Summary
Consumers are increasingly becoming aware of their health and nutritional requirements, and in this context, vitamins produced in situ by microbes may suit their needs and expectations. B groups vitamins are essential components of cellular metabolism and among them riboflavin is one of the vital vitamins required by bacteria, plants, animals and humans. Here, we focus on the importance of microbial production of riboflavin over chemical synthesis. In addition, genetic abilities for riboflavin biosynthesis by lactic acid bacteria are discussed. Genetically modified strains by employing genetic engineering and chemical analogues have been developed to enhance riboflavin production. The present review attempts to collect the currently available information on riboflavin production by microbes in general, while placing greater emphasis on food grade lactic acid bacteria and human gut commensals. For designing riboflavin‐enriched functional foods, proper selection and exploitation of riboflavin‐producing lactic acid bacteria is essential. Moreover, eliminating the in situ vitamin fortification step will decrease the cost of food production.
Introduction
Riboflavin is present in many foods such as green vegetables, dairy products, eggs and meat. The recommended daily intake for riboflavin is 1.3 mg day−1 for men and 1.1 mg day−1 for women (Food and Nutrition Board, 1998). In Western countries, mostly milk and dairy products contribute to the daily intake of riboflavin besides yeast, cereals, meats, fatty fish and green leafy vegetables (Cooperman and Lopez, 1991; Powers et al., 1993). Grain products contain only low amounts of riboflavin because of loss of this vitamin during processing of the grains. Nevertheless, fortification practices make certain breads and cereals very good sources of riboflavin (Hill and Nalubola, 2002; Powers, 2003). In defiance of the presence of most of the vitamins in a variety of foods, human riboflavin deficiency persists in both developing and industrialized countries (O'Brien et al., 2001; Blanck et al., 2002) because of insufficient food intake and unbalanced diet (LeBlanc et al., 2011). In developing nations, its deficiency prevails in populations whose diet lacks dairy products and meat (Combs, 1992; Rohner et al., 2007). A high prevalence of poor riboflavin status has been observed among adolescent girls in the United Kingdom and among the Irish population (O'Brien et al., 2001; Powers, 2003). Riboflavin deficiency is associated with impaired vision, reduced growth rate, increased levels of homocysteine with consequent cardiac risk (Moat et al., 2003), pre‐eclampsia (Wacker et al., 2000), oxidative stress (Ashoori and Saedisomeolia, 2014) and anaemia (Shi et al., 2014). Riboflavin deficiency can lead to liver and skin damage, and changes in cerebral glucose metabolism (LeBlanc et al., 2011) with symptoms like hyperaemia, sore throat, oedema of oral and mucous membranes, cheilosis and glossitis (Wilson, 1983).
Riboflavin has been traditionally synthesized for food and feed fortification by chemicals means, but past decade has witnessed emerging information about commercial completive microbial processes for its production (Stahmann et al., 2000). Riboflavin is synthesized by many bacteria and its biosynthesis pathway has been studied in both Gram‐positive and Gram‐negative bacteria, but it has been extensively studied only in two organisms namely in Bacillus subtilis (Perkins and Pero, 2002) and Escherichia coli (Bacher et al., 1996). Currently, three microorganisms are exploited for riboflavin production: Ashbya gossypii, Candida famata and B. subtilis (Perkins et al., 1999; Stahmann et al., 2000; Schallmey et al., 2004). In recent years, the use of lactic acid bacteria (LAB) was proposed for vitamin synthesis. These microorganisms are able to synthesize B‐group vitamins particularly riboflavin to obtain fermented bio‐enriched food (Capozzi et al., 2011; Laino et al., 2012; Vaesken et al., 2012). The use of LAB is a common practice in the dairy industry, and the addition of the riboflavin‐producing strain into fermented products such as fermented milks, yoghurt, and cheeses increases riboflavin concentrations, which is feasible and economically viable (LeBlanc et al., 2005a). The obvious practical advantages of vitamin‐producing LAB that fortification happens in situ. The in situ fortification advantage of LAB makes them a good choice for bio prospecting bacteria which can act as vitamin supplier to human hosts (Burgess et al., 2009). The adaptability of LAB to fermentation processes, their biosynthetic capability and metabolic versatility are the key features that make them ideal candidates for in situ production of riboflavin in food (Arena et al., 2014b). Gut commensals are able to synthesize vitamin K as well as most of the water‐soluble B‐vitamins, such as biotin, folates, nicotinic acid, panthotenic acid, pyridoxine, riboflavin and thiamine (Hill, 1997). In this review, focus is placed on the LAB and their genetic ability to biosynthesize riboflavin.
Microbes taking place of chemical factories for riboflavin production
Although humans and animals lack the ability to synthesize most of the vitamins, bacteria have inherent potential to produce those metabolites (LeBlanc et al., 2011). With modern lifestyle, consumers are becoming more health conscious and discerned in their food choices (Burgess et al., 2004). In such a situation, riboflavin‐supplying LAB offer a clear advantage over chemical synthesis by increasing the nutritional value of food (LeBlanc et al., 2012). Chemical synthesis of a vitamin is being replaced by fermentation processes because of economic and environmental considerations of the latter. Besides the economic advantages, additional benefits of the microbial synthesis include the use of renewable sources, environmental‐friendly approach and superior quality of the final product (Fig. 1) (Van Loon et al., 1996).
Figure 1.
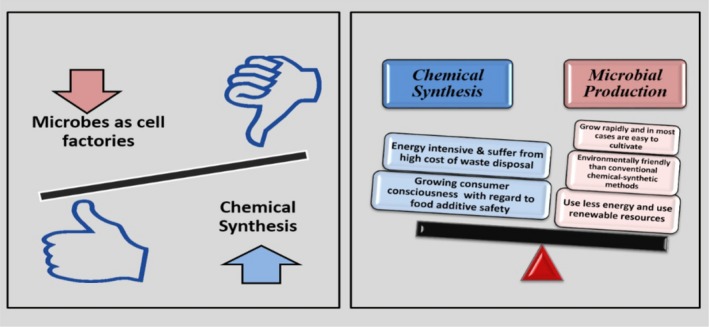
Advantages of microbes as cell factories for vitamin synthesis.
Importance of riboflavin to humans and bacteria
Each B‐group vitamin acts in synergy to maintain the body's homeostasis by playing major roles in metabolic processes (LeBlanc et al., 2011). One of such essential vitamins, i.e. riboflavin, is an obligatory component of cellular metabolism and is responsible for normal development, growth, reproduction, lactation, physical performance of well‐being. Metabolically, riboflavin is the precursor of flavin mononucleotide (FMN) and flavin adenine dinucleotide (FAD), both of which act as electron carriers in oxidation‐reduction reactions (Fischer and Bacher, 2005). They help in the metabolism of carbohydrates, amino acids, energy production and also activate folate and pyridoxine to their respective coenzyme forms (Food and Nutrition Board, 1998; Massey, 2000), which constitute the basis for its clinical applications. Until now, riboflavin has received relatively little attention, but interest is increasing with its recognition as an essential component of cellular biochemistry (Thakur and Tomar, 2015). One study has suggested that dietary vitamin intake leads to a relatively low risk of vitamin deficiency in all age and sex groups (Mensink et al., 2013). Riboflavin is being used for headache (Schetzek et al., 2013) and migraine management (Sherwood et al., 2014). According to Foley and colleagues (2014) riboflavin supplementation can combat the progression of neurodegenerative conditions. Another study by Shi and colleagues (2014) showed that inadequate riboflavin intake was associated with an increased risk of persistent anaemia. Riboflavin can act as a protectant from oxidative injury independently by the conversion of its reduced form to oxidized form, or as a component of glutathione redox cycle (Ashoori and Saedisomeolia, 2014). According to Hassan and colleagues (2013) riboflavin acts as an efficient adjuvant, which is confirmed in many cancer cell lines and animal‐based studies, and it is promising under photodynamic therapy (Hassan et al., 2013. Recently, riboflavin has been shown to improve the efficiency of conventional therapies in different diseases such as Staphylococcus aureus infection and cisplatin‐induced intestinal epithelial cell apoptosis (Bodiga et al., 2012; Mal et al., 2013).
Although requirement for riboflavin may be rare among bacteria, it is known to be an essential growth factor for Enterococcus faecalis, Streptococcus pyogenes, Listeria monocytogenes and some lactobacilli (Koser, 1968). The biosynthetic deficiency correlates with the absence of riboflavin biosynthetic genes in the genomes of these organisms (Vitreschak et al., 2002). The sensitive growth response of Lactobacillus casei to riboflavin was used to develop one of the first microbiological assays for a vitamin (Snell and Strong, 1939) and it is based on the presence of an efficient transport system that allows the uptake of exogenous riboflavin. Riboflavin uptake inversely correlates with the riboflavin concentration present during cell growth and increases in riboflavin‐requiring mutants (Coquard et al., 1997).
Regulation of riboflavin biosynthesis in bacteria
The riboflavin biosynthesis in bacteria was analysed using comparative analysis of genes, operons and regulatory elements (Vitreschak et al., 2002). A model for regulation of riboflavin biosynthesis is based on the formation of alternative RNA structure involving the RFN element (a mononucleotide riboswitch is highly conserved RNA element that is found frequently in the 5′ untranslated region of prokaryotic mRNA that encodes for FMN biosynthesis and transport proteins) (Fig. 2) (Gelfand et al., 1999; Vitreschak et al., 2002). The RFN element can be found on the chromosome of many, but not all, bacterial species (Gelfand et al., 1999; Vitreschak et al., 2002; Wels et al., 2006). In Gram‐positive bacteria, riboflavin metabolism and transport genes are regulated at transcription attenuation, whereas in Gram‐negative bacteria riboflavin biosynthesis genes are regulated on level of translation initiation (Fig. 2) (Vitreschak et al., 2002). The enzymatic activities required to catalyse the biosynthesis of riboflavin from guanosine triphosphate (GTP) and ribulose‐5‐phosphate are encoded by four genes (ribG, ribB, ribA and ribH) as shown in (Fig. 3) (Perkins et al., 1999). According to these authors, these genes are located in an operon, the gene order of which differs from the order of enzymatic reactions. In GTP cyclohydrolase II activity, which catalyses the first step in riboflavin biosynthesis, GTP is encoded by the third gene in the operon, ribA. The RibA gene also contains a second enzymatic function that synthesizes a four‐carbon unit from ribulose‐5‐phosphate (Richter et al., 1992; 1993) that is utilized in a later step (lumazine synthase). The second and third enzymatic steps (deamination of the pyrimidine ring of structure and the subsequent reduction of the ribosyl side‐chain) are controlled by another bi‐functional enzyme encoded by the first gene of the operon ribG (Richter et al., 1997). The penultimate step in riboflavin biosynthesis, is catalysed by lumazine synthase, the product of the last rib gene, ribH (Perkins et al., 1999). Riboflavin synthase, which controls the last step of the pathway, is encoded by the second gene of the operon, ribE (Perkins et al., 1999). Transcription of the four riboflavin genes is primarily controlled by the ribP1 promoter and regulatory region located at the 5′ end of the operon (Perkins et al., 1999). In addition, the last two rib genes in the operon, ribA and ribH, are also transcribed from a second promoter (ribP2) and regulatory region RFN (Perkins et al., 1999).
Figure 2.
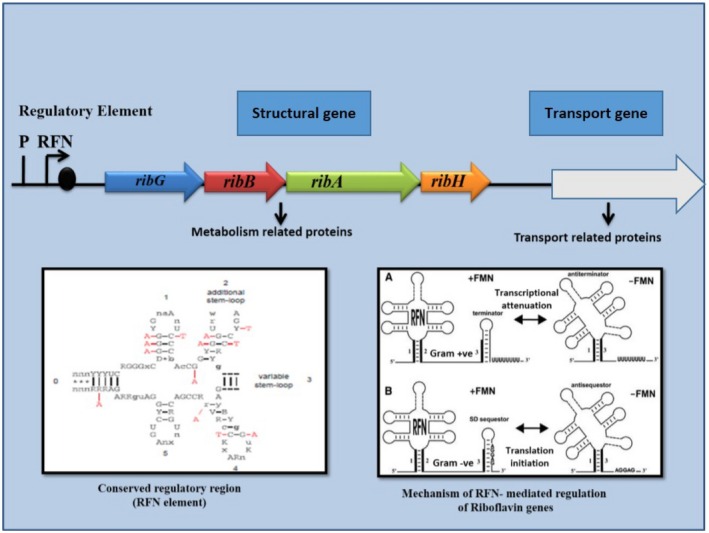
Regulation of riboflavin biosynthesis genes in Gram‐positive and Gram‐negative bacteria.
Figure 3.
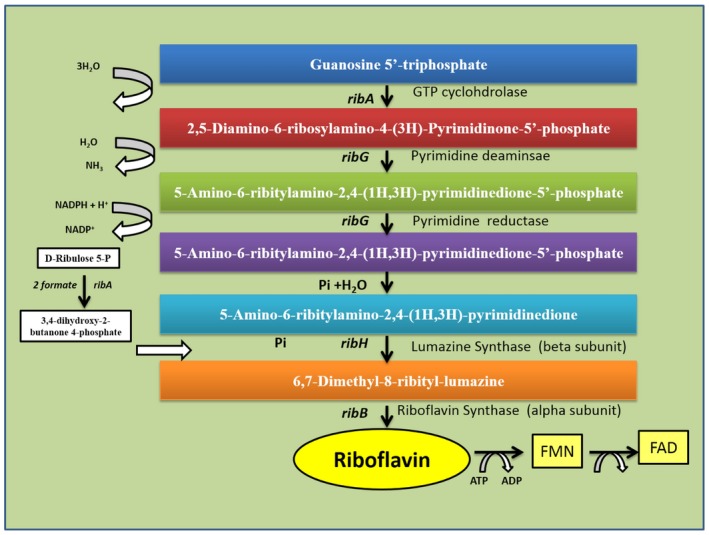
Riboflavin biosynthesis pathway in bacteria.
Genetic ability of riboflavin production in LAB
According to Capozzi and colleagues (2012), the genetic information for riboflavin biosynthesis in LAB is species specific and/or strain specific. It is clear from the previous reports of comparative genome analysis that the ability to synthesize riboflavin is shared by several of the sequenced members of LAB although an interrupted or partially present rib operon is sometimes observed in certain strains (Table 1). According to bioinformatics analysis by Burgess and colleagues (2004), when the first gene (ribG) is absent from the genome, it is more likely that the riboflavin operon will be incomplete. The sequenced genome of Lactobacillus plantarum strain WCFS1 contains an incomplete rib operon, which is devoid of the entire ribG and part of the ribB genes (Kleerebezem et al., 2003). As expected, this strain is unable to grow in the absence of riboflavin (Burgess et al., 2006).
Table 1.
Presence/absence of riboflavin biosynthesis genes among different LAB strains (adapted from Capozzi et al., 2012 and Valle et al., 2014)
| Organism | ribG | ribB | ribA | ribH |
|---|---|---|---|---|
| Lactococcus lactis subsp. cremoris SK11 | + | + | + | + |
| Lactococcus lactis subsp. cremoris NZ9000 | + | + | + | + |
| Lactococcus lactis subsp. lactis KF147 | + | + | + | + |
| Lactococcus lactis subsp. cremoris A76 | + | + | + | + |
| Lactococcus lactis subsp. cremoris MG1363 | + | + | + | + |
| Lactococcus lactis subsp. lactis CV56 | + | + | + | + |
| Lactobacillus brevis ATCC367 | + | + | + | + |
| Lactobacillus plantarum WCFSI | − | − | + | + |
| Lactobacillus plantarum subsp. plantarum ST‐III | + | + | + | + |
| Lactobacillus plantarum JDM1 | + | + | + | + |
| Lactobacillus plantarum CRL725 | + | + | + | + |
| Lactobacillus gasseri ATCC33323 | − | − | − | + |
| Lactobacillus casei ATCC334 | − | − | − | − |
| Lactobacillus bulgaricus ATCC BAA365 | − | − | − | − |
| Lactobacillus delbrueckii subsp. bulgaricus ND02 | + | + | + | + |
| Lactobacillus delbrueckii subsp. bulgaricus 2038 | + | + | + | + |
| Lactobacillus delbrueckii subsp. bulgaricus ATCC 11842 | − | − | − | − |
| Lactobacillus acidophilus NCFM | − | − | − | − |
| Lactobacillus acidophilus 30SC 8293 | − | − | − | + |
| Leuconostoc mesenteroides subsp. mesenteroides J18 | + | + | + | + |
| Leuconostoc mesenteroides subsp. mesenteroides ATCC 8293 | + | + | + | + |
| Lactobacillus amylovorus GRL1118 | − | − | − | + |
| Lactobacillus amylovorus GRL 1112 | − | − | − | + |
| Lactobacillus buchneri NRRL B‐30929 | − | − | − | − |
| Lactobacillus crispatus ST1 | + | + | + | + |
| Lactobacillus fermentum IFO 3956 | + | + | + | + |
| Lactobacillus fermentum MTCC8711 | + | + | + | + |
| Lactobacillus helveticus DPC 4571 | − | − | − | − |
| Lactobacillus helveticus H10 | − | − | − | − |
| Lactobacillus johnsonii NCC 533, DPC 6026, FI9785 | − | − | − | − |
| Lactobacillus kefiranofaciens ZW3 | − | − | − | − |
| Lactobacillus reuteri DSM 20016 | + | + | + | + |
| Lactobacillus reuteri SD2112 | − | − | + | − |
| Lactobacillus reuteri JCM 1112 | + | + | + | + |
| Lactobacillus rhamnosus ATCC 8530, GG | − | − | − | − |
| Lactobacillus salivarius CECT 5713, UCC118 | + | + | + | + |
+, Presence; −, absence.
Human gut commensals and riboflavin biosynthesis ability
According to a recent study of systematic genome assessment of B‐vitamin biosynthesis, a complete riboflavin operon is present in all Bacteroidetes, Fusobacteria and 36 genomes (92%) of Proteobacteria (Magnusdottir et al., 2015). In their study, the authors have placed Firmicutes as more potent producers of riboflavin compared with other examined vitamins. The Actinobacteria phylum contains only two genomes that are publicly available, namely, those of Corynebacterium ammoniagenes DSM 20306 and Bifidobacterium longum ATCC 15697, which have the coding capacity for riboflavin biosynthesis. Interestingly, gut commensals that produce riboflavin are detected by the innate immune system through a metabolic intermediate as riboflavin precursors found in many bacteria and yeast selectively activating mucosal‐associated invariant T cells, an abundant population of innate‐like T cells in humans (Corbett et al., 2014). Riboflavin biosynthesis genes seem to be partially or completely absent from the majority of currently available bifidobacterial genomes (Ventura et al., 2007). Recently, four bifidobacterial species are predicted to possess a complete riboflavin biosynthesis pathway (Milani et al., 2014), which may represent an additional mechanism for microbe–host interactions by stimulation of the host's immune system (Corbett et al., 2014). The possibility of co‐evolution of gut microbes in the human gut makes them suitable for de novo synthesis (LeBlanc et al., 2012; Magnusdottir et al., 2015). The latter study suggests that human gut bacteria actively exchange B‐vitamins among each other, which leads to the survival of organisms that do not synthesize any of these essential cofactors. However, all non‐producing organisms from the human gut contained the riboflavin transporter role, indicating their need for the riboflavin‐derived cofactors FMN and FAD. It was almost completely absent in the Bacteroidetes, Fusobacteria and Proteobacteria, whereas the de novo synthesis pathway was found in nearly all genomes of the three phyla (Magnusdottir et al., 2015). Another study supports the fact that vitamin metabolism pathways are highly represented in all enterotypes, whereas two among all the examined enterotypes are found to be rich in biosynthesis genes for biotin, riboflavin, pantothenate, ascorbate, thiamine and folate production (Arumugam et al., 2011).
Riboflavin production by LAB
LAB are a group of industrially prominent microorganisms used in the food and dairy industry because of their enormous applications for the biosynthesis of a number of compounds as metabolic end‐products or secondary metabolites (LeBlanc et al., 2012). Many LAB and bifidobacteria produce a range of metabolites including B‐vitamins such as riboflavin and folate, low‐calorie sugars such as mannitol and sorbitol, exopolysaccharides, diacetyl and L‐alanine (Hugenholtz et al., 2002). They also accumulate and biotransform inorganic selenium to organic and elemental forms, which are useful for human and animal nutrition. (Pophaly et al., 2014; Saini et al., 2014). There are three reports of riboflavin‐producing lactobacilli from India (Table 2) (Jayashree et al., 2010; Guru and Viswanathan, 2013; Thakur and Tomar, 2015). Thakur and Tomar, (2015) have reported the riboflavin production in Lactobacillus fermentum KTLF1 (2.36 mg) and L. plantarum (and 2.13 mg l−1) in MRS medium (Thakur and Tomar, 2015). According to Jayashree and colleagues (2010) efficient riboflavin‐producing bacterium L. fermentum MTCC 8711 showed 2.29 mgl l−1 of riboflavin in chemically defined media after 24 h. Guru and Viswanathan (2013) have observed that L. acidophilus produces higher riboflavin levels compared with Lactococcus lactis. They have recommended whey as a better fermentation medium compared with skim milk for riboflavin production (Guru and Viswanathan, 2013). Valle and colleagues (2014) have evaluated over 179 strains of LAB to increase the riboflavin levels in soymilk. The development of novel functional foods with enhanced vitamin content has been suggested and it would contribute to an ever‐growing market for these products (Stanton et al., 2005). The production of fermented food products with elevated levels of B‐vitamins increases their commercial and nutritional value and eliminates the need for fortification (Burgess et al., 2009). Different strategies have been applied to improve microbial production of vitamins during fermentation (Sybesma et al., 2006). Riboflavin overproduction can be achieved either by genetic engineering (Perkins et al., 1991) or by exposure to purine analogues and/or the toxic riboflavin analogue roseoflavin (Table 2) (Burgess et al., 2004). The same authors have obtained overproduction of riboflavin up to 24 mg l−1 and up to around 0.9 mg l−1 using nisin induction and roseoflavin respectively (Burgess et al., 2004). Often, the increased riboflavin production phenotype is associated with mutations at the regulatory region (RFN), which increases the transcription of the riboflavin operon (Burgess et al., 2006). Roseoflavin‐resistant strains of Leu. mesenteroides overproduced up to 0.5 mg l−1 of riboflavin, whereas riboflavin‐overproducing L. plantarum and Propionibacterium freudenreichii were able to synthesize up to around 0.6 mg l−1 and 3 mg l−1 respectively (Burgess et al., 2006). The genetic engineering is an interesting way to exploit the industrially important strains that cannot produce riboflavin or in other strains that produce it at low level and are physiologically inactive (Capozzi et al., 2012). These two strategies for riboflavin overproduction have been successfully employed in various LAB and non‐LAB so far.
Table 2.
Various LAB and non‐LAB screened for riboflavin production
| Riboflavin production strategy | Organism | Source | References |
|---|---|---|---|
| Genetic engineering/exposure to purine/toxic riboflavin analogue | Microbes screened for enhanced riboflavin production | ||
| L. lactis | Yoghurt | LeBlanc and colleagues (2005) | |
| L. lactis subsp. cremoris strain NZ9000 | Burgess and colleagues (2004) | ||
| L. fermentum MTCC8711 | Jayashree and colleagues (2011) | ||
| L. plantarum | Burgess and colleagues (2006) | ||
| L. mesenteroids | Burgess and colleagues (2006) | ||
| P. freudenreichii | Burgess and colleagues (2006) | ||
| Exposure to toxic riboflavin analogue | L. plantarum, L. mesenteroides and L. fermentum | Sourdough | Russo and colleagues (2014) |
| L. plantarum CRL725 | Dairy products | Valle and colleagues (2014) | |
| L. plantarum | Durum wheat flour | Capozzi and colleagues (2011) | |
| Natural | Microbes screened for natural riboflavin production | ||
| L. acidophilus | Curd and cheese | Guru and Viswanathan, 2013 | |
| Bacillus clausii, B. subtilis, B. cereus IP 5832, L. rhamnosus ATCC 53103 | Probiotic formulations | Salvetti et al., 2003 | |
| L. fermentum, L. plantarum and L. mucosae | Human faeces and fermented bamboo shoots | Thakur and Tomar, 2015 | |
| Genetic engineering/exposure to purine/toxic riboflavin analogue | Commercial producers | ||
| A. gossypii | Perkins and colleagues (1999) | ||
| Candida famata | Schallmey and colleagues (2004) | ||
| Bacillus subtilis | Stahmann and colleagues (2000) | ||
Riboflavin overproduction by genetic engineering approach
Lactococcus lactis is a commonly used starter strain that can be converted from riboflavin consumer into riboflavin‐producing factory by overexpressing its riboflavin biosynthesis genes (LeBlanc et al., 2005b). These riboflavin‐producing strains were able to eliminate most physiological manifestations of ariboflavinosis such as stunted growth, elevated erythrocyte glutathione reductase activation coefficient values and hepatomegalia in a riboflavin depletion–repletion model. In another study, Burgess and colleagues (2004) carried out genetic analysis of the riboflavin biosynthetic operon in L. lactis subsp. cremoris strain NZ9000. The strain showed enhanced vitamin synthesis because of simultaneous overexpression of riboflavin biosynthetic genes (ribG, ribH, ribB and ribA) in L. lactis (Burgess et al., 2004). In one study, the inactivation of the folE gene, involved in the folate biosynthesis pathway, which could make more GTP available for the riboflavin biosynthesis, resulted in a 50% enhanced level of riboflavin production by L. fermentum, albeit with double generation time. This phenotype was stably maintained because the folE has been disrupted in the genome (Jayashree et al., 2011). While through site‐directed mutagenesis followed by metabolic engineering, Sybesma and colleagues (2004) modified two complicated biosynthetic pathways in L. lactis that resulted in simultaneous overproduction of both folate and riboflavin (Sybesma et al., 2004). Such strategies do not attempt to generate alternative production strains, but rather replacing riboflavin‐consuming strains used in traditional food fermentation processes with riboflavin‐producing counterparts, thereby increasing riboflavin bioavailability in the food product and introducing an added health benefit. Elevated levels of the vitamin, which would be produced in such foods, would not have any negative health implications as no upper limit of intake has been set for riboflavin because of the lack of evidence on adverse effect in humans (Flynn et al., 2003).
Riboflavin overproduction by chemical analogues approach
The isolation of spontaneous roseoflavin‐resistant mutants is a reliable method to obtain natural riboflavin‐overproducing strains of various species commonly used in the food industry, and it is also acceptable from a consumer/regulatory point of view as it does not involve deliberate genetic engineering (Jayashree et al., 2011). With the increase in the availability of genome sequences it is possible not only to identify the potential mutations that cause riboflavin (over) production, but also to determine how stable such mutations are maintained (LeBlanc et al., 2012). The toxic analogue approach has also been successfully employed for L. plantarum, Leuconostoc mesenteroides and P. freudenreichii (Burgess et al., 2006) and a fermented dairy product made with the latter strain was shown to counteract riboflavin deficiency in an animal model (LeBlanc et al., 2006). The riboflavin‐overproducing Lactobacillus strains were selected by exposure to roseoflavin and several overproducing strains were identified and used for bread fermentation, barley and oat‐fermented products (Russo et al., 2012).The riboflavin‐producing LAB strains including L. plantarum, L. mesenteroides and L. fermentum were isolated from a traditional sourdough (Russo et al., 2014). Overproducing strains of L. fermentum and L. plantarum selected after exposure to roseoflavin were investigated for their probiotic attributes by using an in vitro model and they were able to synthesize riboflavin in co‐culture systems with Caco‐2 cells (Arena et al., 2014a). It was reported that β‐Glucans stimulate the growth of these strains when submitted to oro‐gastrointestinal stress with a positive impact on bacterial adhesion (Arena et al., 2014b). Moreover, the adhesion ability of these strains was evaluated by using gnotobiotic zebrafish larvae as in vivo model, reinforcing the suggestion that they could contribute to further increase the riboflavin supply in the gut environment (Russo et al., 2015). The in vitro adhesion on human epithelial cell lines (mucus‐producing HT‐29) was also studied by the use of riboflavin‐producing L. mucosae KTF (Thakur et al., 2015). Russo and colleagues (2014) have used L. fermentum PBCC11.5 and its parental strain to fortify bread, and they have concluded that bread produced using the co‐inoculum yeast and L. fermentum PBCC11.5 led to an approximately twofold increase of final riboflavin content, which opens new perspectives in the field of functional foods based on a cereal matrix (Russo et al., 2014). In one study by LeBlanc and colleagues (2006) the novel fermented product containing P. freudenreichii B2336, with increased levels of riboflavin, eliminated most physiological manifestations of ariboflavinosis using a riboflavin depletion–repletion model, whereas the product fermented with the non‐riboflavin‐producing strain did not show this beneficial effect. Propionibacterium freudenreichii NIZO B2336 is a spontaneous roseoflavin‐resistant mutant derived from P. freudenreichii B374 that produces higher levels of the riboflavin than that produced by the parental stain (LeBlanc et al., 2006). In another study, riboflavin‐producing LAB strains were isolated and used as a convenient biotechnological application for the preparation of fermented sourdough bread and pasta to enrich them with riboflavin (Capozzi et al., 2011). In this study, L. plantarum was selected for roseoflavin‐resistant to acquire natural riboflavin‐overproducing strains. Valle and colleagues (2014) stated that roseoflavin‐resistant strains are capable of synthesizing riboflavin in soymilk and have led to an interesting and economically feasible biotechnology strategy that could easily be adapted to develop novel vitamin bio‐enriched functional foods with enhanced consumer appeal. All these reports (Table 3) of enhanced riboflavin production in various dairy and cereal‐based products pave the way for analysing the effect of similar riboflavin‐overproducing LAB in human trials (LeBlanc et al., 2005).
Table 3.
In vivo manifestations of riboflavin‐enriched fermented products and riboflavin‐overproducing lactobacilli
| Product | Organism used | In vivo effects | Reference |
|---|---|---|---|
| Fermented milk | P. freudenreichii B2336 | Eliminated most physiological manifestation of ariboflavinosis | LeBlanc and colleagues (2006) |
| Fermented milk | L. lactis NZ9000 | Reversing ariboflavinosis in a riboflavin‐deficiency rat model | LeBlanc and colleagues (2005) |
| – | L. lactis CB010 | Elimination of stunted growth, increased EGRAC values and hepatomaglia in animal model riboflavin depletion–repletion rats | – |
| Soya milk | L. plantarum CRL 725 | – | Valle and colleagues (2014) |
| Yoghurt | P. freudenreichii B2336 | – | Burgess and colleagues (2004) |
| Pasta and bread | L. plantarum | – | Capozzi and colleagues (2011) |
EGRAC, erythrocyte glutathione reductase activity coefficient.
Probiotics and B‐vitamin biosynthesis
Besides traditional applications of LAB, some of the members have been reported to elicit probiotic features (Russo et al., 2015). Food‐related LAB as well as human gut commensals such as bifidobacteria make a certain site in dairy and food industry by imparting various health benefits to human host and carries enzymes to de novo synthesize and supply vitamins (LeBlanc et al., 2012). Two sources of riboflavin are available to humans: a dietary source and riboflavin‐producing microflora of the large intestine (Wrong et al., 1981; Hill, 1997). Vitamins produced by microbes get adsorbed in the colon in contrast to dietary vitamins, which are adsorbed in the proximal tract of the small intestine (Ichihashi et al., 1992; Said and Mohammed, 2006). The site of uptake increases the bioavailability of vitamins synthesised by microbes to human host. Moreover riboflavin‐producing gut commensals may overactivate the innate immune system (Corbett et al., 2014), which also presents the limitations of in situ riboflavin production by gut commensals. Commercialized probiotic bacteria have been included as active ingredients in products such as yoghurt, cheese, ice cream, chocolates pharmaceutical tablets, infant formulas and dietary supplements (Tamime et al., 2005). Fermented foods using LAB are advantageous because they have the potential beneficial effects of probiotic properties coupled with enhanced content of vitamins (Jayashree et al., 2011). Intestinal microbiota has also been shown to produce short chain fatty acids, conjugated linoleic acid, essential amino acids, group B‐vitamins and vitamin K, contributing to the well‐being of a host (Marques et al., 2010). According to Magnusdottir and colleagues (2015) gut microbiota is an important source of B‐vitamins, which lead to changes in the gut microbiota composition and ultimately affecting our dietary B‐vitamin requirements. Salvetti and colleagues (2003) have reported eight probiotic strains from five different probiotic formulations containing Bacillus clausii, B. subtilis, Bacillus cereus IP 5832, Lactobacillus rhamnosus ATCC 53103, which were able to produce riboflavin (Salvetti et al., 2003). Guru and Viswanathan (2013) have reported riboflavin‐producing probiotic L. acidophilus obtained from curd and cheese samples.
Concluding comments
The economic and environmental considerations have led the fermentation‐based method as a model of the environmentally friendly white biotechnology with regard to traditional chemical synthesis of riboflavin (Shi et al., 2009). Bacteria producing even small amounts of riboflavin will be a better choice to be used as a starter for the formation of fermented products rather than traditional starters, which consume riboflavin. So far, information available on whole genomes of various microbes has made it clear that riboflavin‐producing ability is recognized to be strain or subspecies specific. Thus, it can be an attractive approach to bioprospect prolific riboflavin‐producing strains from their diversified natural niche and further enhance their ability to produce this essential vitamin by microbiological and biotechnological interventions. The enzymes required for riboflavin biosynthesis may be completely or partially absent in various available genomes of microbes; nevertheless, the behaviour of multiple coexisting microbial species suggests the possibility of de novo synthesis of riboflavin. LAB also known as power house of dairy industry and imparting health benefits as probiotics are endowed with the ability to synthesize essential biomolecules in particular riboflavin. The carefully selected riboflavin‐producing strains holding probiotic attributes could open the way to be potential candidates for in situ production of riboflavin once these strains get colonized to host intestine (Arena et al., 2014a, 2014b). Thus, considering the extensive application of LAB in the food, pharmaceutical and medicine industry, coupled with consumer demand for healthier foods, the use of these food grade microorganisms as riboflavin factories will be of great advantage in the near future.
Conflict of interest
None declared.
Acknowledgements
The authors thankfully acknowledge the support extended by Indian Council of Agricultural Research (ICAR), New Delhi, and by the Director of National Dairy Research Institute (NDRI), Karnal. The authors also wish to thank the reviewers for their meticulous reviews and constructive comments, which led to necessary improvements of manuscript.
Microbial Biotechnology (2016) 9(4), 441–451
Funding Information No funding information provided.
References
- Arena, M.P. , Russo, P. , Capozzi, V. , Lopez, P. , Fiocco, D. , and Spano, G. (2014a) Probiotic abilities of riboflavin‐overproducing Lactobacillus strains: a novel promising application of probiotics. Appl Microbiol Biotechnol 98: 7569–7581. [DOI] [PubMed] [Google Scholar]
- Arena, M.P. , Caggianiello, G. , Fiocco, D. , Russo, P. , Torelli, M. , Spano, G. , and Capozzi, V. (2014b) Barley β‐glucans‐containing food enhances probiotic performances of beneficial bacteria. Int J Mol Sci 15: 3025–3039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arumugam, M. , Raes, J. , Pelletier, E. , Le Paslier, D. , Yamada, T. , Mende, D.R. , et al (2011) Enterotypes of the human gut microbiome. Nature 473: 174–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashoori, M. , and Saedisomeolia, A. (2014) Riboflavin (vitamin B2) and oxidative stress: a review. Br J Nutr 20: 1–7. [DOI] [PubMed] [Google Scholar]
- Bacher, A. , Eberhardt, S. , and Richter, G. (1996) Biosynthesis of riboflavin In Escherichia coli and Salmonella: Cellular and Molecular Biology, 2nd edn Neidhardt F.C., Curtiss R., III, Ingraham J.L., Lin E.C.C., Low K.B., Magasanik B., et al (eds). Washington, DC, USA: ASM Press, pp. 657–664. [Google Scholar]
- Blanck, H.M. , Bowman, B.A. , Serdula, M.K. , Khan, L.K. , Kohn, W. , and Woodruff, B.A. (2002) Angular stomatitis and riboflavin status among adolescent Bhutanese refugees living in southeastern Nepal. Am J Clin Nutr 76: 430–435. [DOI] [PubMed] [Google Scholar]
- Bodiga, V.L. , Bodiga, S. , Surampudi, S. , Boindala, S. , Putcha, U. , Nagalla, B. , et al (2012) Effect of vitamin supplementation on cisplatin‐induced intestinal epithelial cell apoptosis in Wistar/NIN rats. Nutrition 28: 572–580. [DOI] [PubMed] [Google Scholar]
- Burgess, C. , O'Connell‐Motherway, M. , Sybesma, W. , Hugenholtz, J. , and van Sinderen, D. (2004) Riboflavin production in Lactococcus lactis: potential for in situ production of vitamin‐enriched foods. Appl Environ Microbiol 70: 5769–5777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgess, C.M. , Smid, E.J. , Rutten, G. , and van Sinderen, D. (2006) A general method for selection of riboflavin‐overproducing food grade micro‐organisms. Microb Cell Fact 5: 24. doi:10.1186/1475‐2859‐5‐24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgess, C.M. , Smid, E.J. , and van Sinderen, D. (2009) Bacterial vitamin B2, B11 and B12 overproduction: an overview. Int J Food Microbiol 133: 1–7. [DOI] [PubMed] [Google Scholar]
- Capozzi, V. , Menga, V. , Digesu, A.M. , De Vita, P. , van Sinderen, D. , Cattivelli, L. , et al (2011) Biotechnological production of vitamin B2‐enriched bread and pasta. J Agr Food Chem 59: 8013–8020. [DOI] [PubMed] [Google Scholar]
- Capozzi, V. , Russo, P. , Duenas, M.T. , Lopez, P. , and Spano, G. (2012) Lactic acid bacteria producing B‐group vitamins: a great potential for functional cereals products. Appl Microbiol Biotechnol 96: 1383–1394. [DOI] [PubMed] [Google Scholar]
- Combs, G.F. (1992) The Vitamins: Fundamental Aspects in Nutrition and Health. San Diego, CA, USA: Academic Press. [Google Scholar]
- Cooperman, J.M. , and Lopez, R. (1991) Riboflavin In Handbook of Vitamins. Machlin L.J. (ed.). New York, NY, USA: Marcel Dekker. [Google Scholar]
- Coquard, D.1. , Huecas, M. , Ott, M. , van Dijl, J.M. , van Loon, A.P. , and Hohmann, H.P. (1997) Molecular cloning and characterisation of the ribC gene from Bacillus subtilis: a point mutation in ribC results in riboflavin overproduction. Mol Gen Genet 54: 81–84. [DOI] [PubMed] [Google Scholar]
- Corbett, A.J. , Eckle, S.B. , Birkinshaw, R.W. , Liu, L. , Patel, O. , Mahony, J. , et al (2014) T‐cell activation by transitory neo‐antigens derived from distinct microbial pathways. Nature 509: 361–365. [DOI] [PubMed] [Google Scholar]
- Fischer, M. , and Bacher, A. (2005) Biosynthesis of flavocoenzymes. Nat Prod Rep 22: 324–350. [DOI] [PubMed] [Google Scholar]
- Flynn, A. , Moreiras, O. , Stehle, P. , Fletcher, R.J. , Muller, D.J. , and Rolland, V. (2003) Vitamins and minerals: a model for safe addition to foods. Eur J Nutr 42: 118–130. [DOI] [PubMed] [Google Scholar]
- Foley, A.R. , Menezes, M.P. , Pandraud, A. , Gonzalez, M.A. , Al‐Odaib, A. , Abrams, A.J. , et al (2014) Treatable childhood neuronopathy caused by mutations in riboflavin transporter RFVT2. Brain 137: 44–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Food and Nutrition Board (1998) Riboflavin. Dietary Reference Intakes: Thiamin, Riboflavin, Niacin, Vitamin B6, Vitamin B12, Pantothenic Acid, Biotin, Folate and Choline. Washington DC, USA: National Academies Press, pp. 87–122. [PubMed] [Google Scholar]
- Gelfand, M.S. , Mironov, A.A. , Jomantas, J. , Kozlov, Y.I. , and Perumov, D.A. (1999) A conserved RNA structure element involved in the regulation of bacterial riboflavin synthesis genes. Trends Genet 15: 439–442. [DOI] [PubMed] [Google Scholar]
- Guru, V. , and Viswanathan, K. (2013) Riboflavin production in milk whey using probiotic bacteria – Lactobacillus acidophilus and Lactococcus lactis . Ind J Fund Appl Life Sci 3: 169–176. [Google Scholar]
- Hassan, I. , Chibber, S. , and Naseem, I. (2013) Vitamin B2: a promising adjuvant in cisplatin based chemoradiotherapy by cellular redox management. Food Chem Toxicol 59: 715–723. [DOI] [PubMed] [Google Scholar]
- Hill, D.I. , and Nalubola, R. (2002) Fortification strategies to meet micronutrient needs: successes and failures. Proc Nutr Soc 61: 231–241. [DOI] [PubMed] [Google Scholar]
- Hill, M.J. (1997) Intestinal flora and endogenous vitamin synthesis. Eur J Cancer Prev 6: S43–S45. [DOI] [PubMed] [Google Scholar]
- Hugenholtz, J. , Sybesma, W. , Groot, M.N. , Wisselink, W. , Ladero, V. , Burgess, K. , et al (2002) Metabolic engineering of lactic acid bacteria for the production of nutraceuticals. Antonie Van Leeuwenhoek 82: 217–235. [PubMed] [Google Scholar]
- Ichihashi, T. , Takagishi, Y. , Uchida, K. , and Yamada, H. (1992) Colonic absorption of menaquinone‐4 and menaquinone‐9 in rats. J Nutr 122: 506–512. [DOI] [PubMed] [Google Scholar]
- Jayashree, S. , Jayaraman, K. , and Kalaichelvan, G. (2010) Isolation, screening and characterization of riboflavin producing lactic acid bacteria from Katpadi, Vellore district. Recent Res Sci Technol 2: 83–88. [Google Scholar]
- Jayashree, S. , Rajendhran, J. , Jayaraman, K. , Kalaichelvana, G. , and Gunasekaran, P. (2011) Improvement of riboflavin production by Lactobacillus fermentum isolated from yogurt. Food Biotechnol 25: 240–251. [Google Scholar]
- Kleerebezem, M. , Boekhorst, J. , van Kranenburg, R. , Molenaar, D. , Kuipers, O.P. , Leer, R. , et al (2003) Complete genome sequence of Lactobacillus plantarum WCFS1. Proc Natl Acad Sci 100: 1990–1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koser, S.A. (1968) Vitamin Requirements of Bacteria and Yeasts. Springfield, IL, USA: Charles C. Thomas. [Google Scholar]
- Laino, J.E. , Juarez del Valle, M. , Savoy de Giori, G. , and LeBlanc, J.G.J. (2012) Development of a high folate concentration yogurt naturally bio‐enriched using selected lactic acid bacteria. LWT Food Sci Technol 54: 1–5. [Google Scholar]
- LeBlanc, J.G. , Burgess, C. , Sesma, F. , Savoy de Giori, G. , and van Sinderen, D. (2005a) Ingestion of milk fermented by genetically modified Lactococcus lactis improves the riboflavin status of deficient rats. J Dairy Sci 88: 3435–3442. [DOI] [PubMed] [Google Scholar]
- LeBlanc, J.G. , Burgess, C. , Sesma, F. , de Giori, G.S. , and van Sinderen, D. (2005b) Lactococcus lactis is capable of improving the riboflavin status in deficient rats. Br J Nutr 94: 262–267. [DOI] [PubMed] [Google Scholar]
- LeBlanc, J.G. , Rutten, G. , Bruinenberg, P. , Sesma, F. , de Giori, G.S. , and Smid, E.J. (2006) A novel product fermented with Propionibacterium freudenreichii improves the riboflavin status of deficient rats. Nutrition 22: 645–651. [DOI] [PubMed] [Google Scholar]
- LeBlanc, J.G. , Laino, J.E. , del Vall, M.J. , Vannini, V. , van Sinderen, D. , Taranto, M.P. , et al (2011) B‐group vitamin production by lactic acid bacteria – current knowledge and potential applications. J Appl Microbiol 111: 1297–1309. [DOI] [PubMed] [Google Scholar]
- LeBlanc, J.G. , Milani, C. , de Giori, G.S. , Sesma, F. , van Sinderen, D. , and Ventura, M. (2012) Bacteria as vitamin suppliers to their host: a gut microbiota perspective. Curr Opinion Biotechnol 24: 160–168. [DOI] [PubMed] [Google Scholar]
- Magnusdottir, S. , Ravcheev, D. , de Crecy‐Lagard, V. , and Thiele, I. (2015) Systematic genome assessment of B‐vitamin biosynthesis suggests co‐operation among gut microbes. Front Genet 6: 148. doi:10.3389/fgene.2015.00148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mal, P. , Dutta, K. , Bandyopadhyay, D. , Basu, A. , Khan, R. , and Bishayi, B. (2013) Azithromycin in combination with riboflavin decreases the severity of Staphylococcus aureus infection induced septic arthritis by modulating the production of free radicals and endogenous cytokines. Inflammat Res 62: 259–273. [DOI] [PubMed] [Google Scholar]
- Marques, T.M. , Wall, R. , Ross, R.P. , Fitzgerald, G.F. , Ryan, C.A. , and Stanton, C. (2010) Programming infant gut microbiota: influence of dietary and environmental factors. Curr Opinion Biotechnol 21: 149–156. [DOI] [PubMed] [Google Scholar]
- Massey, V. (2000) The chemical and biological versatility of riboflavin. Biochem Soc T 28: 283–296. [PubMed] [Google Scholar]
- Mensink, G.B. , Fletcher, R. , Gurinovic, M. , Huybrechts, I. , Lafay, L. , Serra‐Majem, L. , et al (2013) Mapping low intake of micronutrients across Europe. Br J Nutr 110: 755–773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milani, C. , Lugli, G.A. , Duranti, S. , Turroni, F. , Bottacini, F. , Mangifesta, M. , et al (2014) Genomic encyclopedia of type strains of the genus Bifidobacterium . Appl Environ Microbiol 80: 6290–6302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moat, S.J. , Ashfield‐Watt, P.A. , Powers, H.J. , Newcombe, R.G. , and McDowell, I.F. (2003) Effect of riboflavin status on the homocysteine‐lowering effect of folate in relation to the MTHFR (C677T) genotype. Clin Chem 49: 295–302. [DOI] [PubMed] [Google Scholar]
- O'Brien, M.M. , Kiely, M. , Harrington, K.E. , Robson, P.J. , Strain, J.J. , and Flynn, A. (2001) The North/South Ireland Food Consumption Survey: vitamin intakes in 18–64‐year‐old adults. Public Health Nutr 4: 1069–1079. [DOI] [PubMed] [Google Scholar]
- Perkins, J. , and Pero, J. (2002) Biosynthesis of riboflavin, biotin, folic acid, and cobalamin In Bacillus subtilis and Its Closest Relatives: From Genes to Cells. Sonenshine A., Hoch J., and Losick R. (eds). Washington, DC, USA: ASM Press, pp. 271–276. [Google Scholar]
- Perkins, J.B. , Pero, J.G. , and Sloma, A. (1991) Riboflavin overproducing strains of bacteria. European patent application 0405370.
- Perkins, J.B. , Sloma, A. , Hermann, T. , Theriault, K. , Zachgo, E. , Erdenberger, T. , et al (1999) Genetic engineering of Bacillus subtilis for the commercial production of riboflavin. J Ind Microbiol Biot 22: 8–18. [Google Scholar]
- Pophaly, S.D. , Poonam, Singh, P. , Kumar, H. , Tomar, S.K. , and Singh, R. (2014) Selenium enrichment of lactic acid bacteria and bifidobacteria: a functional food perspective. Trends food sci tech 39: 135–145. [Google Scholar]
- Powers, H.J. (2003) Riboflavin (vitamin B2) and health. Am J Clin Nutr 77: 1352–1360. [DOI] [PubMed] [Google Scholar]
- Powers, H.J. , Weaver, L.T. , Austin, S. , and Beresford, J.K. (1993) A proposed intestinal mechanism for the effect of riboflavin deficiency on iron loss in the rat. Brit J Nutr 69: 553–561. [DOI] [PubMed] [Google Scholar]
- Richter, G. , Volk, R. , Krieger, C. , Lahm, H.W. , Rothlisberger, U. , and Bacher, A. (1992) Biosynthesis of riboflavin: cloning, sequencing, and expression of the gene coding for 3,4‐dihydroxy‐2‐butanone 4‐phosphate of Escherichia coli . J Bacteriol 174: 4050–4056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter, G. , Ritz, H. , Katzenmeier, G. , Volk, R. , Kohnle, A. , Lottspeich, F. , et al (1993) Biosynthesis of riboflavin: cloning, sequencing, and mapping, and expression of the gene coding for GTP cyclohydrolase II of Escherichia coli . J Bacteriol 175: 4045–4051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter, G. , Fischer, M. , Krieger, C. , Eberhardt, S. , Lüttgen, H. , Gerstenschläger, I. , and Bacher, A. (1997) Biosynthesis of riboflavin: characterization of the bifunctional deaminase‐reductase of Escherichia coli and Bacillus subtilis . J Bacteriol 179: 2022–2028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohner, F. , Zimmermann, M.B. , Wegmueller, R. , Tschannen, A.B. , and Hurrell, R.F. (2007) Mild riboflavin deficiency is highly prevalent in school‐age children but does not increase risk for anaemia in Cote d'Ivoire. Brit J Nutr 97: 970–976. [DOI] [PubMed] [Google Scholar]
- Russo, P. , Lopez, P. , Capozzi, V. , de Palencia, P.F. , Duenas, M.T. , Spano, G. , and Fiocco, D. (2012) Beta‐glucans improve growth, viability and colonization of probiotic microorganisms. Int J Mol Sci 13: 6026–6039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russo, P. , Capozzi, V. , Arena, M.P. , Spadaccino, G. , Duenas, M.T. , Lopez, P. , et al (2014) Riboflavin‐overproducing strains of Lactobacillus fermentum for riboflavin‐enriched bread. Appl Microbiol Biotechnol 98: 3691–3700. [DOI] [PubMed] [Google Scholar]
- Russo, P. , Iturria, I. , Mohedano, M.L. , Caggianiello, G. , Rainieri, S. , Fiocco, D. , et al (2015) Zebrafish gut colonization by mCherry‐labelled lactic acid bacteria. Appl Microbiol Biotechnol 99: 3479–3490. [DOI] [PubMed] [Google Scholar]
- Said, H.M. , and Mohammed, Z.M. (2006) Intestinal absorption of water soluble vitamins: an update. Curr Opin Gastroenterol 22: 140–146. [DOI] [PubMed] [Google Scholar]
- Saini, K. , Tomar, S.K. , Sangwan, V. , and Bhushan, B. (2014) Evaluation of Lactobacilli from human sources for uptake and accumulation of selenium. Biol Trace Elem Res 160: 433–436. [DOI] [PubMed] [Google Scholar]
- Salvetti, S. , Celandroni, F. , Ghelardi, E. , Baggiani, A. , and Senesi, S. (2003) Rapid determination of vitamin B2 secretion by bacteria growing on solid media. J App Microbiol 95: 1255–1260. [DOI] [PubMed] [Google Scholar]
- Schallmey, M. , Singh, A. , and Ward, O.P. (2004) Developments in the use of Bacillus species for industrial production. Can J Microbiol 50: 1–17. [DOI] [PubMed] [Google Scholar]
- Schetzek, S. , Heinen, F. , Kruse, S. , Borggraefe, I. , Bonfert, M. , Gaul, C. , et al (2013) Headache in children: update on complementary treatments. Neuropediatrics 44: 25–33. [DOI] [PubMed] [Google Scholar]
- Sherwood, M.D. , Ran, D. , and Goldman, M.D. (2014) Effectiveness of riboflavin in pediatric migraine prevention. Can Fam Physician mars 60: 157–159. [PMC free article] [PubMed] [Google Scholar]
- Shi, S. , Shen, Z. , Chen, X. , Chen, T. , and Zhao, X. (2009) Increased production of riboflavin by metabolic engineering of the purine pathway in Bacillus subtilis . Biochem Eng J 46: 28–33. [Google Scholar]
- Shi, Z. , Zhen, S. , Wittert, G.A. , Yuan, B. , Zuo, H. , and Taylor, A.W. (2014) Inadequate riboflavin intake and anemia risk in a Chinese population: five‐year follow up of the Jiangsu Nutrition Study. PLoS ONE 9: e88862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snell, E.E. , and Strong, F.M. (1939) A microbiological assay for riboflavin. Ind Eng Chem, Anal ed 11: 346–350. [Google Scholar]
- Stahmann, K.P. , Revuelta, J.L. , and Seulberger, H. (2000) Three biotechnical processes using Ashbya gossypii, Candida famata, or Bacillus subtilis compete with chemical riboflavin production. Appl Microbiol Biotechnol 53: 509–516. [DOI] [PubMed] [Google Scholar]
- Stanton, C. , Ross, R.P. , Fitzgerald, G.F. , and Van Sinderen, D. (2005) Fermented functional foods based on probiotics and their biogenic metabolites. Curr Opinion Biotechnol 16: 198–203. [DOI] [PubMed] [Google Scholar]
- Sybesma, W. , Burgess, C. , Starrenburg, M. , van Sinderen, D. , and Hugenholtz, J. (2004) Multivitamin production in Lactococcus lactis using metabolic engineering. Metab Eng 6: 109–115. [DOI] [PubMed] [Google Scholar]
- Sybesma, W. , Hugenholtz, J. , de Vos, W. , and Smid, E.J. (2006) Safe use of genetically modified lactic acid bacteria in food: bridging the gap between consumers, green groups, and industry. Elect J Biotechnol 9: 424–448. [Google Scholar]
- Tamime, D.A. , Tamime, A.Y. , Saarela, M. , Sondergaard, A.K. , Mistry, V.V. , and Shah, N.P. (2005) Production and maintenance of viability of probiotic micro‐organisms in dairy products In Probiotic Dairy Products.UK. Tamine A.Y. (ed.). Oxford, UK: Blackwell Publishing, pp. 39–72. [Google Scholar]
- Thakur, K. , and Tomar, S.K. (2015) Exploring indigenous Lactobacillus species from diverse niches for riboflavin production. J Young Pharmacists 7: 122–127. [Google Scholar]
- Thakur, K. , Gosh, S. , Kadam, J. , and Tomar, S.K. (2015) Adhesion properties of riboflavin producing Lactobacillus mucosae isolated from human faeces. J Cell Tissue Res 15: 5095–5100. [Google Scholar]
- Vaesken, S.M. , Aperte, A.E. , and Moreiras, V.G. (2012) Vitamin food fortification today. Food Nutr Res 56: 5459. doi:10.3402/fnr.v56i0.5459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valle, J.D.M. , Lainoa, J.E.G. , Savoy, D.G.G. , and LeBlanc, J.G. (2014) Riboflavin producing lactic acid bacteria as a biotechnological strategy to obtain bio‐enriched soymilk. Food Res Int 62: 1015–1019. [Google Scholar]
- Van Loon, A.P.G.M. , Hohmann, H.P. , and Bretzel, W.M. (1996) Development of a fermentation process for the manufacture of riboflavin. Chimia (Aarau) 50: 410–412. [Google Scholar]
- Ventura, M. , Canchaya, C. , Fitzgerald, G.F. , Gupta, R.S. , and van Sinderen, D. (2007) Genomics as a means to understand bacterial phylogeny and ecological adaptation: the case of bifidobacteria. Antonie Van Leeuwenhoek 91: 351–372. [DOI] [PubMed] [Google Scholar]
- Vitreschak, A.G. , Rodionov, D.A. , Mironov, A.A. , and Gelfand, M.S. (2002) Regulation of riboflavin biosynthesis and transport genes in bacteria by transcriptional and translational attenuation. Nucleic Acids Res 30: 3141–3151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wacker, J. , Fruhauf, J. , Schulz, M. , Chiwora, F.M. , Volz, J. , and Becker, K. (2000) Riboflavin deficiency and preeclampsia. Obstet Gynecol 96: 38–44. [DOI] [PubMed] [Google Scholar]
- Wels, M. , Francke, C. , Kerkhoven, R. , Kleerebezem, M. , and Siezen, R.J. (2006) Predicting cisacting elements of Lactobacillus plantarum by comparative genomics with different taxonomic subgroups. Nucleic Acids Res 34: 1947–1958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson, J.A. (1983) Disorders of vitamins: deficiency, excess and errors of metabolism In Harrison's Principles of Internal Medicine. Petersdorf R.G., and Harrison T.R. (eds). New York, NY, USA: McGraw‐Hill Book, pp. 461–470. [Google Scholar]
- Wrong, O.M. , Edmonds, C.J. , and Chadwich, V.S. (1981) The Large Intestine; its Role in Mammalian Nutrition and Homeostasis. New York, NY, USA: Wiley & Sons. [Google Scholar]


