Abstract
The ATPase SecA mediates the posttranslational translocation of a wide range of polypeptide substrates through the SecY channel in the cytoplasmic membrane of bacteria. We have determined the crystal structure of a monomeric form of Bacillus subtilis SecA at a 2.2-Å resolution. A comparison with the previously determined structures of SecA reveals a nucleotide-independent, large conformational change that opens a deep groove similar to that in other proteins that interact with diverse polypeptides. We propose that the open form of SecA represents an activated state.
Many polypeptides are exported from bacterial cells after completion of their biosynthesis, employing a posttranslational translocation pathway. N-terminal signal sequences direct polypeptides to the Sec machinery in the cytoplasmic membrane. The major components of the machinery are the transmembrane proteins, SecY, SecE, and SecG, which form the protein-conducting SecY channel, as well as the cytosolic ATPase SecA (1, 2).
SecA is essential and sufficient to move polypeptides through the channel, but how it utilizes cycles of ATP hydrolysis to provide the driving force for translocation is poorly understood. The currently favored model is a membrane insertion and deinsertion mechanism in which SecA binds a polypeptide segment in the cytosol and inserts with it into the channel (3). Upon ATP hydrolysis, SecA moves back into the cytosol, leaving the polypeptide segment in the channel, and grabs the next segment, continuing this process until the polypeptide is across the membrane. Several lines of evidence support insertion of SecA across the membrane, including protease protection of SecA upon interaction with SecY (3, 4) and apparent periplasmic exposure of SecA (5, 6). However, alternative interpretations of these data have been proposed, suggesting that SecA may not insert into the channel (7). Indeed, the x-ray structure of the SecY channel suggests that the channel cannot accommodate a SecA molecule (8). What is clear is that SecA undergoes conformational changes that are coupled to its interaction with ligands, and that domain movements driven by the ATPase cycle are required to move polypeptide chains through the channel.
SecA exists in an equilibrium between dimeric and monomeric states (9–11), with the monomeric state stabilized by interactions with acidic phospholipids or the SecY complex (9, 12–14). A synthetic signal peptide also induces dissociation of the dimer (9, 12). A SecA mutant that fails to dimerize retains significant translocation activity (see ref. 9; E. Or and T.A.R., unpublished data). These data suggest that the active form of SecA may be monomeric. Further experiments indicate that the same ligands, phospholipids, and signal peptides can induce additional conformational changes in SecA (11, 15).
Crystal structures of SecA from Bacillus subtilis and Mycobacterium tuberculosis show that it contains two domains that are similar to the two RecA-like domains that form the ATPase site of superfamily 1 and 2 helicases (16, 17). In helicases, such as PcrA, the two RecA-like domains move relative to one another during the ATPase cycle, generating domain movements that translocate the helicase along nucleic acids (18). Mutagenesis of residues in SecA (9, 19, 20) supports the idea that it uses a similar mechanism to generate domain movements that are coupled to polypeptide translocation.
SecA also contains the following three domains with no similarity to any helicase domains: the preprotein crosslinking domain (PPXD), helical wing domain (HWD), and helical scaffold domain (HSD) (16, 17). How these domains can generate a promiscuous peptide-binding site is not clear from the previous structures. We describe the crystal structure of B. subtilis SecA in a monomeric state in which the PPXD, previously implicated in polypeptide binding, undergoes a dramatic rigid body rotation. This conformational change opens a deep groove in the protein, which is similar to the peptide-binding sites in other proteins that interact with a diverse range of polypeptide substrates. The open conformation may correspond to an activated state of SecA.
Methods
Protein Purification and Crystallization. Full-length B. subtilis SecA was cloned into the plasmid pET19b, incorporating a prescission protease cleavage site after the N-terminal histidine tag, and expressed in BL21 cells. After purification with nickel–nitrilotriacetic acid (Ni-NTA) agarose, the histidine tag was cleaved. SecA was purified further with a MonoQ column and dialyzed against 10 mM Hepes, pH 7.5/0.5 mM Tri(2-carboxyethyl)phosphine. Crystals of SecA were grown by using the hanging-drop method with a reservoir containing 100 mM Hepes (pH 7–7.5), 250 mM magnesium acetate, and 7–12% polyethylene glycol 8000. Crystals were frozen in reservoir solution containing 10% butanediol.
Structure Determination. SecA crystals contain a single molecule in the asymmetric unit and belong to the space group C2 with the following cell dimensions: a = 149.2 Å, b = 107.9 Å, c = 72.1 Å, and β = 95.0°. Data were processed with hkl2000 (21). The structure was determined by molecular replacement using the program amore (22). The previously published structure of SecA (1M6N), with residues 221–354 omitted, was used as a search model. After refinement by using cns, density modification was carried out (23), and it yielded clear density for the region corresponding to residues 221–354. After addition of a model for this region, further cycles of manual rebuilding by using the program o (24) and refinement by using cns were performed. The structure of ADP-bound SecA was also determined from crystals soaked in adenosine 5′-[β,γ-imido]triphosphate. No density for the γ phosphate was seen in the electrondensity map, indicating that ADP (a contaminant in 5′-[β,γ-imido]triphosphate) was bound.
Maleimide Fluorescein Labeling and Chemical Crosslinking. Single cysteine mutations (E234C or K824C) were introduced into N95 Escherichia coli SecA containing a C-terminal histidine tag and lacking cysteines (C98S). Mutants were purified with nickel–nitrilotriacetic acid (Ni-NTA) agarose, followed by a Superdex 200 gel-filtration column. We labeled 0.6 μM SecA in 50 mM Hepes, pH 7/50 mM KCl by addition of 63 μM maleimide fluorescein. After 3 min at 25°C, DTT was added to a concentration of 330 mM. Samples were run on an SDS/PAGE gel and visualized under UV light. Crosslinking with 20 mM 1-ethyl-3-(3-dimethylaminopropyl)carbodiimide was performed under identical conditions for 10 min before quenching with 250 mM glycine and SDS sample buffer. The following detergents were used in labeling and crosslinking experiments: 1-myristoyl-2-hydroxy-sn-glycero-3-[phospho-rac-(1-glycerol)], 1,2-diheptanoyl-sn-glycero-3-phosphocholine, dodecyl maltoside, decyl maltoside, octyl maltoside, octyl glucoside, CYMAL4, CY-MAL6, lauryldimethylamine-N-oxide, digitonin, and SDS. Large unilamellar vesicles were prepared from 70% 1,2-dioleoyl-sn-glycerol-3-phosphoethanolamine and 30% 1,2-dioleoyl-sn-glycerol-3-[phospho-rac-(1-glycerol)] (15) and added to a final concentration of 2 mg/ml.
Results
Crystallization of SecA. Crystals of B. subtilis SecA that diffracted to 2.2 Å were obtained, and the structure was solved by molecular replacement by using parts of the published B. subtilis SecA structure (16) as a search model. Most of the electron density could be readily interpreted, and many regions designated in the previous structure as “unreliable” are well ordered. The C-terminal 61 residues were disordered, although mass spectrometry indicated that they were present in the crystallized protein. The structure of ADP-bound SecA was also determined. Crystallographic statistics are shown in Table 1.
Table 1. Crystallographic statistics.
| Data set | SecA (apo)* | SecA (ADP)* |
|---|---|---|
| Resolution, Å | 2.18 | 2.9 |
| Unique reflections | 58,197 | 25,014 |
| I/Iσ | 28.37 (2.65) | 20.93 (2.08) |
| Completeness, % | 98.8 (97.7) | 99.3 (94.0) |
| Rsym† | 0.063 (0.516) | 0.073 (0.479) |
| Rcryst,‡ % | 24.0 (31.3) | 22.8 (38.6) |
| Rfree,§ % | 27.3 (33.5) | 29.2 (45.2) |
| rms deviation bond angles, ° | 1.1 | 1.3 |
| rms deviation bond length, Å | 0.007 | 0.008 |
| Average B factor | 56.1 | 61.5 |
| Most favored angles,¶ % | 90.5 | 88.3 |
| Allowed angles,¶ % | 8.8 | 10.4 |
| Generously allowed angles,¶ % | 0.7 | 1.3 |
Values in parentheses refer to data in the highest-resolution shell (2.26-2.18 Å and 3-2.9 Å in the apo and ADP datasets, respectively).
Rsym = ΣhklΣi|Ii(hkl) – I(hkl)|/ΣhklΣi|Ii(hkl), where I(hkl) is the average intensity.
Rcryst = Σhkl∥Fobs| – k|Fcalc∥/Σhkl|Fobs|, where Fcalc and Fobs are the calculated and observed structure factor amplitudes, respectively.
Rfree is the same as Rcryst except that the summation is over randomly selected reflections excluded from the refinement.
Defined by procheck.
Description of the Structure. As noted in refs. 16 and 17, the ATPase domain of SecA is made up of two RecA-like folds referred to as nucleotide-binding folds 1 and 2 (NBF1 and NBF2) (Fig. 1). Similar to other RecA-like domains, they contain a sheet of β-strands, flanked on either side by α-helices. The nucleotide-binding site is found at the interface between NBF1 and NBF2, with both domains contributing residues that are essential for catalysis (Fig. 1).
Fig. 1.
Structure of monomeric B. subtilis SecA. Monomeric B. subtilis SecA is presented as a ribbon diagram. NBF1 is shown in yellow, NBF2 is shown in blue, the PPXD is shown in orange, the HSD is shown in green, and the HWD is shown in cyan. ADP is shown in a ball-and-stick representation. The images were prepared by using molscript (40), raster3d (41), or spock (available at http://mackerel.tamu.edu/spock).
The HSD contains a long α-helix extending from the end of NBF2. At its C terminus, it forms part of a three-helix bundle, which forms an interface with NBF1. The HWD is an insertion into the three-helix bundle. It contains two α-helices, but other regions in the domain appear to be less ordered (high B factors). The PPXD, containing α-helices and β-strands, emerges from NBF1 and forms an interface with the HSD. The crystal structure of SecA in the presence of magnesium–ADP is almost identical to the nucleotide-free structure, but it shows some small changes in side-chain conformations around the bound nucleotide.
SecA Is Crystallized as a Monomer. In the previous B. subtilis SecA structure, two SecA monomers pack head to tail in the crystal lattice, likely representing the physiological dimer (16) (Fig. 2a). An antiparallel dimer in solution is supported by fluorescence resonance energy transfer experiments (11). E. coli SecA can be converted to a monomeric species by mutation of residues in the HSD or by deletion of the 11 N-terminal residues (ref. 9; E. Or and T.A.R., unpublished data). These residues are at the dimer interface (shown in color in Fig. 2a), supporting the physiological relevance of the dimer seen in the previous crystal structure. The interface of this dimer buries 5,442 Å2 (determined by using cns and a 1.4-Å probe radius) (23). The largest intermolecular interface in our structure buries a much smaller area than the physiological dimer (2,410 Å2; Fig. 2b), the crystal contacts are different, and residues important for SecA dimerization are in a minor crystal contact. In addition, the intersubunit distances obtained from fluorescence resonance energy transfer experiments in solution are not consistent with our structure being the physiological dimer (11). All of these data suggest that we have crystallized monomeric SecA.
Fig. 2.
Contacts in different SecA crystal forms. (a) Ribbon diagram of the previously determined B. subtilis SecA structure, which is likely to be the physiological dimer (16). (b) The largest contact in the crystal lattice of monomeric B. subtilis SecA. (c) The proposed M. tuberculosis SecA dimer (17). Residues 757–768 in the HSD and the N terminus of SecA that are important for dimerization of E. coli SecA are shown in red and yellow, respectively.
The crystal packing in our structure is also different from that observed for M. tuberculosis SecA. It has been proposed that the M. tuberculosis SecA structure also represents a physiological dimer, but its interface is different from that of the B. subtilis SecA dimer (Fig. 2c) and it buries only 2,822 Å2 (17). Residues that are important for dimerization of E. coli SecA are not localized at this interface (Fig. 2c), suggesting that the M. tuberculosis SecA structure also represents a monomeric state of SecA.
Domain Movements in B. subtilis SecA. Comparison with the previous structures shows that monomeric B. subtilis SecA is essentially identical in the NBF1 and NBF2 regions (Fig. 3); the average difference (rms deviation) for the peptide backbone in the NBFs (including residues 57–217 and 356–540) is 0.7 Å when compared with dimeric B. subtilis SecA, and 0.92 Å when compared with M. tuberculosis SecA (omitting residues in the insert in NBF2 of M. tuberculosis SecA). In contrast, the other domains have undergone large movements.
Fig. 3.
Domain movements in monomeric SecA. Ribbon diagram of monomeric B. subtilis SecA in the open conformation (a) and of a single subunit of dimeric B. subtilis SecA in the closed conformation (b). Color codes are as described for Fig. 1. The first and last helices in the PPXD are represented as cylinders to better visualize the transition between the conformations. The arrows in a indicate the movements that are required to convert the open conformation to the closed conformation. The side chains of residues 232 and 773 are shown in red in stick representation. Corresponding E. coli SecA residue numbers are given in parentheses. These residues were mutated to cysteines in E. coli SecA, and the accessibility of residue 824 to a modification reagent was used to probe the transition from the closed to the open conformation.
The most dramatic domain movement in monomeric B. subtilis SecA is an ≈60° rigid body rotation of the PPXD (Fig. 3 and Movie 1, which is published as supporting information on the PNAS web site). The “hinge point” occurs around residues 226 and 348 in the two strands connecting the PPXD and NBF1. The rms deviation for the α-carbon atoms in the PPXD (including residues 232–343) between monomeric B. subtilis SecA and M. tuberculosis SecA is 1.7 Å. The region is essentially identical to the dimeric B. subtilis domain, with deviations mostly in residues declared as unreliable in the dimer structure. Rotation of the PPXD results in the breaking of its “porous” interface with the HSD and HWD (16) and the formation of a large groove between these three domains (Fig. 3).
Together, the HWD and HSD undergo a rotation of ≈15° relative to the same domains in the dimeric B. subtilis SecA (Fig. 3). Additionally, in the monomeric B. subtilis SecA, the HSD is shifted toward NBF2 by >2 Å. Its overall fold is similar, with an rms deviation of 1.5 Å for superimposing it with the same region of dimeric SecA. There is a short stretch of 3/10 helix between residues 599 and 601 in the monomeric B. subtilis SecA, possibly indicating that this region may be able to accommodate further conformational changes. Given that the drastic conformational changes in monomeric B. subtilis SecA occur in the absence of any changes in NBF1 and NBF2, they are unlikely to represent nucleotide-dependent movements.
Potential Interaction Sites for Substrate and Binding Partners. The movement of PPXD opens a large groove, ≈10 Å deep and 15 Å wide, between the PPXD and the HSD/HWD, which is an excellent candidate for a peptide-binding site (groove 1; red in Fig. 4a). It is similar to peptide-binding grooves in other proteins, such as calmodulin (Fig. 4b) (25) and Hsp70 (Fig. 4c) (26), which interact with diverse substrates. Groove 1 contains numerous charged and polar residues that might interact with the peptide backbone. It also contains two large pockets, one in the HSD and HWD (pocket H) and the other in the PPXD (pocket P; Fig. 4 d and e), which might accommodate side chains of the peptide substrate.
Fig. 4.
Potential ligand-binding sites in monomeric B. subtilis SecA. (a) A surface representation of SecA showing the surface grooves 1 and 2 in red and blue, respectively. (b) Surface representation of calmodulin with bound peptide shown in green as an α-carbon trace (25). (c) Surface representation of Hsp70, with the C terminus occupying the peptide-binding groove shown in green as an α-carbon trace (26). (d and e) Surface representations of groove 1, showing the location of pockets H and P, respectively. The surface is rendered transparent, and the underlying peptide backbone is color-coded as described for Fig. 1.
There is also a shallow groove extending across the HSD and part of NBF1 adjacent to groove 1 (groove 2; blue in Fig. 4a) (16). The upper side of the groove is lined by two β-strands connecting the PPXD and NBF1. In dimeric B. subtilis SecA, this groove was occupied partially by the C-terminal residues, 794–798 (16), but in monomeric B. subtilis SecA, the equivalent region is disordered, and therefore, the groove is accessible.
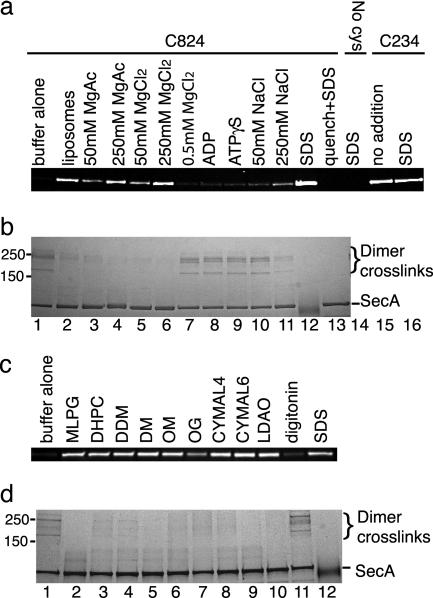
E. coli SecA Can Adopt a Monomeric, Open Conformation. To determine whether E. coli SecA can adopt an open conformation in solution, we tested the accessibility of residues in the interface between the PPXD and HSD to the thiol-modifying reagent maleimide fluorescein. Based on the B. subtilis SecA structures, a cysteine introduced at position 824 of E. coli SecA (773 in B. subtilis SecA) should be buried in the closed, but not the open, conformation (Fig. 3). A cysteine at position 234 of E. coli SecA (232 in B. subtilis SecA) should be accessible in both states. As expected, in the presence of SDS, both cysteines were labeled efficiently by fluorescein maleimide (Fig. 5a, lanes 12 and 16), whereas a cysteine-less mutant (N95 SecA, C98S) was not labeled (lane 14). In buffer alone, the cysteine at position 824 was not labeled (lane 1), indicating that the ground state of SecA is a closed conformation. In the presence of liposomes, this residue was labeled efficiently (lane 2), suggesting that lipid-associated SecA adopts an open conformation. A similar conformational change was induced by the addition of 50–250 mM Mg2+ (lanes 3–6) or various nondenaturing detergents (Fig. 5c, lanes 2–10), an exception being digitonin, which is likely the mildest tested detergent (lane 11). These results indicate that SecA can adopt an open conformation in solution when not constrained in a crystal lattice.
Fig. 5.
E. coli SecA can adopt an open conformation in solution. (a) SecA containing single cysteines at positions 824 or 234, or a mutant lacking cysteines (no cys), were labeled with maleimide fluorescein in the presence of the indicated additions. When nucleotide (0.25 mM) was added, 0.5 mM MgCl2 was also included. The samples were separated by SDS/PAGE and visualized under UV light. In the lane labeled quench+SDS, the quenching reagent, either DTT or glycine, was added before labeling or crosslinking. (b) In parallel, samples were crosslinked with 1-ethyl-3-(3-dimethylaminopropyl)carbodiimide, separated by SDS/PAGE, and stained with Coomassie blue. The position of dimer crosslinks is indicated. (c) Modification reactions with SecA containing a cysteine at position 824 were performed as in a in the presence of 1-myristoyl-2-hydroxy-sn-glycero-3-[phospho-rac-(1-glycerol)] (MLPG, 0.1 mM); 1,2-diheptanoyl-sn-glycero-3-phosphocholine (DHPC, 4.2 mM); dodecyl maltoside (DDM, 0.6 mM); decyl maltoside (DM, 3.6 mM); octyl maltoside (OM, 39 mM); octyl glucoside (OG, 36.4 mM); CYMAL4 (15.2 mM); CYMAL6 (1.12 mM); lauryldimethylamine-N-oxide (LDAO, 2 mM); digitonin (1%); or SDS (0.5%). (d) In parallel, samples were crosslinked with 1-ethyl-3-(3-dimethylaminopropyl)carbodiimide.
In parallel to the experiments described above, we determined the oligomeric state of SecA by chemical crosslinking (9). In buffer alone, crosslinking with 1-ethyl-3-(3-dimethylaminopropyl)carbodiimide led to the appearance of several high-molecular-weight species, diagnostic of the dimeric state of SecA (Fig. 5b, lane 1) (9). The addition of lipid, Mg2+, or nondenaturing detergents, which efficiently converted SecA to an open conformation (Fig. 5 a and c), also caused dissociation of the SecA dimer, as indicated by the loss of high-molecular-weight crosslinks (Fig. 5 b, lanes 2 and 3–6, and d, lanes 2–10). Digitonin was ineffective in dimer dissociation (Fig. 5d, lane 11), and 250 mM NaCl caused partial dissociation of the dimer and only inefficient labeling with maleimide fluorescein. Several of the detergents used, such as 1-myristoyl-2-hydroxy-sn-glycero-3-[phospho-rac-(1-glycerol)] (12) and dodecyl maltoside (9), as well as high salt concentrations have been demonstrated (10, 11) to cause dissociation of the SecA dimer. Thus, there is a general correlation between conversion of SecA into an open conformation and dissociation of the dimer into monomers.
Potential Domain Interactions During the ATPase Cycle. One ADP molecule is bound in the cleft between NBF1 and NBF2 (Fig. 1) (16, 17), and there is no evidence for a second ATP-binding site (proposed in ref. 27). Comparison with the structure of PcrA bound to a nonhydrolysable ATP analogue (18) shows that, for ATP-hydrolysis to occur, NBF2 must move relative to NBF1 such that the essential and conserved arginine residue in helicase motif 6 (9, 28) of SecA can coordinate the γ-phosphate of ATP (Fig. 6a). Relative movement of the two NBFs is supported by experiments showing that the linker between them becomes protease-resistant in the presence of nucleotide (29). The reorientation of NBF1 and NBF2 might be propagated to other domains, resulting in movement of the polypeptide substrate. Conversely, conformational changes in other domains may affect the ATPase cycle. The strand leading from NBF1 to the PPXD emerges from the Walker B motif (Fig. 6b), and the strand leading from the PPXD to NBF1 is connected to the strand between the Walker A and B strands. Both strands connecting NBF1 to the PPXD line groove 2.
Fig. 6.
The ATPase site and mutations affecting SecA function. (a) The Walker A motifs of ADP-bound SecA and ATP-bound PcrA are superimposed. The Walker A and B motifs and motif 6 of monomeric B. subtilis SecA are shown in color, and the equivalent motifs in PcrA are shown in gray (18). Arginine residues in motif 6 are shown in stick representation, and a magnesium ion bound to SecA is shown as a green sphere. The ATP bound to PcrA and the ADP bound to SecA are shown in a stick representation in gray and light blue, respectively. The phosphorus atom of the γ-phosphate of ATP bound to PcrA is shown in red. (b) A view of the SecA ATPase site showing that the Walker B motif (red) is connected to the PPXD via a β-strand (gray). The β-strand (blue) that leads from the PPXD back to NBF1 is connected to the strand between the Walker A (black) and B motifs. NBF2 is omitted for clarity, and the remaining domains are color-coded as described for Fig. 1. The aspartate residue in the Walker B motif and ADP are shown in stick representation. (c) SecA mutations that suppress mutations in SecY channel components are shown in yellow, and PrlD mutations that suppress signal sequence mutations are shown in cyan on a backbone representation of monomeric B. subtilis SecA. Residues identified in both screens are shown in green. ADP is shown as a stick representation.
Many superactive SecA mutations that suppress multiple mutations in SecY channel components (30) and the PrlD mutations that suppress signal peptide defects (31, 32) map to residues that are close to the nucleotide-binding site (Fig. 6c). However, the superactive SecA mutation (H309Y; residue 288 in B. subtilis) is found in pocket P, and the A373V mutation (residue 353 in B. subtilis), identified both as a PrlD and a superactive SecA mutant, is found in groove 2, attesting to the importance of these sites and interdomain communication.
Discussion
We have determined the crystal structure of SecA in a monomeric, open conformation in which movement of the PPXD, HSD and HWD generates a deep groove on the surface of the protein. We demonstrate that E. coli SecA adopts a similar open conformation in solution under various conditions. Most significantly, the interaction of SecA with lipids, which is likely to be an early step during protein translocation, induces the open conformation. Fluorescence resonance energy transfer and Trp fluorescence experiments have also demonstrated movement of the PPXD and HSD upon interaction with lipid (11, 15). Trp-775 in the HSD of E. coli SecA moves from a buried environment to a solvent-exposed environment upon binding lipid or signal peptide. In the B. subtilis dimeric structure, this Trp residue is buried in the hydrophobic core of the HSD, whereas in the monomeric B. subtilis SecA structure, the residue is in a more polar environment in the close proximity of three water molecules.
Lipid binding not only induces an open conformation of SecA but also dissociates the SecA dimer (9, 12, 13). The observations that a monomeric SecA mutant retains significant translocation activity (9) and that acidic phospholipids are essential for efficient protein translocation (33) indicate that the monomeric open conformation represents an important, activated state of SecA. It is unclear whether the SecA dimer could adopt an open conformation, but it has been reported that the dimer can adopt two different conformations (10). However, because most conditions that induce an open conformation also dissociate the dimer, it appears that the monomer can undergo the conformational change to the open state more readily.
The large groove (groove 1) between the PPXD, HSD and HWD is an excellent candidate for a peptide-binding site. Crosslinking experiments indicate that the PPXD is in close proximity to bound preprotein (34), and mutagenesis of residues in the PPXD further supports a role for it in polypeptide binding (35). Groove 1 has similar dimensions to those seen in other proteins that interact with a wide range of peptide substrates, including the Hsp70 family (26), calmodulin (36), and OppA (37). In these cases, the walls of a deep groove embrace the polypeptide, and its side chains are accommodated in large pockets. The two pockets (P and H) in SecA are large enough to accommodate bulky side chains of the substrate. They are lined mostly, but not exclusively, by polar and charged residues. Structural analysis of OppA demonstrates how such pockets can accommodate each of the 20-aa side chains (37): the pockets are filled with water molecules that are displaced, depending on the size of the side chain. A similar principle is also used by the nonanchor positions of the MHC I peptide-binding site (38).
Based on crosslinking experiments, SecA is likely to change its affinity for peptide substrate during the ATPase cycle (34, 39). A high-affinity state could be induced by nucleotide-dependent conformational changes in groove 1 such that, in the high-affinity state, the groove would close around the peptide. Binding and release cycles could be coupled to other conformational changes that push the polypeptide into the SecY channel. It has been proposed (16) that groove 2 may form a polypeptide- or signal sequence-binding site. Alternatively, or in addition, this shallow, conserved groove may bind to the cytoplasmic loops in SecY (8). The structures of SecA now provide a basis to address some of these ideas.
Supplementary Material
Acknowledgments
We thank S. Harrison for advice and critical reading of the manuscript; T. Ellenberger for discussions; B. van den Berg, U. Unligil, E. Toth, D. King, T. Hollis, and M. Groll for advice and reagents; M. Becker, L. Berman, and S. LaMarra for support at beamline X25 (Brookhaven National Laboratory, supported by the U.S. Department of Energy, Division of Materials Sciences and Division of Chemical Sciences, under contract no. DE-AC02-98CH10886); and A. Joachimiak, S. Ginell, and R. Alkire at beamline ID19 (Advanced Photon Source, supported by the U.S. Department of Energy, Office of Science, Office of Basic Energy Sciences, under contract no. W-31-109-ENG-38). We also thank Y. Modis and E. Or for critical reading of the manuscript. The work was supported by a grant from the National Institutes of Health (to T.A.R.). T.A.R. is a Howard Hughes Medical Institute Investigator. W.M.C. was supported by a fellowship from the Damon Runyon Cancer Research Foundation.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: NBF1/2, nucleotide-binding folds 1 and 2; PPXD, preprotein crosslinking domain; HSD, helical scaffold domain; HWD, helical wing domain.
Data deposition: The atomic coordinates have been deposited in the Protein Data Bank, www.pdb.org (PDB ID codes 1TF5 and 1TF2 for the nucleotide-free and ADP-bound forms of SecA, respectively).
References
- 1.Benach, J. & Hunt, J. F. (2004) Nature 427, 24–26. [DOI] [PubMed] [Google Scholar]
- 2.Mori, H. & Ito, K. (2001) Trends Microbiol. 9, 494–500. [DOI] [PubMed] [Google Scholar]
- 3.Economou, A. & Wickner, W. (1994) Cell 78, 835–843. [DOI] [PubMed] [Google Scholar]
- 4.Eichler, J. & Wickner, W. (1998) J. Bacteriol. 180, 5776–5779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ramamurthy, V. & Oliver, D. (1997) J. Biol. Chem. 272, 23239–23246. [DOI] [PubMed] [Google Scholar]
- 6.van der Does, C., den Blaauwen, T., de Wit, J. G., Manting, E. H., Groot, N. A., Fekkes, P. & Driessen, A. J. (1996) Mol. Microbiol. 22, 619–629. [DOI] [PubMed] [Google Scholar]
- 7.van der Does, C., Manting, E. H., Kaufmann, A., Lutz, M. & Driessen, A. J. (1998) Biochemistry 37, 201–210. [DOI] [PubMed] [Google Scholar]
- 8.Van den Berg, B., Clemons, W. M., Jr., Collinson, I., Modis, Y., Hartmann, E., Harrison, S. C. & Rapoport, T. A. (2004) Nature 427, 36–44. [DOI] [PubMed] [Google Scholar]
- 9.Or, E., Navon, A. & Rapoport, T. (2002) EMBO J. 21, 4470–4479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Woodbury, R. L., Hardy, S. J. & Randall, L. L. (2002) Protein Sci. 11, 875–882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ding, H., Hunt, J. F., Mukerji, I. & Oliver, D. (2003) Biochemistry 42, 8729–8738. [DOI] [PubMed] [Google Scholar]
- 12.Benach, J., Chou, Y. T., Fak, J. J., Itkin, A., Nicolae, D. D., Smith, P. C., Wittrock, G., Floyd, D. L., Golsaz, C. M., Gierasch, L. M. & Hunt, J. F. (2003) J. Biol. Chem. 278, 3628–3638. [DOI] [PubMed] [Google Scholar]
- 13.Bu, Z., Wang, L. & Kendall, D. A. (2003) J. Mol. Biol. 332, 23–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Duong, F. (2003) EMBO J. 22, 4375–4384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ding, H., Mukerji, I. & Oliver, D. (2001) Biochemistry 40, 1835–1843. [DOI] [PubMed] [Google Scholar]
- 16.Hunt, J. F., Weinkauf, S., Henry, L., Fak, J. J., McNicholas, P., Oliver, D. B. & Deisenhofer, J. (2002) Science 297, 2018–2026. [DOI] [PubMed] [Google Scholar]
- 17.Sharma, V., Arockiasamy, A., Ronning, D. R., Savva, C. G., Holzenburg, A., Braunstein, M., Jacobs, W. R., Jr., & Sacchettini, J. C. (2003) Proc. Natl. Acad. Sci. USA 100, 2243–2248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Velankar, S. S., Soultanas, P., Dillingham, M. S., Subramanya, H. S. & Wigley, D. B. (1999) Cell 97, 75–84. [DOI] [PubMed] [Google Scholar]
- 19.Sianidis, G., Karamanou, S., Vrontou, E., Boulias, K., Repanas, K., Kyrpides, N., Politou, A. S. & Economou, A. (2001) EMBO J. 20, 961–970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schmidt, M. O., Brosh, R. M., Jr., & Oliver, D. B. (2001) J. Biol. Chem. 276, 37076–37085. [DOI] [PubMed] [Google Scholar]
- 21.Otwinowski, Z. & Minor, W. (1997) Methods Enzymol. 276, 307–326. [DOI] [PubMed] [Google Scholar]
- 22.Collaborative Computational Project Number 4 (1994) Acta Crystallogr. D 50, 760–763.15299374 [Google Scholar]
- 23.Brunger, A. T., Adams, P. D., Clore, G. M., DeLano, W. L., Gros, P., Grosse-Kunstleve, R. W., Jiang, J. S., Kuszewski, J., Nilges, M., Pannu, N. S., et al. (1998) Acta Crystallogr. D 54, 905–921. [DOI] [PubMed] [Google Scholar]
- 24.Jones, T. A., Zou, J. Y., Cowan, S. W. & Kjeldgaard (1991) Acta Crystallogr. A 47, 110–119. [DOI] [PubMed] [Google Scholar]
- 25.Yamauchi, E., Nakatsu, T., Matsubara, M., Kato, H. & Taniguchi, H. (2003) Nat. Struct. Biol. 10, 226–231. [DOI] [PubMed] [Google Scholar]
- 26.Morshauser, R. C., Hu, W., Wang, H., Pang, Y., Flynn, G. C. & Zuiderweg, E. R. (1999) J. Mol. Biol. 289, 1387–1403. [DOI] [PubMed] [Google Scholar]
- 27.Mitchell, C. & Oliver, D. (1993) Mol. Microbiol. 10, 483–497. [DOI] [PubMed] [Google Scholar]
- 28.Koonin, E. V. & Gorbalenya, A. E. (1992) FEBS Lett. 298, 6–8. [DOI] [PubMed] [Google Scholar]
- 29.Karamanou, S., Vrontou, E., Sianidis, G., Baud, C., Roos, T., Kuhn, A., Politou, A. S. & Economou, A. (1999) Mol. Microbiol. 34, 1133–1145. [DOI] [PubMed] [Google Scholar]
- 30.Matsumoto, G., Nakatogawa, H., Mori, H. & Ito, K. (2000) Genes Cells 5, 991–999. [DOI] [PubMed] [Google Scholar]
- 31.Fikes, J. D. & Bassford, P. J., Jr. (1989) J. Bacteriol. 171, 402–409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Huie, J. L. & Silhavy, T. J. (1995) J. Bacteriol. 177, 3518–3526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hendrick, J. P. & Wickner, W. (1991) J. Biol. Chem. 266, 24596–24600. [PubMed] [Google Scholar]
- 34.Kimura, E., Akita, M., Matsuyama, S. & Mizushima, S. (1991) J. Biol. Chem. 266, 6600–6606. [PubMed] [Google Scholar]
- 35.Kourtz, L. & Oliver, D. (2000) Mol. Microbiol. 37, 1342–1356. [DOI] [PubMed] [Google Scholar]
- 36.Hoeflich, K. P. & Ikura, M. (2002) Cell 108, 739–742. [DOI] [PubMed] [Google Scholar]
- 37.Sleigh, S. H., Seavers, P. R., Wilkinson, A. J., Ladbury, J. E. & Tame, J. R. (1999) J. Mol. Biol. 291, 393–415. [DOI] [PubMed] [Google Scholar]
- 38.Jones, E. Y. (1997) Curr. Opin. Immunol. 9, 75–79. [DOI] [PubMed] [Google Scholar]
- 39.van Voorst, F., Vereyken, I. J. & de Kruijff, B. (2000) FEBS Lett. 486, 57–62. [DOI] [PubMed] [Google Scholar]
- 40.Kraulis, P. J. (1991) J. Appl. Crystallogr. 24, 946–950. [Google Scholar]
- 41.Merritt, E. A. & Bacon, D. J. (1997) Methods Enzymol. 277, 505–524. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.