Abstract
In contrast to the traditional biological paradigms focused on ‘specificity’, recent research and theoretical efforts have focused on functional ‘promiscuity’ exhibited by proteins and enzymes in many biological settings, including enzymatic detoxication, steroid biochemistry, signal transduction and immune responses. In addition, divergent evolutionary processes are apparently facilitated by random mutations that yield promiscuous enzyme intermediates. The intermediates, in turn, provide opportunities for further evolution to optimize new functions from existing protein scaffolds. In some cases, promiscuity may simply represent the inherent plasticity of proteins resulting from their polymeric nature with distributed conformational ensembles. Enzymes or proteins that bind or metabolize noncognate substrates create ‘messiness’ or noise in the systems they contribute to. With our increasing awareness of the frequency of these promiscuous behaviors it becomes interesting and important to understand the molecular bases for promiscuous behavior and to distinguish between evolutionarily selected promiscuity and evolutionarily tolerated messiness. This review provides an overview of current understanding of these aspects of protein biochemistry and enzymology.
Keywords: Enzyme promiscuity, Detoxication enzymes, Enzyme evolution, Evolvability, Intrinsically Disordered proteins
1. Introduction
The structural biology revolution that spanned the 1980s–1990s temporarily reinforced the long-held belief that enzymes and receptors were exquisitely specific in their substrate or ligand interactions. An explosion of published X-ray structures seemed to confirm the traditional perspective that receptors and enzymes were ‘special’ because of their specificity. It was easy to visualize directly, based on models derived from crystallography, that enzyme and protein active sites usually exploit all possible ‘handles’ for their interactions with their cognate ligands. Structurally similar ligands can be selectively recognized by different active sites because enzymes or proteins can exploit spatially optimized hydrogen bonds, ionic interactions and hydrophobic contacts, and they can also exclude non cognate ligands via steric clashes or charge repulsion [1–6]. The structural perspective revealed the mechanisms by which enzymes and proteins achieve the molecular recognition that had been heralded for decades.
However, the pendulum has swung. During the past 15 years that perspective has expanded to accommodate the growing realization that most enzymes or proteins are not as ‘ligand specific’ as the textbooks, or crystal structures, suggested, supporting the initial observations made by a few [7,8]. In fact, many enzymes are conspicuously promiscuous in vitro despite their critical roles in core metabolism in vivo. A search of the literature published in the past five years reveals a dramatic increase in the number of publications with ‘promiscuity’ in the title compared to ‘specificity’ (which remains high but more constant), as expected for the increased interest in the subject and the corresponding new insights that result.
More importantly, our growing awareness of promiscuity as a property of proteins has been accompanied by the realization that functional promiscuity or ‘messiness’ has clear roles in biology and biotechnology [9–17]. It is now apparent that promiscuity is as biologically important as specificity and a significant challenge lies in understanding how biological systems achieve controlled promiscuity and how they exploit it or tolerate it. Intuitively, it is a greater challenge to understand promiscuity than specificity, and definitive rules or concepts about promiscuity are still being developed.
My lab has devoted significant effort toward this aspect of protein structure and function. Our efforts with detoxication enzymes have revealed some useful lessons about the origins of molecular promiscuity and the behavior of promiscuous enzymes, which may be applicable to steroid enzymology. Many of the enzymes in steroid biosynthetic pathways are cytochrome P450s (CYPs) related to the highly promiscuous CYPs involved in detoxication, and we have considered the relative promiscuity of individual isoforms within this family and others. Many of our findings are relevant to this edition. This overview of promiscuity extends beyond enzymes to include other proteins and receptors.
2. Definitions
As with all new fields of study, it is critical to define terms. There have been many terms used to describe variations of promiscuous behavior, including terms defined in thoughtful and extensive reviews by Tawfik et al. [9,13]. I will limit the terms here to distinguish a few types of promiscuous behavior that are most well described, and those types of promiscuity most relevant to this edition. The definitions I find most useful are purely operational and less restrictive than those used by others [9,13] and are schematized in Fig. 1. If an enzyme or protein interacts with multiple structurally distinct ligands or substrates at a single binding site, this is promiscuous behavior. In contrast, others prefer to reserve the use of the term ‘promiscuous’ for cases where an enzyme or protein interacts with a ligand other than the ligand or set of ligands it is ‘supposed to’ interact with based on its biological role. With that definition, the term ‘promiscuity’ is applicable when an enzyme or receptor ‘makes a mistake’. With this more restrictive definition, enzymes or proteins that interact with multiple substrates or ligands as part of their normal function would be called ‘multispecific’ rather than promiscuous. Arguably, ‘multispecific’ would be a better term for enzymes that, in accord with their biological function, have clear specificity toward multiple substrates, as many do, rather than the proteins or enzymes that have no clear preference for any ligands. Therefore, to capture adequately the biological scope of the physico–chemical trait wherein enzymes and proteins are not as specific as once described, I apply ‘promiscuity’ to any case where multiple ligands can bind at a common site. Of course, then, essentially all proteins or enzymes are promiscuous to some degree and this demands consideration of how much promiscuity is tolerable vs. useful in different situations, as with steroid metabolism and signaling that are the focus of this edition. Even with these differences in the use of terms by different groups, several definitions are consistent with those depicted in Fig. 1 for both enzymes and receptors. Types of promiscuity that are relevant for enzymes are schematized in Fig. 2.
Fig. 1.
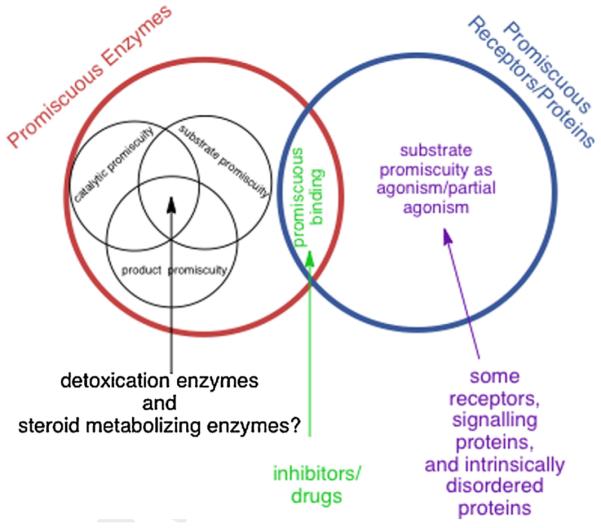
Types of promiscuous behavior exhibited by enzymes, receptors and other proteins. The specific types of promiscuous behavior summarized are described in the text. The relative sizes of the compartments do not reflect their relative frequency or abundance. For example, promiscuous binding is more frequent than catalytic promiscuity, but this is not reflected in this figure.
Fig. 2.
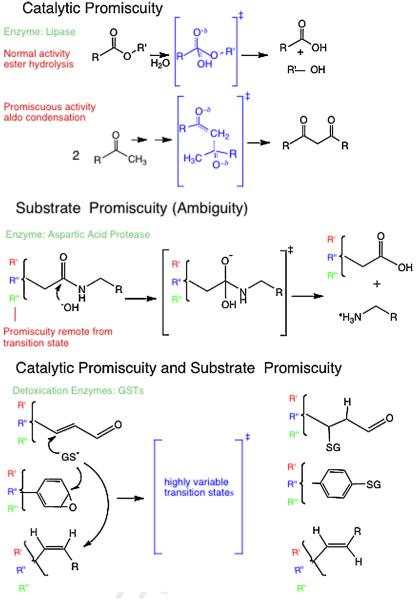
Chemical representation of promiscuous enzyme behaviors. Top: catalytic promiscuity includes structurally distinct transition states (in brackets) within the same active site. Structural differences also naturally occur remote from the transitions state for the different substrates (R vs. R′). Middle: substrate promiscuity refers to similar types of reactions, and hence similar local transition states (in brackets), for a single enzyme with a series of substrates that have structural differences remote from the local transition state (R, R′, R″). Bottom: bottom depicts the combination of catalytic and substrate promiscuity for detoxication enzymes that catalyze multiple types of reactions, with different local transition state structures (in brackets), and with variable structures remote from the transition state for any specific reaction type. For example the GSTs catalyze Michael type additions, addition to aryl epoxides (which subsequently aromatizes via dehydration), and cis-trans isomerization via addition–elimination, each for a wide range of substrate structures.
For enzymes, “catalytic promiscuity” is the ability of a single enzyme isoform to catalyze different types of chemical transformations, such as hydrolysis of esters or lactones vs. structurally distinct phosphotriesters. In this case a single enzyme has the ability to stabilize transition states of different reaction types. Also for enzymes, “substrate ambiguity”, or “substrate promiscuity” refers to their ability to perform the same type of chemical transformation on different substrate structures. For example, some reductases metabolize fatty acyl CoAs of different acyl chain length [18] and some kinases recognize peptide motifs rather than specific peptide sequences [19]. In the case of substrate promiscuity the local transition states for the reaction are very similar or identical, but the structure of the substrate remote from the transition state varies. In addition to catalytic promiscuity and substrate promiscuity of enzymes, ‘product promiscuity’ should be acknowledged. Product promiscuity refers to the situation when a single enzyme converts a single substrate to multiple products in reactions that require different transition states. For example, proteases that cleave a peptide at a single peptide bond generate two product peptides from a single substrate peptide, but this requires a single transition state and does not represent product promiscuity. On the other hand a protease that hydrolyzes a single peptide at multiple peptide bonds requires stabilization of multiple transition states that differ in the neighboring structure. Many proteases exhibit product promiscuity and substrate promiscuity and it is clearly advantageous for their biological plasticity [20–22]. Similarly, bacterial 3α, 20β-hydroxysteroid oxidoreductases are known to oxidize either hydroxy group, yielding two different products, and requiring two different transition states from a single substrate [23]. This could rationally be called product promiscuity. Product promiscuity is likely also to be important in several steroid pathways [24,25]. Furthermore, it is a clear trait of detoxication enzymes, and potential advantages of product promiscuity have been speculated [26].
In some cases individual enzymes exhibit all three types of behavior, as is well documented for CYPs involved in detoxication, wherein a single CYP can oxidize an extraordinary range of functional groups on structurally diverse substrates [27]. A single CYP isoform may catalyze a combination of N-dealkylation, aryl epoxidation, aliphatic hydroxylation and hydrocarbon desaturation and other reactions on a single substrate (catalytic promiscuity), or it may perform a single type of reaction such as N-dealkylation for a wide range of substrates (substrate promiscuity), or it may lack regiospecificity and exhibit product promiscuity. Other detoxication enzymes also exhibit extreme substrate promiscuity and catalytic promiscuity including previously mentioned glutathione transferases (GSTs) [28], UDP-glucuronosyl transferases (UGTs) [29], and sulfotransferases (SULTs) [30]. These enzymes catalyze the reaction of a broad range of electrophilic or nucleophilic functional groups on substrates with the cofactors, glutathione, UDP-glucuronic acid, or 3′-phospho-adenosine-5′phosphosulfate, respectively. A critically important point is that there is a wide range of functional groups on the substrates that reacts with the cofactor for each enzyme; any set of reactions catalyzed by an individual enzyme can include very different local transitions states as well as very different substrate structures remote from the transition state. Furthermore, examples of product promiscuity are common with detoxication enzymes, particularly with CYPs [31–33]. Interestingly, steroid metabolizing CYPs also exhibit catalytic, substrate, and product promiscuity.
For some detoxication enzymes the situation is even more complex. For example, the efflux transporter P-glycoprotein (ABCB1) [34] exports an enormous range of structurally distinct xenobiotics, at the expense of very specific ATP hydrolysis. Arguably, the enzyme is substrate promiscuous with respect to export but not catalytically promiscuous with respect to ATP hydrolysis, even though many different transported substrates activate the hydrolysis. Examples of this type amplify the need for rigorous terminology when discussing promiscuity. These labels imply that catalytic, substrate, and product promiscuity result from reactions within a single active site on the enzyme, although different subsites within a large contiguous active site may contribute differentially to binding of different substrates or different activities. Importantly, the behavior of detoxication enzymes has been suggested by others to be an example of “multispecificity” rather than promiscuity per se because of the clear biological advantage in metabolizing many substrates [9,13]. My perspective is different. Because the CYPs, GSTs, UGTs, and other detoxication enzymes require stabilization of very different local transition states, and often have no clear preference for any specific substrates, they are defined here as catalytically promiscuous, rather than ‘multispecific’, even though there is a clear functional advantage to this type of behavior. In this view, the degree to which an enzyme accommodates many substrates vs. a single ‘cognate’ substrate reflects its degree of promiscuity, regardless of whether the promiscuity has clear purpose or is related to a specific biochemical pathway. Here, a ‘cognate’ ligand is the one for which the binding site has specifically evolved, and a noncognate ligand binds due to the promiscuity of the site.
Upon moving from enzymes to receptors or proteins we must acknowledge the distinction between catalysis and activation of other biological processes. Although there is a fundamental distinction between a kinetic promiscuity of enzymes resulting from stabilization of multiple transition states and a thermodynamic promiscuity resulting from multiple ligands stabilizing an active receptor conformation, this has yet to be considered with any formalism and is beyond the scope of this review. However, some noncognate ligands promote receptor activation or partial activation including recent examples and ‘classic’ behavior of G protein-coupled receptors (partial agonists) [35,36], which is analogous to substrate promiscuity for enzymes; the receptor performs its ‘normal function’ with noncognate ligands at lower efficiency. For these promiscuous interactions the noncognate ligand is able to shift the conformational equilibrium of the receptor or protein toward an active conformation that can, at least partially, perform its function.
A notable cross section of overlap between receptors and enzymes is the binding of inhibitors. Both can passively bind noncognate ligands that prevent catalysis or signaling simply by competition with the cognate ligands. It is interesting that this behavior has provided the conceptual basis for much of traditional drug discovery; the drug discovery processes assumes that noncognate ligands exist and that they will bind and inhibit their target. This assumption further predicts that a high degree of inhibitory promiscuity makes a target more ‘druggable’ because a larger fraction of chemical space will yield inhibitory drugs. Speculatively, the relative promiscuity of a drug target could be predictive of the relative effort required to develop a drug.
A special case of promiscuity may occur with intrinsically disordered proteins (IDPs), which have minimal secondary or tertiary structure until they interact with ligands or other proteins [37,38]. Different ligands or partners can select different conformations from the disordered ensemble and induce different conformations for different functions. It is currently unclear how to characterize their behavior within the spectrum of promiscuity. If each functional conformation was induced by a specific ligand this might be multispecificity. Alternatively if multiple ligands induce a conformation required for one function, then this could be considered as promiscuous agonism or promiscuous partial agonism. Further studies with IDPs may clarify the best terminology to describe their promiscuity.
In contrast “multifunctional enzymes” catalyze multiple distinct chemical reactions with a clear metabolic purpose, but they utilize different catalytic sites, residues, or cofactors to achieve these different reactions [39–41]. For example, some Type I restriction enzymes methylate DNA using a site distinct from the site that hydrolyzes the DNA [42]. In many cases multifunctional enzymes are multisubunit complexes with the different catalytic activities distributed among the different subunits.
3. Roles of promiscuity in biology and biotechnology
Because several of the reviews referenced above have focused on the roles of promiscuity in biology, I will only summarize their main points. Although only few examples are highlighted here, many biochemical, physiological, and medical studies are now elucidating the importance of this ‘messy’ biology.
3.1. Evolution of new protein function and in vitro evolution
Divergent evolution of new protein function from an existing pool of structural scaffolds likely occurs by sampling promiscuous enzyme variants resulting from mutation with relaxed substrate specificity, enroute to an optimized protein with new function [9–14]. In effect, a spontaneous mutation occurs in one copy of a gene that encodes an altered protein that catalyzes a reaction on a substrate not metabolized by the wild type enzyme. If this reaction, either as catalytic promiscuity, substrate promiscuity, or product promiscuity has any survival advantage or confers additional ‘fitness,’ then this mutant will be retained and further optimized by additional mutation. Importantly, this optimization of new function occurs without forfeiting the original function, as long as only one copy of the gene is mutated.
Of course, the survival value of any promiscuous intermediate depends also on whether there is a disadvantage, or negative trade-off, that accompanies the new activity. Thus the evolutionary trajectory of the gene is not two dimensional, moving simply from old function to new function. Instead, the evolutionary intermediates may have new properties in addition to their relaxed specificity, such as altered stability or expression, and the evolutionary trajectory will be affected to avoid negative trade-offs. As a result the evolutionary trajectory is multidimensional and the other behaviors acquired in the promiscuous intermediates will contribute to the evolutionary trajectory [9,43]. For example if the mutant protein was hypersensitive to proteolysis or aggregation then the mutation will either not be retained or the negative trade off must be resolved with further evolution; promiscuity will only be useful as an evolutionary intermediate if the corresponding loss in stability or other property is not too disadvantageous.
An important concept is that the promiscuous intermediates not only provide an opportunity for divergent evolution, but they are also are more easily optimized than specific enzymes to yield new efficient and specific enzymes with a small number of mutations [44–48]. Promiscuous intermediates are highly ‘evolvable’ and it has been suggested that promiscuity is actually selected as an advantageous trait within the entire proteome in order to ensure evolutionary adaptability. In fact, significant experimental evidence suggests that protein/enzyme promiscuity per se is a trait that is required to optimize evolutionary efficiency, because fewer mutations may be required when starting from a promiscuous template than from a previously optimized enzyme with high specificity. Notably, this idea has been incorporated into in vitro evolution strategies to obtain proteins with new functions. Several groups have considered, and even demonstrated, the advantage of starting directed evolution toward a defined target function from a promiscuous template [44–48].
3.2. Detoxication and immunity
Perhaps the most obvious role for functional promiscuity in biology is in chemical detoxication, wherein several enzyme systems contribute to the conversion of hydrophobic xenobiotics to more water soluble metabolites that are more efficiently eliminated. These enzyme systems include those mentioned above, the hepatic CYPs, UGTs, cytosolic SULTs, GSTs, and others. In addition, various transporters, such as P-glycoprotein, efflux exogenous compounds out of cells to facilitate their clearance. The extraordinary catalytic and substrate promiscuity of these enzymes and transporters is an obvious advantage for their role in detoxication, wherein they promote the elimination of a diverse range of chemicals, including many we have never been exposed to before. It is unrealistic to have an enzyme or transporter to eliminate every possible environmental toxin to which we could be exposed, so catalytic promiscuity is the next best solution. There are, however, less well appreciated manifestations of promiscuity regarding detoxication enzymes, such as the product promiscuity defined above, which may have an advantage in detoxication [26]. It is interesting to consider this in the context of the definitions summarized above.
A similar advantage of promiscuity may be envisioned for the immune system. The maturation of promiscuous germ line IgGs via somatic mutation to afford higher affinity, more specific, IgGs directed against a specific antigen has an obvious functional role in the immune response. This process provides a mechanism for maximal diversification of antibody–antigen recognition from a fixed pool of germ line IgGs [49,50]. Interestingly, the somatic mutations occur specifically in the IgG hypervariable regions. As with detoxification enzymes, it is unrealistic to have a gene encoding an antibody for every possible antigen to which we might be exposed, so the maturation to a highly specific antibody from a pool of promiscuous templates has obvious utility.
3.3. Signal transduction
Several signal transduction pathways that mediate the transfer of information from the cell surface to the genome include promiscuous scaffold proteins that interact with many different components of specific pathways. These are examples of substrate promiscuity, rather than catalytic promiscuity, in as much as multiple binding partners are able to elicit a biological function from the promiscuous protein. For example, individual SH3 domains mediate a wide range of signaling response by recognition of varying peptide sequences. As a result, biochemical pathways comprise a highly complex network rather than discrete parallel pathways. It appears that in some cases intrinsic disorder of individual proteins contributes to their promiscuous function, wherein different binding partners induce different structures from the disordered state [51–54]. A similar situation occurs with the unfolded protein response and stress responses in the endoplasmic reticulum [55,56]. Here also, multiple interconnected pathways mediate the clearance of misfolded proteins, and these pathways depend on promiscuous interactions between, in some cases, disordered proteins. These pathways appear to have clear roles in the development of several diseases states.
The resulting redundancy and interconnectedness of pathways allows for complex regulation and adaptive responses. The promiscuity of the interacting components also introduces ‘noise’ in various phenotypes of biochemical markers and it is fascinating to consider how the noise can be tolerated, muted or amplified, as considered below in Section 5.
4. Methods for measuring promiscuity
4.1. Comparison of methods available to date
As a result of the realization that promiscuity is an important biological property, there have been some attempts to quantify it or compare its relative degree among different proteins. Such metrics would enable our understanding of the physical–chemical properties that optimize promiscuity or different intermediates along evolutionary pathways, in order to optimize protein engineering or directed evolution. In addition, a quantitative index would facilitate hypothesis driven experiments aimed to understand mechanistic aspects of promiscuous function or evolution. To date, several of the proposed methods for comparing relatively, or quantifying, promiscuity have some merit and each has some limitations. Each can be applied in useful ways if the ‘user’ understands what is actually being measured.
One approach to compare relative promiscuity is based on comparison of enzymes across different functional labels as defined by the Enzyme Commission of the International Union of Biochemists and Molecular Biologists, which assigns ‘EC numbers.’ If a candidate enzyme metabolizes a substrate assigned to enzymes with a different EC number then the candidate enzyme may be catalytically promiscuous, because it appears competent to catalyze different types of reactions [9,43]. The greater the distance between reactions within the EC phylogeny, the more promiscuous the structure is. For example, in considering an enzyme as a candidate for catalytic promiscuity, if EC numbers for two reactions catalyzed by an the enzyme differ only in the last digit, then the types of reactions are the same and only the substrate is different, so reaction with the noncognate substrate represents substrate promiscuity but not catalytic promiscuity. Conversely, if the EC numbers are different in the first few digits then this could be catalytic promiscuity because they potentially are different ‘classes’ of reactions catalyzed by the candidate enzyme. This approach has been validated qualitatively by historical data concerning the catalytic properties of some enzymes and it nicely predicts the ‘intuitively’ high promiscuity of some enzymes and the low promiscuity of others.
The marriage of this approach with structural databases may have utility in identifying enzymes for directed evolution of new function when the target enzyme does not need to be in the same structural family as its parent [57,58]. This method essentially identifies structurally related active sites that are represented in separate regions of ‘EC space.’ The combination of the EC phylogeny with structural considerations has nice predictive features for in silico directed evolution from promiscuous protein scaffolds [59]. However, this approach obviously requires a crystal structure to assign a promiscuity score for any enzyme, or strictly speaking its mutants. In addition it is not highly quantitative in as much as it does not address the issue of how similar or how different the transition states are that are associated with reactions denoted by different EC numbers; there is no accounting for chemical similarity across different reaction types. Because the transition states for all enzymes of different EC families are not equidistant in reaction space, this method provides no basis for assigning a quantitative degree of promiscuity or a difference in promiscuity for pairs of enzymes. On the other hand, the advantage of computationally comparing large sets of proteins to find those likely to have a predefined promiscuous function without any experimental effort is significant.
A related approach uses functional annotation from the KEGG database and graphical methods to assign similarity of the reactions assigned to individual enzymes [60]. The similarity or difference in the reactions catalyzed by the enzymes is scored and this can be correlated with structural features or sequence signatures of the enzymes. As much as the KEGG database is conceptually analogous to the EC categorization of function, this method focuses on catalytic promiscuity. The approach has been used to understand design and evolution of metabolic networks and provides a metric to examine the distribution of promiscuity or specificity across species or temporally through evolution [61]. For example, the method predicts that enzymes involved in amino acid and lipid metabolism are among the most promiscuous and therefore are among the oldest enzymes, which supported the types of reactions required for the broadest range of life forms. Both of these methods have potential utility in understanding catalytic promiscuity as it relates to in vitro or in vivo evolution.
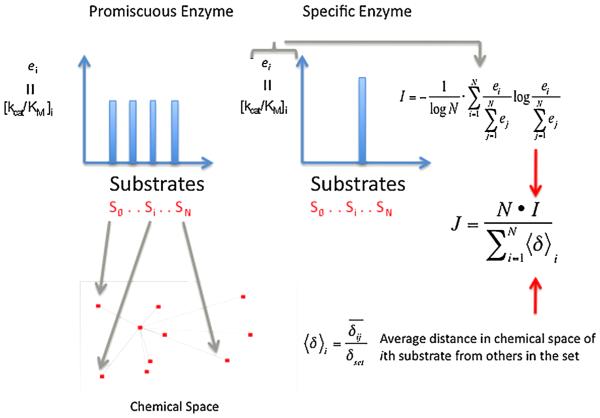
A few years ago my lab developed a quantitative index of promiscuity, designated “J,” that is independent of structure and is based on parameter entropy as schematized in Fig. 3 [62,63]. The method is highly versatile and allows one, for example, to query the promiscuity of either the enzyme toward different substrates or reactions, or the promiscuity of the substrate toward different enzymes. The approach accommodates both catalytic promiscuity and substrate promiscuity, either separately or as a single behavior [62], in contrast to the misconception [6,57] that it only is useful for substrate promiscuity or multispecificity. We have applied it successfully to both catalytically promiscuous (multispecific as defined by others) detoxication enzymes and to substrate promiscuous biosynthetic enzymes. Notably, we have not yet quantified product promiscuity with this index, but this would be possible. Although we first developed the approach for catalytic behavior, a strength of the method is its adaptability to any functional parameter such as inhibitory promiscuity, or binding promiscuity [62]. For example in order to compare enzyme promiscuity with regard to catalytic efficiency, we utilize the distribution of kcat/KM values for a given enzyme with a basis set of structurally diverse substrates. In effect, the distribution of kcat/KM values will be broad and flat (nearly identical values) if the enzyme has minimal preference for any individual substrates within the basis set. In contrast if the enzyme has strong preference for one substrate, such as its physiologic cognate substrate, the distribution of kcat/KM parameter will be narrow, with high values only for the cognate substrate or close analogs and low values for all other substrates. Specifically, the fractional kcat/KM of each substrate divided by the summed kcat/KMS for the entire ensemble is determined for each substrate. This ratio represents the probability that an enzyme will metabolize any particular substrate when presented with all of the substrates in the set at a concentration equal to their KM value. This probability is a measure of the ‘disorder’ of the system. High disorder corresponds to high promiscuity and low predictability of which substrate the enzyme will choose. Low disorder corresponds to low promiscuity, and a high degree of certainty about which substrate will be metabolized. The degree of disorder is normalized for the degree of chemical space sampled by the substrate basis set, so that the final parameter is not dependent on the substrate set used, as long as an adequate fraction of chemical space is sampled. A detailed description of the calculation of the chemical space represented by the substrate set is found in reference [62]. Briefly, The chemical diversity within a basis set of substrates is assessed with a value <δ>i which is the average distance in chemical space of each compound from the others in the substrate set, normalized to the greatest distance within the set. A <δ>i near 1 indicates that the substrates are, on average, far from one another, and the set is chemically diverse. A value near 0 indicates the substrates are chemically very similar. A commonly used chemometric approach to assign relative positions of compounds in n-dimensional chemical space is to score a binary string, or a keyset, of ‘n’ chemical properties that can be scored with a ‘1’ (yes) or a ‘0’ (no). For example, the keyset may include the queries: is there an aromatic ring? Are there two aromatic rings? Are there more than three aromatic rings? Is there an aldehyde? Is there an alcohol? Is there an alkyl nitrogen? Is there a nitrogen heterocycle? Is there a positive charge? Are there two positive charges? etc. A well known and validated keyset is the MDL set, which is widely used for similarity searches and for comparison of substructures in lead compounds in drug design [64]. From the pairwise comparison of each bit in the keyset for each pair of substrates, the relative location of each substrate in n-dimensional space is determined and the distance between them can be calculated, and a normalized mean value for the set of distances, <δ>i can be determined [62,64]. Diversity of the substrate set is best ensured by using a large set of chemical properties to calculate <δ>i, and demanding a value of <δ>i near 1. Obviously, there is some human judgment required; a set of substrates could appear diverse if the chemical properties in the keyset were chosen to emphasize their differences, rather than to sample a large part of chemical space. Although a detailed discussion of these methods is outside the scope of this review, it is generally accepted that even 50 well chosen chemical properties can provide useful relative measures of chemical diversity or similarity.
Fig. 3.
Schematized summary for calculating the promiscuity index, J. For a series of substrates the enzyme efficiency, ei = (kcat/KM) is determined and the term ‘I’ is calculated. I reflects the normalized probability that any given substrate will be chosen by an enzyme, when the enzyme is exposed simultaneously to low concentrations of each substrate, or the relative preference for any substrate. ‘I’ reflects the distribution of these preferences; a promiscuous enzyme with little preference (e1 = e2 = en) yields an I = 1 and a specific enzyme with a large e for only one substrate yields I = 0. It may be necessary to scale I to account for the chemical similarity or difference among the substrates used; if an enzyme has the same ei for all substrates that are far from one another in chemical space (chemically very different) than it is more promiscuous than an enzyme that has the same ei for substrates that sample a narrow range of chemical space (chemically similar). Substrates far away from others in chemical space, large <δ>i, add more weight to the calculated promiscuity, J. J = 1 for a perfectly promiscuous enzyme (no preference for any substrate) and J = 0 for a completely specific enzyme.
A hypothetical example based on steroids is relevant here. If several steroid compounds differ only in a substituent at, say position 17, the same enzyme may metabolize them all. If the keyset only addresses properties of the substituent at position 17, and does not include properties of the steroid nucleus, the substrate set will appear to be diverse based on <δ>i and the resulting ‘J’ value will be artificially high. This could still be useful if the goal is to compare the promiscuity of a series of steroid metabolizing enzymes with respect to their ability to tolerate substitution at position 17. On the other hand, if the goal is to determine the overall promiscuity of a steroid metabolizing enzyme to determine how susceptible it is to inhibition by non steroid compounds, then a keyset with a much wider range of chemical properties not focused on substituents at position 17 would be more powerful and would reveal that the substrate set is inadequate to answer that question. Thus, the quality of any specific substrate set depends on the goal when determining ‘J’. Conversely, the degree of chemical space that needs to be sampled depends on the purpose of the measurement. Therefore, a comparison of ‘J’ values for a set enzymes, or correspondingly a set proteins, should probably only be done when the same substrate sets are used for each enzyme. The diversity within this data set that is required depends on the comparison that the user wants to perform. If the user has a good understanding of these issues, then very robust and informative comparison can be based on ‘J’ values, and even ‘I’ values in some cases [62,63].
We have enumerated specific criteria to benchmark the chemical similarity of substrates within the basis set to ensure the robustness of the final promiscuity value. Similar chemometric methods could easily be used to benchmark chemical similarity of transition states and thus further understand catalytic promiscuity. This promiscuity index has been a useful parameter that allows for testing hypotheses about promiscuity in quantitative ways.
Each of these approaches to quantify promiscuity is imperfect but they are useful tools. Each has utility in considering different questions related to promiscuity [57,58,60]. The effort required to develop these approaches reflects the growing interest in promiscuity as an important property of proteins. The utility of a quantitative index is best appreciated with few examples, which reflect the types of questions that become addressable when such an index is applied.
4.2. Examples of quantitation of promiscuity
Many investigators have suggested the intuitive hypothesis that catalytic promiscuity (or multispecificity as defined by others) is promoted by flexibility or structural plasticity; enzymes that can adapt their conformations easily would be expected to catalyze reactions with more substrates than substrate specific enzymes. In effect, functional flexibility requires structural flexibility. This hypothesis is difficult to address without tools to quantify promiscuity and ‘flexibility’. Honaker et al. [65,66] used a series of mutants of the highly promiscuous GST enzymes to determine their substrate promiscuity against a highly diverse basis set of substrates. Fortuitously, some A-class GSTs exhibited a reversible low temperature, barrierless transition in differential scanning calorimetry (DSC) experiments that corresponds to rearrangement of the active site and its C-terminal ‘lid.’ From a specialized analysis of the DSC transition it was possible to estimate the conformational breadth or diversity of the active sites for the series of GSTs with a thermodynamic treatment. The catalytic promiscuity measured by the method outlined in Fig. 3 correlated well with the conformational heterogeneity, supporting the expectation that the most promiscuous enzymes are the most ‘flexible’ [67]. In fact the DSC data indicated that the active site included an ensemble of different locations of the C-terminal helix, wherein secondary structure was maintained with different degrees of tertiary contact, as suggested for ‘molten globule’ states initially described for protein folding intermediates. This is in contrast to the case of intrinsically disordered proteins in signal transduction cascades mentioned above, wherein secondary structure is induced, differentially, by different substrates or binding partners.
In another example, Foti et al. [68] used this promiscuity index and considered the relationship between catalysis and inhibition. Hypothetically, catalysis requires highly specific alignment of relevant functional groups on the enzyme and the substrate, whereas competitive inhibition merely requires some favorable interactions compared to solvent–substrate and solvent–enzyme interactions. Therefore, enzymes would, on average, be more promiscuous with respect to inhibition than catalysis. This hypothesis has important implications for evolution because susceptibility to inhibition (inhibitory promiscuity) could be a significant negative trade off of substrate promiscuity or catalytic promiscuity of evolutionary intermediates that are sampled en route to new enzymes. A quantitative understanding of this relationship could elaborate mechanistic evolutionary models. In fact, the hypothesis is the basis for most small molecule drug design; typically, there are many more inhibitory drugs than substrates for therapeutic targets.
When this hypothesis was considered for a series of CYPs, including isoforms involved in either detoxication or in specific biosynthetic pathways, it was found that that all of the CYPs in the series were highly promiscuous with respect to inhibition, with a higher average inhibitory promiscuity than catalytic promiscuity. Interestingly, susceptibility to inhibition does not appear to be a significant disadvantage of high substrate promiscuity among CYPs collectively because even relatively specific isoforms already exhibited high promiscuity toward inhibition; in general little additional susceptibility to inhibition was incurred upon increasing catalytic promiscuity across isoforms. However within the CYP family, the specific isoforms with lower promiscuity (more substrate specific) would likely have a larger negative trade off of inhibitory promiscuity if they mutated to promiscuous intermediates in an evolutionary process. It is important to emphasize that this analysis did not strictly test any evolutionary hypothesis. Rather it tested the possibility that physico–chemical differences between evolutionarily optimized detoxication enzymes and their more catalytically specific counterparts resulted in differences in their susceptibility to inhibition. A more direct query about the evolutionary relationship between ‘binding’ promiscuity and catalytic promiscuity was considered by Khersonsky et al., who suggest that substrate promiscuity is more abundant than catalytic promiscuity, using enzymes encoded by the Escherichia coli genome as a model with a small number of model substrates and reaction types [69].
The important point is that access to a quantitative index for substrate promiscuity or catalytic promiscuity allows for the interrogation of hypotheses about promiscuous behavior that are otherwise intractable. Optimistically, such tools could provide additional insight into physical and chemical features associated with promiscuity or into evolutionary roles for promiscuity. Improvements to each of the methods described here could further improve our understanding of promiscuity in quantitative ways.
5. Next steps for understanding promiscuity
With our current appreciation for the pervasiveness of functional promiscuity in many biological contexts, we can refine our inquiry concerning its roles. Again, Tawfik has elegantly summarized many of the ideas that naturally emerge [13], but it is useful to reiterate them here. In some cases it is easy to rationalize the biological advantage of functional promiscuity in vivo, such as chemical detoxication or the immune response, and the role of promiscuity in natural and in vitro evolution is well established [70]. However, some inherent promiscuity encoded by individual enzymes that participate in specific pathways could translate into noise or ‘messiness’ at the system level without any clear purpose. Metabolic networks naturally include some protein-protein or protein–ligand interactions between noncognate partners and the enzymatic production of ‘mistakes’. The impact of these mistakes or the resulting noise will be context dependent. Under many biological circumstances this noise will be biologically irrelevant with no detriment to the cell or organism. More interestingly, it is likely that some noise in biological systems is actually advantageous, but this is difficult to demonstrate directly [71–73].
Focusing only on the potential disadvantage of noise, it is obvious that the noise resulting from promiscuity only becomes threatening under circumstances where the incorrect interactions or incorrect reactions are effectively competing with the ‘correct’ ones [74]. This will depend on the concentrations of the ligands and enzymes and their relative affinities and functional parameters. Therefore, even high levels of promiscuity for an individual enzyme or several enzymes may have no negative impact on the system as a whole. Alternatively a modest negative impact may be preferable to the cost of achieving higher levels of fidelity. The key point is that perfection is expensive. If it is possible for biological systems to create perfectly specific enzymes and proteins, and this is an open question, then it would likely come at some cost. Therefore, we are left wondering whether the widespread messiness we have come to appreciate is, instead, a reflection of biological genius. It is tempting to suggest that the promiscuity inherent in biology is tolerated with minimal detriment rather than corrected at high cost. Even that suggestion, possibly, falls short of the potential clever exploitation of promiscuity. In light of the role that promiscuity plays in facilitating evolution [43–46], it is likely that the messiness is advantageous on the evolutionary time scale even if modestly disadvantageous on the time scales of an individual’s life span. These aspects of biological promiscuity remain to be clarified, in general, but specifically for the steroid systems included in this volume. The steroid pathways may uniquely frame the questions that emerge about biochemical systems with the realization that the participating enzymes and receptors are promiscuous. Steroid receptors exhibit promiscuous agonism and steroid metabolizing enzymes demonstrate catalytic promiscuity, substrate promiscuity and product promiscuity. Some of these promiscuous reactions are tolerated by ‘gating mechanisms’ wherein a receptor is exposed to only a single sterol ligand due to expression of the enzyme that generates it, without expression of the enzyme that generates its potential, promiscuous, competitor ligand. The promiscuity of the receptor has no cost in the context of its localization. Similarly, for example, estrogen receptor is ‘protected’ from promiscuous interactions with 5α-androstane-3β,17β-diol, to which it binds in vitro, by expression in the same cells of CYP7B1, which metabolizes it via 7α-hydroxlyation. The metabolite has much weaker affinity for the receptor. This type of ‘cross talk’ between steroid ligands and receptors and enzymes is common yet it remains uncertain how much noise is tolerated. Additional work is needed to distinguish which of these reactions is noise, to understand mechanism for controlling the noise, and to identify examples of useful promiscuity within these steroid pathways.
6. Summary
Several ‘flavors’ of promiscuous behavior by enzymes and proteins have become widely appreciated. Although some difference of opinion remains concerning the best definitions for these types of behavior, our collective appreciation for promiscuity has matured. In many cases the biological utility of promiscuity is apparent. In other cases promiscuity has no obvious function and it introduces ‘biological noise’ that may simply be tolerated by organisms. Some steroid metabolizing enzymes exhibit catalytic, substrate, and product promiscuity, as do several detoxication enzymes. Continued effort with steroid systems is needed to distinguish between biologically useful promiscuity vs. biologically tolerated promiscuity in pathways that exhibit crosstalk between steroid metabolizing enzymes and receptors.
Acknowledgments
WMA thanks Professor Abhinav Nath for helpful discussion. This work was supported by the Department of Medicinal Chemistry, University of Washington.
Footnotes
This article is part of a Special Issue entitled ‘Steroid/Sterol signaling’.
References
- [1].Neurath H, Walsh KA. Role of proteolytic enzymes in biological regulation (a review) Proc. Natl. Acad. Sci. U. S. A. 1976;73(11):3825–3832. doi: 10.1073/pnas.73.11.3825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Fersht A, Winter G. Protein engineering. Trends Biochem. Sci. 1992;17(8):292–295. doi: 10.1016/0968-0004(92)90438-f. [DOI] [PubMed] [Google Scholar]
- [3].Russell AJ, Fersht AR. Rational modification of enzyme catalysis by engineering surface charge. Nature. 1987;328(6130):496–500. doi: 10.1038/328496a0. [DOI] [PubMed] [Google Scholar]
- [4].Fersht AR, Shi JP, Knill-Jones J, Lowe DM, Wilkinson AJ, Blow DM, Brick P, Carter P, Waye MM, Winter G. Hydrogen bonding and biological specificity analysed by protein engineering. Nature. 1985;314(6008):235–238. doi: 10.1038/314235a0. [DOI] [PubMed] [Google Scholar]
- [5].Sinha N, Smith-Gill SJ. Electrostatics in protein binding and function. Curr. Protein Pept. Sci. 2002;3(6):601–614. doi: 10.2174/1389203023380431. [DOI] [PubMed] [Google Scholar]
- [6].Perona JJ, Hadd A. Structural diversity and protein engineering of the aminoacyl-tRNA synthetases. Biochemistry. 2012;51(44):8705–8729. doi: 10.1021/bi301180x. [DOI] [PubMed] [Google Scholar]
- [7].Pocker Y, Guilbert LJ. Catalytic versatility of erythrocyte carbonic anhydrase. Kinetic studies of the enzyme-catalyzed hydrolysis of methyl pyridyl carbonates, Biochemistry. 1972;11(2):180–190. doi: 10.1021/bi00752a007. [DOI] [PubMed] [Google Scholar]
- [8].Pocker Y, Meany JE. The catalytic versatility of erythrocyte carbonic anhydrase. II. Kinetic studies of the enzyme-catalyzed hydration of pyridine aldehydes, Biochemistry. 1967;6(1):239–246. doi: 10.1021/bi00853a037. [DOI] [PubMed] [Google Scholar]
- [9].Khersonsky O, Tawfik DS. Enzyme promiscuity: a mechanistic and evolutionary perspective. Annu. Rev. Biochem. 2010;79:471–505. doi: 10.1146/annurev-biochem-030409-143718. [DOI] [PubMed] [Google Scholar]
- [10].Tokuriki N, Tawfik DS. Protein dynamism and evolvability. Science. 2009;324(5924):203–207. doi: 10.1126/science.1169375. [DOI] [PubMed] [Google Scholar]
- [11].Nobeli I, Favia AD, Thornton JM. Protein promiscuity and its implications for biotechnology. Nat. Biotechnol. 2009;27(2):157–167. doi: 10.1038/nbt1519. [DOI] [PubMed] [Google Scholar]
- [12].Basu MK, Poliakov E, Rogozin IB. Domain mobility in proteins: functional and evolutionary implications. Brief Bioinform. 2009;10(3):205–216. doi: 10.1093/bib/bbn057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Tawfik DS. Messy biology and the origins of evolutionary innovations. Nat. Chem. Biol. 2010;6(10):692–696. doi: 10.1038/nchembio.441. [DOI] [PubMed] [Google Scholar]
- [14].Glasner ME, Gerlt JA, Babbitt PC. Mechanisms of protein evolution and their application to protein engineering. Adv. Enzymol Relat. Areas Mol. Biol. 2007;75:193–239. doi: 10.1002/9780471224464.ch3. [DOI] [PubMed] [Google Scholar]
- [15].Noy N. Ligand specificity of nuclear hormone receptors: sifting through promiscuity. Biochemistry. 2007;46(47):13461–13467. doi: 10.1021/bi7018699. [DOI] [PubMed] [Google Scholar]
- [16].Giuseppe M, Luigia M, Elena P, Yan F, Luigi M. Enzyme promiscuity in the hormone-sensitive lipase family of proteins. Protein Pept. Lett. 2012;19(2):144–154. doi: 10.2174/092986612799080400. [DOI] [PubMed] [Google Scholar]
- [17].Mohamed MF, Hollfelder F. Efficient, crosswise catalytic promiscuity among enzymes that catalyze phosphoryl transfer. Biochim. Biophys. Acta. 2013;1834(1):417–424. doi: 10.1016/j.bbapap.2012.07.015. [DOI] [PubMed] [Google Scholar]
- [18].Lin F, Das D, Lin XN, Marsh EN. Aldehyde-forming fatty acyl-CoA reductase from cyanobacteria: expression, purification and characterization of the recombinant enzyme. FEBS J. 2013;280(19):4773–4781. doi: 10.1111/febs.12443. [DOI] [PubMed] [Google Scholar]
- [19].Roskoski R., Jr. ERK1/2 MAP kinases: structure, function, and regulation. Pharmacol. Res. 2012;66(2):105–143. doi: 10.1016/j.phrs.2012.04.005. [DOI] [PubMed] [Google Scholar]
- [20].Di Cera E. Serine proteases. IUBMB Life. 2009;61(5):510–515. doi: 10.1002/iub.186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Guy JL, Lambert DW, Warner FJ, Hooper NM, Turner AJ. Membrane-associated zinc peptidase families: comparing ACE and ACE2. Biochim. Biophys. Acta. 2005;1751(1):2–8. doi: 10.1016/j.bbapap.2004.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Huntington JA. Thrombin plasticity. Biochim. Biophys. Acta. 2012;1824(1):246–252. doi: 10.1016/j.bbapap.2011.07.005. [DOI] [PubMed] [Google Scholar]
- [23].Liu Y, Lv T, Ren J, Wang M, Wu Q, Zhu D. The catalytic promiscuity of a microbial 7α-hydroxysteroid dehydrogenase. Reduction of non-steroidal carbonyl compounds. Steroids. 2011;76(10–11):1136–1140. doi: 10.1016/j.steroids.2011.05.001. [DOI] [PubMed] [Google Scholar]
- [24].Eick GN, Colucci JK, Harms MJ, Ortlund EA, Thornton JW. Evolution of minimal specificity and promiscuity in steroid hormone receptors. PLoS Genet. 2012;8(11):e1003072. doi: 10.1371/journal.pgen.1003072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Lathe R, Kotelevtsev Y. Steroid signaling: ligand-binding promiscuity, molecular symmetry, and the need for gating. Steroids. 2014;82:14–22. doi: 10.1016/j.steroids.2014.01.002. [DOI] [PubMed] [Google Scholar]
- [26].Cook DL, Atkins WM. Enhanced detoxication due to distributive catalysis and toxic thresholds: a kinetic analysis. Biochemistry. 1997;36(36):10801–10806. doi: 10.1021/bi971284b. [DOI] [PubMed] [Google Scholar]
- [27].Guengerich FP. Mechanisms of cytochrome P450 substrate oxidation: MiniReview. J. Biochem. Mol. Toxicol. 2007;21(4):163–168. doi: 10.1002/jbt.20174. [DOI] [PubMed] [Google Scholar]
- [28].Armstrong RN. Structure, catalytic mechanism, and evolution of the glutathione transferases. Chem. Res. Toxicol. 1997;10(1):2–18. doi: 10.1021/tx960072x. [DOI] [PubMed] [Google Scholar]
- [29].Bock KW. Human UDP-glucuronosyltransferases: feedback loops between substrates and ligands of their transcription factors. Biochem. Pharmacol. 2012;84(8):1000–1006. doi: 10.1016/j.bcp.2012.07.009. [DOI] [PubMed] [Google Scholar]
- [30].Allali-Hassani A, Pan PW, Dombrovski L, Najmanovich R, Tempel W, Dong A, Loppnau P, Martin F, Thornton J, Edwards AM, Bochkarev A, Plotnikov AN, Vedadi M, Arrowsmith CH. Structural and chemical profiling of the human cytosolic sulfotransferases. PLoS Biol. 2007;5(5):e97. doi: 10.1371/journal.pbio.0050097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Roberts AG, Yang J, Halpert JR, Nelson SD, Thummel KT, Atkins WM. The structural basis for homotropic and heterotropic cooperativity of midazolam metabolism by human cytochrome P450 3A4. Biochemistry. 2011;50(50):10804–11018. doi: 10.1021/bi200924t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Harrelson JP, Atkins WM, Nelson SD. Multiple-ligand binding in CYP2A6: probing mechanisms of cytochrome P450 cooperativity by assessing substrate dynamics. Biochemistry. 2008;47(9):2978–2988. doi: 10.1021/bi702020y. [DOI] [PubMed] [Google Scholar]
- [33].Darbyshire JF, Iyer KR, Grogan J, Korzekwa KR, Trager WF. Substrate probe for the mechanism of aromatic hydroxylation catalyzed by cytochrome P450. Drug Metab. Dispos. 1996;24(9):1038–1045. [PubMed] [Google Scholar]
- [34].Senior AE. Two ATPases. J. Biol. Chem. 2012;287(36):30049–30062. doi: 10.1074/jbc.X112.402313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Schmitt KC, Rothman RB, Reith ME. Nonclassical pharmacology of the dopamine transporter: atypical inhibitors, allosteric modulators, and partial substrates. J. Pharmacol. Exp. Ther. 2013;346(1):2–10. doi: 10.1124/jpet.111.191056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Jasper JR, Insel PA. Evolving concepts of partial agonism. The beta-adrenergic receptor as a paradigm. Biochem. Pharmacol. 1992;43(2):119–130. doi: 10.1016/0006-2952(92)90268-n. [DOI] [PubMed] [Google Scholar]
- [37].Mark WY, Liao JC, Lu Y, Ayed A, Laister R, Szymczyna B, Chakrabartty A, Arrowsmith CH. Characterization of segments from the central region of BRCA1: an intrinsically disordered scaffold for multiple protein–protein and protein–DNA interactions? J. Mol. Biol. 2005;345(2):275–287. doi: 10.1016/j.jmb.2004.10.045. [DOI] [PubMed] [Google Scholar]
- [38].Uversky VN, Oldfield CJ, Dunker AK. Showing your ID: intrinsic disorder as an ID for recognition, regulation and cell signaling. J. Mol. Recognit. 2005;18(5):343–384. doi: 10.1002/jmr.747. [DOI] [PubMed] [Google Scholar]
- [39].Kirschner K, Bisswanger H. Multifunctional proteins. Annu. Rev. Biochem. 1976;45:143–166. doi: 10.1146/annurev.bi.45.070176.001043. [DOI] [PubMed] [Google Scholar]
- [40].Cheng XY, Huang WJ, Hu SC, Zhang HL, Wang H, Zhang JX, Lin HH, Chen YZ, Zou Q, Ji ZL. A global characterization and identification of multifunctional enzymes. PLoS One. 2012;7(6):e38979. doi: 10.1371/journal.pone.0038979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Kivirikko KI, Myllylä R, Pihlajaniemi T. Protein hydroxylation: prolyl 4-hydroxylase, an enzyme with four cosubstrates and a multifunctional subunit. FASEB J. 1989;3(5):1609–1617. [PubMed] [Google Scholar]
- [42].Loenen WA, Dryden DT, Raleigh EA, Wilson GG. Type I restriction enzymes and their relatives. Nucleic Acids Res. 2014;42(1):20–44. doi: 10.1093/nar/gkt847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Khersonsky O, Tawfik DS, Mander L, Lui H-W, editors. Vol. 8. Oxford; Elsevier: 2010. Comprehensive Natural Products II Chemistry and Biology; pp. 48–90. [Google Scholar]
- [44].Dellus-Gur E, Toth-Petroczy A, Elias M, Tawfik DS. What makes a protein fold amenable to functional innovation? Fold polarity and stability trade-offs. J. Mol. Biol. 2013;425(14):2609–2621. doi: 10.1016/j.jmb.2013.03.033. [DOI] [PubMed] [Google Scholar]
- [45].Tokuriki N, Jackson CJ, Afriat-Jurnou L, Wyganowski KT, Tang R, Tawfik DS. Diminishing returns and tradeoffs constrain the laboratory optimization of an enzyme. Nat. Commun. 2012;3:1257. doi: 10.1038/ncomms2246. [DOI] [PubMed] [Google Scholar]
- [46].Chakraborty S. An automated flow for directed evolution based on detection of promiscuous scaffolds using spatial and electrostatic properties of catalytic residues. PLoS One. 2012;7(7):e40408. doi: 10.1371/journal.pone.0040408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Williams GJ, Zhang C, Thorson JS. Expanding the promiscuity of a natural-product glycosyltransferase by directed evolution. Nat. Chem. Biol. 2007;3(10):657–662. doi: 10.1038/nchembio.2007.28. [DOI] [PubMed] [Google Scholar]
- [48].Díaz Arenas C, Cooper TF. Mechanisms and selection of evolvability: experimental evidence. FEMS Microbiol. Rev. 2013;37(4):572–582. doi: 10.1111/1574-6976.12008. [DOI] [PubMed] [Google Scholar]
- [49].Willis JR, Briney BS, DeLuca SL, Crowe JE, Jr., Meiler J. Human germline antibody gene segments encode polyspecific antibodies. PLoS Comput. Biol. 2013;9(4):e1003045. doi: 10.1371/journal.pcbi.1003045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].James LC, Roversi P, Tawfik DS. Antibody multispecificity mediated by conformational diversity. Science. 2003;299(5611):1362–1367. doi: 10.1126/science.1079731. [DOI] [PubMed] [Google Scholar]
- [51].Castagnoli L, Costantini A, Dall'Armi C, Gonfloni S, Montecchi-Palazzi L, Panni S, Paoluzi S, Santonico E, Cesareni G. Selectivity and promiscuity in the interaction network mediated by protein recognition modules. FEBS Lett. 2004;567(1):74–79. doi: 10.1016/j.febslet.2004.03.116. [DOI] [PubMed] [Google Scholar]
- [52].Dunker AK, Cortese MS, Romero P, Iakoucheva LM, Uversky VN. Flexible nets. The roles of intrinsic disorder in protein interaction networks. FEBS J. 2005;272(20):5129–5148. doi: 10.1111/j.1742-4658.2005.04948.x. [DOI] [PubMed] [Google Scholar]
- [53].Peggs KS, Allison JP. Co-stimulatory pathways in lymphocyte regulation: the immunoglobulin superfamily. Br. J. Haematol. 2005;130(6):809–824. doi: 10.1111/j.1365-2141.2005.05627.x. [DOI] [PubMed] [Google Scholar]
- [54].Patil A, Kinoshita K, Nakamura H. Domain distribution and intrinsic disorder in hubs in the human protein-protein interaction network. Protein Sci. 2010;19(8):1461–1468. doi: 10.1002/pro.425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Pollice A, Vivo M, La Mantia G. The promiscuity of ARF interactions with the proteasome. FEBS Lett. 2008;582(23–24):3257–3262. doi: 10.1016/j.febslet.2008.09.026. [DOI] [PubMed] [Google Scholar]
- [56].Lee J, Ozcan U. Unfolded protein response signaling and metabolic diseases. J. Biol. Chem. 2014;289(3):1203–1211. doi: 10.1074/jbc.R113.534743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Chakraborty S, Rao BJ. A measure of the promiscuity of proteins and characteristics of residues in the vicinity of the catalytic site that regulate promiscuity. PLoS One. 2012;7(2):e32011. doi: 10.1371/journal.pone.0032011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Chakraborty S, Ásgeirsson B, Rao BJ. A measure of the broad substrate specificity of enzymes based on ‘duplicate' catalytic residues. PLoS One. 2012;7(11):e49313. doi: 10.1371/journal.pone.0049313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Chakraborty S, Minda R, Salaye L, Dandekar AM, Bhattacharjee SK, Rao BJ. Promiscuity-based enzyme selection for rational directed evolution experiments. Methods Mol. Biol. 2013;978:205–216. doi: 10.1007/978-1-62703-293-3_15. [DOI] [PubMed] [Google Scholar]
- [60].Carbonell P, Faulon JL. Molecular signatures-based prediction of enzyme promiscuity. Bioinformatics. 2010;26(16):2012–2019. doi: 10.1093/bioinformatics/btq317. [DOI] [PubMed] [Google Scholar]
- [61].Carbonell P, Lecointre G, Faulon JL. Origins of specificity and promiscuity in metabolic networks. J. Biol. Chem. 2011;286(51):43994–44004. doi: 10.1074/jbc.M111.274050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Nath A, Atkins WM. A quantitative index of substrate promiscuity. Biochemistry. 2008;47(1):157–166. doi: 10.1021/bi701448p. [DOI] [PubMed] [Google Scholar]
- [63].Nath A, Zientek MA, Burke BJ, Jiang Y, Atkins WM. Quantifying and predicting the promiscuity and isoform specificity of small-molecule cytochrome P450 inhibitors. Drug Metab. Dispos. 2010;38(12):2195–2203. doi: 10.1124/dmd.110.034645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Durant JL, Leland BA, Henry DR, Norse JG. Reoptimization of MDL keys foruse in drug discovery. J. Chem. Inform. Comput. Sci. 2002;42:1273–1280. doi: 10.1021/ci010132r. [DOI] [PubMed] [Google Scholar]
- [65].Honaker MT, Acchione M, Zhang W, Mannervik B, Atkins WM. Enzymatic detoxication, conformational selection, and the role of molten globule active sites. J. Biol. Chem. 2013;288(25):18599–18611. doi: 10.1074/jbc.M112.445767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Honaker MT, Acchione M, Sumida JP, Atkins WM. Ensemble perspective for catalytic promiscuity: calorimetric analysis of the active site conformational landscape of a detoxification enzyme. J. Biol. Chem. 2011;286(49):42770–42776. doi: 10.1074/jbc.M111.304386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Hou L, Honaker MT, Shireman LM, Balogh LM, Roberts AG, Ng KC, Nath A, Atkins WM. Functional promiscuity correlates with conformational heterogeneity in A-class glutathione S-transferases. J. Biol. Chem. 2007;282(32):23264–23274. doi: 10.1074/jbc.M700868200. [DOI] [PubMed] [Google Scholar]
- [68].Foti RS, Honaker M, Nath A, Pearson JT, Buttrick B, Isoherranen N, Atkins WM. Catalytic versus inhibitory promiscuity in cytochrome P450s: implications for evolution of new function. Biochemistry. 2011;50(13):2387–2393. doi: 10.1021/bi1020716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Khersonsky O, Malitsky S, Rogachev I, Tawfik DS. Role of chemistry versus substrate binding in recruiting promiscuous enzyme functions. Biochemistry. 2011;50(13):2683–2690. doi: 10.1021/bi101763c. [DOI] [PubMed] [Google Scholar]
- [70].Gerlt JA, Babbitt PC. Enzyme (re)design: lessons from natural evolution and computation. Curr. Opin. Chem. Biol. 2009;13(1):10–18. doi: 10.1016/j.cbpa.2009.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Viney M, Reece SE. Adaptive noise. Proc. Biol. Sci. 2013;280(1767):20131104. doi: 10.1098/rspb.2013.1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Simpson ML, Cox CD, Allen MS, McCollum JM, Dar RD, Karig DK, Cooke JF. Noise in biological circuits. Wiley Interdiscip. Rev. Nanomed. Nanobiotechnol. 2009;1(2):214–225. doi: 10.1002/wnan.22. [DOI] [PubMed] [Google Scholar]
- [73].Ladbury JE, Arold ST. Noise in cellular signaling pathways: causes and effects. Trends Biochem. Sci. 2012;37(5):173–178. doi: 10.1016/j.tibs.2012.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Schreiber G, Keating AE. Protein binding specificity versus promiscuity. Curr. Opin. Struct. Biol. 2011;21(1):50–61. doi: 10.1016/j.sbi.2010.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]