Abstract
Apoptosis is known as programmed cell death. Some non-apoptotic cell death is increasingly recognized as genetically controlled, or ‘regulated’. However, the full extent and diversity of these alternative cell death mechanisms remains uncharted. Here, we surveyed the landscape of pharmacologically-accessible cell death mechanisms. Of 56 caspase-independent lethal compounds, modulatory profiling revealed ten inducing three types of regulated non-apoptotic cell death. Lead optimization of one of the ten resulted in the discovery of FIN56, a specific inducer of ferroptosis. Ferroptosis occurs when the lipid repair enzyme GPX4 is inhibited. We found that FIN56 promotes degradation of GPX4. We performed chemoproteomics to reveal that FIN56 also binds to and activates squalene synthase, an enzyme involved in the cholesterol synthesis, in a manner independent of GPX4 degradation. These discoveries reveal that dysregulation of lipid metabolism is associated with ferroptosis. This systematic approach is a means to discover and characterize novel cell death phenotypes.
Introduction
Cells die not merely as a consequence of catastrophic failure of homeostasis, but also when regulated or programmed cell death is activated1. Apoptosis s a well understood form of regulated cell death. The biological importance of apoptosis is highlighted by the fact that it is evolutionarily conserved in multicellular organisms2, and that it is utilized in developmental programs. The unique morphological and biochemical changes associated with apoptosis are due to the executioner caspase proteases. However, apoptosis is not the only form of regulated cell death. For example, cells commit to die upon tumor necrosis factor-alpha (TNF-α) treatment, even when catalytic activity of the executioner caspase-3 is inhibited3. Intriguingly, non-apoptotic cell death can be blocked by small molecules, and this concept is now appreciated as a type of regulated cell death, which has been implicated in several physiological contexts and pathological conditions1,4,5. Cells actively commit to die through non-apoptotic cell death in some physiological contexts, such as infection: damage- or pathogen-associated molecular patterns (DAMP or PAMP) are released from dying cells and trigger inflammatory responses of neighboring cells and immune cells6.
Ferroptosis is a non-apoptotic form of regulated cell death. Its relevance to certain pathological conditions has been reported, such as periventricular leukomalacia, nephrotic tubular death, and Huntington’s disease7,8. It is distinct from other regulated cell death phenotypes, such as apoptosis and necroptosis4,9. Ferroptosis is characterized by extensive lipid peroxidation, which can be suppressed by iron chelators or lipophilic antioxidants. Mechanistically, ferroptosis inducers are divided into two classes: (1) inhibitors of cystine import via system xc− (e.g., erastin)9,10, which subsequently causes depletion of glutathione (GSH)11, and (2) covalent inhibitors (e.g., (1S, 3R)-RSL3) of glutathione peroxidase 4 (GPX4)12. Since GPX4 reduces lipid hydroperoxides using GSH as a co-substrate13, both compound classes ultimately result in loss of GPX4 activity, followed by elevated levels of lipid reactive oxygen species (ROS) and consequent cell death.
There is some crosstalk among distinct regulated cell death phenotypes4,14. Each proposed cell death phenotype has generally been studied using different models15. However, a universal comparison of different cell death phenotypes would be highly beneficial to understand the mechanisms governing cell death.
We hypothesized that evaluating diverse pharmacologically-accessible cell death mechanisms would help map the landscape of cell death. Of 3,169 compounds evaluated as cell death probes, we found that 451 compounds were lethal without activating caspases. Of these, most activated unregulated necrotic death, but a technique termed modulatory profiling revealed that a subset of lethal compounds induced three types of regulated non-apoptotic cell death: metal-ion-dependent cell death, necrostatin-1-dependent cell death, and ferroptosis. Further optimization of one probe resulted in the discovery of FIN56, a new and specific inducer of ferroptosis. Ferroptosis has been reported to be induced when the lipid repair enzyme glutathione peroxide 4 (GPX4) is directly inhibited, or indirectly inactivated by depletion of glutathione12. We found that, in contrast to these mechanisms, FIN56 induced ferroptosis by inducing degradation of GPX4. We performed a chemoproteomic analysis to find that FIN56 binds to and activates squalene synthase, an enzyme involved in the cholesterol synthesis, to suppress non-steroidogenic metabolites – most likely coenzyme Q10 – in the mevalonate pathway, which enhances sensitivity to FIN56-induced ferroptosis, in a manner independent of GPX4 degradation. These discoveries together reveal that dysregulation of lipid metabolism is associated with ferroptosis.
Results
Modulatory profiling revealed three kinds of cell death
Towards mapping the landscape of cell death, we sought small molecule inducers of regulated, non-apoptotic cell death (Fig. 1a). We tested 3,169 lethal compounds for induction of caspase-independent cell death in HT-1080 fibrosarcoma cells and BJeLR engineered transformed fibroblasts, the cell lines used for the modulatory profiling experiment16. We found that 451 compounds (14%) triggered cell death without activation of caspases 3/7, detected using a fluorogenic substrate. These compounds were defined as ‘caspase-3/7-independent lethals’ (CILs) (Supplementary Results, Supplementary Fig. 1, Supplementary Note 1). Thus, while most lethal compounds activate caspase activity (irrespective of whether caspase activity is required for their lethality), a significant number of compounds were lethal without activating cleavage of this fluorogenic caspase activity probe.
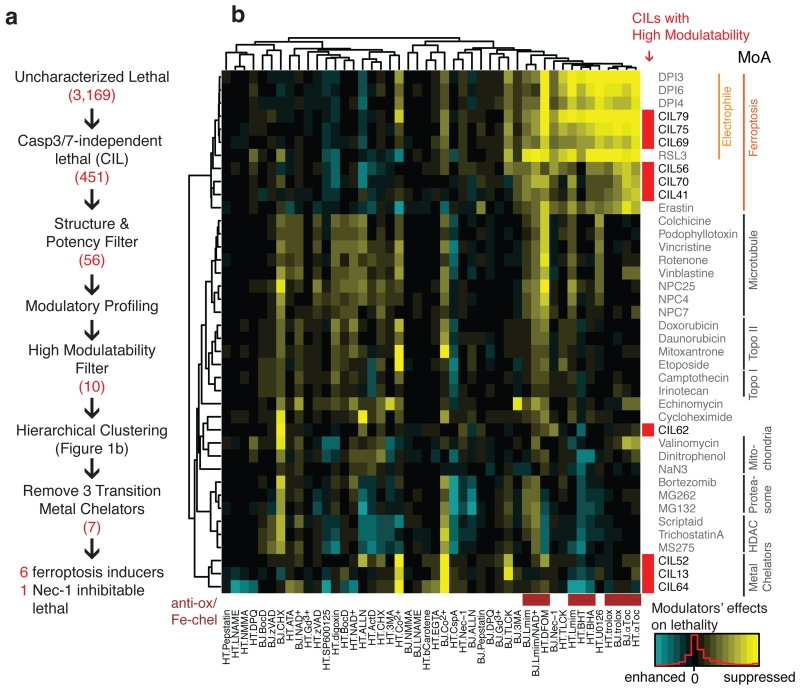
Figure 1. Modulatory profiling revealed three types of regulated non-apoptotic cell death.
a. Experimental scheme to identify regulated non-apoptotic cell death inducers with high modulatability. The numbers in red are the number of compounds satisfying each criterion. b. Hierarchical clustering of modulatory profiles of 10 CILs with high modulatability and 30 characterized lethal compounds from several classes of lethal mechanisms. Lethal compounds are shown on the right. 10 CILs are indicated in red. 46 modulators are shown on the bottom (28 death modulators in 2 cell lines, HT-1080 or BJeLR). Antioxidants and iron-chelators are indicated in brown. A detailed list of modulators is shown in the Supplementary Table 2. Supplementary Fig. 1–4 show additional data on the CIL screening scheme, modulatory profiling scheme, and structures and characterization of ten regulated non-apoptotic cell death inducers.
We examined in more detail 56 structurally diverse and potent (EC80 < 2.8 μg/mL) CILs using a modulatory profiling strategy. In a previous study, the clustering of modulatory profiles, or changes in potency and efficacy of a lethal compound induced by co-treatment with chemical death modulators (Supplementary Table 2), revealed that compounds with the same mechanism of action share similar modulatory profiles17. Toward evaluation of diverse regulated cell death programs, modulatory profiling should facilitate grouping of pharmacological agents by their induction of specific regulated cell death programs. We discovered ten compounds inducing three types of regulated non-apoptotic cell death. We further discovered a specific ferroptosis inducer through lead optimized and uncovered its mechanism of action. In this analysis, ten of the 56 CIL compounds exhibited “high modulatability”, meaning their lethality was significantly suppressed or enhanced by specific pharmacological or genetic agents17 (Supplementary Fig. 2; Supplementary Data Set 1); high modulatability correlates with activity through a specific lethal pathway. When analyzed along with other well-characterized lethal compounds, these CILs fell into three classes (Fig. 1b, Supplementary Fig. 3a). The first class (CIL13, 52, 64) acted via metal chelation, as these compounds were inhibited by cobalt (II), and bind to cobalt (II) in vitro (Supplementary Fig. 3b,c). The second class (CIL62) induces cell death that is suppressed by necrostatin-118 (Supplementary Fig. 3d,e; note that this does not necessarily imply necroptosis, as necrostatin-1 has necroptosis-independent effects19); and the third class (CIL41, 56 (1), 69, 70, 75, 79) comprises ferroptosis inducers, as suggested by their suppression by canonical ferroptosis inhibitors (iron chelators and lipophilic antioxidants; Supplementary Table 3), and clustering with known ferroptosis inducers. We focused further studies on the six CIL compounds that clustered with ferroptosis inducers.
Of the six ferroptotic CILs, three (CIL69, 75, 79) are putative electrophiles and clustered most closely with known electrophilic ferroptosis inducers, such as (1S, 3R)-RSL3 (Supplementary Fig. 4a). The remaining CILs consisted of two novel scaffolds: CIL41/70 and CIL56. CIL41/70 induced ROS accumulation, detected using H2-DCFDA (Supplementary Fig. 4b), and cell death that was strongly suppressed by ferroptosis inhibitors (the lipophilic antioxidant α−tocopherol, and the iron chelator deferoxamine) (Supplementary Fig. 4c).
All known ferroptosis inducers induce selective lethality in a BJ engineered cell line series, namely BJeH, BJeHLT, DRD, and BJeLR. These cells were initially created to demonstrate that normal human fibroblasts can be transformed into tumor cells by introducing defined genetic elements (human telomerase, SV40 small and large T antigens, and oncogenic HRASG12V)16. BJeLR and DRD, which overexpress oncogenic-Ras, were found to be more sensitive to ferroptosis inducers than BJeHLT and BJeH, which do not express oncogenic-Ras. CIL41/70, unlike all other ferroptosis-inducing compounds reported to date, did not exhibit oncogenic RAS selectivity in the BJ engineered cell line series20 (Supplementary Fig. 4d). Moreover, 203 commercially available structural analogs of CIL41/70 were tested and found to be less potent than CIL56 itself (Supplementary Note, Supplementary Fig. 4e). Given that CIL56 was the most potent compound, and that CIL56 retained some degree of selectivity towards oncogenic-RAS-expressing cells in the BJ series (Fig. 2a), we speculated that CIL56 was more likely than CIL41/70 to yield a potent and selective probe of ferroptosis for further mechanistic analysis. Thus, we selected CIL56 for more detailed characterization.
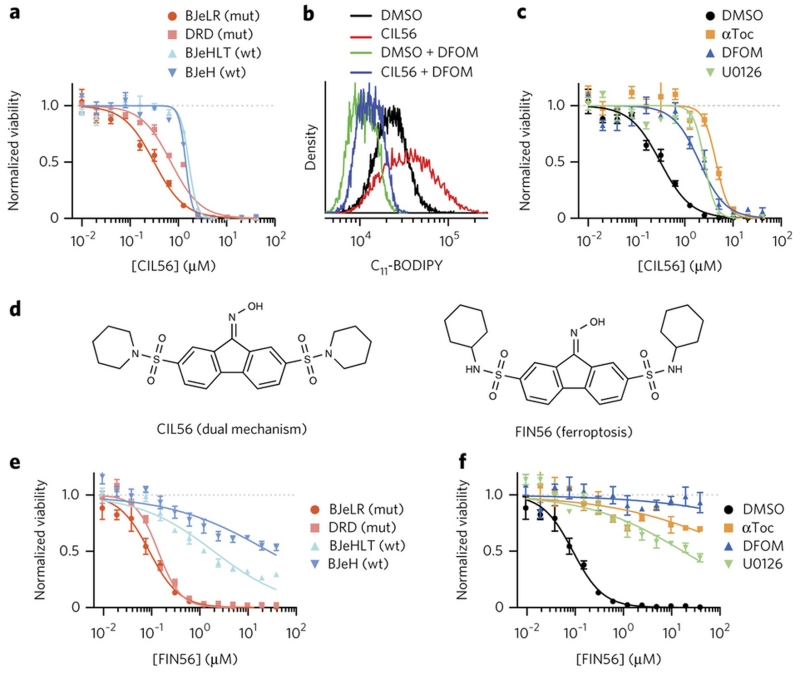
Figure 2. Optimization of CIL56 revealed a potent and selective ferroptosis inducer.
a, e. HRASG12V selectivity. Viability of four engineered BJ cell lines treated with (a) CIL56 or (e) FIN56 for 48 hrs. mut: cells tumor-transformed due to HRASG12V overexpression, wt: isogenic cells without HRASG12V. b. Lipid ROS generation. Flow cytometry analysis with BODIPY-581/591 C11 staining in HT-1080 cells incubated with test compounds for six hours. DFOM: 152 μM deferoxamine. c, f. Effects of ferroptosis inhibitors on viability of HT-1080 cells co-treated with (c) CIL56 or (f) FIN56 for 48 hrs. αToc: 100 μM α-tocopherol, U0126: 3.8 μM. d. Chemical structures of CIL56 and FIN56. See Supplementary Fig. 5 for structure activity relationship around the CIL56 scaffold. Experiments in a-f were performed in biological triplicates, and single representative results were shown; error-bars indicate s.e.m. of technical triplicates.
Probe optimization led to a specific ferroptosis inducer
CIL56 induces iron-dependent ROS (Fig. 2b). Antioxidants and iron chelators only suppressed the lethality of low concentrations of CIL56 (Fig. 2c). We thus hypothesized that CIL56 was capable of engaging two independent death pathways: ferroptosis at low concentrations, and a necrotic, non-suppressible phenotype at higher concentrations. We sought to identify a more selective analog of CIL56 that retained the ability to induce ferroptosis, but lacked the ability to induce the other form of death. Structure activity relationship analysis of the CIL56 scaffold determined that the oxime moiety was crucial to induce ferroptosis, and that hydrophobicity of the piperidine moieties of CIL56 correlated with potency (Supplementary Fig. 5). Ultimately, we discovered an analog (SRS7–34) that has cyclohexylamine moieties in place of the piperidine moieties of CIL56 (Fig. 2d). This new compound, termed FIN56 (2) (Ferroptosis inducer derived from CIL56), exhibited greater potency as well as greater oncogenic RAS selectivity in the BJ cell line series than CIL56 (Fig. 2e); it was also fully suppressed by the ferroptosis inhibitors deferoxamine and α-tocopherol (Fig. 2f), indicating that it engages only ferroptosis and does not have the ability to engage the second death mechanism activated by CIL56.
FIN56-induced ferroptosis involves decreased GPX4 protein
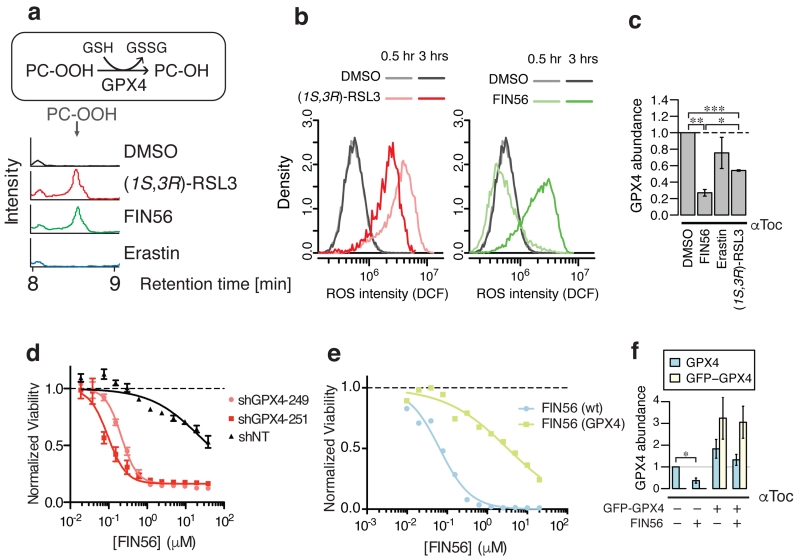
Previously reported ferroptosis inducers either deplete glutathione (caused by inhibiting cystine uptake) or are covalent GPX4 inhibitors. In the NCI60 cell line panel21, we found that GPX4 inhibitors were more cell-line selective than compounds inducing glutathione depletion (Supplementary Fig. 6a)22. In this regard, FIN56 was more similar to other GPX4 inhibitors than to GSH synthesis inhibitors. Supporting this, we found that FIN56 did not deplete glutathione, suggesting that it also did not block cystine import; however, FIN56 did cause the loss of GPX4 activity in cell lysates (Fig. 3a, Supplementary Fig. 6b). Intriguingly, compared to the covalent GPX4 inhibitor (1S, 3R)-RSL3, FIN56 was slower to induce the accumulation of ROS (Fig. 3b), suggesting that it did not cause loss of GPX4 activity via direct inhibition of enzymatic activity. Indeed, we determined that the abundance of GPX4, but not that of the related selenoprotein glutathione peroxidase 1 (GPX1), was substantially decreased following FIN56 treatment (5 μM, 10 hr) (Fig. 3c, Supplementary Fig 6c,d). Neither erastin nor (1S, 3R)-RSL3 affected the levels of GPX4 to the extent of FIN56, demonstrating that this effect was specific to FIN56. While GPX4 knockdown enhanced FIN56 lethality (Fig. 3d), FIN56-induced cell death was suppressed by GFP-GPX4 fusion protein overexpression (Fig. 3e,f, Supplementary Fig. 6e,f), suggesting that the loss of GPX4 protein was critical for FIN56-induced ferroptosis. We also confirmed that selenite supplementation, which is known to upregulate expression of selenoproteins, including GPX423, also suppressed FIN56 lethality (Supplementary Fig. 6g,h). The decrease in GPX4 protein was not inhibited by α-tocopherol, indicating that it was not a downstream consequence of lipid ROS generation. Together, these results suggested that FIN56 triggers ferroptosis through a mechanism involving the regulation of GPX4 protein abundance.
Figure 3. FIN56-induced ferroptosis decreases GPX4 expression.
a. GPX4 enzymatic activity in BJeLR cells upon ferroptosis inducer treatment. b. Kinetics of ROS generation upon 0.5 μM (1S, 3R)-RSL3 or 5 μM FIN56 treatment detected with 25 μM H2-DCFDA staining in BJeLR cells. c. GPX4 protein abundance in BJeLR cells upon co-treatment with 100 μM αToc and ferroptosis inducers for ten hours. d. The effect of shGPX4 on FIN56-induced ferroptosis in BJeLR cells. e. The effects of GFP-GPX4 fusion protein overexpression on sensitivity to FIN56 in BJeLR cells. f. The effects of GFP-GPX4 fusion protein overexpression on endogenous and exogenous GPX4 protein abundance upon FIN56 treatment. Supplementary Fig. 6 supports more connection between FIN56 and GPX4 as well as corresponding westerns to c and f. Statistical significance (paired two-tailed t-test) – *: p < 0.05, **: p < 0.005, ***: p < 0.0005, †: p < 0.01, ††: p < 0.001, n.s.: not significant. Experiments in a-f were done in biological triplicates. Single representative results are shown for a,b,d,e; error-bars in d indicate s.e.m. of technical triplicates.
We further investigated how the decrease of GPX4 protein is caused by FIN56. We found that the GPX4 transcript level increased rather than decreased upon FIN56 treatment (Supplementary Fig. 6i), which suggests that FIN56-induced depletion of GPX4 protein is not mediated by transcriptional changes in GPX4 mRNA. Observations that cells treated with cycloheximide, which inhibit ribosome function, did not substantially decrease GPX4 protein levels compared to a housekeeping gene α-tubulin (Supplementary Fig. 6j), and that knockdown of tRNA isopentenyltransferase 1 (TRIT1) 24, which has been reported to be required for selenoprotein synthesis including GPX4, did not dramatically affect GPX4 protein abundance (Supplementary Fig. 6k-m), suggested that FIN56 did not inhibit GPX4 protein synthesis, but rather it induced post-translational GPX4 protein degradation. It is not clear, however, how GPX4 degradation is regulated, because a proteosome inhibitor, MG132, did not inhibit GPX4 degradation significantly, nor protect cells from FIN56 lethality. It is of note that GPX4 was reported to decrease when the proto-oncogene serine/threonine-protein (Pim) kinases were inhibited25; however, this is not relevant to FIN56 lethality because pan-Pim kinase inhibitors did not induce ferroptosis, as these compounds were not suppressed by α-tocopherol (Supplementary Fig. 6n).
SQS encoded by FDFT1 gene as a target protein of FIN56
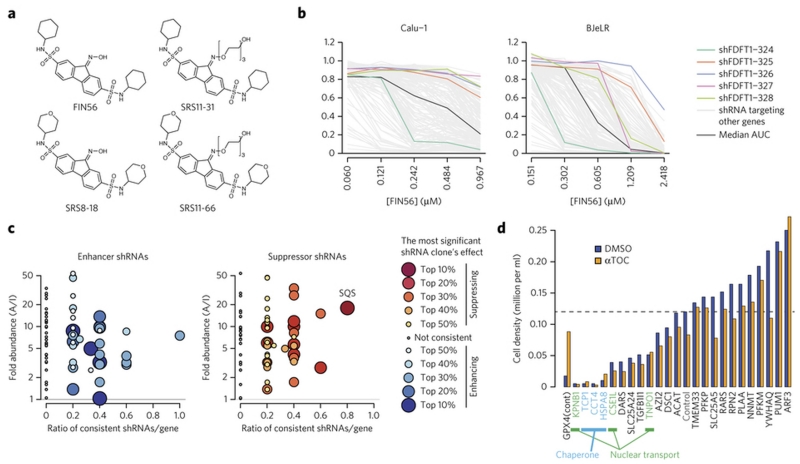
To better understand the mechanism of action of FIN56, we decided to seek direct binding proteins of FIN56 using a chemoproteomic approach. First, we explored structural analogs of FIN56. This resulted in creation of SRS11-31, an analog with a polyethylene glycol (PEG) moiety, which induces ferroptosis at 10-fold higher EC50 than FIN56 (Fig. 4a, Supplementary Fig. 7). On the other hand, substitution of the cyclohexyl moiety in FIN56 with a 4-tetrahydropyran (SRS8-18 (3)) or its PEG-conjugate (SRS11-66) (4) resulted in complete loss of activity. Next, both SRS11-31 (5) (active, or A) and SRS11-66 (inactive, or I) were conjugated to Profinity epoxide resin through an epoxy ring-opening reaction, and the resins were incubated with HT-1080 whole cell lysates. The pull-down proteins found with each probe were identified and quantified by mass spectrometry. 70 proteins excluding universally expressed proteins (actins, tubulins and ribosome subunits) were found to be more abundant on the resin conjugated with the active probe.
Figure 4. Squalene synthase (SQS) encoded by FDFT1 as FIN56’s target protein.
a. Active and inactive FIN56 analogs with PEG linkers. Supplementary Fig. 7 shows their potency in HT-1080 cells. b. Effects of five shRNAs against FDFT1 on FIN56. The results in two of the four cell lines were shown. Five shRNA clones targeting FDFT1 are shown in polychromatic lines. A black line indicates a shRNA which gives no effect (median AUC among tested shRNAs) in each cell line. Grey lines indicate shRNAs targeting other genes. c. Summary of proteomic target identification and shRNA screening targeting 70 identified genes on FIN56. Each dot summarizes the result of multiple shRNAs targeting a gene. Each shRNA is considered ‘consistent’ when it exerts indicated effect (enhancing or suppressing FIN56). X-axis shows the ratio: the number of consistent shRNAs inducing indicated (i.e., enhancing or suppressing FIN56) effects to the total number of shRNAs targeting the gene. Y-axis shows fold-enrichment of protein abundance on active versus inactive probe-beads in pull-down assay. See Supplementary Fig. 8 for more description. d. Effect of siRNAs against ‘loss-of-function’ candidates on BJeLR cell viability. Cells were grown under DMSO or ATOC (α-tocopherol)’s existence. shRNA screens in b,c were performed once in four cell lines. siRNA experiment in d was performed in BJeLR twice and mean of biological replicates.
We tested whether these candidate target proteins were inhibited (loss-of-function) or activated (gain-of-function) by FIN56 to induce ferroptosis using RNA interference (RNAi). RNAi-mediated knockdown of the relevant target should enhance or suppress FIN56 sensitivity, depending on FIN56’s mechanism of action (Supplementary Fig. 8a). Expression of many genes is affected by off-target effects of RNAi, whose phenotype may differ from cell line to cell line; however, on-target effects should be more likely consistent among different cell lines. Therefore, we examined the effects of up to five shRNA clones per gene encoding 70 candidate target proteins in four independent ferroptosis-susceptible cell lines, i.e., HT-1080, BJeLR, Calu-1 lung adenocarcinoma and 143B osteosarcoma (Fig. 4b, Supplementary Fig.8b,c). Candidate FIN56 targets responsible for ferroptosis were sought using two criteria: (i) high selective abundance on the active vs. inactive probe resins, and (ii) high proportion of consistently performing shRNAs in all four cell lines subjected to RNAi-mediated silencing of each gene (Fig. 4c, Supplementary Fig.8d). Among the 70 tested proteins, we found proteins, such as chaperones and nuclear transport proteins, whose knockdown not only enhanced FIN56 lethality, but also induced toxicity by itself. However, their lethality was not suppressed by α-tocopherol, unlike GPX4 knockdown, indicating that these cell death phenotypes were distinct from ferroptosis. Perturbation of these essential proteins by FIN56 might contribute to its potency, but they are not the primary targets of FIN56 (Fig. 4d).
Validating the functional relevance of the target
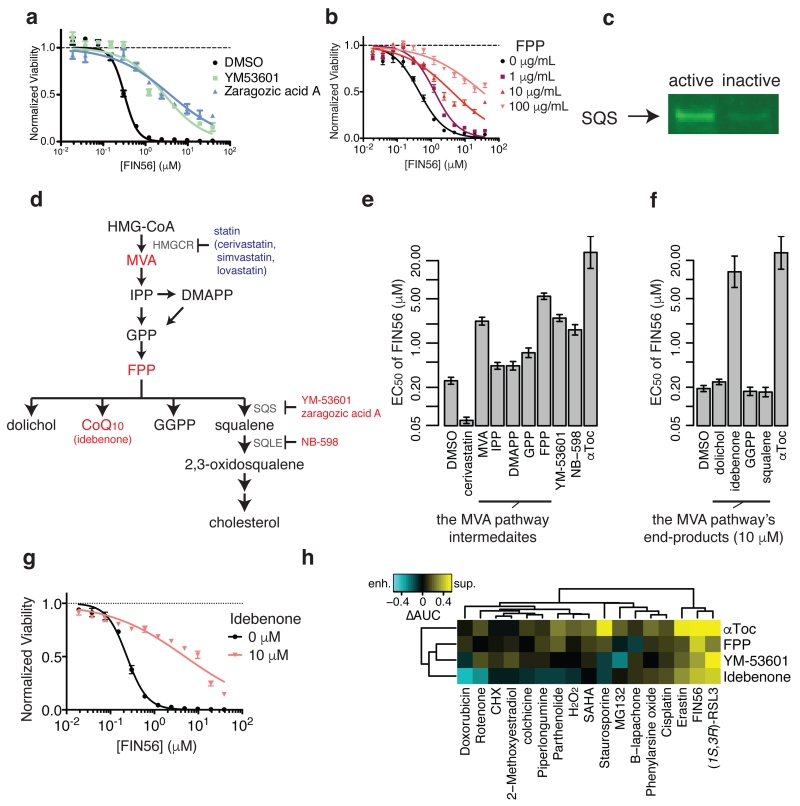
We found that four of the five shRNAs against FDFT1 mRNA (which encodes SQS protein) suppressed FIN56 consistently in all four cell lines tested, indicating that FIN56 activates, rather than inhibits, SQS (a ‘gain-of-function’ model). Therefore, we explored this possibility and studied how the FIN56-SQS interaction is relevant to FIN56’s lethality. We confirmed that not only shRNAs targeting FDFT1, but also small molecule inhibitors of the SQS activity (YM-53601 and zaragozic acid A), suppress FIN56 lethality (Fig. 5a). SQS is an enzyme acting downstream of 3-hydroxy-3-methylglutaryl-CoA (HMG-CoA) reductase in the mevalonate pathway. SQS couples two farnesyl pyrophosphate (FPP) molecules to form squalene. Inhibition of SQS consequently increases the pool of FPP. FPP is essential for multiple processes, including protein prenylation and metabolite synthesis (e.g., sterols, coenzyme Q10 (CoQ10), dolichol and heme) 26, some subset of which may be relevant to the modulatory effect on ferroptosis sensitivity. Supplementation with FPP indeed suppressed the lethality of FIN56 (Fig. 5b). We also examined SQS-FIN56 binding, by confirming that SQS from HT-1080 whole cell lysate binds selectively to active probes versus an inactive probe (Fig. 5c). Moreover, bacterially-expressed truncated human SQS protein27 (with the lipophilic N and C termini removed) binding to an active affinity probe was efficiently suppressed by pre-incubation of purified SQS protein with FIN56, suggesting that SQS and FIN56 directly interact (Supplementary Fig. 9).
Figure 5. Validating SQS as the functionally relevant target to FIN56’s lethality.
a. Effects of chemical inhibitors of SQS on FIN56’s lethality. b. Effects of Farnesyl-PP on FIN56’s lethality. c. Detection of SQS using pull-down assay from HT-1080 whole cell lysate with active or inactive probes. Note that the probes are the same as what were used for chemoproteomic target identification. d. Schematics of the mevalonate pathway. Large letters are metabolites, small letters are enzymes responsible for the reactions or small molecules. Red and blue letters indicate the molecules (inhibitors or metabolites) suppressed or enhanced FIN56’s lethality. The detailed results are shown in e and f. e. Perturbation of the mevalonate pathway and their effects on FIN56’s lethality. Concentrations: cerivastatin (1 μM), metabolites (100 μM), YM-53601 (5 μM), NB-598 (25 μM), αToc (100 μM). f. Supplementation of 10 μM end-products of the MVA pathway and their effects on FIN56. g. Effect of 10 μM idebenone on FIN56 in HT-1080. h. Modulatory profiling between the modulators of the MVA pathway and various lethal compounds inducing oxidative stress. See Supplementary Fig. 9-–10 for SQS pull-down with competition and effect of statins on FIN56 lethality. a,b,g were performed in biological replicates and error-bars are s.e.m. of technical triplicates; e,f in biological duplicates and error-bars are standard errors of EC50 estimation from sigmoidal curve-fitting; h in singlicate. Full image of c is in Supplementary Figure 14.
We focused on the role of upstream processes from FPP in the mevalonate pathway. We found that statins, chemical inhibitors of HMG-CoA reductase, enhanced FIN56 lethality (Supplementary Fig. 10a). HMG-CoA reductase synthesizes mevalonic acid (MVA). Supplementation of MVA reversed the effect of cerivastatin, the most potent and selective statin tested (Supplementary Fig. 10b,c), demonstrating that the HMG-CoA-reductase-inhibiting effect of cerivastatin is responsible for its enhancement of FIN56 lethality. However, we also realized that their effects did not affect GPX4 abundance (Supplementary Fig. 10d).
More extensive investigation of the mevalonate pathway revealed that supplementation of FPP suppresses FIN56 more significantly than other isoprenoid pyrophosphates, which suggests that FPP is functionally relevant to regulating the sensitivity to FIN56 (Fig. 5d). We also found that an inhibitor of squalene monooxygenase (SQLE), which is a rate-limiting step downstream of SQS in cholesterol synthesis28, as well as an SQS inhibitor, suppressed FIN56, supporting the notion that non-steroidogenic products of the mevalonate pathway contribute to suppression of FIN56 (Fig. 5e). Among these metabolites derived from FPP, we found that idebenone, a hydrophilic analog of CoQ10, was the only suppressor of FIN56-induced ferroptosis (Fig. 5f,g), suggesting that CoQ10 is the connection between the mevalonate pathway and regulation of sensitivity to FIN56. Note that supplementation of CoQ10 is known to be ineffective due to its extreme hydrophobicity29. To further assess the specificity of the three mevalonate pathway modulators (FPP, YM-53601, and idebenone) as well as α-tocopherol, we utilized a modulatory profiling scheme in HT-1080 cells with lethal compounds, including ones inducing oxidative stress: we found that both GPX4 inhibitors, FIN56 and (1S,3R)-RSL3, were potently suppressed by all the compounds (Fig. 5h). Erastin, a GSH depletor, was also suppressed somewhat by the modulators, while the rest of compounds showed more distinct patterns. These results suggest that the mevalonate pathway modulators are more specific inhibitors of ferroptosis, rather than of lethality in general.
CoQ10 is an electron carrier in the mitochondrial respiratory chain and an endogenous antioxidant. However, it does not function in regulating ferroptosis via utilizing either of these properties. 143B cells with or without mitochondrial DNA (ρ+ and ρ0 cells, respectively) were both sensitive to FIN56-induced ferroptosis (Supplementary Fig. 11a,b) suggesting no involvement of the respiratory chain in ferroptosis; moreover, while the lipophilic anti-oxidant α-tocopherol suppressed staurosporine-induced cell death, idebenone enhanced it. Finally, although both α-tocopherol and idebenone inhibited lipid ROS generation upon FIN56 treatment, idebenone did not change basal lipid ROS level, unlike α-tocopherol (Supplementary Fig. 11c). Thus, exactly how CoQ10 protects cells from FIN56 remains elusive, but may involve reprogramming lipid metabolism in a way that is not conducive to the execution of ferroptosis.
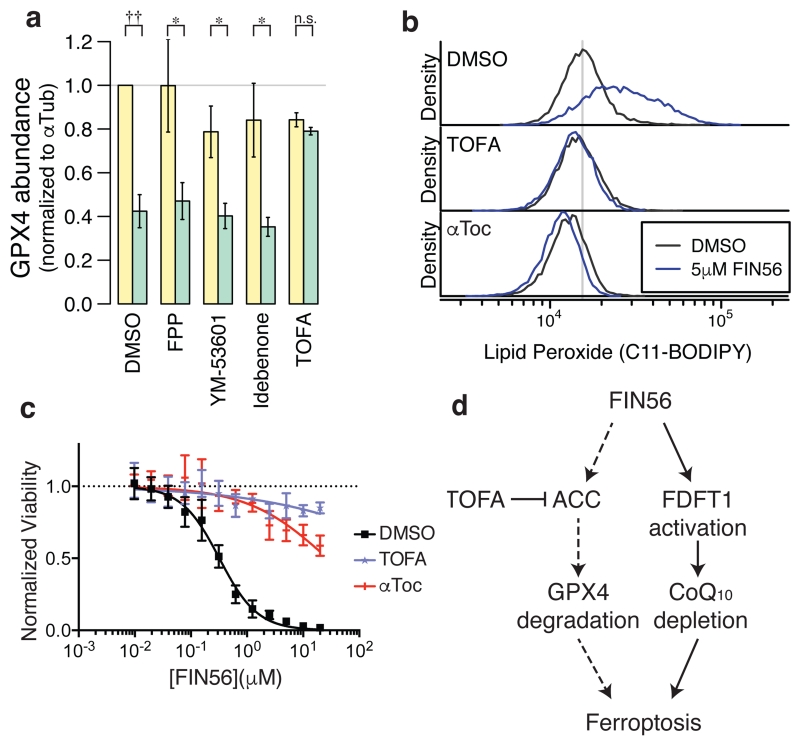
An ACC inhibitor prevents GPX4 protein degradation
Modulators of the mevalonate pathway, such as idebenone, are potent suppressors of ferroptosis, particularly of direct and indirect GPX4 inhibitors (FIN56 and (1S,3R)-RSL3) (Fig. 5h); however, these compounds did not block the decrease of GPX4 protein upon FIN56 treatment or induce its overexpression (Fig. 6a, Supplementary Fig. 12), indicating there may be an additional pathway that regulates GPX4 protein level in response to FIN56 treatment. We discovered that 5-(tetradecyloxy)-2-furoic Acid (TOFA), an inhibitor of acetyl-CoA carboxylase (ACC), inhibited the loss of GPX4. TOFA was also found to be a potent suppressor of FIN56 and suppressed lipid ROS generation upon FIN56 treatment (Fig. 6b,c). ACC is an enzyme involved in fatty acid synthesis. However, ACC itself was not identified as a direct FIN56 target and the detailed mechanism linking FIN56 to ACC remains to be understood.
Figure 6. ACC inhibitor prevents GPX4 protein degradation.
a. Effects of the mevalonate pathway modulators and ACC inhibitor on GPX4 abundance with or without FIN56. The corresponding gel is shown in Supplementary Fig. 12. b. Lipid peroxide levels upon TOFA or αToc treatments. c. Effects of TOFA and αToc on FIN56 lethality. d. Model of FIN56’s mechanism of action. Dotted arrows indicate the mechanistic details are still elusive. Statistical significance (paired two-tailed t-test) – *: p < 0.05, ††: p < 0.001, n.s.: not significant. Supplementary Fig. 11d also supports that the mevalonate pathway modulators did not change GPX4 abundance. a–c were performed in biological triplicates. mean and s.e.m. are shown in a; single representative results were shown in b and c. error-bars are s.e.m. of technical triplicates in c.
In conclusion, characterization of FIN56 revealed that its mechanism involves two distinct pathways (Fig. 6d): one pathway is degradation of GPX4, which requires the enzymatic activity of ACC; the other pathway is activation of SQS, which leads to coenzyme Q10 depletion. Together these effects result in potent induction of ferroptosis.
Discussion
Initially, ferroptosis inducers were discovered through screens for oncogenic-RAS synthetic lethality in BJ engineered cells. It may not be a coincidence that HRASG12V not only induces malignant transformation, but it also sensitizes cells to ferroptosis inducers in these cells, as overexpression of oncogenic Ras was reported to modulate lipid metabolism30–32. Indeed, our studies suggest that fatty acid synthesis and the mevalonate pathway both regulate sensitivity to ferroptosis through distinct mechanisms. These and other studies of regulated non-apoptotic cell death and its intersection with lipid metabolism are still at an early stage.
This systematic discovery and characterization of different cell death phenotypes has suggested that ferroptosis is a significant form of pharmacologically accessible cell death. In the future, additional pharmacological cell death stimuli examined with modulatory profiling may reveal yet additional cell death mechanisms22. The inventory of regulated cell death discovered by these systematic approaches may provide insight into a global view of the possibilities of cell death.
Methods
Chemicals
3,169 uncharacterized lethal compounds, as well as structural analogs of CIL41/70, were purchased from Asinex, ChemBridge, ChemDiv, Enamine, InterBioScreen, MayBridge, TimTec, Vitas M Labs, and Zelinsky. Chemical modulators (Supplementary Note) used in modulatory profiling were obtained as described before17. U-0126 was obtained from LC Laboratories. Deferoxamine mesylate, α-tocopherol, sodium selenite, zaragozic acid A, mevalonolactone and simvastatin were purchased from Sigma-Aldrich. Cerivastatin was obtained from Waterstone Technology. Lovastatin were obtained from Santa Cruz. were purchased from Cayman Chemical. Isoprenoid pyrophopsphates (isopentenyl-PP, dimethylallyl-PP, geranyl-PP, farnesyl-PP and geranylgeranyl-PP) ammonium salt were purchased from Isoprenoids. Idebenone was purchased from Tocris Bioscience. Pim kinase inhibitors, CX-6258 and AZD-1208, were purchased from Selleck Chemicals. Erastin and (1S, 3R)-RSL3 were synthesized as described previously (Yagoda et al, 2007, Yang et al., 2014) 12,33. CIL56, FIN56 and their structural analogs were synthesized as described (#WO2008140792A1)34. Building blocks for these compounds were purchased from Matrix Scientific and Sigma Aldrich. Purity of the purchased compounds were not assessed. Structures of 56 CILs as well as CIL41/70 analogs are in Supplementary Note 1. Total synthesis of CIL56 analogs and their 1H NMR data are described in Supplementary Note 2.
Cell Lines and Media
Four engineered BJ cell lines (BJeLR/DRD/BJeHLT/BJeH) were obtained from Robert Weinberg (Whitehead Institute). 143B cells (osteosarcoma) were from Eric Schon (Columbia University). Calu-1 (lung adenocarcinoma) and HT-1080 (fibrosarcoma) cells were from American Type Culture Collection. The four BJ cell lines were grown in DMEM High-Glucose media (Life Technologies), 20% Medium 199 (Sigma), and 15% heat-inactivated fetal bovine serum (FBS). HT-1080 cells were grown in DMEM High-Glucose media with 1% non-essential amino acids (Life Technologies) and 10% FBS. 143B cells were grown in DMEM High-Glucose media with 1% glutamine and 10% FBS. Calu-1 cells were grown in McCoy’s 5A media (Life Technologies) supplemented with 10% FBS. All the cell lines were grown at 37°C under 5% CO2.
Cell Viability Assay
1000 cells/36 μl were seeded in each well in 384-well plates. Lethal compounds were dissolved and a 2-fold, 12-point dilution series were prepared in DMSO. Compound solutions were further diluted with media at 1:25 and 4 μl/well of the diluted solutions were added to cell cultures immediately after cells were seeded. When ferroptosis inhibitors (100 μM α-tocopherol, 152 μM deferoxamine, or 10 μM U-0126) were co-treated with lethal inducers, they were supplemented to cell culture at the same time as lethal compounds were added, and the cells were incubated for 24 hrs. When other cell death modulating compounds (100 nM sodium selenite, 1 μM cerivastatin, 100 μg/mL mevalonic acid) were co-treated, they were first supplemented to cell culture for 24 hrs before lethal compounds were added to cell culture and further incubated for 24 hrs at 37°C under 5% CO2. On the day of the viability measurement, 10 μl/well of 50% Alamar Blue diluted in media (Life Technologies) was added and further incubated at 37°C for 6 hrs. Fluorescence intensity (ex/em: 530/590) was measured with a Victor 3 plate reader (Perkin Elmer) and the normalized viability was calculated by VL = (IL – I0)/(IV – I0), where VL, I0, IV, and IL are the normalized viability, raw fluorescence intensities from the wells containing media, cells treated with a vehicle (negative control), and cells with the lethal compound (L), respectively. When the effect of a chemical modulator (M) on L was calculated, we instead used the equation: VL|M = (IM,L – I0)/(IM,V – I0), where VL|M, IM,L and IM,V were the normalized viability, and fluorescence intensity from cells treated with M and V, and from cells with M and L. respectively. The viability was typically measured in biological triplicates unless otherwise specified. A representative dose-response curve, the mean and standard error of normalized viability from one replicate were plotted. HT-1080 viability upon modulator treatments corresponding to Fig. 3c, 5e, 5f are available in Supplementary Fig. 13a–c.
Caspase-3/7 Activation Assay
Apo-ONE Homogeneous Caspase-3/7 Assay (Promega) was used according to manufacturers’ protocol with a minor modification. First, we optimized the assay (Supplementary Fig. 1a,b). HT-1080 cells were seeded at 1000 cells/40 μl in each well of a 384-well plate, incubated for one hour and treated with test compounds for different durations (from 3.5 to 48 hrs). 15 μl culture media was aspirated from each well and 5 μl of a mixture of lysis buffer and Caspase-3/7 fluorogenic substrate from the kit was added. Plates were kept in the dark at room temperature for 16 hrs, and the fluorescence (ex/em: 490/535) of each well was measured using a Victor 3 plate reader. In the optimization, the fluorescence of apoptosis inducer-treated cells started increasing after 6 to 12 hrs and lethal compounds that induce strong positive signals in the end (i.e., apoptosis inducers) were distinguished from non-apoptotic inducers as early as after 18 hrs. So we treated cells with lethal compounds for 18 hrs in further analysis. In the screening mode, we incubated HT-1080 cells with screening molecules for 18 hrs, processed cells, and measured the fluorescence. Compounds inducing the fluorescence with similar intensity as vehicle (DMSO) treatments were defined as Caspase-Independent Lethals (CILs).
Discovery of Novel Ferroptosis Inducers Using Modulatory Profiling
Collection of Caspase Independent Lethals (CILs)
We first collected uncharacterized synthetic compounds from various vendors for multiple different screening purposes. Of them, we found 3,169 compounds to be lethal in BJeLR cells. We first sought compounds that induce non-apoptotic cell death in two ferroptosis susceptible cell lines, HT-1080 and BJeLR. 451 compounds were tested at 5.3 μg/mL in these two cell lines, and 451 compounds showed (i) an EC80 < 2.8 μg/mL in both cell lines and (ii) no activation of caspase-3/7 at 5.3 μg/mL. Cell viability and caspase-3/7 activity assays were performed as described before. 95 percentile of Apo-ONE fluorescence from 0.13% DMSO treated cells was set as threshold of caspase-independent lethality, and the raw fluorescent value of each well was divided by the threshold for normalization; lethal compounds were considered caspase-3/7-independent when the normalized fluorescent values were less than one. These compounds were defined as caspase-3/7 independent lethals (CILs). Next, we computed the structural similarity among the 451 compounds based on Pubchem’s fingerprint35, and removed structurally similar compounds (cutoff: Tanimoto coefficient is 0.9). We also removed compounds that did not satisfy Lipinski’s rule of five with a minor modification (no more than five hydrogen bond donors, no more than ten hydrogen bond acceptors, molecular weight ranging from 250 to 500, a partition coefficient log P not greater than five), and compounds whose biological activities were known. Finally, we retested the selected compounds in 2-fold dilution series in the two cell lines, and closely examined 56 compounds with low EC50 in modulatory profiling.
Modulatory Profiling of CILs
56 CILs were examined using modulatory profiling (Supplementary Fig. 2)17. HT-1080 and BJeLR cells were seeded at 1000 cells/40μl in each well of a 384 well plate. They were co-treated with a lethal compound (L) and a death modulator (M) in technical triplicates. 56 CILs were added to cells in a 2-fold, 14-point dilution series. Death modulators were reagents known to perturb cell death signaling pathways, and were treated at a single concentration, as described before17. Cells were incubated with lethal compounds and modulators for 48 hrs and the normalized viability was measured as described above. For each combination of L and M, the area under the dose-response curve (AUC) was computed. An effect of M on L (EM|L) was represented by the difference of AUCs between the modulator (AUCM,L) and the vehicle (AUCV,L), or EM|L=AUCM,L-AUCV,L (Supplementary Fig. 2b). When M suppresses or enhances the effect of L, EM|L>0 or EM|L<0, respectively.
The modulatability of each L (mL) was an average of the absolute value of the effect of all M’s on L, or , where nM is the number of modulators, 46. Large mL indicates that L induces a selective cell death phenotype17. We computed mL for each CIL and each known lethal compound from different classes of mechanism of action (i.e., HDAC inhibitors, proteosomal inhibitors, mitochondrial uncouplers, topoisomerase I and II inhibitors, microtubule destabilizers, and ferroptosis inducers). mL of CILs larger than the median value of known lethal compounds’ mL were considered ‘high’; we identified high-modulatability CILs. For generating hierarchical clustering of the modulatory profiles, the distance between modulatory profiles (P) of two compounds Pi and Pj was defined by dij=1–corrPerason(Pi,Pj) and hierarchical clustering with average-linkage method was used to generate dendrograms.
Compounds’ Cobalt (II) Binding Assay in vitro
For each cluster I compounds (CIL13, 52, 64), 2-fold dilution series were prepared with DMSO. 30 μL/well of either water or 10 μM cobalt (II) chloride solution were added to each well of a 384 well plate. Dilution series of each compound in DMSO were added by 10 μL/well. After solutions were mixed by shaking the plate using Victor 3 plate reader (Perkin Elmer), absorbance was scanned between 300 and 700 nm for each compound. Finally the following ΔΔAbsorbance for each compound was plotted in Supplementary Figure 3b:
where Abs is an Absorbance at each wavelength.
Search of Commercially Available Structural Analogs of CIL41/70
Structural analogs possessing the core scaffold of CIL41/70 (SMILES: C(=O)([*])O/N=C([*])/N ) were searched for in eMolecules (http://www.emolecules.com) and purchased from the vendors described above. Their lethality was tested in HT-1080 cells in triplicate.
Analysis of Reactive Oxygen Species Generation
200,000 HT-1080 or BJeLR cells were grown in 6-well plates at 37°C for 16 hrs. Cells were treated with test compounds for the indicated time, then trypsinized, pelleted and washed once with PBS. For lipophilic or aqueous ROS detection, cells were resuspended in Hanks Balanced Salt Solution (HBSS, Life Technologies) containing test compounds as well as C11-BODIPY(581/591) (2 μM) or H2-DCFDA (25 μM), respectively (Life Technologies), and incubated for 10 mins at 37°C. Cells were then pelleted, resuspended in 500 uL HBSS, strained through a 40 μm cell strainer (BD Falcon), and analyzed using BD Accuri C6 flow cytometer (BD Biosciences). Both dyes were measured in the FL1 channel. Experiments were done in biological triplicates and representative results are shown.
Glutathione Quantification Assay
500,000 HT-1080 cells were seeded in 10 cm dish. Cells were grown at 37°C for 16 hrs. On the day of the analysis, cells were co-treated with 100 μM α-tocopherol and either vehicle (DMSO) or a ferroptosis inducer (10 μM erastin, 0.5 μM (1S,3R)-RSL3, or 5 μM FIN56) and incubated for 10 hrs. Cells were then trypsinized, pelleted, washed once with 400 μL ice-cold PBS containing 1 mM EDTA and sonicated. After the cell debris was pelleted and removed, both oxidized and reduced glutathione in 120 μL of sample was quantified in technical triplicates using QuantiChrome glutathione assay kit (BioAssay Systems). The glutathione quantity was normalized to protein concentration measured with Bradford assay (Bio-Rad).
GPX4 Enzymatic Activity assay
17 million BJeLR cells were seeded in 225 cm2 tissue culture flasks (Corning). Vehicle (0.1 % DMSO for 11 hrs) or test compounds were added (10 μM erastin for 11 hrs, 0.5 μM (1S,3R)-RSL3 for 2 hrs, or 5 μM FIN56 for 10 hrs). GPX4 enzymatic activity assay was performed as described previously12. Briefly, ten million cells were resuspended in cell lysis buffer. Cells were sonicated to make cell lysates followed by centrifugation at 14,000 rpm for 10 mins to make cleared cell lysates. Protein concentration of the cleared cell lysates was determined using Bradford protein assay. 200 μg of cellular protein was mixed with phosphatidyl choline hydroperoxide (PC-OOH), a GPX4 specific substrate, and reduced glutathione, a GPX4 co-factor. The mixture was incubated at 37°C for 30 mins followed by lipid extraction using chloroform:methanol (2:1) solution. The lipid extract was evaporated using a Rotavap, and re-dissolved in 100% ethanol before injecting into LC-MS instrument for PLPC quantitation.
Reverse Transcription-quantitative PCR
0.2 to 1 million cells grown in 6 well dishes were trypsinized, pelleted, resuspended in Buffer RLT (Qiagen), and homogenized with QIAshredder (Qiagen). RNA was further extracted using RNeasy Mini Kit (Qiagen). 2 μg of extracted RNA from each sample was converted to cDNA using TaqMan reverse transcription reagents (Life Technologies). qPCR primers were designed to detect all splicing variants using Primer Express 2.0 (Applied Biosystems). The designed primers were confirmed to amplify only the designated gene transcripts using in-silico PCR (http://genome.ucsc.edu/cgi-bin/hgPcr). For qPCR reactions, primers, cDNA and Power SYBR Green PCR Master Mix (Applied Biosystems) was mixed, and quantitation was performed using a StepOnePlus real-rime PCR system (Applied Biosystems). Experiments were done in biological triplicates, and the mean and the standard error of a representative result was shown.
Western Blot
300,000 cells (HT-1080 or BJeLR) were seeded per well in 6 well plates. When cells were co-treated with ferroptosis inducers (10 μM erastin for 11 hrs, 0.5 μM (1S,3R)-RSL3 for 2 hrs, or 5 μM FIN56) and 100 μM α-tocopherol, they were added to cell culture at the same time and incubated for 10 hrs (Fig. 2e). When other death modulating compounds (100 nM sodium selenite, 1 μM cerivastatin, 100 μg/mL mevalonic acid, 30 μM C75, or 2 μM cerulenin) were co-treated, cell were pre-incubated with the modulators for 24 hrs before treated with ferroptosis inducers for 6 hrs. Cell lysis, SDS-PAGE and protein transfer to PVDF membrane were performed as previously described9. Antibodies used: anti-human α-tubulin antibody (Santa Cruz, sc-32293, 1:10,000 dilution), anti-human GPX4 antibody (Abcam, ab41787, 1:2,000 dilution), anti-human GPX1 antibody (R&D Systems, AF3798, 1:1,000 dilution), anti-human SQS antibody (Abcam, ab109723 for full length, ab195046 for truncated, both at 1:1000 dilution). Secondary antibodies were from LI-COR (1:3,000 dilution). The PVDF membrane labeled with primary and secondary antibodies were scanned using an Odyssey Imaging System (LI-COR). Experiments were done in biological triplicates, and the mean and the standard error of intensity was plotted. Full gel images are shown in Supplementary Figure 14.
Gene Knockdown Experiment
shRNAs were designed by the RNAi consortium were used and gene knockdown was performed as described previously9. For siRNA mediated gene knockdown experiments, HT-1080 cells were reverse transfected with 5 nM siRNA. siRNA targeting GPX4 (Dharmacon, #L-011676-00), TRIT1 (Dharmacon, #L-018831-02) or non-targeting (Qiagen) were mixed with 2 μL Lipofectamine RNAiMAX (Invitrogen) in a well of a 12-well plate. After incubation for 30 mins at room temperature, 30,000 cells were added to each well and knockdown was allowed to proceed for 48 hrs. Cells were then harvested and re-seeded for RT-qPCR, viability assay, and western blots. For RT-qPCR, cells were re-seeded into 12 well plates and harvested the following day as described above. For viability assay, 1000 cells/well were re-seeded into 384 well plates for 24 hrs before compound addition for another 24 hrs before addition of Alamar Blue. Compounds were added in a 12-point, 2-fold dilution series, with the highest concentration of compound being 36.5 μM (erastin), 10 μM ((1S,3R)-RSL3), 38.7 μM (FIN56). For western blots, cells were harvested 48 hrs after knockdown (no re-seeding).
FIN56 Target Identification
Conjugation of active and inactive probes with Profinity Epoxide Resin
6 μmol of Active (SRS11-31) or inactive (SRS11-66) probes dissolved in 500 μL DMSO and 300mg of Profinity™ Epoxide Resin (Bio-Rad) were incubated in saturated sodium bicarbonate at 45 °C for 3 days. The conjugation reaction was ended by adding 120 μL of 1 M ethanolamine to the reaction mixture. The conjugated probe-beads were used for further protein pull-down assay.
Affinity Chromatography with active and inactive probe beads
8 million HT-1080 cells were seeded in two 15 cm polystyrene tissue culture dishes and grown overnight. Culture media was removed from dishes and cells were washed five times with cold PBS. After PBS was completely removed, the cells were treated with 2 mL per plate of lysis buffer (25 mM MOPS (pH 7.2), 15 mM EGTA, 15 mM MgCl2, 2 mM DTT, 1 mM sodium orthovanadate, 1 mM sodium fluoride, 0.5% NP-40, 60 mM β-glycerophosphate, protease inhibitor cocktail (Sigma-Aldrich P8340)) were added, scraped, and collected. After cells were agitated at 4 °C for 15 minutes, insoluble components were precipitated at 14,000 × g at 4 °C for 10 minutes and the supernatant was removed and the protein concentration was measured using Bradford assay.
250 μg protein (up to 400 μL of the whole cell lysate) each was incubated with active and inactive probe-beads, filled with 20 mL of bead buffer (50 mM Tris-HCl (pH 7.4), 250 mM NaCl, 5 mM EDTA, 5 mM EGTA, 5 mM NaF, 0.1% NP-40) and incubated at 4 °C for 12 hours. Beads were then collected using Poly-prep chromatography columns (Bio-Rad). Beads were further washed with 8 mL bead buffer for three times, and finally the beads were transferred to eppendorf tubes.
Bacterial expression and purification of truncated human SQS for competition assay
The plasmid encoding the truncated human squalene synthase (SQS 31-370) in pET28a expression vector, kindly provided by Chia-I Liu and Andrew H.J. Wang (Academia Sinica, Taiwan), was confirmed by DNA sequencing (Gene Wiz, Inc.) and then used to transform Escherichia coli BL21-Gold (DE3) competent cells (Agilent Technologies). The cells with the construct were grown in LB media supplemented with 100 μg/mL ampicillin at 37° C until OD600nm reached 1. Protein expression was induced with 0.5mM isopropyl β-D-thiogalactoside (IPTG) at 17° C overnight (12-13 hr). Cells were pelleted (4,000 × g, 20 min, 4° C) and lysed by sonication in SQS Buffer (50 mM Tris, pH 7.4, 250 mM NaCl, 5 mM imidazole, 5 mM MgCl2, 1 mM TCEP). Cell lysate was then centrifuged at 15,000 × g, 45 min, 4° C. The supernatant was loaded onto a chromatography column containing Ni Sepharose 6 Fast Flow beads (GE Life Sciences) equilibrated with SQS Buffer. After two washes and one non-specific wash of the beads, the bound SQS was eluted with 250 mM imidazole in the same buffer. The purity of eluted fractions was verified by SQS-PAGE as more than 90% pure. The fractions containing SQS were concentrated, flash frozen, and stored at −80° C. Protein concentration was determined using absorbance at 280 nm with molar extinction coefficient (ε) 42,860 M−1 cm−1 (for reduced SQS with N-terminal His12 tag as calculated from amino acid sequence by ExPASy ProtParam).
SQS competition assay with purified truncated SQS, FIN56 and active probe
Purified truncated SQS was prepared as described above. The protein solution was diluted with bead buffer (50 mM Tris-HCl, pH 7.4, 250 mM NaCl, 5 mM EDTA, 5 mM EGTA, 5 mM NaF, 0.1% NP-40, 1 mM TCEP). 190 μL of 1 or 10 ng/mL SQS solution was mixed with 10 μL of DMSO or competitor solution (2mM FIN56 solution in DMSO), where DMSO and/or FIN56 were at the final concentrations of 5% and 100 μM, respectively. This mixture was rotated and incubated at 4 °C for 2 hrs. 15 uL of active probe-beads solution (33% slurry) was added to the solution and incubated for another 2 hrs. The beads was spun down (500 rpm, 4°C, 1 min), supernatant was removed and washed with 500 μL bead buffer for three times totally. After the third wash, 20 μL of 3× SDS sample loading buffer was added and boiled at 95 °C for 5 mins. Samples were spun for 1 min at 13,000 rpm in the table top centrifuge and 10 μL of each was loaded on SDS-PAGE. SQS was confirmed using immunoblotting.
Proteomic Analysis for Target identification
At Quantitative Proteomics Core at Columbia University, the proteins in pull-down samples with active (SRS11-31) and inactive (SRS11-66) beads were eluted at 80 °C in 50 mM ammonium bicarbonate with 0.1% Rapigest detergent with a protease inhibitors cocktail P8340 (Sigma-Aldrich). Biological triplicate culture and affinity pulldowns for bound active and inactive compound were prepared. Cysteines in the protein samples were reduced with dithiothreitol and alkylated with iodoacetamide, and proteins were digested with trypsin (6 ng/μL Promega Corp, #V511A in 50 mM ammonium bicarbonate). A digest of yeast alcohol dehydrogenase (50 fmol) was added as an internal detection control.
Three chromatograms were recorded for each of six biological replicates (three active, three inactive), yielding 12 chromatograms. Analytical separation was on a NanoAcquity UPLC (Waters Corp.), with a 120-minute chromatogram on a 75 μm ID × 25 cm HSS T3 1.8 μm particle diameter reverse phase C18 column at a flowrate of 300 nL/min with an acetonitrile/formic acid gradient at 45°C. Identification and quantitation of proteins bound to the beads was performed by label-free proteomic profiling on a Synapt G2 HDMS (quadrupole-time-of-flight) mass spectrometer (Waters Corp.) using data-independent scanning (MSE) as described previously12 except that spectra were recorded in positive ion sensitivity mode without ion mobilty. Spectra were searched against a human Uniprot complete proteome with ProteinLynx Global Server version 2.5 RC9 (Waters Corp.) and post-processed with Elucidator software Ver. 3.3.0.1.SP3_CRE52.21 (Ceiba Solutions, Inc.) as described previously36.
Target validation using custom shRNA library
Custom lentiviral shRNA library were generated using plasmids encoding the first generation the RNAi consortium (TRC) shRNAs targeting 70 genes identified in chemoproteomics following the RNAi consortium’s instruction. These lentiviral shRNAs were laid out in an arrayed format using 384 well plates, and infected in four cell lines (HT-1080, BJeLR, Calu-1, and 143B). Our rational of the screening design was that if shRNAs target the bona fide FIN56 targets, the shRNAs should show ‘consistent’ FIN56-enhancing or suppressing effects in all four cell lines, depending on how FIN56 acts on the target protein (Supplementary Fig. 8a). If the shRNAs act on off-targets to change sensitivity to FIN56, their effects should be less consistent. Gene knockdown was performed as described above; 400 cells per well in 384 well plates were seeded and incubated for a day, lenvirus containing shRNAs were infected in them on the next day, and puromycin was added two days after infection. Cells were treated with 2-fold 8-point dilution seires of FIN56 after 24 hrs and incubated for another 48 hrs before alamar blue was added and incubated for 6 hrs.
Data analysis for discovering functionally-relevant targets
Alamar blue fluorescence intensity data were normalized as described above. For each cell line, dose-curves of FIN56 for each shRNA treatment were plotted and overlaid on a single plot first. Of eight tested concentrations of FIN56, extreme (low or high) concentrations that did not kill or completely kill cells treated with any shRNAs were removed; eventually 4 or 5 more informative concentrations were used for further analysis, as in Fig. 4b and Supplementary Fig. 8b. For each shRNA treatment, area under dose-curve (AUC) upon FIN56 treatment in dilution series was computed, rank-ordered across all tested shRNAs in each cell line. Based on its ‘consistency’ across four cell lines and the magnitude of effects, shRNAs were classified into 11 categories (five consistently suppressing groups (top 10%, top 20%, etc. five consistently enhancing groups (top 10%, etc.), and non-consistent). Note that ‘non-consistent’ indicates that the shRNA induced FIN56-enhancing effects in some cell lines and FIN56-suppressing effects in others. A gene is ranked based on the consistent effects of shRNAs targeting it. When at least one of the shRNAs targeting a gene shows consistent FIN56-enhancing or –suppressing effect, the gene was considered potentially functionally relevant target of FIN56, through ‘loss-of-function’ or ‘gain-of-function’ scenarios, respectively.
Validation of ‘loss-of-function’ targets using pooled-siRNAs
Candidates of ‘loss-of-function’ target of FIN56 was further targeted by pooled-siRNAs. Previously, pooled-siRNA against GPX4, target of RSL3, was shown to phenocopy the RSL3; siGPX4 induced ferroptosis that was suppressed by α-tocopherol as well as siGPX4 induce oncogenic-Ras selectivity across BJ series. Expecting that siRNAs against bona fide ‘loss-of-function’ target induce ferroptosis, we treated siRNAs against the candidates as well as GPX4 as a positive control. The transfection of siRNAs in BJeLR cells was performed in BJeLR cells. 1 mL Opti-MEM (Life Technologies) mixed with 20 pmol of pooled-siRNA and 5 μL RNAiMAX (Life Technologies) was incubated to form complex for 15 mins, and aliquoted 500 μL/well in 6 well dishes. Next, 120,000 BJeLR/1.5 mL in each well were seeded and grown for two days. Cells were then trypsinized and seeded again at 120,000 per well and grown with or without supplementation of α-tocopherol. After two days, cells were trypsinized and cell density was measured using an automated cell counter (Vicell, Beckman Coulter).
Statistical Analysis and Data Visualization
Dose-response curve plotting and EC50 computation were performed with Prism 5.0c. p-values between the differences in EC50’s were computed using Akaike’s information criterion (AIC). The rest of the statistics and plotting were performed using R language and the following R Packages and functions: ChemmineR package for Pubchem’s fingerprint and Tanimoto coefficient computation, heatmap.2 function in gplots package for plotting heatmap, flowCore and flowViz packages were modified for plotting fcs files in flow cytometry. Statistical significance of protein expression (on western blot) was calculated using paired two-tailed t-test.
Supplementary Material
Acknowledgements
We thank Hai Li at Columbia University and Christopher Henderson at Biogen Idec for providing mevalonic acid and farnesyl pyrophosphate, Chia-I Liu and Andrew H.J. Wang at Academia Sinica in Taiwan for the truncated human squalene synthase construct, Eric Lee for assistance with ACC experiments, and Vasanthi Viswanathan for helpful discussions. This research was funded by the Howard Hughes Medical Institute, National Institute of Health (5R01CA097061, 5R01GM085081, R01CA161061), and New York Stem Cell Science (C026715) to B.R.S. and a K99 Pathway to Independence Award from the National Cancer Institute to S.J.D. (1K99CA166517-01).
Footnotes
Competing financial interests
The authors declare to have no competing interests as defined by Nature Publishing Group, or other interests that might be perceived to influence the results and/or discussion reported in this article.
Author Contributions
K.S. and B.R.S. conceived of the project, designed the experiments, analyzed the data and wrote the manuscript. K.S. performed all experiments and analyses except: R.S. and C.A.V. synthesized CIL56 analogs (Supplementary Fig. 5, 7, Fig. 2d, 4a), A.K. expressed and purified SQS protein, and performed confirmatory SQS pull-down experiment and competition assays (Supplementary Fig. 9), W.S.Y. performed the GPX4 enzymatic assay (Fig. 3a), M.H. performed confirmatory siRNA experiments (Supplementary Fig. 6k–m). L.M.B. performed proteomic analysis for target identification (Fig. 4), S.J.D. participated in characterization of TOFA’s effect on FIN56 (Fig. 6) and A.J.W. assisted in modulatory profiling experiments (Fig. 1b). S.J.D. assisted in writing the manuscript.
References
- 1.Fuchs Y, Steller H. Programmed Cell Death in Animal Development and Disease. Cell. 2011;147:742–758. doi: 10.1016/j.cell.2011.10.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aravind L, Dixit VM, Koonin EV. The domains of death: evolution of the apoptosis machinery. Trends Biochem. Sci. 1999;24:47–53. doi: 10.1016/s0968-0004(98)01341-3. [DOI] [PubMed] [Google Scholar]
- 3.Degterev A, et al. Identification of RIP1 kinase as a specific cellular target of necrostatins. Nat. Chem. Biol. 2008;4:313–321. doi: 10.1038/nchembio.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Berghe TV, Linkermann A, Jouan-Lanhouet S, Walczak H, Vandenabeele P. Regulated necrosis: the expanding network of non-apoptotic cell death pathways. Nat. Rev. Mol. Cell Biol. 2014;15:135–147. doi: 10.1038/nrm3737. [DOI] [PubMed] [Google Scholar]
- 5.Kaczmarek A, Vandenabeele P, Krysko DV. Necroptosis: The Release of Damage-Associated Molecular Patterns and Its Physiological Relevance. Immunity. 2013;38:209–223. doi: 10.1016/j.immuni.2013.02.003. [DOI] [PubMed] [Google Scholar]
- 6.Kono H, Rock KL. How dying cells alert the immune system to danger. Nat. Rev. Immunol. 2008;8:279–289. doi: 10.1038/nri2215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Skouta R, et al. Ferrostatins Inhibit Oxidative Lipid Damage and Cell Death in Diverse Disease Models. J. Am. Chem. Soc. 2014;136:4551–4556. doi: 10.1021/ja411006a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Linkermann A, et al. Synchronized renal tubular cell death involves ferroptosis. Proc. Natl. Acad. Sci. 2014;111:16836–16841. doi: 10.1073/pnas.1415518111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dixon SJ, et al. Ferroptosis: An Iron-Dependent Form of Nonapoptotic Cell Death. Cell. 2012;149:1060–1072. doi: 10.1016/j.cell.2012.03.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dixon SJ, et al. Pharmacological inhibition of cystine–glutamate exchange induces endoplasmic reticulum stress and ferroptosis. eLife. 2014;3:e02523. doi: 10.7554/eLife.02523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hayano M, Yang WS, Corn CK, Pagano NC, Stockwell BR. Loss of cysteinyl-tRNA synthetase (CARS) induces the transsulfuration pathway and inhibits ferroptosis induced by cystine deprivation. Cell Death Differ. 2015 doi: 10.1038/cdd.2015.93. doi:10.1038/cdd.2015.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yang WS, et al. Regulation of Ferroptotic Cancer Cell Death by GPX4. Cell. 2014;156:317–331. doi: 10.1016/j.cell.2013.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Imai H, Nakagawa Y. Biological significance of phospholipid hydroperoxide glutathione peroxidase (PHGPx, GPx4) in mammalian cells. Free Radic. Biol. Med. 2003;34:145–169. doi: 10.1016/s0891-5849(02)01197-8. [DOI] [PubMed] [Google Scholar]
- 14.Linkermann A, Stockwell BR, Krautwald S, Anders H-J. Regulated cell death and inflammation: an auto-amplification loop causes organ failure. Nat. Rev. Immunol. 2014;14:759–767. doi: 10.1038/nri3743. [DOI] [PubMed] [Google Scholar]
- 15.Hitomi J, et al. Identification of a Molecular Signaling Network that Regulates a Cellular Necrotic Cell Death Pathway. Cell. 2008;135:1311–1323. doi: 10.1016/j.cell.2008.10.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hahn WC, et al. Creation of human tumour cells with defined genetic elements. Nature. 1999;400:464–468. doi: 10.1038/22780. [DOI] [PubMed] [Google Scholar]
- 17.Wolpaw AJ, et al. Modulatory profiling identifies mechanisms of small molecule-induced cell death. Proc. Natl. Acad. Sci. 2011;108:E771–E780. doi: 10.1073/pnas.1106149108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Degterev A, et al. Chemical inhibitor of nonapoptotic cell death with therapeutic potential for ischemic brain injury. Nat. Chem. Biol. 2005;1:112–119. doi: 10.1038/nchembio711. [DOI] [PubMed] [Google Scholar]
- 19.Takahashi N, et al. Necrostatin-1 analogues: critical issues on the specificity, activity and in vivo use in experimental disease models. Cell Death Dis. 2012;3:e437. doi: 10.1038/cddis.2012.176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yang WS, Stockwell BR. Synthetic lethal screening identifies compounds activating iron-dependent, nonapoptotic cell death in oncogenic-RAS-harboring cancer cells. Chem. Biol. 2008;15:234–245. doi: 10.1016/j.chembiol.2008.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shoemaker RH. The NCI60 human tumour cell line anticancer drug screen. Nat. Rev. Cancer. 2006;6:813–823. doi: 10.1038/nrc1951. [DOI] [PubMed] [Google Scholar]
- 22.Shimada K, Hayano M, Pagano NC, Stockwell BR. Cell-Line Selectivity Improves the Predictive Power of Pharmacogenomic Analyses and Helps Identify NADPH as Biomarker for Ferroptosis Sensitivity. Cell Chem. Biol. 2016;23:225–235. doi: 10.1016/j.chembiol.2015.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Romanowska M, et al. Effects of selenium supplementation on expression of glutathione peroxidase isoforms in cultured human lung adenocarcinoma cell lines. Lung Cancer. 2007;55:35–42. doi: 10.1016/j.lungcan.2006.09.007. [DOI] [PubMed] [Google Scholar]
- 24.Fradejas N, et al. Mammalian Trit1 is a tRNA [Ser]Sec -isopentenyl transferase required for full selenoprotein expression. Biochem. J. 2013;450:427–432. doi: 10.1042/BJ20121713. [DOI] [PubMed] [Google Scholar]
- 25.Song JH, et al. Deletion of Pim kinases elevates the cellular levels of reactive oxygen species and sensitizes to K-Ras-induced cell killing. Oncogene. 2015;34:3728–3736. doi: 10.1038/onc.2014.306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tansey TR, Shechter I. Structure and regulation of mammalian squalene synthase. Biochim. Biophys. Acta BBA - Mol. Cell Biol. Lipids. 2000;1529:49–62. doi: 10.1016/s1388-1981(00)00137-2. [DOI] [PubMed] [Google Scholar]
- 27.Liu C-I, et al. Structural insights into the catalytic mechanism of human squalene synthase. Acta Crystallogr. D Biol. Crystallogr. 2014;70:231–241. doi: 10.1107/S1399004713026230. [DOI] [PubMed] [Google Scholar]
- 28.Chugh A, Ray A, Gupta JB. Squalene epoxidase as hypocholesterolemic drug target revisited. Prog. Lipid Res. 2003;42:37–50. doi: 10.1016/s0163-7827(02)00029-2. [DOI] [PubMed] [Google Scholar]
- 29.Gueven N, Woolley K, Smith J. Border between natural product and drug: Comparison of the related benzoquinones idebenone and coenzyme Q10. Redox Biol. 2015;4:289–295. doi: 10.1016/j.redox.2015.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hirsch HA, et al. A Transcriptional Signature and Common Gene Networks Link Cancer with Lipid Metabolism and Diverse Human Diseases. Cancer Cell. 2010;17:348–361. doi: 10.1016/j.ccr.2010.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Santos CR, Schulze A. Lipid metabolism in cancer. FEBS J. 2012;279:2610–2623. doi: 10.1111/j.1742-4658.2012.08644.x. [DOI] [PubMed] [Google Scholar]
- 32.Kamphorst JJ, Fan J, Lu W, White E, Rabinowitz JD. Liquid Chromatography–High Resolution Mass Spectrometry Analysis of Fatty Acid Metabolism. Anal. Chem. 2011;83:9114–9122. doi: 10.1021/ac202220b. [DOI] [PMC free article] [PubMed] [Google Scholar]
Methods-only references
- 33.Yagoda N, et al. RAS-RAF-MEK-dependent oxidative cell death involving voltage-dependent anion channels. Nature. 2007;447:865–869. doi: 10.1038/nature05859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cholody WM, et al. Derivatives of fluorene, anthracene, xanthene, dibenzosuberone and acridine and uses thereof. 2008 at < http://www.google.com/patents/WO2008140792A1>.
- 35.Backman TWH, Cao Y, Girke T. ChemMine tools: an online service for analyzing and clustering small molecules. Nucleic Acids Res. 2011:gkr320. doi: 10.1093/nar/gkr320. doi:10.1093/nar/gkr320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Alegre-Aguarón E, et al. Growth Factor Priming Differentially Modulates Components of the Extracellular Matrix Proteome in Chondrocytes and Synovium-Derived Stem Cells. PLoS ONE. 2014;9:e88053. doi: 10.1371/journal.pone.0088053. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.