Abstract
Eosinophilic oesophagitis (EoE) is characterized by oesophageal dysfunction and oesophageal eosinophilia refractory to proton-pump-inhibitor treatment. EoE is a food allergy, as elimination of food trigger(s) abrogates the disease, while trigger(s) reintroduction causes recurrence. The allergic mechanism of EoE involves both IgE and non-IgE processes. There is a break in oral tolerance, the immune mechanism allowing enteric exposure to food and micro-organisms without causing deleterious immune responses. Changes in life-style, alterations in gut flora and use of antibiotics may be increasing disease prevalence. Mouse models of EoE and human studies revealed the role of regulatory T-cells and iNKT-cells in the pathogenesis. Th2-cytokines like IL-4, IL-5 and IL-13, and other cytokines like TGFβ and TSLP are involved. Perhaps no one cytokine is critically important for driving the disease. Control of EoE may require a pharmaceutical approach that blocks more than one target in the Th2-inflammatory pathway.
Keywords: eosinophilic oesophagitis, allergic mechanism, antigen sensitization, oral tolerance, pathogenesis, t-helper lymphocyte type 2 immunity, food allergy, barrier dysfunction
Introduction
The concept of eosinophilic oesophagitis (EoE) as a food allergy seems foreign to many patients, and even physicians, since EoE does not exhibit the typical symptoms associated with allergic reactions, like hives, swelling, pruritus, wheezing or anaphylaxis. Instead, it causes symptoms of oesophageal dysfunction such as dysphagia, food impaction, vomiting and pain. The National Institute of Allergy and Infectious Disease expert panel defines food allergy as “an adverse health effect arising from a specific immune response that occurs reproducibly on exposure to a given food” [1]. According to this definition, EoE is unambiguously a food allergy because elimination of the food trigger abrogates the disease, while reintroduction causes disease recurrence [2,3]. Histologically, EoE is characterized by significant oesophageal eosinophilia (>15 eos/HPF) refractory to treatment with proton-pump-inhibitors (PPIs) [4].
Oesophageal eosinophilia was first described by Dobbins and colleagues in 1977 in a patient with concurrent eosinophilic gastroenteritis and a normal pH study [5]. Landres and co-workers, in 1978, reported the first case of eosinophilic infiltration isolated to the oesophagus with associated oesophageal dysfunction [6]. The absence of acid reflux, based on a negative pH study, suggested factors other than gastroesophageal reflux disease (GERD) were operative. Two case series, published in 1982 and 1985, concluded GERD was the underlying cause of EoE [7,8].
This paradigm of oesophageal eosinophilia as a result of GERD shifted in 1993 when Attwood and colleagues retrospectively compared patients with oesophageal eosinophilia who had normal and abnormal 24-hour pH studies [9]. They concluded that patients with significant oesophageal eosinophilia, dysphagia, and a normal pH study represented a distinct clinical group caused by factors other than GERD. This non-GERD trigger of oesophageal eosinophilia remained unknown until Kelly and colleagues, in 1995, published convincing evidence that food proteins caused the disease [2]. They showed elimination of food proteins from the diet by administrating amino-acid based elemental diets significantly reduced PPI-refractory oesophageal eosinophilia and improved clinical symptoms. Moreover, re-introduction of the food proteins caused recurrence of the symptoms. Markowitz and colleagues replicated these observations in 2003 with a larger cohort [10]. Recent clinical trials with empirical elimination diet [11] and skin testing-directed elimination diet [3] further support EoE as a food allergen-driven disease. Although there is overwhelming evidence that food allergens drive EoE, not all patients achieve histological and/or symptomatic remission with elimination diets. This discrepancy may result from non-compliance to treatment or other allergic triggers (e.g. aeroallergens).
This review discusses our current understanding of the allergic mechanism involved in EoE. Also delineated are potential therapeutic targets and opportunities for future research.
IgE or non-IgE mediated?
Food allergy is classified based on the mechanism of antigen recognition: IgE-mediated (immediate type) or non-IgE-mediated (delayed type). Food allergy is usually caused by IgE-mediated reactions. IgE-mediated food allergy is characterized by a reproducible, rapid onset of symptoms after ingesting the offending food. The classic example is the immediate development of hives, swelling, and wheezing after consuming peanuts in an individual with IgE-mediated peanut allergy. To trigger an IgE-mediated food allergic reaction, an individual must first be exposed to the food antigen and become sensitized by producing antigen-specific IgE antibodies. During sensitization, activated antigen-presenting cells (APCs) prime the native T-cells to differentiate into Th2-cells, which provide signals necessary to induce B-cell class-switching to generate IgE. These antigen-specific IgE molecules attach to the surface of mast cells. Upon re-exposure to the offending substance, the allergen crosslinks the IgE molecules on the mast cells causing rapid release of their preformed inflammatory mediators. This response is followed by subsequent de novo synthesis and release of lipid mediators, cytokines and chemokines that orchestrate a late phase immune response in which inflammatory cells such as eosinophils infiltrate tissue.
Although EoE patients do not exhibit the typical IgE-mediated food allergic reactions; their oesophageal lining, and only their oesophageal lining, acquires the characteristic elements of an IgE-mediated response such as dendritic cells (DCs), antigen-specific IgE, class-switched B-cells [12], tryptase-positive mast cells [13] and Th2-cytokines [14,15]. Yet serum levels of allergen-specific IgE and the results from skin prick testing (SPT) correlate poorly with the food trigger(s). Furthermore, systemic blockage of IgE-antibodies fails to eradicate oesophageal eosinophilia both in murine models [16] and in human clinical trials [4]. IgE-deficient (Igh-7−/−) and B-cell deficient mice develop experimental EoE [16,17], suggesting IgE may not be required to induce or maintain EoE. Collectively, these observations from human and mice studies suggest there are other non-IgE-mediated pathways important in the EoE pathogenesis. This is consistent with the categorization of EoE as a mixed IgE- and non-IgE-mediated food allergy in the recent guidelines [1].
Oral tolerance
We encounter millions of foreign antigens through our skin, gastrointestinal and respiratory tract. One of the key functions of our immune system is to distinguish pathogenic from non-pathogenic foreign antigens allowing an appropriate response. We do not develop allergic reactions to foods that we consume under normal circumstances because our immune system is tolerant to these foreign, but non-pathogenic food antigens. This is called oral tolerance [18].
Food allergy results from loss of oral tolerance. Clinical studies point towards antibiotic use during infancy, caesarean delivery, pre-term birth and lack of breast-feeding as early-life risk factors predisposing people to EoE [19]. Early-life exposures help shape our microbiome, which plays a critical role in development of oral tolerance [20,21].
Foxp3+ regulatory T-cells (Tregs) help maintain oral tolerance [22,23]. They regulate the pro-inflammatory pathway and maintain immune homeostasis. EoE patients have fewer Tregs in their oesophageal tissue compared to healthy controls, and corticosteroid treatment does not correct this deficiency [24]. This imbalance between Tregs and effector T-cells may be partially responsible for breaking oral tolerance in susceptible individuals. How to restore oral tolerance is an area of active research.
Sensitization
Antigen sensitization is a prerequisite for breaking oral tolerance. Various murine models of EoE show allergic sensitization can occur via the airway [25], skin [16,26] or gastrointestinal tract [27,28]. The route of allergic sensitization in human EoE is largely unknown, but clinical observations provided some insight. Multiple reports describe new-onset EoE in paediatric and adult patients undergoing oral immunotherapy (OIT) for IgE-mediated food allergies [29]. During OIT, increasing doses of food antigens are reintroduced to restore oral tolerance. These patients have IgE-mediated food allergy, but do not have EoE prior to starting OIT. While undergoing desensitization, they become susceptible to EoE. This observation suggests that allergic sensitization for EoE can occur orally, and that mechanisms of antigen sensitization in EoE may differ from that of IgE-mediated food allergy.
Other studies suggest that sensitization to food allergens in EoE results from aeroallergen inhalation [30]. While airway and skin are routes of sensitization to trigger experimental EoE in mice [16,25,26], there is limited human data supporting this hypothesis. Yet, the majority of EoE patients have respiratory allergies [4]. It is proposed that food allergen sensitization in EoE patients mostly results from cross-reactivity to birch pollens via PR-10 proteins (based on component-resolved diagnostics from a preliminary human study) [30].
Epithelial production of IL-33 and thymic stromal lymphopoietin (TSLP) have become key areas of interest in allergic sensitization [22]. TSLP is a key cytokine in antigen sensitization that promotes Th2-cell development [16]. Genetic studies link EoE susceptibility to TSLP variants [31,32], and an ex-vivo study showed TSLP enhances the basophil response in oesophageal biopsies of human subjects with EoE [16]. Furthermore, TSLP is necessary for development of EoE in mice, which can be prevented with antibody-mediated TSLP neutralization [16].
Genomic studies are elucidating critical mechanisms of allergic sensitization in EoE. A large genome-wide association study (GWAS) correlated EoE susceptibility to 9 of the 22 antigen sensitization loci [32]. However, how these specific genes are involved in antigen sensitization in EoE has yet to be explored.
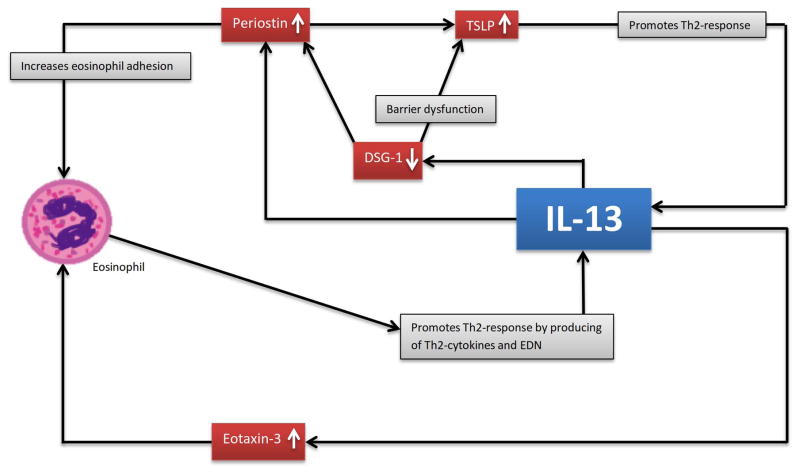
Sensitization may be facilitated by dysfunction in tight junctions that allow antigen penetration via the epithelial barrier [33]. EoE patients have reduced expression of desmosomal cadherin desmoglein-1 (DSG-1) in oesophageal tissues [34]. Reduction of DSG-1 weakens the oesophageal epithelial integrity and barrier function, potentiating allergic sensitization [35]. This in turn drives a Th2-response producing IL-13, which strongly down-regulates the expression of DSG-1 and propagates the local inflammatory process (figure 1).
FIGURE 1. Self-perpetuated inflammatory cycle mediated by IL-13.
IL-13 up-regulates periostin, while down-regulating DSG-1. Down-regulation of DSG-1 enhances the production of TSLP and periostin. Up-regulation of periostin further enhances the expression of TSLP, which is a strong driver of the Th2-response that propels the IL-13 inflammatory cycle. IL-13 stimulates the oesophageal epithelial cells to express eotaxin-3. Both eotaxin-3 and periostin recruit eosinophils to the site of inflammation; eotaxin-3 as a chemo-attractant, while periostin increases eosinophil adhesion. Eosinophils produce IL-13 and release EDN to activate DCs to prime Th2-differentiation.
Although the exact mechanism by which antigens are sensitized to cause EoE is unknown; genetic, epidemiologic, translational, human and animal studies are shedding light on this complicated process.
Lessons from human oesophageal biopsies
Assessment of treatment response is heavily dependent on histological evaluation of oesophagus biopsies. Patients treated with an elimination diet undergo multiple endoscopies and biopsies to identify the food(s) triggering the disease. These biopsies are taken during both disease and remission states, providing researchers the opportunity to study the allergic mechanism at tissue level.
From histological analysis of these biopsies, researchers have learned that active EoE displays a prominent eosinophilic infiltration with increased density of mast cells, basophils and lymphocytes [36]. Immunohistochemistry and/or immunofluorescence analysis further characterized these cells, which include tryptase- and TGF-β1-positive mast cells [37], CD8+ T-cells [38], IgG4+ plasma cells [39] and Vα24Jα18+ T-cells (a cell marker for invariant natural killer T (iNKT)-cells) [40].
However, histological analysis provides no information on cell function at the site of inflammation. Using unbiased whole-genome-wide transcript expression profile analysis of oesophageal tissues with Affymetric DNA chip, Blanchard et al uncovered 574 genes uniquely expressed in EoE [13]. The gene encoding eotaxin-3 was most highly induced. This landmark study provided a framework for investigation of inflammatory pathways critical for driving EoE. Yet, these results did not reveal the whole story, since genome-wide expression chips have modest sensitivity and cannot detect expression of potentially critical immunological cytokines produced at low levels. For example, IL-13 was not part of the initial EoE transcriptome identified by Affymetrix DNA microarray analysis. In subsequent years, higher resolution mRNA analysis techniques uncovered additional transcripts. Using human inflammatory cytokine and receptor PCR array, IL-13 and IL-5 were found to be markedly up-regulated [14]. Another key set of mRNAs, not identified by the Affymetrix DNA chip, is associated with the iNKT pathway [41]. Oesophageal tissue from patients with EoE display heightened expression of chemokine ligand 16 (CXCL16), iNKT-cell-associated cell marker Vα24, and CD1d compared to healthy control oesophageal tissue. This up-regulated gene expression was more pronounced in patients aged <6 years at diagnosis and correlated with the expression of eotaxins and periostin.
In 2010, a GWAS based on 181 patients with EoE and 1,974 controls identified an EoE susceptibility locus at 5q22, which is a gene region encoding TSLP. The association of TLSP to EoE was confirmed in 2014 with a larger cohort GWAS [32]. This GWAS also identified another EoE susceptibility locus at 2p23, encoding CAPN14, which is significantly more expressed in the oesophagus. However, the role of CAPN14 in EoE is unknown [40].
Micro-RNAs (miRNAs) are regulators of mRNA expression and translation. The first miRNA profile in EoE was reported in 2012 [42]. Lu et al found that EoE was associated with 32 regulated miRNAs and was distinguished from non-eosinophilic forms of oesophagitis. The exact functions of these regulated miRNAs also remain unknown.
Lessons from animal models
In 2001, Mishra et al developed the first murine model of EoE [25]. Mice were challenged intranasally with Aspergillus fumigatus and they developed the characteristic features of EoE. Rayapudi et al demonstrated indoor insect allergens could induce experimental EoE [43]. These studies raised awareness that aeroallergens may have an etiological role in EoE.
Several immune cell types and cytokines are proved important for expression of EoE in murine models. Using lymphocyte-deficient mice, Mishra et al found that T-cells, but not B-cells, are required for the development of experimental EoE [17]. By analysing mice overexpressing IL-5, either by genetic manipulation or by IL-5 administration, researchers established the central role of IL-5 in trafficking eosinophils to the oesophagus [44]. Intra-tracheal administration of IL-13 was shown to be sufficient to induce experimental EoE in a STAT6-dependent pathway [45]. Noti et al developed an IgE-independent murine model and showed that TSLP-regulated basophils contributed to the pathogenesis of experimental EoE [16]. In a food allergen sensitization model, Rajavelu et al found eotaxins and iNKT-cells were important for EoE development. Taken together, these mice studies have elucidated the potential routes of antigen sensitization (oral, respiratory and cutaneous), the role of different lymphocyte subsets and the importance of Th2 adaptive immunity in driving EoE. Key mouse models and their findings are summarized in table 1.
TABLE 1.
Lessons learned from mouse models.
| Key findings | Reference |
|---|---|
| First murine model of EoE. EoE induced by intratracheal administration of Aspergillus fumigatus. | Mishra 2001 [25] |
| EoE induced by intratracheal IL-13. Mechanism is dependent on IL-5, eotaxin-1, and STAT6. | Mishra 2003 [45] |
| Mice were sensitized epicutaneously to develop experimental EoE. First evidence that antigens could be sensitized via the skin in EoE. | Akei 2005 [26] |
| Mice deficient in the eotaxin-3 receptor (CCR3−/−) could not develop EoE. | Blanchard 2006 [13] |
| T-cells, but not B-cells, are required to trigger EoE. | Mishra 2007 [17] |
| IL-5-deficient mice or eosinophil-lineage deficient mice had significantly less tissue remodelling. | Mishra 2008 [82] |
| EoE was induced intranasally by cockroach and dust mite allergens in an eotaxin and IL-5 mediated pathway. Mice deficient in eotaxin-1/2, CCR3 or IL-5 failed to develop EoE. | Rayapudi 2010 [43] |
| Administration of anti-Siglec-F antibodies significantly decreased eosinophilic inflammation. | Rubinstein 2011 [28] |
| Mice were sensitized intraperitoneally, and then challenged intranasally or intragastrically with corn or peanut to induce EoE. Disease process is dependent on eotaxin and iNKT-cells. | Rajavelu 2012 [83] |
| IgE is not required to generate experimental EoE. EoE is TSLP and basophil dependent. | Noti 2013 [16] |
| Mast cell-deficient mice model had reduced hyperplasia and hypertrophy, but showed no difference on eosinophil recruitment to the oesophagus. | Niranjan 2013 [84] |
Putting the Puzzle Together
Based on the findings in human and mice, we can postulate on the allergic mechanism involved in EoE (see figure 2).
FIGURE 2. Proposed allergic mechanisms of EoE.
Antigen is presented by APCs, which promotes a Th2-response in EoE. Th2-cytokines are primarily produced by Th2-cells, but can be produced by ILC2, iNKT-cells, eosinophils and mast cells. IL-5 increases the proliferation and differentiation of eosinophils in the bone marrow, maintains their survival and facilitates their migration into the blood. IL-13 stimulates eotaxin-3 production from the epithelial cells, which recruits circulating eosinophils to the oesophagus. Eosinophils produce Th2-cytokines propagating the inflammatory cycle. They release cytotoxic granules causing tissue injury. They activate DCs via EDN to prime Th2-cells and possibly iNKT-cells. Eosinophils activate mast cells via MBP. Mast cells can also be activated by antigens cross-linking their surface IgE. Activated mast-cells release IL-13, IL-5 and TFGβ-1. Together, mast cells and eosinophils cause fibrosis, remodelling and dysmotility in a TGFβ-1 mediated pathway.
Several mechanisms have been proposed to tip the balance from tolerance to sensitization. One of them is impaired barrier function as EoE patients have barrier dysfunction of their oesophagus. This is supported by (i) histological findings of dilated intercellular spaces [46], (ii) increased oesophageal permeability [34] and (iii) decreased DSG-1 expression [47]. DSG-1 is important in maintaining barrier integrity. However, barrier disruption alone is not sufficient to induce EoE, as most patients with disorders of barrier disruption, such as inflammatory bowel disease and psoriasis, do not have higher than expected incidence of food allergy. Tregs, tolergenic DCs and IL-10 producing macrophages provide additional protection [22]. Several lines of evidence suggested that disturbance in the microbial flora (associated with early risk factors such as C-section and early antibiotic use) can impair normal development of this regulatory mechanism, leading to loss of oral tolerance [19–21]. Th2 propensity in susceptible individuals also contributes to tipping the balance from tolerance to sensitization. Th2-cytokine expression is markedly elevated in EoE [14].
Milk, wheat and eggs are the most common food triggers of EoE [48]. We do not know why these foods are more apt to trigger EoE than others, though there is evidence that certain foods process intrinsic immunological properties that can directly induce innate immune responses. For example, milk sphingomyelin can activate iNKT promoting Th2-response [49], and peanut allergen Ara h1 can directly bind to CD209 on DCs [50]. Given the limited number of foods that can trigger EoE, it is proposed there are certain intrinsic immunological properties of these food proteins that can confer allergenicity, although this remains controversial [51]. Taken together, the intrinsic properties of certain food proteins, barrier dysfunction, Th2 propensity and composition of microbiota influenced by early life events predisposing to dysregulation of the regulatory circuit, all contribute to sensitization.
APCs engulf, process and present peptides coupled to MHC class II molecules on their surface. APCs are classified into “professional” and “atypical” [52]. The professional APCs in the oesophagus are DCs, which are considered the most important APCs in EoE. DC levels are increased in the oesophagus of children with EoE compared to healthy controls [53]. Professional APCs constitutively express MHC class II structural proteins and antigen-processing machinery. By contrast, atypical APCs such as epithelial cells and eosinophils do not constitutively express MHC class II molecules, but can up-regulate the expression of MHC class II under pathological conditions [52]. Mulder et al showed that epithelial cells from EoE biopsies expressed the MHC class II protein HLA-DR. Using a human oesophageal cell line HET-1A, they demonstrated epithelial cells can engulf, process and present antigen in an IFN-γ-dependent manner. This data supports the potential role of oesophageal epithelial cells in presenting antigen in EoE [54].
Multiple investigators have shown eosinophils from EoE biopsies express HLA-DR [55]. Le-Carlson et al demonstrated the presence of costimulatory markers on eosinophils (CD40 and CD80) and activation markers on T-cells (CD28 and CD69) within the oesophageal epithelia of patients with EoE [56]. Collectively, these data imply, but do not prove that eosinophils can present antigen to T-cells. This concept remains controversial since there is no compelling evidence eosinophils can present antigen and activate naive T-cells in an antigen-specific manner [52].
Antigens are processed and presented to naive T-cells, which are primed to mature into antigen-specific Th2-cells under the influence of Th2-cytokines. Oesophageal T-cells of EoE patients are characterized by Th2-defining cell surface markers such as CCR8 and CRTH2 [15]. The most extensively studied Th2-cytokines are IL-13, IL-5 and IL-4 (see table 2).
TABLE 2.
Cytokines and cells involved in EoE mechanism.
| Cytokines and cells | Origin | Key function(s) in EoE |
|---|---|---|
| TSLP | Epithelial cells |
|
| Th2-cells | Lymphocyte progenitor cells in the bone marrow |
|
| B-cells | Lymphocyte progenitor cells in the bone marrow |
|
| Mast cells | Myeloid progenitor cells in the bone marrow |
|
| Eosinophils | Myeloid progenitor cells in the bone marrow |
|
| Basophils | Myeloid progenitor cells in the bone marrow |
|
| ILC2 | Lymphocyte progenitor cells in the bone marrow |
|
| IL-4 | Origin unclear; basophils, Th2-cells |
|
| IL-5 | Primarily produced by Th2 cells, also by eosinophils and mast cells |
|
| IL-9 | Origin unclear; eosinophils, ILC2 cells |
|
| IL-13 | Primarily produced by Th2 cells, also by eosinophils and mast cells |
|
| IL-33 | Epithelial cells |
|
| Eotaxin-3 | Epithelial cells |
|
| Periostin | Epithelial cells |
|
IL-13 is produced by Th2-cells and recruits eosinophils via an eotaxin-3 driven pathway [47]. It appears to activate the local tissue inflammatory response in Th2-associated diseases. Levels of IL-13 are increased 16-fold in the oesophagus of EoE patients [47]. It induces eotaxin-3 (CCL26) production from the oesophageal epithelial cells [13,57], which recruits eosinophils [26,45,57]. In addition, it recruits eosinophils by promoting fibroblasts to produce periostin [58], which increases eosinophil adhesion and TSLP levels [59]. IL-13 reduces barrier integrity by down-regulating DSG-1; decreased levels of DSG-1 increases periostin and TSLP [33,34,60].
TSLP is an epithelial-derived cytokine associated with multiple allergic disorders [16] and is known to promote the Th2-response [61]. TSLP levels are elevated in oesophageal tissues of patients with EoE [31], and it is required to induce experimental EoE [16]. There is evidence TSLP can activate DCs to drive the Th2-response via OX40 ligands [62], and its level is associated with heightened basophil responses in human [16]. Taken together, IL-13 perpetuates the local inflammatory process by recruiting IL-13-producing eosinophils, via stimulation of eotaxin-3 and periostin production, as well as by inducing TSLP secretion which promotes a strong Th2-response (see figure 1).
IL-5 is expressed by Th2-cells [14], eosinophils [63], basophils [64], innate lymphoid type 2 cells (ILC2) and iNKT-cells [65,66]. Its level is increased in the oesophagus of EoE patients [67]. IL-5 is involved in the differentiation and maturation of eosinophils in the bone marrow [65]. It promotes eosinophil survival [68] and migration into blood [69]. Epithelial cell-derived eotaxin-3, induced by IL-13, recruits the circulating eosinophils from the blood to accumulate in the oesophagus. In addition, IL-5 plays a role in the development, metabolism and function of basophils [70], but the exact role of IL-5-basophils axis in EoE is not defined.
IL-4 is overexpressed in oesophageal biopsies from EoE patients [12], but its exact role is not established. Although the source of IL-4 is controversial, basophils contribute significantly in initiating the Th2-inflammatory response [61]. Once the Th2-response is initiated, Th2-cells also produce IL-4 [71]. A key function of IL-4 is to induce B-cell class-switching [12], but it also plays a role in eosinophil recruitment by stimulating eotaxin-3 production from the epithelial cells [57,72].
Eosinophils are the end-effector cells of EoE. They are responsible for the injury in the oesophagus. Recruitment and migration of eosinophils from the bone marrow to the oesophagus is orchestrated by a complex network of cytokines, chemokines and lipid mediators [73]. Activated eosinophils at the site of inflammation produce a wide array of pro-inflammatory cytokines including IL-4, IL-5, IL-13, TGFβ, and TNF. These cytokines modulate the allergic response and contribute to tissue damage [14,63]. Tissue remodelling in EoE patients is largely controlled by TGFβ, which causes fibrosis and influences epithelial growth [74]. The toxic proteins, such as eosinophil peroxidase, cationic protein, neurotoxin (EDN) and major basic protein (MBP) damage the epithelium [63]. EDN skews the adaptive immunity to a Th2-response by activating DCs via the Toll-like receptor 2-myeloid differentiation factor 88 signalling pathway [75]. Leukotriene C4 increases vascular permeability, smooth muscle contraction and may therefore contribute to oesophageal dysmotility [36,63]. However, leukotriene inhibitors are ineffective in resolving inflammation in human EoE [4].
Mast cells are also increased in EoE [76] and are derived from the same CD34+ progenitor cell type as eosinophils. While the oesophagus is devoid of eosinophils at baseline, mast cells normally reside in the oesophagus. Their recruitment to the oesophagus is dependent on the SCF-SCFR axis [76]. MCTC (mast cells with tryptase and chymase) are the predominant mast cell-type in the oesophageal epithelium. They store and release TGFβ-1, IL-4, IL-5, IL-13, eotaxin, histamine, leukotrienes, proteases and lipid mediators when activated. The best-characterized mechanism of mast cell activation is cross-linking of their surface IgE molecules in an antigen-specific manner. Mast cells with antigen-specific IgE bound to their Fc-receptors are present in the oesophagus of EoE patients [12,38], but whether mast cells are activated in an IgE-dependent manner in EoE remains controversial, as discussed previously. Among the non-IgE mechanisms of mast cell activation, eosinophil-derived MBP seems the most relevant, although not yet proven in EoE [76]. Mast cells, through the production of TGFβ-1, play a central role in tissue remodelling and promoting aberrant smooth muscle contractility in EoE [37,77].
Conclusion, and Future Clinical and Research Directions
Variable symptoms and responses to treatments suggest EoE represents a group of heterogeneous diseases of the oesophagus, rather than a single disease. Three key points are known about EoE pathogenesis: it (1) is Th2-mediated, (2) requires loss of oral tolerance and (3) is characterized by eosinophil recruitment to the oesophagus.
Due to engagement of multiple compensatory inflammatory pathways in EoE, therapeutic trials using agents designed to block multiple components of the Th2 pathway are likely to display efficacy. Biologics neutralizing single agents like IL-5, IL-13, CRTH2 and IgE show limited efficacy in EoE [78]. Therapeutics targeting multiple pathways are necessary and finding these is an active and on-going field of research. A clinical trial with Dupilumab, targeting both IL-4 and IL-13, is underway. Recently, JAK1/3-inhibitors have shown promising effects in a murine model of asthma [79]. This finding may present a novel therapeutic approach for EoE.
Strengthening oral tolerance also may be an effective strategy in preventing and/or treating EoE. The rapid increase in the prevalence of EoE cannot be explained by changes in genetic susceptibility [80]. Risk factors that may predispose to EoE include use of antibiotics during infancy, caesarean delivery, premature birth and lack of breast-feeding. These circumstances correlated with changes in the gut microbiota and with dysregulation of Tregs and iNKT-cells. Mice studies show that manipulating the microbiota can restore oral tolerance [20,21]. Changes in microbiota are associated with various human diseases, but its role in EoE remains to be elucidated. Successful modulation of the microbiota to treat human disease is feasible as demonstrated by the efficacy of faecal transplant in the treatment of refractory Clostridium difficile [81]. Future research should explore the role of the microbiota in the pathogenesis of EoE, and whether manipulation of microbiota can restore oral tolerance, prevent and/or treat EoE.
Eosinophilic infiltration in the oesophagus is the hallmark histologic feature of EoE. Eosinophils cause tissue damage, remodelling and fibrosis. They produce various mediators that propel the Th2-pathway to perpetuate the local inflammatory cycle. The fact that eosinophils only affect the oesophagus in EoE suggests there are oesophagus-specific chemotactic signals that may offer opportunities for therapeutic intervention.
Research agenda.
Restoring oral tolerance via modulation of microbiota may prevent/treat EoE. We need a better understanding of how the microbiota contributes to the risk of EoE development and loss of oral tolerance.
Simultaneous blockade of more than a single target in the Th2 pathways may be required to abrogate the disease, due to the compensatory inflammatory processes in EoE.
Targeted disruption of eosinophil trafficking to the oesophagus may abate EoE without systemic side effects. Oesophagus-specific chemotactic signalling pathways needs to be better defined.
Acknowledgments
We thank Joel V. Weinstock, MD and Peter A. Bonis, MD for their valuable input on this work. This work is supported in part by the Consortium of Eosinophilic Gastrointestinal Disease Researchers (CEGIR). CEGIR (U54 AI117804) is part of the Rare Diseases Clinical Research Network (RDCRN), an initiative of the Office of Rare Diseases Research (ORDR), NCATS, and is funded through collaboration between NCATS, NIAID and NIDDK.
Footnotes
Conflict of Interest: None
References
- 1.Sampson HA, Aceves S, Bock SA, James J, Jones S, Lang D, et al. Food allergy: A practice parameter update—2014. J Allergy Clin Immunol. 2014 Nov;134(5):1016–25. e43. doi: 10.1016/j.jaci.2014.05.013. [DOI] [PubMed] [Google Scholar]
- 2.Kelly KJ, Lazenby AJ, Rowe PC, Yardley JH, Perman JA, Sampson HA. Eosinophilic Esophagitis Attributed to Gastroesophageal Reflux: Improvement With an Amino Acid-Based Formula. Gastroenterology. 1995;109(1):1503–12. doi: 10.1016/0016-5085(95)90637-1. [DOI] [PubMed] [Google Scholar]
- 3.Spergel JM, Andrews T, Brown-Whitehorn TF, Beausoleil JL, Liacouras CA. Treatment of eosinophilic esophagitis with specific food elimination diet directed by a combination of skin prick and patch tests. Ann Allergy Asthma Immunol. 2005 Oct;95(1):336–43. doi: 10.1016/S1081-1206(10)61151-9. [DOI] [PubMed] [Google Scholar]
- *4.Dellon ES, Gonsalves N, Hirano I, Furuta GT, Liacouras CA, Katzka DA. ACG Clinical Guideline: Evidence Based Approach to the Diagnosis and Management of Esophageal Eosinophilia and Eosinophilic Esophagitis (EoE) Am J Gastroenterol. 2013 Apr;108(1):679–92. doi: 10.1038/ajg.2013.71. [DOI] [PubMed] [Google Scholar]
- 5.Dobbins JW, Sheahan DG, Behar J. Eosinophilic Gastroenteritis with Esophageal Involvement. Gastroenterology. 1977;72(6):1312–6. [PubMed] [Google Scholar]
- 6.Landres RT, Kuster GG, Strum WB. Eosinophilic esophagitis in a patient with vigorous achalasia. Gastroenterology. 1978;74(6):1298–301. [PubMed] [Google Scholar]
- 7.Winter HS, Madara JL, Stafford RJ, Grand RJ, Quinlan J-E, Goldman H. Intraepithelial eosinophils: a new diagnostic criterion for reflux esophagitis. Gastroenterology. 1982;83:818–23. [PubMed] [Google Scholar]
- 8.Lee RG. Marked eosinophilia in esophageal mucosal biopsies. Am J Surg Pathol. 1985 Jul;9(7):475–9. doi: 10.1097/00000478-198507000-00002. [DOI] [PubMed] [Google Scholar]
- 9.Attwood SE, Smyrk TC, Demeester TR, Jones JB. Esophageal eosinophilia with dysphagia. Dig Dis Sci. 1993;38(1):109–16. doi: 10.1007/BF01296781. [DOI] [PubMed] [Google Scholar]
- 10.Markowitz J. Elemental diet is an effective treatment for eosinophilic esophagitis in children and adolescents. Am J Gastroenterol. 2003 Apr;98(4):777–82. doi: 10.1111/j.1572-0241.2003.07390.x. [DOI] [PubMed] [Google Scholar]
- 11.Gonsalves N, Yang G, Doerfler B, Ritz S, Ditto AM, Hirano I. Elimination Diet Effectively Treats Eosinophilic Esophagitis in Adults; Food Reintroduction Identifies Causative Factors. Gastroenterology. 2012 Jun;142(7):1451–9. e1. doi: 10.1053/j.gastro.2012.03.001. [DOI] [PubMed] [Google Scholar]
- 12.Vicario M, Blanchard C, Stringer KF, Collins MH, Mingler MK, Ahrens A, et al. Local B cells and IgE production in the oesophageal mucosa in eosinophilic oesophagitis. Gut. 2010 Jan 1;59(01):12–20. doi: 10.1136/gut.2009.178020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *13.Blanchard C. Eotaxin-3 and a uniquely conserved gene-expression profile in eosinophilic esophagitis. J Clin Invest. 2006 Jan 19;116(2):536–47. doi: 10.1172/JCI26679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Blanchard C, Stucke EM, Rodriguez-Jimenez B, Burwinkel K, Collins MH, Ahrens A, et al. A striking local esophageal cytokine expression profile in eosinophilic esophagitis. J Allergy Clin Immunol. 2011 Jan;127(1):208–17. e7. doi: 10.1016/j.jaci.2010.10.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Islam SA, Ling MF, Leung J, Shreffler WG, Luster AD. Identification of human CCR8 as a CCL18 receptor. J Exp Med. 2013 Sep 23;210(10):1889–98. doi: 10.1084/jem.20130240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *16.Noti M, Wojno EDT, Kim BS, Siracusa MC, Giacomin PR, Nair MG, et al. Thymic stromal lymphopoietin–elicited basophil responses promote eosinophilic esophagitis. Nat Med. 2013 Jul 21;19(8):1005–13. doi: 10.1038/nm.3281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mishra A, Schlotman J, Wang M, Rothenberg ME. Critical role for adaptive T cell immunity in experimental eosinophilic esophagitis in mice. J Leukoc Biol. 2007 Jan 29;81(4):916–24. doi: 10.1189/jlb.1106653. [DOI] [PubMed] [Google Scholar]
- 18.Burks AW, Laubach S, Jones SM. Oral tolerance, food allergy, and immunotherapy: Implications for future treatment. J Allergy Clin Immunol. 2008 Jun;121(6):1344–50. doi: 10.1016/j.jaci.2008.02.037. [DOI] [PubMed] [Google Scholar]
- 19.Jensen ET, Kappelman MD, Kim HP, Ringel-Kulka T, Dellon ES. Early life exposures as risk factors for pediatric eosinophilic esophagitis. J Pediatr Gastroenterol Nutr. 2013 Jul;57(1):67–71. doi: 10.1097/MPG.0b013e318290d15a. [DOI] [PubMed] [Google Scholar]
- 20.Oyoshi MK, Oettgen HC, Chatila TA, Geha RS, Bryce PJ. Food allergy: Insights into etiology, prevention, and treatment provided by murine models. J Allergy Clin Immunol. 2014 Feb;133(2):309–17. doi: 10.1016/j.jaci.2013.12.1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Noverr MC, Huffnagle GB. The “microflora hypothesis” of allergic diseases. Clin Exp Allergy. 2005 Dec;35(12):1511–20. doi: 10.1111/j.1365-2222.2005.02379.x. [DOI] [PubMed] [Google Scholar]
- 22.Johnston LK, Chien KB, Bryce PJ. The Immunology of Food Allergy. J Immunol. 2014 Mar 15;192(6):2529–34. doi: 10.4049/jimmunol.1303026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hadis U, Wahl B, Schulz O, Hardtke-Wolenski M, Schippers A, Wagner N, et al. Intestinal Tolerance Requires Gut Homing and Expansion of FoxP3+ Regulatory T Cells in the Lamina Propria. Immunity. 2011 Feb;34(2):237–46. doi: 10.1016/j.immuni.2011.01.016. [DOI] [PubMed] [Google Scholar]
- 24.Stuck MC, Straumann A, Simon H-U. Relative lack of T regulatory cells in adult eosinophilic esophagitis - no normalization after corticosteroid therapy. Allergy. 2011 May;66(5):705–7. doi: 10.1111/j.1398-9995.2010.02525.x. [DOI] [PubMed] [Google Scholar]
- 25.Mishra A, Hogan SP, Brandt EB, Rothenberg ME. An etiological role for aeroallergens and eosinophils in experimental esophagitis. J Clin Invest. 2001 Jan 1;107(1):83–90. doi: 10.1172/JCI10224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Akei HS, Mishra A, Blanchard C, Rothenberg ME. Epicutaneous Antigen Exposure Primes for Experimental Eosinophilic Esophagitis in Mice. Gastroenterology. 2005 Sep;129(3):985–94. doi: 10.1053/j.gastro.2005.06.027. [DOI] [PubMed] [Google Scholar]
- 27.Hogan SP, Mishra A, Brandt EB, Royalty MP, Pope SM, Zimmermann N, et al. A pathological function for eotaxin and eosinophils in eosinophilic gastrointestinal inflammation. Nat Immunol. 2001;2(4):353–60. doi: 10.1038/86365. [DOI] [PubMed] [Google Scholar]
- 28.Rubinstein E, Cho JY, Rosenthal P, Chao J, Miller M, Pham A, et al. Siglec-F Inhibition Reduces Esophageal Eosinophilia and Angiogenesis in a Mouse Model of Eosinophilic Esophagitis: J Pediatr Gastroenterol Nutr. 2011 Oct;53(4):409–16. doi: 10.1097/MPG.0b013e3182182ff8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lucendo AJ, Arias Á, Tenias JM. Relation between eosinophilic esophagitis and oral immunotherapy for food allergy: a systematic review with meta-analysis. Ann Allergy Asthma Immunol. 2014 Dec;113(6):624–9. doi: 10.1016/j.anai.2014.08.004. [DOI] [PubMed] [Google Scholar]
- 30.Van Rhijn BD, van Ree R, Versteeg SA, Vlieg-Boerstra BJ, Sprikkelman AB, Terreehorst I, et al. Birch pollen sensitization with cross-reactivity to food allergens predominates in adults with eosinophilic esophagitis. Allergy. 2013 Nov;68(11):1475–81. doi: 10.1111/all.12257. [DOI] [PubMed] [Google Scholar]
- 31.Rothenberg ME, Spergel JM, Sherrill JD, Annaiah K, Martin LJ, Cianferoni A, et al. Common variants at 5q22 associate with pediatric eosinophilic esophagitis. Nat Genet. 2010 Apr;42(4):289–91. doi: 10.1038/ng.547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *32.Kottyan LC, Davis BP, Sherrill JD, Liu K, Rochman M, Kaufman K, et al. Genome-wide association analysis of eosinophilic esophagitis provides insight into the tissue specificity of this allergic disease. Nat Genet. 2014 Aug;46(8):895–900. doi: 10.1038/ng.3033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Samuelov L, Sarig O, Harmon RM, Rapaport D, Ishida-Yamamoto A, Isakov O, et al. Desmoglein 1 deficiency results in severe dermatitis, multiple allergies and metabolic wasting. Nat Genet. 2013 Aug 25;45(10):1244–8. doi: 10.1038/ng.2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sherrill JD, Kc K, Wu D, Djukic Z, Caldwell JM, Stucke EM, et al. Desmoglein-1 regulates esophageal epithelial barrier function and immune responses in eosinophilic esophagitis. Mucosal Immunol. 2014 May;7(3):718–29. doi: 10.1038/mi.2013.90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Groschwitz KR, Hogan SP. Intestinal barrier function: Molecular regulation and disease pathogenesis. J Allergy Clin Immunol. 2009 Jul;124(1):3–20. doi: 10.1016/j.jaci.2009.05.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *36.Rothenberg ME. Molecular, Genetic, and Cellular Bases for Treating Eosinophilic Esophagitis. Gastroenterology. 2015 Feb; doi: 10.1053/j.gastro.2015.02.002. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Aceves SS, Chen D, Newbury RO, Dohil R, Bastian JF, Broide DH. Mast cells infiltrate the esophageal smooth muscle in patients with eosinophilic esophagitis, express TGF-β1, and increase esophageal smooth muscle contraction. J Allergy Clin Immunol. 2010 Dec 1;126(6):1198–204. e4. doi: 10.1016/j.jaci.2010.08.050. [DOI] [PubMed] [Google Scholar]
- 38.Lucendo AJ, Navarro M, Comas C, Pascual JM, Burgos E, Santamaría L, et al. Immunophenotypic characterization and quantification of the epithelial inflammatory infiltrate in eosinophilic esophagitis through stereology: an analysis of the cellular mechanisms of the disease and the immunologic capacity of the esophagus. Am J Surg Pathol. 2007 Apr;31(4):598–606. doi: 10.1097/01.pas.0000213392.49698.8c. [DOI] [PubMed] [Google Scholar]
- 39.Clayton F, Fang JC, Gleich GJ, Lucendo AJ, Olalla JM, Vinson LA, et al. Eosinophilic Esophagitis in Adults Is Associated With IgG4 and Not Mediated by IgE. Gastroenterology. 2014 Sep;147(3):602–9. doi: 10.1053/j.gastro.2014.05.036. [DOI] [PubMed] [Google Scholar]
- 40.Rayapudi M, Rajavelu P, Zhu X, Kaul A, Niranjan R, Dynda S, et al. Invariant natural killer T-cell neutralization is a possible novel therapy for human eosinophilic esophagitis. Clin Transl Immunol. 2014 Jan 10;3(1):e9. doi: 10.1038/cti.2013.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lexmond WS, Neves JF, Nurko S, Olszak T, Exley MA, Blumberg RS, et al. Involvement of the iNKT cell pathway is associated with early-onset eosinophilic esophagitis and response to allergen avoidance therapy. Am J Gastroenterol. 2014 May;109(5):646–57. doi: 10.1038/ajg.2014.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lu TX, Sherrill JD, Wen T, Plassard AJ, Besse JA, Abonia JP, et al. MicroRNA signature in patients with eosinophilic esophagitis, reversibility with glucocorticoids, and assessment as disease biomarkers. J Allergy Clin Immunol. 2012 Apr 1;129(4):1064–75. e9. doi: 10.1016/j.jaci.2012.01.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rayapudi M, Mavi P, Zhu X, Pandey AK, Abonia JP, Rothenberg ME, et al. Indoor insect allergens are potent inducers of experimental eosinophilic esophagitis in mice. J Leukoc Biol. 2010 Aug 1;88(2):337–46. doi: 10.1189/jlb.0110025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mishra A, Hogan SP, Brandt EB, Rothenberg ME. IL-5 Promotes Eosinophil Trafficking to the Esophagus. J Immunol. 2002 Mar 1;168(5):2464–9. doi: 10.4049/jimmunol.168.5.2464. [DOI] [PubMed] [Google Scholar]
- 45.Mishra A, Rothenberg ME. Intratracheal IL-13 induces eosinophilic esophagitis by an IL-5, eotaxin-1, and STAT6-dependent mechanism. Gastroenterology. 2003;125(5):1419–27. doi: 10.1016/j.gastro.2003.07.007. [DOI] [PubMed] [Google Scholar]
- 46.Mueller S, Neureiter D, Aigner T, Stolte M. Comparison of histological parameters for the diagnosis of eosinophilic oesophagitis versus gastro-oesophageal reflux disease on oesophageal biopsy material. Histopathology. 2008 Dec;53(6):676–84. doi: 10.1111/j.1365-2559.2008.03187.x. [DOI] [PubMed] [Google Scholar]
- 47.Blanchard C, Mingler MK, Vicario M, Abonia JP, Wu YY, Lu TX, et al. IL-13 involvement in eosinophilic esophagitis: transcriptome analysis and reversibility with glucocorticoids. J Allergy Clin Immunol. 2007 Dec;120(6):1292–300. doi: 10.1016/j.jaci.2007.10.024. [DOI] [PubMed] [Google Scholar]
- 48.Lucendo AJ, Arias Á, González-Cervera J, Yagüe-Compadre JL, Guagnozzi D, Angueira T, et al. Empiric 6-food elimination diet induced and maintained prolonged remission in patients with adult eosinophilic esophagitis: a prospective study on the food cause of the disease. J Allergy Clin Immunol. 2013;131(3):797–804. doi: 10.1016/j.jaci.2012.12.664. [DOI] [PubMed] [Google Scholar]
- 49.Jyonouchi S, Smith CL, Saretta F, Abraham V, Ruymann KR, Modayur-Chandramouleeswaran P, et al. Invariant natural killer T cells in children with eosinophilic esophagitis. Clin Exp Allergy. 2014 Jan;44(1):58–68. doi: 10.1111/cea.12201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shreffler WG, Castro RR, Kucuk ZY, Charlop-Powers Z, Grishina G, Yoo S, et al. The major glycoprotein allergen from Arachis hypogaea, Ara h 1, is a ligand of dendritic cell-specific ICAM-grabbing nonintegrin and acts as a Th2 adjuvant in vitro. J Immunol. 2006 Sep 15;177(6):3677–85. doi: 10.4049/jimmunol.177.6.3677. [DOI] [PubMed] [Google Scholar]
- 51.Masilamani M, Commins S, Shreffler W. Determinants of food allergy. Immunol Allergy Clin North Am. 2012 Feb;32(1):11–33. doi: 10.1016/j.iac.2011.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kambayashi T, Laufer TM. Atypical MHC class II-expressing antigen-presenting cells: can anything replace a dendritic cell? Nat Rev Immunol. 2014 Oct 17;14(11):719–30. doi: 10.1038/nri3754. [DOI] [PubMed] [Google Scholar]
- 53.Teitelbaum JE, Fox VL, Twarog FJ, Nurko S, Antonioli D, Gleich G, et al. Eosinophilic esophagitis in children: immunopathological analysis and response to fluticasone propionate. Gastroenterology. 2002 May;122(5):1216–25. doi: 10.1053/gast.2002.32998. [DOI] [PubMed] [Google Scholar]
- 54.Mulder DJ, Pooni A, Mak N, Hurlbut DJ, Basta S, Justinich CJ. Antigen Presentation and MHC Class II Expression by Human Esophageal Epithelial Cells. Am J Pathol. 2011 Feb;178(2):744–53. doi: 10.1016/j.ajpath.2010.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Davis BP, Rothenberg ME. Antigen Presentation by Eosinophils in Eosinophilic Esophagitis?: J Pediatr Gastroenterol Nutr. 2013 Mar;56(3):242. doi: 10.1097/MPG.0b013e31827ab8d3. [DOI] [PubMed] [Google Scholar]
- 56.Le-Carlson M, Seki S, Abarbanel D, Quiros A, Cox K, Nadeau KC. Markers of Antigen Presentation and Activation on Eosinophils and T Cells in the Esophageal Tissue of Patients With Eosinophilic Esophagitis. J Pediatr Gastroenterol Nutr. 2013 Mar;56(3):257–62. doi: 10.1097/MPG.0b013e3182758d49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Blanchard C, Durual S, Estienne M, Emami S, Vasseur S, Cuber J-C. Eotaxin-3/CCL26 gene expression in intestinal epithelial cells is up-regulated by interleukin-4 and interleukin-13 via the signal transducer and activator of transcription 6. Int J Biochem Cell Biol. 2005 Dec;37(12):2559–73. doi: 10.1016/j.biocel.2005.06.010. [DOI] [PubMed] [Google Scholar]
- 58.Blanchard C, Mingler MK, McBride M, Putnam PE, Collins MH, Chang G, et al. Periostin facilitates eosinophil tissue infiltration in allergic lung and esophageal responses. Mucosal Immunol. 2008 Jul;1(4):289–96. doi: 10.1038/mi.2008.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Masuoka M, Shiraishi H, Ohta S, Suzuki S, Arima K, Aoki S, et al. Periostin promotes chronic allergic inflammation in response to Th2 cytokines. J Clin Invest. 2012 Jul 2;122(7):2590–600. doi: 10.1172/JCI58978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Blanchard C, Stucke EM, Burwinkel K, Caldwell JM, Collins MH, Ahrens A, et al. Coordinate Interaction between IL-13 and Epithelial Differentiation Cluster Genes in Eosinophilic Esophagitis. J Immunol. 2010 Apr 1;184(7):4033–41. doi: 10.4049/jimmunol.0903069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Siracusa MC, Saenz SA, Hill DA, Kim BS, Headley MB, Doering TA, et al. TSLP promotes interleukin-3-independent basophil haematopoiesis and type 2 inflammation. Nature. 2011 Aug 14;477(7363):229–33. doi: 10.1038/nature10329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ito T, Wang Y-H, Duramad O, Hori T, Delespesse GJ, Watanabe N, et al. TSLP-activated dendritic cells induce an inflammatory T helper type 2 cell response through OX40 ligand. J Exp Med. 2005 Oct 31;202(9):1213–23. doi: 10.1084/jem.20051135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hogan SP, Rosenberg HF, Moqbel R, Phipps S, Foster PS, Lacy P, et al. Eosinophils: Biological Properties and Role in Health and Disease. Clin Exp Allergy. 2008 May;38(5):709–50. doi: 10.1111/j.1365-2222.2008.02958.x. [DOI] [PubMed] [Google Scholar]
- 64.Phillips C, Coward WR, Pritchard DI, Hewitt CRA. Basophils express a type 2 cytokine profile on exposure to proteases from helminths and house dust mites. J Leukoc Biol. 2003 Jan;73(1):165–71. doi: 10.1189/jlb.0702356. [DOI] [PubMed] [Google Scholar]
- 65.Sakuishi K, Oki S, Araki M, Porcelli SA, Miyake S, Yamamura T. Invariant NKT cells biased for IL-5 production act as crucial regulators of inflammation. J Immunol. 2007 Sep 15;179(6):3452–62. doi: 10.4049/jimmunol.179.6.3452. [DOI] [PubMed] [Google Scholar]
- 66.Mirchandani AS, Salmond RJ. Innate Lymphoid Cells in Type 2 Immune Responses. Arch Immunol Ther Exp (Warsz) 2014 Dec 20; doi: 10.1007/s00005-014-0327-5. in press. [DOI] [PubMed] [Google Scholar]
- 67.Gupta SK, Fitzgerald JF, Kondratyuk T, HogenEsch H. Cytokine expression in normal and inflamed esophageal mucosa: a study into the pathogenesis of allergic eosinophilic esophagitis. J Pediatr Gastroenterol Nutr. 2006;42(1):22–6. doi: 10.1097/01.mpg.0000188740.38757.d2. [DOI] [PubMed] [Google Scholar]
- 68.Ochiai K, Kagami M, Matsumura R, Tomioka H. IL-5 but not interferon-gamma (IFN-gamma) inhibits eosinophil apoptosis by up-regulation of bcl-2 expression. Clin Exp Immunol. 1997 Jan;107(1):198–204. doi: 10.1046/j.1365-2249.1997.d01-884.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Menzies-Gow A, Ying S, Phipps S, Kay AB. Interactions between eotaxin, histamine and mast cells in early microvascular events associated with eosinophil recruitment to the site of allergic skin reactions in humans. Clin Exp Allergy J Br Soc Allergy Clin Immunol. 2004 Aug;34(8):1276–82. doi: 10.1111/j.1365-2222.2004.02014.x. [DOI] [PubMed] [Google Scholar]
- 70.Gauvreau GM, Ellis AK, Denburg JA. Haemopoietic processes in allergic disease: eosinophil/basophil development. Clin Exp Allergy J Br Soc Allergy Clin Immunol. 2009 Sep;39(9):1297–306. doi: 10.1111/j.1365-2222.2009.03325.x. [DOI] [PubMed] [Google Scholar]
- 71.Zhu X, Wang M, Mavi P, Rayapudi M, Pandey AK, Kaul A, et al. Interleukin-15 Expression Is Increased in Human Eosinophilic Esophagitis and Mediates Pathogenesis in Mice. Gastroenterology. 2010 Jul;139(1):182–93. e7. doi: 10.1053/j.gastro.2010.03.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Cheng E, Zhang X, Huo X, Yu C, Zhang Q, Wang DH, et al. Omeprazole blocks eotaxin-3 expression by oesophageal squamous cells from patients with eosinophilic oesophagitis and GORD. Gut. 2013 Jun;62(6):824–32. doi: 10.1136/gutjnl-2012-302250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zhang S, Wu X, Yu S. Prostaglandin D2 receptor-type prostanoid receptor 2 mediates eosinophil trafficking into the esophagus: PGD2 in esophageal eosinophil trafficking. Dis Esophagus. 2014 Aug;27(6):601–6. doi: 10.1111/dote.12118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Aceves SS, Newbury RO, Dohil R, Bastian JF, Broide DH. Esophageal remodeling in pediatric eosinophilic esophagitis. J Allergy Clin Immunol. 2007 Jan;119(1):206–12. doi: 10.1016/j.jaci.2006.10.016. [DOI] [PubMed] [Google Scholar]
- 75.Yang D, Chen Q, Su SB, Zhang P, Kurosaka K, Caspi RR, et al. Eosinophil-derived neurotoxin acts as an alarmin to activate the TLR2-MyD88 signal pathway in dendritic cells and enhances Th2 immune responses. J Exp Med. 2008 Jan 21;205(1):79–90. doi: 10.1084/jem.20062027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Arias Á, Lucendo AJ, Martínez-Fernández P, González-Castro AM, Fortea M, González-Cervera J, et al. Dietary Treatment Modulates Mast Cell Phenotype, Density, and Activity in Adult Eosinophilic Esophagitis. Clin Exp Allergy. 2015 Feb 1; doi: 10.1111/cea.12504. in press. [DOI] [PubMed] [Google Scholar]
- 77.Beppu LY, Anilkumar AA, Newbury RO, Dohil R, Broide DH, Aceves SS. TGF-β1-induced phospholamban expression alters esophageal smooth muscle cell contraction in patients with eosinophilic esophagitis. J Allergy Clin Immunol. 2014 Nov;134(5):1100–7. e4. doi: 10.1016/j.jaci.2014.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Sawas T, Dhalla S, Sayyar M, Pasricha PJ, Hernaez R. Systematic review with meta-analysis: pharmacological interventions for eosinophilic oesophagitis. Aliment Pharmacol Ther. 2015 Feb;:1–10. doi: 10.1111/apt.13147. [DOI] [PubMed] [Google Scholar]
- 79.Ashino S, Takeda K, Li H, Taylor V, Joetham A, Pine PR, et al. Janus kinase 1/3 signaling pathways are key initiators of TH2 differentiation and lung allergic responses. J Allergy Clin Immunol. 2014 Apr;133(4):1162–74. e4. doi: 10.1016/j.jaci.2013.10.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *80.Bonis PAL. Putting the puzzle together: epidemiological and clinical clues in the etiology of eosinophilic esophagitis. Immunol Allergy Clin North Am. 2009 Feb;29(1):41–52. viii. doi: 10.1016/j.iac.2008.09.005. [DOI] [PubMed] [Google Scholar]
- 81.Weingarden A, González A, Vázquez-Baeza Y, Weiss S, Humphry G, Berg-Lyons D, et al. Dynamic changes in short- and long-term bacterial composition following fecal microbiota transplantation for recurrent Clostridium difficile infection. Microbiome. 2015 Dec; doi: 10.1186/s40168-015-0070-0. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Mishra A, Wang M, Pemmaraju VR, Collins MH, Fulkerson PC, Abonia JP, et al. Esophageal Remodeling Develops as a Consequence of Tissue Specific IL-5-Induced Eosinophilia. Gastroenterology. 2008 Jan;134(1):204–14. doi: 10.1053/j.gastro.2007.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Rajavelu P, Rayapudi M, Moffitt M, Mishra A, Mishra A. Significance of para-esophageal lymph nodes in food or aeroallergen-induced iNKT cell-mediated experimental eosinophilic esophagitis. Am J Physiol - Gastrointest Liver Physiol. 2012 Apr 1;302(7):G645–54. doi: 10.1152/ajpgi.00223.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Niranjan R, Mavi P, Rayapudi M, Dynda S, Mishra A. Pathogenic role of mast cells in experimental eosinophilic esophagitis. AJP Gastrointest Liver Physiol. 2013 Jun 15;304(12):G1087–94. doi: 10.1152/ajpgi.00070.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]