Abstract
Incorporation of strain-specific synthetic DNA tags into yeast Saccharomyces cerevisiae gene-deletion strains has enabled identification of gene functions by massively parallel growth rate analysis. However, it is important to confirm the sequences of these tags, because mutations introduced during construction could lead to significant errors in hybridization performance. To validate this experimental system, we sequenced 11,812 synthetic 20-mer molecular bar codes and adjacent sequences (>1.8 megabases synthetic DNA) by pyrosequencing and Sanger methods. At least 31% of the genome-integrated 20-mer tags contain differences from those originally synthesized. However, these mutations result in anomalous hybridization in only a small subset of strains, and the sequence information enables redesign of hybridization probes for arrays. The robust performance of the yeast gene-deletion dual oligonucleotide bar-code design in array hybridization validates the use of molecular bar codes in living cells for tracking their growth phenotype.
Disruption of gene function by deletion allows elucidation of both qualitative and quantitative functions of genes. The yeast Saccharomyces cerevisiae is a particularly powerful experimental system, because multiple deletion strains can easily be pooled for parallel growth assays. Individual deletion strains have recently been created for 5,918 ORFs, representing nearly all of the estimated 6,000 genetic loci (1). Tagging of each deletion strain with one or two unique 20-nt sequences allows identification of genes affected by specific growth conditions without prior knowledge of gene functions (2). Hybridization of bar-code DNA to oligonucleotide arrays can be used to measure the growth rate of each strain over a period of several cell division generations, which represents an index of strain fitness.
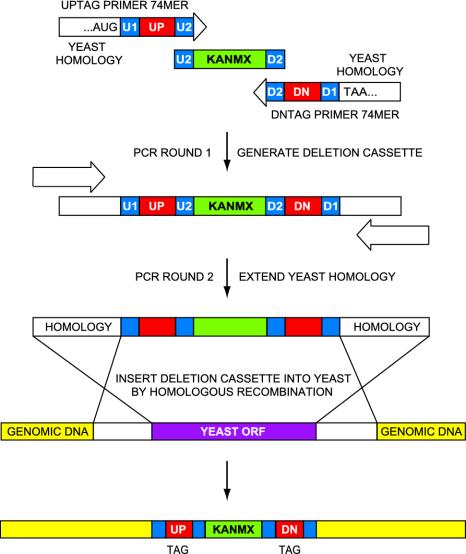
For each strain, the ORF was replaced and tagged by mitotic recombination with the selectable resistance gene KanMX cassette, which is linked to one or two unique 20-mer sequence tags (UPTAG and DNTAG) that are flanked on both sides by common primer sequences (U1 and U2 for UPTAG, D1 and D2 for DNTAG) that vary in length by design from 17 to 19 nt (2) (Fig. 1). A tag and its associated primers are collectively referred to as a “bar code.” Although a large majority of strains have two bar codes, some ORFs were replaced with only a single bar code in an earlier feasibility study. The 11,812 different 20-mer oligonucleotide tags were computationally selected for uniqueness of each sequence (2) (see Table 1, which is published as supporting information on the PNAS web site).
Fig. 1.
Construction of the KanMX deletion cassette and the gene-deletion molecular barcoding strategy. A PCR-based deletion strategy was used to systematically replace each ORF with a cassette containing a kanamycin-(geneticin) resistance marker along with a unique pair of oligonucleotide (20-mer) molecular tags (2). In the first round of PCR, upstream and downstream 74-mer primers were used to amplify the KanMX gene (from pF-kanMX4) and to incorporate the two bar-code sequences into the deletion cassette. These primers begin at the 5′-end with 18 (or 17) bp of genomic sequence, followed by a bar code comprised of 18 bp of sequence (U1 or D1) common to all deletion constructs, a unique 20-mer tag, and 18 (or 19) bp of sequence (U2 or D2) homologous to the KanMX gene. In the second round of PCR, the ORF-specific homology of the deletion construct was extended to 45 bp by using two upstream ORF-specific primers (UP45 or DN45). This was necessary to increase the ORF-targeting specificity during mitotic recombination, promoting efficient insertion of the cassette into the desired strain. Finally, the deletion cassette was integrated into the yeast chromosome by homologous recombination. Proper incorporation of each cassette was verified by PCR by using primers selected from the gene-specific region, 200–400 bp upstream from the ORF start codon, along with a common primer from the KanMX region. As shown, the red (tag) and blue (flanking) segments of synthetic DNA are critical to bar-code amplification and discrimination by hybridization probes.
Although ORF replacement was previously verified for each strain by several PCR amplifications (1), the identity of the 20-mer tag identifiers (see also the Yeast Deletion Project web site, http://sequence-www.stanford.edu/group/yeast_deletion_project), 17- to 19-mer common primer sequences and the sequence of both recombination junctions in the yeast genome of each gene-deletion strain were not previously confirmed by direct sequencing. Moreover, although the tags were computationally designed to have similar hybridization properties, it has been observed that there are some tags that hybridize with greater intensities than others, and some tags that do not hybridize at all. This is likely due to a combination of factors, including specific and unpredictable hybridization properties of a tag sequence, or mutation of the tags and/or primers during construction of the deletion. Each of the three steps in the production of the yeast gene deletion (i.e., chemical synthesis of oligonucleotides, PCR amplification, and introduction of the KanMX deletion cassette into the yeast genome by mitotic recombination at specific genome locus) is prone to stochastic mutations with variable error frequencies that may alter the designed sequence of the 20-mer tag identifier sequences, 17- to 19-mer common primer sequences, and/or genome integration coordinates. In anticipation of such variation, two different bar codes were incorporated into most strains so that quantitative growth phenotype could be obtained with either or both bar codes. To assay the prevalence of such errors, we directly sequenced all incorporated bar codes. The effects of errors on hybridization performance could then be studied by using hybridization data from a previous study (3). The results of this work not only validate the identities of the yeast gene-deletion tags but also reveal the robustness of the yeast gene-deletion dual oligonucleotide bar-code design in array hybridization performance. Most of the yeast gene-deletion strains are unaffected in hybridization performance for the identified corresponding changes (or “defects”) in bar-code sequence, because as many as two or more nucleotide defects per bar code are required to reduce array hybridization performance.
Methods
DNA Samples. Yeast gene-deletion strains were obtained in 96-well plates from Research Genetics (Huntsville, AL). Cultures were grown in 200 μl of yeast extract/peptone/dextrose media in covered plates for 3 days under ambient conditions (yield ≈108 cells). Genomic DNA from yeast was extracted by washing cells with 100 μl of H2O and then resuspending in 50 μl of H2O. Washed and resuspended cells (1 μl, ≈2 × 106 cells) were used as templates for PCR.
Oligonucleotides. Synthetic DNA oligonucleotides were obtained from Qiagen (Alameda, CA) or MWG Biotech (High Point, NC). A complete set of 11,812 yeast ORF-specific PCR primers was obtained from Illumina (San Diego) or from the oligonucleotide synthesis facility at the Stanford Genome Technology Center. Yeast deletion primer sequences for the complete set of strains are listed at http://www-sequence.stanford.edu/group/yeast_deletion_project/Deletion_primers_PCR_sizes.txt. Outer PCR (common)primersare:UPTAG,TAG1_FPCR2(5′-TCATGCCCCTGAGCTGCGCACGT-3′); DNTAG, TAG2_FPCR2 (5′-TCGCCTCGACATCATCTGCCCAGA-3′). Inner PCR primers: UPTAG, TAG1_FPCR3 (5′-Biotin-GAGCTGCGCACGTCAAGACTGTC-3′) and TAG1_RPCR1 (5′-GATGTCCACGAGGTCTCT-3′); DNTAG, TAG2_FPCR3 (5′-Biotin-GACATCATCTGCCCAGATGCGAAG-3′) and TAG2_RPCR1 (5′-ACGGTGTCGGTCTCGTAG-3′). Sequencing primers are: UPTAG, TAG1_RSEQ1 (5′-GATGTCCACGAGGTCT-3′); DNTAG, TAG2_RSEQ1 (5′-ACGGTGTCGGTCTCGT-3′).
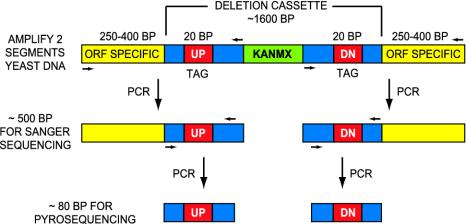
DNA Amplification. Amplification of DNA for bar-code pyrosequencing was a two-stage process (Fig. 2). Initial amplification of each of the two bar-code-containing segments (UPTAG and DNTAG) was conducted separately by using one common primer from the gene-deletion cassette ≈150 nt downstream of the tag (UPTAG TAG1_FPCR2, or DNTAG TAG2_FPCR2, 20 pmol per 15-μl reaction), and one strain-specific primer from a region ≈300 nt upstream of the tag (yeast ORF-specific UPTAG primer A or DNTAG primer D, 20 pmol). Each 15-μl PCR contained 3.3 mM MgCl2, 0.27 mM dNTPs, and 1.5 units Taq polymerase. PCR was run for 35 cycles with extension at 55°C. The actual sizes of the amplicons varied between 400 and 500 nt, depending on the strain. These products (5 μl of a 1,000-fold dilution, 2–5 ng of DNA) were used as templates for nested PCR along with two common primers, one 40–45 nt downstream from the tag (UPTAG TAG1_FPCR1, or DNTAG TAG2_FPCR1, 5 pmol), and the other located in an 18-nt common region immediately upstream of the tag (UPTAG TAG1_RPCR1, or DNTAG TAG2_RPCR1, 5 pmol). One inner PCR primer (FPCR1) was 5′-biotinylated to enable subsequent strand separation. Each 50-μl PCR contained 2.5 mM MgCl2, 0.2 mM dNTPs, 1.5 units Taq polymerase, and 0.25 μl of tetramethylenesulfone. The inner PCR was run with a touchdown program, 65–57°C over 14 cycles, then 26 cycles at 57°C. The resulting products were either 78 or 82 nt in length. Approximately 1 pmol of DNA was obtained per reaction. After removal of 10 μl for gel analysis, the remaining 40-μl product volume was used for pyrosequencing.
Fig. 2.
Schematic diagram of the yeast gene-deletion cassette sequencing strategy. Amplification of molecular bar-code-containing DNA for sequencing required two rounds of PCR for two different sequencing methods. Yeast deletion strains were grown from frozen cell stocks in 96-well microtiter plates. From cellular genomic DNA, an initial round of PCR using one ORF-specific primer (in a region 400–500 bases upstream of the deleted ORF) and one common primer (from the cassette region) were used to generate two amplicons suitable for Sanger (dideoxy terminator) sequencing, each containing one of the two 20-base tags (UPTAG or DNTAG) as well as DNA from the adjacent homologous recombination junction. A second inner round of PCR using two common primers, one from the 18-base region immediately upstream of the tag and the other from a region in the deletion cassette (≈60 bp downstream from the tag) was used to generate two short (78- to 82-bp) bar-code-containing segments for pyrosequencing. One of the inner PCR primers was 5′ biotinylated to facilitate DNA cleanup and strand separation before pyrosequencing. Sequencing primers were selected to read on opposite strands, with the Sanger primer reading upstream, starting within the deletion cassette and reading the bar code and into the recombination junction (gene-specific region). The pyrosequencing primer is complementary to the 18-bp common region immediate upstream of the tag, enabling reading of the 20-mer tag and several bases from the common flanking region downstream (cassette region).
Pyrosequencing Sample Preparation. The biotinylated PCR products (1 pmol) were each immobilized onto 15 μl of streptavidin-coated superparamagnetic beads (Dynabeads M-280 streptavidin, Dynal, Oslo). The beads were then passed although a series of wash plates by using a 96-pin magnetic tool (Dynal). The first wash contained 10 mM Tris, 2 M NaCl, 1 mM EDTA, and 0.1% Tween 20, pH 7.6. The second wash contained water. Next, single-stranded DNA template was obtained by treatment of the immobilized PCR duplex in a plate containing 0.10 M NaOH for 5 min. The beads containing immobilized single-stranded DNA were moved to a final wash plate containing 20 mM Tris and 2 mM MgCl2, pH 7.6, then were transferred to a plate containing sequencing primer (UPTAG TAG1_RSEQ1 or DNTAG TAG2_RSEQ1, 5 pmol) in 45 μl of buffer (20 mM Tris and 2 mM MgCl2, pH 7.6). Each sequencing primer was identical to the corresponding PCR primer immediately upstream of the tag except for lacking the last two nucleotides before the start of the tag. The primer was annealed to template at 70°C for 3 min, 50°C for 5 min, and 25°C for at least 5 min. Samples were used immediately for pyrosequencing.
Pyrosequencing. Primed DNA templates were placed in a pyrosequencing 96-well microtiter plate, and pyrosequencing substrate and enzyme mixtures were dispensed by using the fully automated plate-based PSQ96 pyrosequencing instrument (Pyrosequencing, Uppsala, Sweden). The progress of sequencing was followed in real time by using Pyrosequencing SQA software. The first two nucleotides sequenced were common to either all of the UPTAG or the DNTAG samples and thus provided an internal standard for single-nucleotide incorporation by polymerase. Nucleotide triphosphates were delivered sequentially at 1-min intervals. Run time was 52 min (13 cycles of the four dNTPs). Standard nucleotide and reagent concentrations are described (10).
The identity and quantity of nucleotide extension were determined by automated measurement of the amount of light generated after addition of a dNTP. The actual length of each tag was determined by sequencing several common nucleotides beyond the expected 20-mer tag. Raw data were interpreted either with Pyrosequencing evaluation software or manually.
Sanger Dideoxy Sequencing. The universal primer approach for tag amplicons was used in Sanger DNA sequencing on an ABI 3700 DNA Analyzer (Applied Biosystems) by using the BigDye terminator chemistry (Ver. 3.0) according to the manufacturer's manual. The FPCR3_TAG1 (UPTAG) and FPCR3_TAG2 (DNTAG) general sequencing primers were used in cycle sequencing in separate reactions (Fig. 2).
Hybridization Signal Data Analysis. Hybridization intensities were analyzed from 14 replicate control chip hybridizations derived from a pool of heterozygous yeast deletion strains, as described (3). The data set consists of hybridizations that were performed with an aliquot of the heterozygous yeast gene-deletion pool after removal from –80°C and thawing. The scanned intensities were scaled to yield the same overall intensity for all chips. The same control chip data set was used to analyze the log base 10 ratio of UPTAG/DNTAG. For each position on the chip, the average intensity was calculated from 36 control chips, discarding the maximum and minimum intensities at each position. Each deletion strain has four associated bar codes on a chip: UPTAG sense, UPTAG antisense, DNTAG sense, and DNTAG antisense. The same bar codes on different strands are different in hybridization performance. The better hybridizations from UPTAG and DNTAG were used for a ratio analysis.
Results
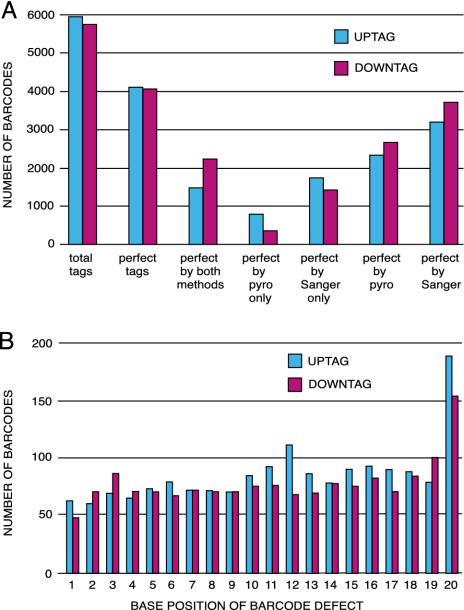
Identification of Sequence Mutations in the Yeast Gene-Deletion Bar Codes. The fidelity of yeast gene-deletion strain construction was characterized by sequencing the mitotic recombination junctions by using the standard Sanger dideoxy chain termination sequencing method (Fig. 2). Pyrosequencing (8) was also used for identification of mutations in the 20-mer tags for all 5,918 different heterozygous yeast gene-deletion strains (Fig. 2). Tag analysis by the two different sequencing methods is compared in Fig. 3A. Readable traces produced by either of the two sequencing methods were in agreement by producing identical sequence results. Examination of sequences reveals that 31% of bar-code sequences feature differences from the original design (or “defects”) in the 20-mer tag sequences. Deletions account for 14%, substitutions 16%, and insertions 1%, 31%, 18%, 28%, and 17% of the associated common primer sequences U1, U2, D1, and D2, respectively, contain defects. (Fig. 1 and Table 2, which is published as supporting information on the PNAS web site). The difference in the frequency of sequence variation between U1 and U2 and between D1 and D2 is consistent with a decrease in the fidelity of chemical synthesis of DNA with distance from the first position at the 3′-end of each oligonucleotide (Fig. 3B). U2 and D2 common sequences are synthesized at the 3′-end of the 74-mer oligonucleotide, whereas U1 and D1 common sequences are synthesized closer to the 5′ end (Fig. 1). However, <10% of the nucleotide sequence defects in the common primer sequences would be expected to affect PCR amplification of the bar codes, because the occurrence of substitutions in the first three nucleotides adjacent to the tag is 3%, 9%, 2%, and 3% for U1, U2, D1, and D2, respectively (Table 2). To our knowledge, this is the largest collection of synthetic DNA sequenced (>1.8 megabases) and the most comprehensive analysis of sequencing results generated by pyrosequencing and Sanger sequencing methods.
Fig. 3.
Tag sequence defects. (A) Comparison of tag analysis by pyrosequencing and Sanger sequencing. Blue bars represent UPTAGs, and purple bars represent DNTAGs. Nondefective 20-mer sequences were obtained by pyrosequencing or Sanger sequencing for >8,000 (≈69%) of the 11,812 unique tags. Sanger sequencing was performed to 2-fold coverage relative to pyrosequencing. Both methods were required to complete unambiguous identification of all tags. (B) Occurrence of sequence variation by position within tags. Summary of nucleotide sequence defects in 10,889 Sanger-sequenced UPTAGs (blue) and DNTAGs (purple) by position within the tag. The occurrence of nucleotide deletion at a given position in the molecular tag is similar at most positions (0.6–0.8%), except at position 20, which is more variable (1.5–2.0%).
The actual sequences for the entire UPTAG and DNTAG primers, including the tag, found by Sanger sequencing, as well as the putative sequences for each ORF in the collection, can be found in http://www-deletion.stanford.edu/deletion_sequences/deletion_sequences.html. These files list all of the information pertaining to the Sanger sequencing, including annotation of the base errors, with their quality scores. This site also contains a link to a blast site where an experimenter can check a putative sequence against the primers sequenced by Sanger sequencing.
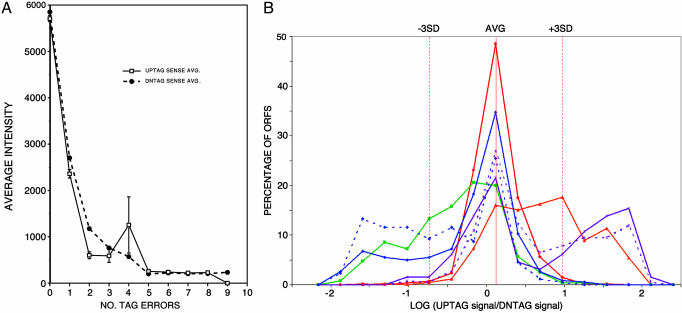
Variation in Hybridization Performance of Mutated Bar Codes. With sequence information for individual bar codes, the relationship between defects and hybridization performance can be characterized by using hybridization signals from a previous quantitative phenotypic study (3). In this previous study, all strains were pooled and grown under standard conditions, followed by PCR amplification of tag sequences and hybridization to arrays. The oligonucleotide arrays were fabricated with the original computationally designed sequences, rather than the actual sequences determined in the present work. Average absolute hybridization intensity is plotted vs. number of tag defects in Fig. 4A, and it is clear that defects lead in general to a reduction in hybridization. On average, the array hybridization intensities are above background (>1,000 average intensity units) only for perfect tags or for those tags that have one tag error. Nevertheless, there are many individual cases where a tag with two or more tag errors can yield hybridization signals above background. However, hybridization intensity is a complex function of several factors in this experiment, including the starting amount of each strain and the effect of gene deletion on growth rate. For strains with two tags, the ratio of signals for the two tags is a better measure of anomalous hybridization, because in principle, the DNTAG and UPTAG that correspond to a single ORF are represented in the pool of all tags with the same number of copies, and the ratio of the signals should be constant. Defects in one of the two tags, however, would be expected to alter this ratio. The log base 10 of UPTAG signal/DNTAG signal (LR) of each ORF was calculated to determine the effect of bar-code sequence errors on hybridization performance (Fig. 4B).
Fig. 4.
Tag hybridization performance. (A) Defects in bar-code sequences decrease absolute hybridization intensity. For each of 5,918 different tags, the hybridization signal from a previous quantitative phenotypic study (16) was plotted vs. the number of sequence defects. The solid line with square points represents UPTAG sense average intensity, and the dotted line with circle points represents DNTAG sense average intensity. The error bars are shown at each point in the graph except for points whose error bar is too small or that have a limited size of population. The total number of tags, UPTAG;DNTAG, containing no error, is: 3,326;3,734, 808;838, 122;153, 41;38, 20;15, 7;5, 4;8, 1;9, 1;2, and 0;3 tags contain 1, 2, 3, 4, 5, 6, 7, 8, or 9 errors in a UPTAG;DNTAG, respectively. (B) Ratio effect of sequence defects on UPTAG/DNTAG ratio. The relative hybridization performance for each bar code was analyzed as described in Methods for hybridization signal data analysis. The histograms were constructed with bins of width = 1 SD and were plotted by using the center of each bin to represent the bin. The red solid line with circle points ( ) represents the LR distribution of reference set of ORF population with no defect in deletion strains. The orange in DNTAGs only. The purple solid line with star points (
) represents the LR distribution of reference set of ORF population with no defect in deletion strains. The orange in DNTAGs only. The purple solid line with star points ( ) solid line with triangle points (
) solid line with triangle points ( ) represents the distribution represents the distribution of the population with defects in D1/D2s only, whereas the purple dotted line with star points (
) represents the distribution represents the distribution of the population with defects in D1/D2s only, whereas the purple dotted line with star points ( ) represents the distribution of the population with defects of the population with defects in D1(3)/D2(3)s only. The green solid line with square points (
) represents the distribution of the population with defects of the population with defects in D1(3)/D2(3)s only. The green solid line with square points ( ) represents the distribution of the population with defects in UPTAGs only. The blue solid line with diamond points (
) represents the distribution of the population with defects in UPTAGs only. The blue solid line with diamond points ( ) represents the distribution of the population with defects in U1/U2s only, and the blue dotted line with diamond points (
) represents the distribution of the population with defects in U1/U2s only, and the blue dotted line with diamond points ( ) represents the distribution of the population with defects in U1(3)/U2(3)s only. The graphs show that the defect(s) in the bar code decreases the hybridization intensity of the tag compared to the other error-free tag of the same ORF, making the log ratio abnormally high or low.
) represents the distribution of the population with defects in U1(3)/U2(3)s only. The graphs show that the defect(s) in the bar code decreases the hybridization intensity of the tag compared to the other error-free tag of the same ORF, making the log ratio abnormally high or low.
Average LR (0.1186) and the SD (= 0.2852) were calculated from a reference population of 818 ORFs with totally error-free (or “perfect”) bar codes. A histogram of this population is shown in red in Fig. 4B (where percent of total population is plotted vs. binned LR), and the distribution is relatively normal. The distribution of 425 ORFs with defects in the DNTAG only (average LR = 0.7246) is skewed to the right of the reference data, presumably because defects in the DNTAG decrease hybridization of the tag, making the log ratios abnormally high. On the other hand, the distribution of 526 ORFs with defects in the UPTAG only (average LR = –0.4102) is skewed to the left of the reference data, because defects in UPTAGs decrease their hybridization signals, making the log ratios abnormally low.
Defects in primers (U1, U2, D1, and D2) outside tags can contribute to the decrease in hybridization intensities. The distribution of 368 ORFs with defects in D1/D2 only (average LR = 0.6544) is skewed to the right of the reference data, presumably because defects in the D1/D2 decrease PCR performance of the bar codes, making the log ratios abnormally high. On the other hand, the distribution of 564 ORFs with defects in the U1/U2 only (average LR =–0.2673) is skewed to the left of the reference data, because defects in the U1/U2 decrease PCR performance of the bar codes, making the log ratios abnormally low. The effect of the defects in primers is more obvious for defects that are located near the tags. This is presumably due to a decreased priming efficiency by common primer with mismatched 3′ end, which results in lower PCR yield of the corresponding bar codes. The distribution of 65 ORFs with defects in three-base adjacent to the tag of D1/D2 [D1(3), D2(3)] only (average LR = 0.7245) is skewed further to the right of the reference data, whereas the distribution of 173 ORFs with defects in three-base adjacent to the tag of U1/U2 [U1(3), U2(3)] only (average LR =–0.5900) is skewed further to the left of the reference data than the distributions of ORFs with defects in D1/D2 and U1/U2.
In summary, 98% of the reference population has an LR value within ± 3 SD of the average, and this range was therefore chosen to define “normal” hybridization. ORFs (n = 760) with LR values below this range are considered to have anomalous UPTAG hybridization signals and ORFs (n = 845) with LR values greater than this range are considered to have anomalous DNTAG hybridization. The list of all such tags is found in Table 3, which is published as supporting information on the PNAS web site. The percentage of ORF population with defects that have LR outside the normal range can be found in Table 4, which is published as supporting information on the PNAS web site.
Some of the sequence defects can cause hybridization of the tags below background (<1,000 average intensity units). In Table 5, which is published as supporting information on the PNAS web site, there are 182 ORFs that would be excluded from analysis because the hybridization intensities of both tags are below background (or one tag is below background if the deletion strain has only one bar code). One hundred forty-four (79%) of these ORFs have defects in both bar codes, or could not be confirmed to have zero defects.
Discussion
The overall occurrence of defects in bar codes was unexpectedly high (Fig. 3B). The observed frequency of nucleotide substitution at a given position in the 20-mer tags is approximately the same as the frequency for nucleotide deletion (≈1% at each position). These sequence errors are well above the predicted error frequencies from raw Sanger sequencing data (5, 6). Nucleotide insertion, however, is found to occur with a frequency that is an order of magnitude lower than that for either deletion or substitution. Tag sequence defects are confirmed by two independent sequencing methods (Sanger and pyrosequencing) and thus cannot be due to software errors in base calling (7). Although a single-nucleotide deletion might be expected to occur randomly during the solid-phase oligonucleotide synthesis of the tag-containing 74-mer primer (from a periodic failure to deblock the 5′-terminal nucleotide for one coupling cycle), substitution is less likely to arise during chemical synthesis, either from a failure in reagent delivery or from a sequence programming mistake, because no keyed input of the oligonucleotide sequence is required. Some intrinsic contamination of the reagent deoxynucleotide phosphoramite monomers used in automated synthesis (e.g., 0.05% each of the other three amidites is typical) would give rise to a substitution rate of <0.2% per bar-code base position, or ≈4% occurrence within a 20-mer tag. The high substitution rate (16%) cannot be easily explained by nucleotide incorporation errors during PCR amplification of the bar codes before sequencing, because Taq polymerase typically misincorporates only one nucleotide in 103–104 for native DNA. However, chemical damage to synthetic DNA might increase the mutation. Nucleotide substitutions in the bar-code sequence could also arise during integration of the deletion cassette into the yeast deletion strain by homologous recombination, although the mechanism is unclear.
Although the standard method for DNA mutation scanning is still automated Sanger sequencing, the expense and labor involved make it difficult to scan a large population of short (e.g., 20-mer tags) DNA samples. To accelerate the process and reduce the cost of finding DNA sequence variations, the pyrosequencing method has been developed. Pyrosequencing is a real-time sequencing-by-synthesis method in which base extension on a primed DNA template is monitored via chemiluminescent detection of the inorganic pyrophosphate released after incorporation of a dNTP by DNA polymerase (8). Compared to the Sanger method, the pyrosequencing technique has an inherently shorter read length of up to 50 bases. Pyrosequencing has found wide application in detection of single-nucleotide polymorphisms (9, 10) and recently for examination of 10–20 base sequence variations in human papillomavirus strains (11). Although limited in read length, pyrosequencing has been used to overcome certain difficulties encountered in Sanger sequencing, such as poor electrophoretic resolution of dye-terminated DNA fragments, which arise from DNA sequence-dependent secondary structures (9). In this work, pyrosequencing provided complementary DNA sequence information as well as independent confirmation of the bar-code Sanger sequencing results.
Defects in bar codes generally result in a decreased hybridization signal. Defects in the sequence of tags presumably decrease hybridization performance of the tags, whereas defects in the sequence of the common primers, especially three bases adjacent to the tags on both 5′ and 3′ ends of the tags, presumably decrease PCR amplification of the bar codes. Some of these defects can cause the hybridization of the tags to be below background intensity. ORFs that contain defects in both bar codes might be excluded from data analysis, although the deletion strains were included in the pool. Further study is needed to evaluate possible defects in growth rate calculation.
The yeast S. cerevisiae provides a useful model system, because it is one of the most genetically tractable eukaryotic organisms. Whole-genome functional analysis studies using the complete collection of yeast gene-deletion strains not only can address assignment of function to genes but can also elucidate biochemical metabolic pathways and reveal quantitative drug and natural product interactions with gene products. By sequencing all synthetic tags from the nearly complete collection of heterozygous diploid yeast gene-deletion strains, verification for each bar code in phenotypic analysis has now been provided. Identification of all specific defects in bar codes suggests that future growth rate experiments and data analysis can be designed to avoid or compensate for the effects of the defects in the hybridization signal. Although identified defects in tags or primers can alter hybridization in a subset of strains, the built-in redundancy of having two bar codes for one deletion strand can be used to avoid the effect of defects in data analysis. Future oligonucleotide arrays can now be redesigned to match actual tags if desired. The results show that the range of detection in the quantitative phenotypic analysis depends on the precise design of tag complements on the surface of an oligonucleotide array. This may lead to more complete and comprehensive yeast functional genomic studies in the future using redesigned oligonucleotide arrays and may facilitate more accurate analysis of results taking into consideration the potential for crosshybridization among tags. There are at least 2,060 tags that can be corrected by array redesign. Yeast gene-deletion strains with large deletions in tags or defects in common primers may be remade by recombination of a new deletion cassette into the yeast genome.
One powerful application of the yeast gene-deletion strain collection is the identification of biologically active natural products (12, 13). Natural products are extensively used by microorganisms, plants, and animals for chemical communication and competition among cells to obtain selective growth advantages (see http://sequence-www.stanford.edu/group/yeast_deletion_project). Like genetic mutations, natural products can potentially generate conditional phenotype of each yeast gene, thus these compounds can be used to elucidate gene function with specificity that is superior to conditional genetic mutations (14, 15). Because conventional genetic methods are not feasible in many organisms, the use of natural products as genetic tools is of great value in the study of gene functions. The compounds identified by the use of the tagged yeast gene-deletion strains could be useful as tools in studies of genetically less tractable organisms such as the pathogenic yeast Candida albicans and potentially in the treatment of human diseases such as cancer, because many genes and biochemical pathways are universally conserved. Incorporation of molecular bar codes into other microorganisms is a useful method for identifying gene functions by growth rate.
Supplementary Material
Acknowledgments
We thank the following people for technical help: C. Komp, D. Faulkner, D. Bruno, and F. Aviles for ABI 3700 sequencing support; M. Karhanek and T. Jones for assistance with data interpretation and analysis; J. Kumm (Stanford Genome Technology Center) for providing the normalized hybridization intensities used in this analysis; and M. Brock for critical reading of the manuscript. This work was supported by National Aeronautics and Space Association (NASA) Center for Nanotechnology, NASA Fundamental Biology Program, and NASA Biomolecular Systems Research Program, NASA contract NAS2-99092 and National Institutes of Health Grant 5PO1HG00205. K.A. and O.J. were supported by the NASA Administrator's Fellowship Program.
Abbreviation: LR, log base 10 of UPTAG signal/DNTAG signal.
References
- 1.Winzeler, E. A., Shoemaker, D. D., Astromoff, A., Liang, H., Anderson, K., Andre, B., Bangham, R., Benito, R., Boeke, J. D., Bussey, H., et al. (1999) Science 285, 901–906. [DOI] [PubMed] [Google Scholar]
- 2.Shoemaker, D. D., Lashkari, D. A., Morris, D., Mittmann, M. & Davis R. W. (1996) Nat. Genet. 14, 450–456. [DOI] [PubMed] [Google Scholar]
- 3.Giaever, G., Flaherty, P., Kumm, J., Proctor, M., Nislow, C., Jaramillo, D. F., Chu, A. M., Jordan, M. I., Arkin, A. P., et al. (2004) Proc. Natl. Acad. Sci. USA 101, 793–798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Giaever, G., Chu, A. M., Ni, L., Connelly, C., Riles, L., Veronneau, S., Dow, S., Lucau-Danila, A., Anderson, K., Andre, B., et al. (2002) Nature 418, 387–391. [DOI] [PubMed] [Google Scholar]
- 5.Richterich, P. (1998) Genome Res. 8, 251–259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lawrence, C. B. & Solovyev V. V. (1994) Nucleic Acids Res. 22, 1272–1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ewing, B. & Green, P. (1998) Genome Res. 8, 186–194. [PubMed] [Google Scholar]
- 8.Ronaghi, M., Uhlen, M. & Nyren, P. (1998) Science 281, 363–365. [DOI] [PubMed] [Google Scholar]
- 9.Ronaghi, M., Nygren, M., Lundeberg, J. & Nyren, P. (1999) Anal. Biochem. 267, 65–71. [DOI] [PubMed] [Google Scholar]
- 10.Ronaghi, M. (2003) Methods Mol. Biol. 212, 189–195. [DOI] [PubMed] [Google Scholar]
- 11.Gharizadeh, B., Ghaderi, M., Donnelly, D., Amini, B., Wallin, K. L. & Nyren, P. (2003) Electrophoresis 24, 1145–1151. [DOI] [PubMed] [Google Scholar]
- 12.Steinmetz, L. M., Scharfe, C., Deutschbauer, A. M., Mokranjac, D., Herman, Z. S., Jones, T, Chu, A. M., Giaever, G., Prokisch, H., Oefner, P. J., et al. (2002) Nat. Genet. 31, 400–404. [DOI] [PubMed] [Google Scholar]
- 13.Giaever, G., Shoemaker, D. D., Jones, T. W., Liang, H., Winzeler, E. A., Astromoff, A. & Davis, R. W. (1999) Nat. Genet. 21, 278–283. [DOI] [PubMed] [Google Scholar]
- 14.Hung, D. T., Jamison, T. F. & Schreiber, S. L. (1996) Chem. Biol. 8, 623–639. [DOI] [PubMed] [Google Scholar]
- 15.Schreiber, S. L. (1992) Chem. Eng. News 70, 22–32. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.