Abstract
Recent studies have shown that the range of affinities of T cell receptors (TCRs) against non-mutated cancer peptide/class I complexes are lower than TCR affinities for foreign antigens. Raising the affinity of TCRs for optimal activity of CD8 T cells, and for recruitment of CD4 T cell activity against a class I antigen, provides opportunities for more robust adoptive T cell therapies. However, TCRs with enhanced affinities also risk increased reactivity with structurally related self-peptides, and off-target toxicities. Careful selection of tumor peptide antigens, in silico proteome screens, and in vitro peptide specificity assays will be important in the development of the most effective, safe TCR-based adoptive therapies.
Introduction
Most of the reported tumor-associated epitopes for CD8+ T cells represent self-peptides presented by class I products [1]. While an immune response to upregulated self-antigens can be generated, the process of central tolerance in the thymus has evolved to eliminate T cells that express TCRs that react too strongly with these pepMHC [2,3]. The absence of peripheral T cells that might have reacted with a specific antigen has been referred to as the “hole in the repertoire”. Recent studies with viral antigens, including HIV, raised the question of whether the collection of self-peptides that have structural homology to viral peptides might operate during negative selection to diminish the response to such foreign antigens [4,5]. When the target antigen is a self-peptide, a “hole in the repertoire” is even more likely. The ability to manipulate the affinities of TCRs, and to introduce TCRs into T cells for adoptive T cell therapies provides opportunities to overcome these limitations associated with negative selection against potential self-cancer antigens. Here we discuss the issues associated with raising the affinities of TCRs in order to drive robust T cell activity, and the potential risks this involves with TCR cross-reactivities and toxicities.
TCR affinities for self-protein epitopes
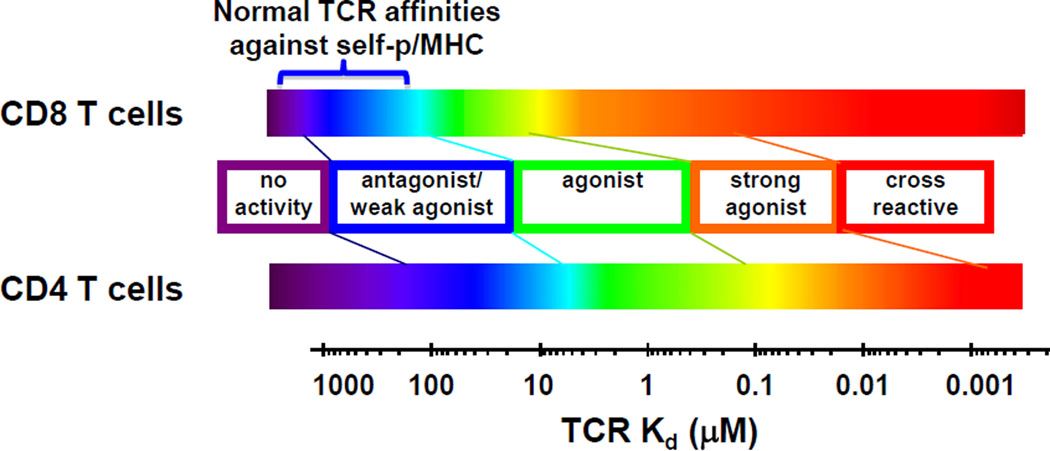
Although some peripheral T cells against tumor-associated self-peptides escape negative selection, it has been shown that their TCRs exhibit lower average affinities than typical foreign antigens (mean Kd values of about 100 µM, compared to mean Kd values of about 10 µM for viral antigens)(Fig. 1) [6]. Nevertheless, naturally occurring TCRs that bind to tumor-associated pepMHC can mediate some clinical responses when introduced in a gene-modified, adoptive transfer setting [7]. TCRs with even modestly higher affinity can yield significant increases in potency [8], as it is believed that the affinities of wild-type TCRs for self-pepMHC tumor-associated epitopes may be too low for optimal CD8+ T cell activity. Importantly, these wild-type TCRs do not mediate CD4 T cell activity [9–11], due to their absolute requirement for CD8 [9–15] (Fig. 1).
Figure 1. Relationship between TCR affinity and T cell activity for CD8 and CD4 T cells.
TCR affinity for class I pep/MHC impacts T cell activity for CD8 (top) and CD4 (bottom) T cells. CD8 T cells are more sensitive at weaker affinities, due to the contribution of the CD8 co-receptor to class I MHC binding, regardless of bound peptide. At higher affinity ranges, peptides that are structurally similar to the cognate peptide (tumor antigen in the present review) are capable of functionally cross-reacting with the T cell.
The importance of CD4 T cell activity in anti-tumor immunity has been established, but the ability to use transduced class I-restricted TCRs in establishing long-term CD4 T cell immunity remains to be shown. Nevertheless, re-direction of CD4+ T cell effector activities against class I-restricted epitopes has been shown [12,14–16]. Higher affinity TCRs (e.g. KD values of 1 µM or less) are required to mediate activation in CD4+ T cells as compared to CD8+ T cells, where CD8 can co-engage the class I MHC molecule. Effector CD4+ T cells with higher affinity TCRs have mediated significant tumor control, and in some cases rejections, in mouse models [11,17–19]. TCRs with increased affinity against MART1/HLA-A2 [8], NY-ESO-1/HLA-A2 [20], MAGE-A3/HLA-A2 [21], and MAGE-A3/HLA-A1 [22], and ability to mediate in vitro CD4+ T cell activity, have been evaluated in adoptive T cell transfer studies. While good persistence of TCR+CD4+ T cells has been reported in some patients [8,20], the in vivo activity of the CD4+ T cells bearing these receptors has not been fully characterized; thus, the extent of anti-tumor response, or even reactivity against normal tissues, that are mediated by CD4+ compared to CD8+ T cells is not clear. In summary, it remains to be determined if CD4+ T cells with a class I-restricted T cells will persist, and/or be capable of recruiting the activity of CD8+ T cells against other potential tumor antigens.
Despite their low affinity, wild-type TCRs can serve as leads to engineer mutations that confer higher affinity. For modest increases in affinity (probably only a few fold), individual site-directed mutations can be screened for T cell activity [14] or TCRs can be derived from allogeneic or xenogenic T cell clones [21,23,24]. For more significant increases in affinity, directed evolution strategies including yeast [25,26] or phage display [27] have been employed. As the TCRs generated by any of these strategies have not been through negative selection, appropriate screening strategies are necessary to assess specificity. This is especially important since CD8 contributes significantly to the ability of T cells to recognize class I complexes, even if the affinity of the TCR for a structurally similar self-peptide is as low as 300 µM [6,28].
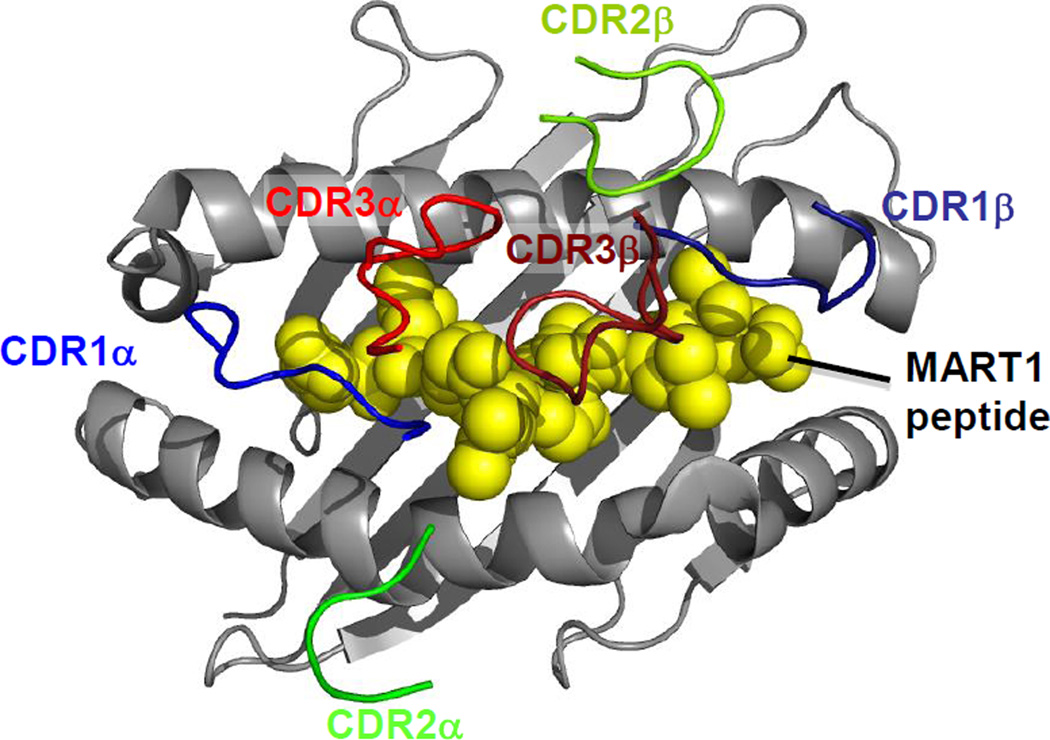
The conserved docking orientation of TCRs on the pepMHC ligand [29,30] (Fig. 2) suggests that peptide specificity might best be retained by mutating CDR3 loops, which typically dock over the peptide. In contrast, CDR2 loops dock over the MHC helices and it is possible that mutations in these residues could promote non-peptide associated increases in binding affinity, and thus reduced peptide specificity. While these concepts are attractive in principle, in practice the binding of a TCR is complex, and the different CDR loops often act cooperatively. For example, CDR3 residues can engage MHC, either directly or through their action on other CDR loops. Furthermore, CDR1α often contacts the N-termini of peptides [31–33]), and thus it could be argued the CDR1α residues have the same potential to confer peptide specificity as CDR3 loops. While there are no strict rules for engineering, avoiding affinity-enhancing modifications of CDR2 loops (Fig. 2) may be wise (e.g. although anecdotal, two patients treated with T cells expressing a CDR2-modified, MAGE-A3/HLA-A1-targeted TCR exhibited lethal effects apparently due to off-tumor off-target cross-reaction with a structurally-related titin peptide [22]).
Figure 2. Conserved TCR docking onto pepMHC.
CDR loops of the DMF5 TCR (PDB ID=3QDG) are shown binding to MART1/HLA-A2 as an example of the highly conserved docking geometry observed between TCRs and MHCs. HLA-A2 is shown in gray; MART1 peptide is shown in yellow. CDR3 loops (CDR3α in red, CDR3β in brick red) dock predominately over peptide, while CDR2 loops (CDR2α in green, CDR3β in lime green) predominately contact MHC. CDR1 loops (CDR1α in blue, CDR1β in navy) can contact both the MHC and the peptide; the CDR1α loop in particular frequently makes specific contacts with the N-terminus of the peptide.
Even when affinity mutations are limited to TCR regions that are predicted to contact the peptide, off-target cross-reactivity was observed in the first high-affinity TCR engineered in a mouse system [16], and in a more recent human TCR against the MAGE-A3/HLA-A2 complex, which cross-reacted with a peptide from MAGE-A12 [34]. To reduce affinities of engineered TCRs to an optimal level for recruitment of both CD8 and CD4 T cell activity, but minimizing the risk of peptide cross-reactivity, we have shown that key CDR2 residues that make energetically important contacts with MHC residues [35] can be mutated to easily generate a panel of TCRs with a range of reduced affinities [36,37].
Thresholds for efficacy and toxicity for on-target reactions
T cells with wild-type affinity TCRs can be extremely sensitive to foreign pepMHC, requiring only a few pepMHC complexes to initiate T cell triggering [38–40]. As indicated earlier, this sensitivity for a class I antigen is in part due to TCR synergy with the co-receptor CD8. The optimal TCR affinity range for safety and efficacy in adoptive therapies appears to depend to some extent on both the TCR and the target, but there are some general rules to follow based on a limited set of mouse and human studies.
Mouse studies
To study aspects of HLA-A2-restricted, tumor-associated epitopes in vivo, mice that express transgenic chimeric human-mouse class I MHC molecules, either A2-Kb [41] or AAD [42] have been used. Chimeric A2-Kb and AAD molecules both consist of human HLA-A2 α1 and α2 domains fused to mouse class I α3 domains from H2-Kb or H2-Db, respectively. The mouse α3 domain allows binding of mouse CD8 and thus the full recognition and activation properties of mouse CD8+ T cells. The chimeric molecules have been shown to present human HLA-A2-restricted epitopes, to bind to mouse CD8, and to elicit CTL responses in transgenic mice, allowing in vivo studies of responses to the HLA-A2-restrcited peptides [41,42].
A panel of gp100/HLA-A2-specific TCRs from vaccinated melanoma patients, spanning a Kd range from 1–100 µM were transduced into CD8+ T cells from HLA-A2-Kb (α3 domain) transgenic mice [41,43]. The A2-Kb strain allows mouse CD8 to engage the class I HLA-A2/Kb molecule. When these T cells were used to treat tumors in A2-Kb mice, both control of tumor and ocular autoimmunity was enhanced with increasing TCR affinity, plateauing at a Kd value of 10 µM [43]. Unfortunately, the same TCRs were not tested in CD4 T cells to determine if the same higher affinity TCRs would mediate improved activity without the ocular effects.
Tyrosinase-specific CTL lines [44,45] and TCR-transgenic mice [46,47] were generated to study efficacy and safety. The high-avidity TCR called FH [42] recognizes human and mouse versions of a tyrosinase368–377 (which vary by a single Y to F mutation at the N-terminus) in the context of the human/mouse chimeric AAD MHC [41,42]. While transfer of the tyrosinase-specific CTL lines delayed the outgrowth of tyrosinase+ AAD+ tumors [45], on-target/off-tumor vitiligo caused by CD8 T cells was observed, and this was accelerated by depletion of CD25+ CD4+ T cells [44,46]. While the precise affinity of the FH TCR is not known, like the gp100 system enhanced binding properties that conferred good activity were correlated with more widespread on-target/off-tumor responses. By contrast, two TCRs that bound with enhanced affinity and conferred high functional avidity to a WT1 peptide presented by Db did not shown signs of toxicity in vivo [48], despite reports that there are low levels of WT1 protein expression in lung and kidney tissues [49]. Hence, determination of an affinity threshold for on-target/off-tumor response is influenced by the distribution and no doubt expression levels of the target peptide.
Human studies
TCRs with various affinities have been used in human adoptive T cell trials with variable efficacy and safety. Notably, T cells expressing the DMF4 (Kd 170 µM) and DMF5 (Kd 40 µM) TCRs against MART-1/HLA-A2 have been used clinically to treat melanoma patients [7,8,31]. The higher affinity DMF5 TCR showed more robust in vitro and clinical antitumor response, but coincident with DMF5's improved anti-tumor response were toxicities due to on-target/off-tumor activity against MART-1 expressed in the eye, ear and skin [8]. Another anti-cancer TCR targeting the cancer-testis antigen NY-ESO-1 used a TCR (1G4-α95:LY) that had a 50-fold higher affinity for its target (Kd=730 nM) than the DMF5 TCR [14]. To date, no on-target/off-tumor effects have been reported in this trial [20]. The NY-ESO-1 antigen has a very restricted expression profile and is thought to be largely absent in normal adult tissues outside of testis. Again, the potential for on-target/off-tumor responses will rely heavily on the selection of target epitope.
Cross-reactivities mediating off-target toxicities
Relationships of TCR affinity and peptide cross-reactivity
In addition to on-target/off-tumor toxicities, TCR candidates for adoptive T cell therapies can exhibit off-target responses as described for the MAGE-A3 systems [22,34]. Both on-target and off-target activities are dependent on the potency of the T cell response, with higher affinity class I-restricted TCRs having the potential to mediate optimal CD8 T cell activity and also CD4 T cell activity. Off-target responses are impacted directly by TCR affinity due to the low affinity threshold necessary to mediate CD8-dependent T cell activity [28]. For example, as affinity for the tumor peptide is increased, affinities for structurally related peptides can also increase [16]. If such structurally related peptides are presented by the class I product on normal tissue at sufficient levels, there is the risk of off-target toxicities.
The issue of peptide cross-reactions by normal TCRs has long been considered important to allow responses to the diverse repertoire of potential epitopes [50,51]. It has been suggested that this cross-reactivity is due in part to the conformational plasticity of the CDRs of a TCR, combined with the low affinities required for CD8 T cell activity. Increasing the affinity of a TCR by mutation of the CDRs can actually reduce this conformational flexibility, and thus can reduce cross-reactivity with structurally unrelated peptides [52]. However, increasing the affinity of the TCR enhances the affinity for structurally related self-peptide variants, as first reported with a nanomolar affinity-matured mouse TCR 2C [16]. It is this feature that is associated with a corresponding increased risk of activation by structurally related self-peptides [53]. Such off-target reactivity with structurally related self-peptides has been seen with engineered human TCRs [15,21] and led to the serious adverse effects in clinical adoptive T cell transfers [22,34].
In summary, the low TCR affinity threshold of CD8-dependent T cell activity dictates that extensive pre-clinical peptide analyses be performed with TCR candidates. The analyses should include activity screening against a battery of peptides and tissues: cognate peptide variants, proteome searches for similar peptides (see below), and tissue-representative cell lines [9,34,54]. Eventually it may be possible to include all peptides that have been validated as members of the immunopeptideome for a class I allele [55].
Proteome searches
Strategies to predict and screen potential self-peptides are crucial in the testing of TCRs that have optimal potential in adoptive T cell therapies. To err on the side of identifying such cross-reactivity, it may be useful to examine a panel of related TCRs with a range of affinities, including those with affinities even higher than the proposed final TCR candidate. As described above, TCR affinity can be easily reduced by mutating conserved CDR2 residues for the eventual clinical candidates [36,37].
Retrospective in silico proteome scanning based on the known reactivity of the MAGE-A3/HLA-A1-specific TCR revealed a peptide from titin that was recognized in vitro and, likely, in vivo [22,54]. In a prospective effort, we performed an in-depth, in silico analysis of over fifty tumor-associated, HLA-A2-restricted, 9-mer peptides from the Cancer Immunity Peptides Database (http://cancerimmunity/org/peptide/). To illustrate the analysis, Table 1 presents features of 19 selected tumor-associated peptides that are identical in humans and mice and that were isolated originally as HLA-A2 restricted antigens. Since MHC binding has been shown to be a critical parameter in judging the fitness of a T cell target peptide [56], each peptide was evaluated for predicted MHC binding via five algorithms (Table 1). The results showed general agreement among the HLA-A2 binding algorithms.
Table 1.
Binding predictions and frequency of similar peptides for tumor-associated epitopes.a
| Tumor Associated Protein |
peptide sequenceb |
SYFPEITHIc | BIMASd half-life (s) |
ANN bindinge IC50 (nM) |
SMM bindingf IC50 (nM) |
NetMHCpang (nM) |
Compiled binding prediction scoreh |
# unique binding peptide matchesi |
% peptide matches in mousej |
|---|---|---|---|---|---|---|---|---|---|
| glypican-3 | FVGEFFTDV | 18 | 830 | 30 | 80 | 40 | 6.5 | 3 | 67 |
| TRP-2 | SVYDFFVWL | 21 | 970 | 20 | 60 | 20 | 6.5 | 4 | 75 |
| RAB38 / NY-MEL-1 | VLHWDPETV | 23 | 60 | 50 | 110 | 100 | 5 | 4 | 75 |
| EZH2 | FMVEDETVL | 21 | 120 | 20 | 40 | 20 | 6 | 9 | 44 |
| cyclin-A1 | FLDRFLSCM | 20 | 80 | 40 | 70 | 40 | 5.5 | 11 | 82 |
| HER-2/neu | RLLQETELV | 24 | 130 | 30 | 80 | 40 | 6 | 11 | 55 |
| HER-2/neu | TLEEITGYL | 23 | 4 | 220 | 430 | 230 | 2.5 | 12 | 42 |
| KIF20A | CIAEQYHTV | 25 | 60 | 100 | 200 | 80 | 5 | 30 | 57 |
| WT1 | CMTWNQMNL | 17 | 20 | 650 | 480 | 400 | 2 | 35 | 34 |
| HER-2/neu | HLYQGCQVV | 23 | 3 | 110 | 130 | 100 | 4 | 41 | 34 |
| HER-2/neu | KIFGSLAFL | 28 | 480 | 10 | 60 | 20 | 6.5 | 42 | 38 |
| gp100 | ITDQVPFSV | 18 | 4 | 70 | 150 | 200 | 3.5 | 48 | 44 |
| WT1 | RMFPNAPYL | 22 | 320 | 7 | 30 | 10 | 6.5 | 50 | 56 |
| ENAH (hMena) | TMNGSKSPV | 21 | 50 | 280 | 290 | 430 | 3 | 55 | 47 |
| Proteinase3 (PR1) | VLQELNVTV | 28 | 490 | 10 | 30 | 10 | 7.5 | 82 | 44 |
| CPSF | KVHPVIWSL | 23 | 110 | 50 | 130 | 60 | 4.5 | 86 | 57 |
| cyclin D1 | LLGATCMFV | 22 | 650 | 20 | 30 | 20 | 7 | 266 | 37 |
| PAX5 | TLPGYPPHV | 24 | 70 | 20 | 40 | 10 | 6 | 341 | 37 |
| PAP | TLMSAMTNL | 21 | 180 | 10 | 60 | 10 | 6 | 818 | 40 |
Shaded cells indicate the strength of HLA-A2 binding predicted for each algorithm: Strong binding = orange; Moderate binding = gold; weak binding = pale yellow; predicted not to bind = no shading.
Selected 9-mer tumor-associated, HLA-A2-restricted peptides from the Cancer Immunity Peptide Database (http://cancerimmunity.org/peptide/).
SYFPEITHI binding scores from online server (http://www.syfpeithi.de/).
BIMAS binding predictions from BioInformatics and Molecular Analysis Section (NIH) online server (http://www-bimas.cit.nih.gov/molbio/hla_bind/).
ANN binding predictions from the Internet Epitope Database (IEDB) Analysis Resource (http://tools.immuneepitope.org/mhci/).
SMM binding predictions from the Internet Epitope Database (IEDB) Analysis Resource (http://tools.immuneepitope.org/mhci/).
NetMHCpan binding predictions from NetMHCpan2.8 Server (www.cbs.dtu.dk/services/NetMHCpan/).
Binding score derived from describing other predictions as not binding (0), weak binding (0.5), moderate binding (1), or strong binding (1.5). The sum of these descriptive scores from each prediction was calculated for each target.
Search for all non-redundant structurally similar 9-mer peptides in the complete Homo sapeins proteome from The Universal Protein Knowledgebase (UniProtKB). Number indicates unique matches predicted to bind at least weakly to HLA-A2 by ANN and SMM.
Percentage of peptides from the human proteome structurally similar to the target (above) which have identical sequences in the Mus musculus proteome.
Each tumor peptide was also examined for sequences in the human and mouse proteomes that showed structural similarities. The scan was guided by a set of heuristics allowing conservative substitutions at every peptide position, plus allowing preferred MHC anchor residues at positions 2 and 9. Using these criteria, the number of unique, identified human peptides that were also predicted to bind to HLA-A2 varied greatly among these tumor epitopes, from as few as 3 to over 800 (Table 1). The number of identical mouse peptides, in anticipation of testing in mouse models, ranged from 2 to 330 (34 to 82%, depending on the cancer self-peptide). This suggests that testing candidate TCRs in models such as the AAD mouse will be useful as one pre-clinical approach to reveal cross-reactivity [11,17,47], especially for targets that are, themselves, identical in the mouse proteome. Of course, the quantitative presence of these peptides as class I complexes will depend also on the transcription and translation of the genes, and on the processing pathways involved in antigen presentation [55].
Conclusions
Engineering TCR affinities against class I cancer peptide antigens provides an opportunity to optimize CD8 T cell activity, but also to recruit CD4 T cells against the cancer. The class I-restricted CD4 T cells can potentially facilitate induction of endogenous T cell responses against other cancer antigens, including patient-specific, mutated peptide antigens [57,58]. However, enhanced affinity of a TCR is associated with a higher risk of off-target peptide reactivity. The uniqueness of a cancer peptide (i.e. fewer structurally related peptides) may be a useful parameter to consider with peptide targets, along with tissue-expression profiles, tumor-expression levels, and links to oncogenicity [1]. Pre-clinical screens for cross-reactivity should include the panel of in silico identified structurally related peptides. As other factors also determine processing and presentation of peptides by class I MHC products, it will eventually be useful to focus in vitro screens on peptides that have been validated as class I complexes, which would allow non-structurally related peptides to be included in a much broader screen [55,59].
Highlights.
Engineered TCRs can overcome a “hole in the T cell repertoire”.
Affinity tuning can yield optimal TCR candidates for adoptive T cell therapy.
On-target/off-tumor toxicity can be revealed by more potent T cells.
Higher affinity TCRs can result in off-target reactivity with related peptides.
Pre-clinical screening methods for off-target T cell activity are critical.
Acknowledgments
We thank past and present members of the lab for their contributions, and many excellent collaborators over the years. This work was supported by NIH grant R01 CA178844 (D.M.K), the Melanoma Research Alliance (D.M.K.)
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Cheever MA, Allison JP, Ferris AS, Finn OJ, Hastings BM, Hecht TT, Mellman I, Prindiville SA, Viner JL, Weiner LM, et al. The prioritization of cancer antigens: a national cancer institute pilot project for the acceleration of translational research. Clin Cancer Res. 2009;15:5323–5337. doi: 10.1158/1078-0432.CCR-09-0737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jameson SC, Hogquist KA, Bevan MJ. Positive selection of thymocytes. Annu Rev Immunol. 1995;13:93–126. doi: 10.1146/annurev.iy.13.040195.000521. [DOI] [PubMed] [Google Scholar]
- 3.Vrisekoop N, Monteiro JP, Mandl JN, Germain RN. Revisiting thymic positive selection and the mature T cell repertoire for antigen. Immunity. 2014;41:181–190. doi: 10.1016/j.immuni.2014.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kosmrlj A, Read EL, Qi Y, Allen TM, Altfeld M, Deeks SG, Pereyra F, Carrington M, Walker BD, Chakraborty AK. Effects of thymic selection of the T-cell repertoire on HLA class I-associated control of HIV infection. Nature. 2010;465:350–354. doi: 10.1038/nature08997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Walker BD, Yu XG. Unravelling the mechanisms of durable control of HIV-1. Nat Rev Immunol. 2013;13:487–498. doi: 10.1038/nri3478. • Although not directly related to cancer immunotherapy, this review contains a highly relevant discussion of how holes in the T cell repertoire can affect responses to a given pepMHC target, or mutations of the target.
- 6. Aleksic M, Liddy N, Molloy PE, Pumphrey N, Vuidepot A, Chang KM, Jakobsen BK. Different affinity windows for virus and cancer-specific T-cell receptors: implications for therapeutic strategies. Eur J Immunol. 2012;42:3174–3179. doi: 10.1002/eji.201242606. •• A compilation of many different TCR affinity measurements revealing that, on average, cancer-specific TCRs bind their targets with lower affinity than TCRs specific for foreign, viral-derived targets.
- 7.Morgan RA, Dudley ME, Wunderlich JR, Hughes MS, Yang JC, Sherry RM, Royal RE, Topalian SL, Kammula US, Restifo NP, et al. Cancer regression in patients after transfer of genetically engineered lymphocytes. Science. 2006;314:126–129. doi: 10.1126/science.1129003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Johnson LA, Morgan RA, Dudley ME, Cassard L, Yang JC, Hughes MS, Kammula US, Royal RE, Sherry RM, Wunderlich JR, et al. Gene therapy with human and mouse T-cell receptors mediates cancer regression and targets normal tissues expressing cognate antigen. Blood. 2009;114:535–546. doi: 10.1182/blood-2009-03-211714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stone JD, Kranz DM. Role of T cell receptor affinity in the efficacy and specificity of adoptive T cell therapies. Front Immunol. 2013;4:244. doi: 10.3389/fimmu.2013.00244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Perez-Diez A, Joncker NT, Choi K, Chan WF, Anderson CC, Lantz O, Matzinger P. CD4 cells can be more efficient at tumor rejection than CD8 cells. Blood. 2007;109:5346–5354. doi: 10.1182/blood-2006-10-051318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Soto CM, Stone JD, Chervin AS, Engels B, Schreiber H, Roy EJ, Kranz DM. MHC-class I-restricted CD4 T cells: a nanomolar affinity TCR has improved anti-tumor efficacy in vivo compared to the micromolar wild-type TCR. Cancer Immunol Immunother. 2013;62:359–369. doi: 10.1007/s00262-012-1336-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chervin AS, Stone JD, Bowerman NA, Kranz DM. Cutting edge: inhibitory effects of CD4 and CD8 on T cell activation induced by high-affinity noncognate ligands. J Immunol. 2009;183:7639–7643. doi: 10.4049/jimmunol.0901664. [DOI] [PubMed] [Google Scholar]
- 13.Chervin AS, Stone JD, Holler PD, Bai A, Chen J, Eisen HN, Kranz DM. The impact of TCR-binding properties and antigen presentation format on T cell responsiveness. J Immunol. 2009;183:1166–1178. doi: 10.4049/jimmunol.0900054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Robbins PF, Li YF, El-Gamil M, Zhao Y, Wargo JA, Zheng Z, Xu H, Morgan RA, Feldman SA, Johnson LA, et al. Single and dual amino acid substitutions in TCR CDRs can enhance antigen-specific T cell functions. J Immunol. 2008;180:6116–6131. doi: 10.4049/jimmunol.180.9.6116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhao Y, Bennett AD, Zheng Z, Wang QJ, Robbins PF, Yu LY, Li Y, Molloy PE, Dunn SM, Jakobsen BK, et al. High-affinity TCRs generated by phage display provide CD4+ T cells with the ability to recognize and kill tumor cell lines. J Immunol. 2007;179:5845–5854. doi: 10.4049/jimmunol.179.9.5845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Holler PD, Chlewicki LK, Kranz DM. TCRs with high affinity for foreign pMHC show self-reactivity. Nat Immunol. 2003;4:55–62. doi: 10.1038/ni863. [DOI] [PubMed] [Google Scholar]
- 17.Engels B, Chervin AS, Sant AJ, Kranz DM, Schreiber H. Long-term persistence of CD4(+) but rapid disappearance of CD8(+) T cells expressing an MHC class I-restricted TCR of nanomolar affinity. Mol Ther. 2012;20:652–660. doi: 10.1038/mt.2011.286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Frankel TL, Burns WR, Peng PD, Yu Z, Chinnasamy D, Wargo JA, Zheng Z, Restifo NP, Rosenberg SA, Morgan RA. Both CD4 and CD8 T cells mediate equally effective in vivo tumor treatment when engineered with a highly avid TCR targeting tyrosinase. J Immunol. 2010;184:5988–5998. doi: 10.4049/jimmunol.1000189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stone JD, Harris DT, Soto CM, Chervin AS, Aggen DH, Roy EJ, Kranz DM. A novel T cell receptor single-chain signaling complex mediates antigen-specific T cell activity and tumor control. Cancer Immunol Immunother. 2014;63:1163–1176. doi: 10.1007/s00262-014-1586-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Robbins PF, Morgan RA, Feldman SA, Yang JC, Sherry RM, Dudley ME, Wunderlich JR, Nahvi AV, Helman LJ, Mackall CL, et al. Tumor regression in patients with metastatic synovial cell sarcoma and melanoma using genetically engineered lymphocytes reactive with NY-ESO-1. J Clin Oncol. 2011;29:917–924. doi: 10.1200/JCO.2010.32.2537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chinnasamy N, Wargo JA, Yu Z, Rao M, Frankel TL, Riley JP, Hong JJ, Parkhurst MR, Feldman SA, Schrump DS, et al. A TCR targeting the HLA-A*0201-restricted epitope of MAGE-A3 recognizes multiple epitopes of the MAGE-A antigen superfamily in several types of cancer. J Immunol. 2011;186:685–696. doi: 10.4049/jimmunol.1001775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Linette GP, Stadtmauer EA, Maus MV, Rapoport AP, Levine BL, Emery L, Litzky L, Bagg A, Carreno BM, Cimino PJ, et al. Cardiovascular toxicity and titin cross-reactivity of affinity-enhanced T cells in myeloma and melanoma. Blood. 2013;122:863–871. doi: 10.1182/blood-2013-03-490565. •• Results from a clinical trial revealing a previously uncharacterized cross-reactivity of a MAGE-A3/HLA-A1 specific TCR used for adoptive therapy, associated with the death of two patients. The reactivity with a titin/HLA-A1 complex was identified by retrospective proteome scanning for structurally-related peptides; pre-clinical screening of T cell activity in response to representative tissue lines in vitro had not revealed this activity.
- 23.de Witte MA, Coccoris M, Wolkers MC, van den Boom MD, Mesman EM, Song JY, van der Valk M, Haanen JB, Schumacher TN. Targeting self-antigens through allogeneic TCR gene transfer. Blood. 2006;108:870–877. doi: 10.1182/blood-2005-08-009357. [DOI] [PubMed] [Google Scholar]
- 24.Theobald M, Biggs J, Dittmer D, Levine AJ, Sherman LA. Targeting p53 as a general tumor antigen. Proc Natl Acad Sci U S A. 1995;92:11993–11997. doi: 10.1073/pnas.92.26.11993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Richman SA, Kranz DM. Display, engineering, and applications of antigen-specific T cell receptors. Biomol Eng. 2007;24:361–373. doi: 10.1016/j.bioeng.2007.02.009. [DOI] [PubMed] [Google Scholar]
- 26.Stone JD, Chervin AS, Aggen DH, Kranz DM. T cell receptor engineering. Methods Enzymol. 2012;503:189–222. doi: 10.1016/B978-0-12-396962-0.00008-2. [DOI] [PubMed] [Google Scholar]
- 27.Ashfield R, Jakobsen BK. Making high-affinity T-cell receptors: a new class of targeted therapeutics. IDrugs. 2006;9:554–559. [PubMed] [Google Scholar]
- 28.Bowerman NA, Crofts TS, Chlewicki L, Do P, Baker BM, Christopher Garcia K, Kranz DM. Engineering the binding properties of the T cell receptor:peptide:MHC ternary complex that governs T cell activity. Mol Immunol. 2009;46:3000–3008. doi: 10.1016/j.molimm.2009.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Garcia KC, Adams JJ, Feng D, Ely LK. The molecular basis of TCR germline bias for MHC is surprisingly simple. Nat Immunol. 2009;10:143–147. doi: 10.1038/ni.f.219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rudolph MG, Stanfield RL, Wilson IA. How TCRs bind MHCs, peptides, and coreceptors. Annu Rev Immunol. 2006;24:419–466. doi: 10.1146/annurev.immunol.23.021704.115658. [DOI] [PubMed] [Google Scholar]
- 31.Borbulevych OY, Santhanagopolan SM, Hossain M, Baker BM. TCRs used in cancer gene therapy cross-react with MART-1/Melan-A tumor antigens via distinct mechanisms. J Immunol. 2011;187:2453–2463. doi: 10.4049/jimmunol.1101268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cole DK, Yuan F, Rizkallah PJ, Miles JJ, Gostick E, Price DA, Gao GF, Jakobsen BK, Sewell AK. Germ line-governed recognition of a cancer epitope by an immunodominant human T-cell receptor. J Biol Chem. 2009;284:27281–27289. doi: 10.1074/jbc.M109.022509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Garboczi DN, Ghosh P, Utz U, Fan QR, Biddison WE, Wiley DC. Structure of the complex between human T-cell receptor, viral peptide and HLA-A2. Nature. 1996;384:134–141. doi: 10.1038/384134a0. [DOI] [PubMed] [Google Scholar]
- 34. Morgan RA, Chinnasamy N, Abate-Daga D, Gros A, Robbins PF, Zheng Z, Dudley ME, Feldman SA, Yang JC, Sherry RM, et al. Cancer regression and neurological toxicity following anti-MAGE-A3 TCR gene therapy. J Immunother. 2013;36:133–151. doi: 10.1097/CJI.0b013e3182829903. •• Results from a small clinical trial with a MAGE-A3/HLA-A2 specific TCR used for adoptive therapy, associated with the death of two patients. In-depth retrospective analysis revealed a low frequency cell type in the central nervous tissue with a previously uncharacterized expression of MAGE-A12 protein and a derived peptide that cross-reacted with the MAGE-A3 TCR.
- 35.Marrack P, Scott-Browne JP, Dai S, Gapin L, Kappler JW. Evolutionarily conserved amino acids that control TCR-MHC interaction. Annu Rev Immunol. 2008;26:171–203. doi: 10.1146/annurev.immunol.26.021607.090421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Chervin AS, Stone JD, Soto CM, Engels B, Schreiber H, Roy EJ, Kranz DM. Design of T-cell receptor libraries with diverse binding properties to examine adoptive T-cell responses. Gene Ther. 2013;20:634–644. doi: 10.1038/gt.2012.80. • This study shows that a panel of TCRs with diverse affinities can be engineered by a single codon manipulation in the CDR2 loop, allowing one to reduce the affinity of interaction between a high-affinity TCR and the MHC helices.
- 37.Smith SN, Sommermeyer D, Piepenbrink KH, Blevins SJ, Bernhard H, Uckert W, Baker BM, Kranz DM. Plasticity in the contribution of T cell receptor variable region residues to binding of peptide-HLA-A2 complexes. J Mol Biol. 2013;425:4496–4507. doi: 10.1016/j.jmb.2013.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Irvine DJ, Purbhoo MA, Krogsgaard M, Davis MM. Direct observation of ligand recognition by T cells. Nature. 2002;419:845–849. doi: 10.1038/nature01076. [DOI] [PubMed] [Google Scholar]
- 39.Purbhoo MA, Irvine DJ, Huppa JB, Davis MM. T cell killing does not require the formation of a stable mature immunological synapse. Nat Immunol. 2004;5:524–530. doi: 10.1038/ni1058. [DOI] [PubMed] [Google Scholar]
- 40.Sykulev Y, Joo M, Vturina I, Tsomides TJ, Eisen HN. Evidence that a single peptide-MHC complex on a target cell can elicit a cytolytic T cell response. Immunity. 1996;4:565–571. doi: 10.1016/s1074-7613(00)80483-5. [DOI] [PubMed] [Google Scholar]
- 41.Vitiello A, Marchesini D, Furze J, Sherman LA, Chesnut RW. Analysis of the HLA-restricted influenza-specific cytotoxic T lymphocyte response in transgenic mice carrying a chimeric human-mouse class I major histocompatibility complex. J Exp Med. 1991;173:1007–1015. doi: 10.1084/jem.173.4.1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Newberg MH, Smith DH, Haertel SB, Vining DR, Lacy E, Engelhard VH. Importance of MHC class 1 alpha2 and alpha3 domains in the recognition of self and non-self MHC molecules. J Immunol. 1996;156:2473–2480. [PubMed] [Google Scholar]
- 43.Zhong S, Malecek K, Johnson LA, Yu Z, Vega-Saenz de Miera E, Darvishian F, McGary K, Huang K, Boyer J, Corse E, et al. T-cell receptor affinity and avidity defines antitumor response and autoimmunity in T-cell immunotherapy. Proc Natl Acad Sci U S A. 2013;110:6973–6978. doi: 10.1073/pnas.1221609110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Colella TA, Bullock TN, Russell LB, Mullins DW, Overwijk WW, Luckey CJ, Pierce RA, Restifo NP, Engelhard VH. Self-tolerance to the murine homologue of a tyrosinase-derived melanoma antigen: implications for tumor immunotherapy. J Exp Med. 2000;191:1221–1232. doi: 10.1084/jem.191.7.1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mullins DW, Bullock TN, Colella TA, Robila VV, Engelhard VH. Immune responses to the HLA-A*0201-restricted epitopes of tyrosinase and glycoprotein 100 enable control of melanoma outgrowth in HLA-A*0201-transgenic mice. J Immunol. 2001;167:4853–4860. doi: 10.4049/jimmunol.167.9.4853. [DOI] [PubMed] [Google Scholar]
- 46.Gregg RK, Nichols L, Chen Y, Lu B, Engelhard VH. Mechanisms of spatial and temporal development of autoimmune vitiligo in tyrosinase-specific TCR transgenic mice. J Immunol. 2010;184:1909–1917. doi: 10.4049/jimmunol.0902778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nichols LA, Chen Y, Colella TA, Bennett CL, Clausen BE, Engelhard VH. Deletional self-tolerance to a melanocyte/melanoma antigen derived from tyrosinase is mediated by a radio-resistant cell in peripheral and mesenteric lymph nodes. J Immunol. 2007;179:993–1003. doi: 10.4049/jimmunol.179.2.993. [DOI] [PubMed] [Google Scholar]
- 48. Schmitt TM, Aggen DH, Stromnes IM, Dossett ML, Richman SA, Kranz DM, Greenberg PD. Enhanced-affinity murine T-cell receptors for tumor/self-antigens can be safe in gene therapy despite surpassing the threshold for thymic selection. Blood. 2013;122:348–356. doi: 10.1182/blood-2013-01-478164. • This study shows the safety and persistence of a adoptively transferred murine T cells carrying TCRs engineered for increased affinity for a WT1/H2-Db complex. The engineered TCRs, unlike the wild-type TCR precursor, were able to bind and be triggered by specific pepMHC in the absence of CD8 co-receptor, suggesting improved functional avidity.
- 49.Inoue K, Ogawa H, Sonoda Y, Kimura T, Sakabe H, Oka Y, Miyake S, Tamaki H, Oji Y, Yamagami T, et al. Aberrant overexpression of the Wilms tumor gene (WT1) in human leukemia. Blood. 1997;89:1405–1412. [PubMed] [Google Scholar]
- 50.Mason D. A very high level of crossreactivity is an essential feature of the T- cell receptor. Immunol Today. 1998;19:395–404. doi: 10.1016/s0167-5699(98)01299-7. [DOI] [PubMed] [Google Scholar]
- 51.Petrova G, Ferrante A, Gorski J. Cross-reactivity of T cells and its role in the immune system. Crit Rev Immunol. 2012;32:349–372. doi: 10.1615/critrevimmunol.v32.i4.50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jones LL, Colf LA, Stone JD, Garcia KC, Kranz DM. Distinct CDR3 conformations in TCRs determine the level of cross-reactivity for diverse antigens, but not the docking orientation. J Immunol. 2008;181:6255–6264. doi: 10.4049/jimmunol.181.9.6255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Holler PD, Kranz DM. Quantitative analysis of the contribution of TCR/pepMHC affinity and CD8 to T cell activation. Immunity. 2003;18:255–264. doi: 10.1016/s1074-7613(03)00019-0. [DOI] [PubMed] [Google Scholar]
- 54. Cameron BJ, Gerry AB, Dukes J, Harper JV, Kannan V, Bianchi FC, Grand F, Brewer JE, Gupta M, Plesa G, et al. Identification of a Titin-derived HLA-A1-presented peptide as a cross-reactive target for engineered MAGE A3-directed T cells. Sci Transl Med. 2013;5:197ra103. doi: 10.1126/scitranslmed.3006034. •• This study described responses to the renal cell cancer vaccine trial with IMA901, containing multiple peptides identified through the immatics XPRESIDENT technology, which combines comparisons between normal and tumor tissues from patients for both their mRNA expression levels and their mass spectrometry-characterized, MHC-presented peptides. This strategy results in extensive knowledge of the normal and cancerous immunopeptidome that should be very useful in target identification and safety screening.
- 55.Walter S, Weinschenk T, Stenzl A, Zdrojowy R, Pluzanska A, Szczylik C, Staehler M, Brugger W, Dietrich PY, Mendrzyk R, et al. Multipeptide immune response to cancer vaccine IMA901 after single-dose cyclophosphamide associates with longer patient survival. Nat Med. 2012;18:1254–1261. doi: 10.1038/nm.2883. [DOI] [PubMed] [Google Scholar]
- 56. Engels B, Engelhard VH, Sidney J, Sette A, Binder DC, Liu RB, Kranz DM, Meredith SC, Rowley DA, Schreiber H. Relapse or eradication of cancer is predicted by peptide-major histocompatibility complex affinity. Cancer Cell. 2013;23:516–526. doi: 10.1016/j.ccr.2013.03.018. • This study detailed a panel of six mouse tumor lines with different peptide-associated epitopes that were targeted by adoptively transferred T cells. The ability of T cell therapy to prevent relapses of the tumors correlated with the affinity of the peptide for the MHC.
- 57.Matsushita H, Vesely MD, Koboldt DC, Rickert CG, Uppaluri R, Magrini VJ, Arthur CD, White JM, Chen YS, Shea LK, et al. Cancer exome analysis reveals a T-cell-dependent mechanism of cancer immunoediting. Nature. 2012;482:400–404. doi: 10.1038/nature10755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Tran E, Turcotte S, Gros A, Robbins PF, Lu YC, Dudley ME, Wunderlich JR, Somerville RP, Hogan K, Hinrichs CS, et al. Cancer immunotherapy based on mutation-specific CD4+ T cells in a patient with epithelial cancer. Science. 2014;344:641–645. doi: 10.1126/science.1251102. •• In this study, whole exome sequencing of a patient with metastatic cholangiocarcinoma identified a mutant peptide epitope restricted by HLA-DQ which stimulated a set of CD4+ tumor infiltrating lymphocytes. These CD4 T cells, which didn't respond to the nonmutated sequence, were expanded in vitro and re-introduced into the patient, resulting in a 30% regression in target lesions, lasting 13 months after T cell infusion.
- 59.Weinschenk T, Gouttefangeas C, Schirle M, Obermayr F, Walter S, Schoor O, Kurek R, Loeser W, Bichler KH, Wernet D, et al. Integrated functional genomics approach for the design of patient-individual antitumor vaccines. Cancer Res. 2002;62:5818–5827. [PubMed] [Google Scholar]