes | 16 |
PD-L1 expression correlated with driver genes
The PD-L1 membrane expression was detected in 186 of the 385 lung adenocarcinoma patients (48.3 %) (Figs. 1, 2). The relationships between clinical parameters or gene characteristics and PD-L1 expression are shown in Table 3. PD-L1 expression was not significantly associated with any clinicopathologic parameters. Patients with PD-L1 positive expression more frequently presented with coexisting mutations (18/24, 75 %), followed by single-gene mutation (145/271, 53.5 %) and pan-negative (23/90, 25.6 %) genes. Differences in PD-L1 expression were found among the coexisting mutations, single-gene mutations and pan-negative genes (P < 0.001).
Fig. 1.
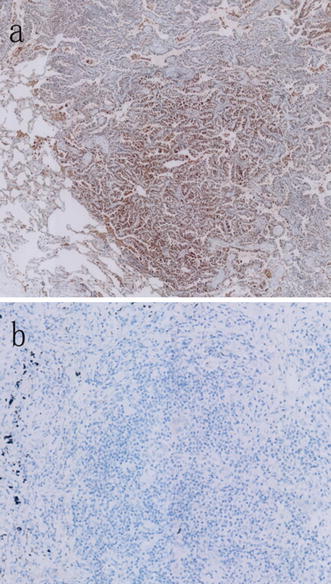
a Positive programmed cell death-ligand 1 (PD-L1) immunohistochemical staining in a patient with adenocarcinoma. b Negative PD-L1 immunohistochemical staining in another patient with adenocarcinoma
Fig. 2.
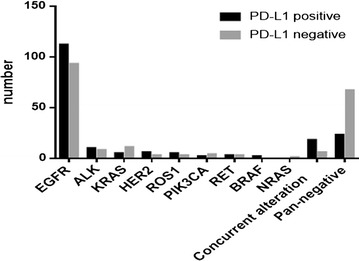
Relationship between PD-L1 expression and driver genes
Table 3.
Clinical characteristics comparison between PD-L1 positive and negative expression in NSCLC patients
| Variables | PD-L1 positive (n = 186) | PD-L1 negative (n = 199) | P |
|---|---|---|---|
| Gender | 0.07 | ||
| Male | 87 | 111 | |
| Female | 99 | 88 | |
| Age | 0.68 | ||
| <60 | 102 | 105 | |
| ≥60 | 84 | 94 | |
| Smoking status | 0.12 | ||
| Never | 121 | 114 | |
| Former/current | 65 | 85 | |
| Pathologic stage | 0.09 | ||
| I + II | 105 | 95 | |
| III | 81 | 104 | |
| EGFR | 0.008 | ||
| Yes | 112 | 93 | |
| No | 74 | 106 | |
| ALK | 0.53 | ||
| Yes | 10 | 8 | |
| No | 176 | 191 | |
| KRAS | 0.16 | ||
| Yes | 5 | 11 | |
| No | 181 | 188 | |
| HER2 | 0.44 | ||
| Yes | 6 | 3 | |
| No | 180 | 196 | |
| ROS1 | 0.65 | ||
| Yes | 5 | 3 | |
| No | 181 | 196 | |
| PIK3CA | 0.74 | ||
| Yes | 2 | 4 | |
| No | 184 | 195 | |
| RET | 0.74 | ||
| Yes | 3 | 3 | |
| No | 183 | 196 | |
| BRAF | 0.45 | ||
| Yes | 2 | 0 | |
| No | 184 | 199 | |
| NRAS | 0.97 | ||
| Yes | 0 | 1 | |
| No | 186 | 198 | |
| Concurrent alteration | 0.01 | ||
| Yes | 18 | 6 | |
| No | 168 | 193 | |
| Pan-negative | <0.01 | ||
| Yes | 23 | 67 | |
| No | 163 | 132 | |
Another antibody (5H1, Cell Signaling Technology, Beverly, MA, USA) was used in 102 patients to detect the PD-L1 expression. The same trend of PD-L1 expression difference existed in patients with different gene abnormality. The PD-L1 positive patitnets was more frequently carried coexisting mutations (5/8, 62.5 %), followed by single-gene positive (32/66, 48.5 %) and pan-negative mutations (10/28, 25.6 %)(P = 0.337).
Survival analysis
The median DFS and OS were 48.3 and 58.1 months, respectively. Patients with positive PD-L1 expression had shorter DFS than those with negative PD-L1 expression (38.0 vs. 50.4 months, P = 0.014) (Fig. 3), but the OS between the two groups showed no significant difference (52.9 vs. 68.2 months, P = 0.069) (Fig. 4; Table 4).
Fig. 3.
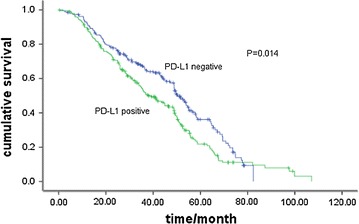
Disease free survival curves in patients with positive or negative programmed cell death-ligand 1 (PD-L1) staining (38.0 vs. 50.4 months, P = 0.014)
Fig. 4.
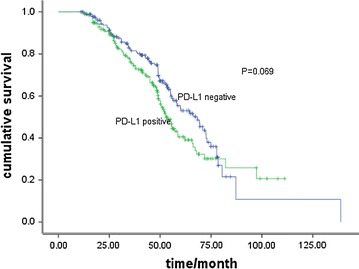
Overall survival curves in patients with positive or negative programmed cell death-ligand 1 (PD-L1) staining (52.9 vs. 68.2 months, P = 0.069)
Table 4.
Univariate analysis for disease-free survival and overall survival
| Variables | Median DFS | P | Median OS | P |
|---|---|---|---|---|
| Gender | 0.74 | 0.44 | ||
| Male | 44.6 | 55.6 | ||
| Female | 48.9 | 59.9 | ||
| Age | 0.23 | 0.39 | ||
| <60 | 49.3 | 59.3 | ||
| ≥60 | 42.9 | 55.2 | ||
| Smoking status | 0.16 | 0.59 | ||
| Never | 49.0 | 58.6 | ||
| Former/current | 41.3 | 56.0 | ||
| Pathologic stage | <0.001 | <0.001 | ||
| I + II | 52.5 | 66.2 | ||
| III | 30.2 | 45.0 | ||
| Adjuvant treatment | 0.54 | 0.76 | ||
| Yes | 49.7 | 59.2 | ||
| No | 46.5 | 56.5 | ||
| Driver genes | 0.23 | 0.24 | ||
| Positive | 48.9 | 58.7 | ||
| Negative | 42.0 | 50.4 | ||
| PD-L1 expression | 0.014 | 0.069 | ||
| Yes | 38.0 | 52.9 | ||
| No | 50.4 | 62.0 | ||
In univariate analysis, early stage (stage I and II versus III) and PD-L1 expression negative were significantly risk factors for tumor recurrence or metastasis (Figs. 3, 4), while only early stage was a favorable prognostic factor of OS (Table 4).
In multivariate analysis, only early stage suggested lower risk for DFS, while PD-L1 expression was not correlated with recurrence or metastasis. Early stage was an independent and favorable prognostic factor for OS (Table 5).
Table 5.
Multivariate survival analysis for disease-free survival and overall survival
| Variables | DFS | OS | ||||
|---|---|---|---|---|---|---|
| HR | 95 % CI | P | HR | 95 % CI | P | |
| Smoking status (smokers vs. non-smokers) | 0.84 | 0.66–1.07 | 0.16 | 1.12 | 0.81–1.53 | 0.48 |
| Stage (III vs. I + II) | 1.71 | 1.32–2.21 | 0.00 | 1.16 | 0.84–1.58 | 0.00 |
| PD-L1 expression (positive vs. negative) | 1.17 | 0.91–1.51 | 0.22 | 1.79 | 1.30–2.46 | 0.37 |
Discussion
This study shows that PD-L1 is overexpressed in 48.3 % (186/385) of lung adenocarcinoma patients and this overexpression is more frequently seen in patients with coexisting mutations, but less frequently in patients with pan-negative genes. The PD-L1 overexpression is not a prognostic factor for overall survival. To the best of our knowledge, this is the first study with the largest number of patients correlating the nine common driver genes in lung adenocarcinoma and PD-L1 expression.
Several studies have reported the association between PD-L1 expression and driver genes [23, 24]. The results of the correlation were controversial. Azuma et al. [14] observed that PD-L1 positive status was significantly associated with EGFR mutations . Mu et al. observed no significant correlation between PD-L1 expression and EGFR/ALK status in stage I NSCLC patients [25]. Similarly, Zhang et al. found that no association between PD-L1 expression and EGFR status in lung adenocarcinoma [25]. Therefore, the role of inhibition of PD-1/PD-L1 pathway and driver genes based on the results of existing studies is inconclusive, due to several reasons. First, most of the samples in previous studies were relatively small. Second, most of the studies focused on EGFR mutations or ALK rearrangements, and other driver genes were not well investigated. Last but not least, racial differences may play an important role in the controversial results.
In the present study, PD-L1 overexpression was more frequent in patients with coexisting mutations than in pan-negative patients. One explanation is that the genetic differences affected epigenetics, which may alter the expression of tumor-associated self-antigens, which in turn, affected tumor antigenicity. Increased number of driver genes reflects a higher level of neoantigens, which alters the immune microenvironment and increases the PD-L1 expression [26].
Because of heterogeneity of tumors, the efficacy of chemotherapy or molecular targeted treatment is relatively limited, combination treatment with different anti-cancer mechanisms drugs hold much potential in this area. Previous studies demonstrated that EGFR and ALK genes could induce PD-L1 expression to facilitate evasion of the host anti-tumour immune response, suggesting an active role for these genes in remodelling the immune microenvironment [27, 28]. In this way, combination of PD-1/PD-L1 blockade with targeted inhibitor or other drugs may be a promising therapeutic strategy to increase the duration of treatment response and delay development of drug resistance.
The role of PD-L1 in predicting the prognosis of NSCLC was controversial in previous studies [20]. Some studies found that negative PD-L1 expression led to superior OS in NSCLC patients compared with positive PD-L1 expression [14, 29], while Yang et al. [30] concluded that PD-L1 expression had no significant correlation with OS. In the present cohort, we found no association between the PD-L1 expression and overall survival in NSCLC patients. However, PD-L1 expression was related to shorter DFS. The results may contribute to the treatment after recurrence or metastasis.
Our study limitations are as follows. One major limitation was its retrospective nature. Second, only 24 patients with coexisting mutations were included, and the small sample size may influence the results of the current study. Third, different antibodies were used in different anti-PD-1 or PD-L1 therapies in clinical trials currently. The choice of antibody and the threshold for positivity might influence the results of different studies. Only one antibody and 5 % threshold were used in the present study. Different anti-PD-L1 antibodies may need to be validated in the same sample in future studies.
Conclusions
In conclusion, we demonstrated the expression of PD-L1 in over 48 % of lung adenocarcinoma patients and the expression was associated with coexisting driver genes. PD-L1 expression is not associated with overall survival in patients with completely resected NSCLC.
Authors’ contributions
CG carried out the molecular genetic studies and carried out the immunoassays. ZY and SZ participated in the design of the study and performed the statistical analysis. ZY and YX conceived of the study, and participated in its design and coordination and helped to draft the manuscript. All authors read and approved the final manuscript.
Acknowledgements
None declared.
Competing interests
The authors declare that they have no competing interests.
Availability of data
All of the histology slides have beeb scanned. It is available if the reader or reviewers required.
Ethics approval
The Ethics Committee of Zhejiang Cancer Hospital approved this study and written informed consent was obtained from each participant.
Abbreviations
- PD-1
programmed death-1
- NSCLC
non-small cell lung cancer
- DFS
disease free survival
- OS
overall survival
Contributor Information
Zhengbo Song, Email: songzhengbo83@163.com.
Xinmin Yu, Email: zjch1971@163.com.
Guoping Cheng, Email: zjzlyy11@163.com.
Yiping Zhang, Phone: +8657188122082, Email: zjzlyy16@163.com.
References
- 1.Fang JY, Dong HL, Wu KS, Du PL, Xu ZX, Lin K. Characteristics and prediction of lung cancer mortality in China from 1991 to 2013. Asian Pac J Cancer Prev. 2015;16(14):5829–5834. doi: 10.7314/APJCP.2015.16.14.5829. [DOI] [PubMed] [Google Scholar]
- 2.Han JY, Park K, Kim SW, Lee DH, KimHY Kim HT, et al. First-SIGNAL:first-line single-agent iressa versus gemcitabine and cisplatin trial innever-smokers with adenocarcinoma of the lung. J Clin Oncol. 2012;30(10):1122–1128. doi: 10.1200/JCO.2011.36.8456. [DOI] [PubMed] [Google Scholar]
- 3.Zhou C, Wu YL, Chen G, Feng J, Liu XQ, Wang C, et al. Erlotinib versus chemotherapy as first-line treatment for patients with advanced EGFR mutationpositive non-small-cell lung cancer (OPTIMAL, CTONG-0802): a multicentre, open-label, randomised, phase 3 study. Lancet Oncol. 2011;12(8):735–742. doi: 10.1016/S1470-2045(11)70184-X. [DOI] [PubMed] [Google Scholar]
- 4.Mitsudomi T, Morita S, Yatabe Y, Negoro S, Okamoto I, Tsurutani J, et al. West Japan Oncology Group. Gefitinib versus cisplatin plus docetaxel in patients with non-small-cell lung cancer harbouring mutations of the epidermal growth factor receptor (WJTOG3405): an open label, randomised phase 3 trial. Lancet Oncol. 2010;11(2):121–128. doi: 10.1016/S1470-2045(09)70364-X. [DOI] [PubMed] [Google Scholar]
- 5.Mok TS, Wu YL, Thongprasert S, Yang CH, Chu DT, Saijo N, et al. Gefitinib or carboplatin-paclitaxel in pulmonary adenocarcinoma. N Engl J Med. 2009;361(10):947–957. doi: 10.1056/NEJMoa0810699. [DOI] [PubMed] [Google Scholar]
- 6.Ciuleanu T, Brodowicz T, Zielinski C, Kim JH, Krzakowski M, Laack E, et al. Maintenance pemetrexed plus best supportive care versus placebo plus best supportive care for non-small-cell lung cancer: a randomised, double-blind, phase 3 study. Lancet. 2009;374(9699):1432–1440. doi: 10.1016/S0140-6736(09)61497-5. [DOI] [PubMed] [Google Scholar]
- 7.Paz-Ares L, de Marinis F, Dediu M, Thomas M, Pujol JL, Bidoli P, et al. Maintenance therapy with pemetrexed plus best supportive care versus placebo plus best supportive care after induction therapy with pemetrexed plus cisplatin for advanced non-squamous non-small-cell lung cancer (PARAMOUNT): a double-blind, phase 3, randomised controlled trial. Lancet Oncol. 2012;13(3):247–255. doi: 10.1016/S1470-2045(12)70063-3. [DOI] [PubMed] [Google Scholar]
- 8.Sequist LV, Waltman BA, Dias-Santagata D, Digumarthy S, Turke AB, Fidias P, et al. Genotypic and histological evolution of lung cancers acquiring resistance to EGFR inhibitors. Sci Transl Med. 2011;3(75):75. doi: 10.1126/scitranslmed.3002003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Temel JS, Greer JA, Muzikansky A, Gallagher ER, Admane S, Jackson VA, et al. Early palliative care for patients with metastatic non-small-cell lung cancer. N Engl J Med. 2010;363(8):733–742. doi: 10.1056/NEJMoa1000678. [DOI] [PubMed] [Google Scholar]
- 10.Zimmermann C, Swami N, Krzyzanowska M, Hannon B, Leighl N, Oza A, et al. Early palliative care for patients with advanced cancer: a cluster-randomised controlled trial. Lancet. 2014;383(9930):1721–1730. doi: 10.1016/S0140-6736(13)62416-2. [DOI] [PubMed] [Google Scholar]
- 11.Muenst S, Schaerli AR, Gao F, Daster S, Trella E, Droeser RA, et al. Expression of programmed death ligand 1 (PD-L1) is associated with poor prognosis in human breast cancer. Breast Cancer Res Treat. 2014;146(1):15–24. doi: 10.1007/s10549-014-2988-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schalper KA, Velcheti V, Carvajal D, Wimberly H, Brown J, Pusztai L, Rimm DL. In situ tumor PD-L1 mRNA expression is associated with increased TILs and better outcome in breast carcinomas. Clin Cancer Res. 2014;20(10):2773–2782. doi: 10.1158/1078-0432.CCR-13-2702. [DOI] [PubMed] [Google Scholar]
- 13.Ali HR, Glont SE, Blows FM, Provenzano E, Dawson SJ, Liu B, et al. PD-L1 protein expression in breast cancer is rare, enriched in basal-like tumours and associated with infiltrating lymphocytes. Ann Oncol. 2015;26(7):1488–1493. doi: 10.1093/annonc/mdv192. [DOI] [PubMed] [Google Scholar]
- 14.Azuma K, Ota K, Kawahara A, Hattori S, Iwama E, Harada T, et al. Association of PD-L1 overexpression with activating EGFR mutations in surgically resected non-small cell lung cancer. Ann Oncol. 2014;25(10):1935–1940. doi: 10.1093/annonc/mdu242. [DOI] [PubMed] [Google Scholar]
- 15.Brahmer JR, Tykodi SS, Chow LQ, Hwu WJ, Topalian SL, Hwu P, et al. Safety and activity of anti-PD-L1 antibody in patients with advanced cancer. N Engl J Med. 2012;366(26):2455–2465. doi: 10.1056/NEJMoa1200694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Herbst RS, Soria JC, Kowanetz M, Fine GD, Hamid O, Gordon MS, et al. Predictive correlates of response to the anti-PD-L1 antibody MPDL3280A in cancer patients. Nature. 2014;515(7528):563–567. doi: 10.1038/nature14011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wolfle SJ, Strebovsky J, Bartz H, Sahr A, Arnold C, Kaiser C, et al. PD-L1 expression on tolerogenic APCs is controlled by STAT-3. Eur J Immunol. 2011;41(2):413–424. doi: 10.1002/eji.201040979. [DOI] [PubMed] [Google Scholar]
- 18.Konishi J, Yamazaki K, Azuma M, Kinoshita I, Dosaka-Akita H, Nishimura M. B7-H1 expression on non-small cell lung cancer cells and its relationship with tumor-infiltrating lymphocytes and their PD-1 expression. Clin Cancer Res. 2004;10(15):5094–5100. doi: 10.1158/1078-0432.CCR-04-0428. [DOI] [PubMed] [Google Scholar]
- 19.Hirahara K, Ghoreschi K, Yang XP, Takahashi H, Laurence A, Vahedi G, et al. Interleukin-27 priming of T cells controls IL-17 production in trans via induction of the ligand PD-L1. Immunity. 2012;36(6):1017–1030. doi: 10.1016/j.immuni.2012.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang A, Wang HY, Liu Y, Zhao MC, Zhang HJ, Lu ZY, et al. The prognostic value of PD-L1 expression for non-small cell lung cancer patients: a meta-analysis. Eur J Surg Oncol. 2015;41(4):450–456. doi: 10.1016/j.ejso.2015.01.020. [DOI] [PubMed] [Google Scholar]
- 21.Travis WD, Brambilla E, Muler-Hermelink HK, et al (eds). WHO classification of tumours. Pathology and genetics of tumours of the lung, pleura, thymus and heart. Lyon: IARC Press; 2004. p. 145–47
- 22.Wu C, Zhao C, Yang Y, He Y, Hou L, Li X, et al. High discrepancy of driver mutations in patients with NSCLC and synchronous multiple lung ground-glass nodules. J Thorac Oncol. 2015;10(5):778–783. doi: 10.1097/JTO.0000000000000487. [DOI] [PubMed] [Google Scholar]
- 23.Akbay EA, Koyama S, Carretero J, Altabef A, Tchaicha JH, Christensen CL, et al. Activation of the PD-1 pathway contributes to immune escape in EGFR-driven lung tumors. Cancer Discov. 2013;3(12):1355–1363. doi: 10.1158/2159-8290.CD-13-0310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang Y, Wang L, Li Y, Pan Y, Wang R, Hu H, et al. Protein expression of programmed death 1 ligand 1 and ligand 2 independently predict poor prognosis in surgically resected lung adenocarcinoma. Onco Targets Ther. 2014;7:567–573. doi: 10.2147/OTT.S59959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mu CY, Huang JA, Chen Y, Chen C, Zhang XG. High expression of PD-L1 in lung cancer may contribute to poor prognosis and tumor cells immune escape through suppressing tumor infiltrating dendritic cells maturation. Med Oncol. 2011;28(3):682–688. doi: 10.1007/s12032-010-9515-2. [DOI] [PubMed] [Google Scholar]
- 26.Le DT, Uram JN, Wang H, Bartlett BR, Kemberling H, Eyring AD, et al. PD-1 blockade in tumors with mismatch-repair deficiency. N Engl J Med. 2015;372(26):2509–2520. doi: 10.1056/NEJMoa1500596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Akbay EA, Koyama S, Carretero J, et al. Activation of the PD-1 pathway contributes to immune escape in EGFR-driven lung tumors. Cancer Discov. 2013;3:1355–1363. doi: 10.1158/2159-8290.CD-13-0310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ota K, Azuma K, Kawahara A, et al. Induction of PD-L1 expression by the EML4–ALK oncoprotein and downstream signaling pathways in non-small cell lung cancer. Clin Cancer Res. 2015;21(17):4014–4021. doi: 10.1158/1078-0432.CCR-15-0016. [DOI] [PubMed] [Google Scholar]
- 29.Pardoll DM. The blockade of immune checkpoints in cancer immunotherapy. Nat Rev Cancer. 2012;12(4):252–264. doi: 10.1038/nrc3239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yang CY, Lin MW, Chang YL, Wu CT, Yang PC. Programmed cell death-ligand 1 expression in surgically resected stage I pulmonary adenocarcinoma and its correlation with driver mutations and clinical outcomes. Eur J Cancer. 2014;50(7):1361–1369. doi: 10.1016/j.ejca.2014.01.018. [DOI] [PubMed] [Google Scholar]


