Abstract
Detection of viral DNA is essential for eliciting mammalian innate immunity. However, viruses have acquired effective mechanisms for blocking host defense. Indeed, in this issue of Cell Host & Microbe, Wu et al. (2015) discover a herpesviral strategy for inhibiting the prominent host sensor of viral DNA, cGAS.
For all metazoa, continual surveillance of the intracellular milieu is essential for both detecting and responding to viral infections. To this end, cells utilize specialized receptor proteins termed “sensors” that recognize pathogen-derived moieties, such as viral DNA, to subsequently initiate antiviral responses. In mammals, these events are required for mobilizing the innate and adaptive arms of the immune system and, ultimately, for pathogen clearance. Consequently, these cellular mechanisms exert extreme selective pressures on viruses, driving the evolution of unique viral gene products that target and disable important host defenses. Understanding virus immune evasion strategies used to inhibit host sensors of viral DNA can accelerate the design of effective therapeutics that directly hinder pathogen fitness.
In this issue, Wu et al. (2015) make a substantial leap forward in this research area by discovering a viral strategy for inhibiting one of the most prominent sensors of viral DNA, cGAS (cyclic GMP-AMP synthase). To place this study in its biological context, recent work has demonstrated that cGAS directly binds to foreign DNA in the cytoplasm, triggering a cascade of events that culminates in the expression of antiviral cytokines (Figure 1, steps 1–6) (Sun et al., 2013; Wu et al., 2013). Specifically, cGAS catalyzes the production of cGAMP (cyclic guanosine monophosphate–adenosine monophosphate) from cellular ATP and GTP pools. In turn, the cGAMP second messenger binds to the ER transmembrane adaptor protein STING (stimulator of interferon genes), triggering activation of the protein kinase TBK-1 and IRF3 (interferon regulatory factor 3) (Ablasser et al., 2013). Subsequently, IRF3 translocates into the nucleus where it orchestrates the expression of immune and inflammatory genes, such as interferons (ifn). Underscoring the significance of this sensor in recognizing multiple pathogens, cGAS was shown to be required for triggering immune responses during infection with several DNA viruses and bacterial pathogens. Interestingly, however, cGAS (also known as C6ORF150 and Mab-21 domain containing 1, MB21D1) was initially found as a potent inhibitor of several RNA viruses in a screen of over 380 interferon-stimulated genes (Schoggins et al., 2011). This suggests that cGAS may possess additional broad-acting antiviral activities. Along these lines, cGAS was also recently demonstrated to interact with and stabilize another DNA sensor, the interferon inducible protein IFI16 (Orzalli et al., 2015). Initially identified as a cytoplasmic sensor, several groups have later demonstrated that IFI16 also acts as a nuclear DNA sensor, being required for STING-dependent IFN expression in response to infections with the nuclear-replicating viruses herpes simplex virus 1 (HSV-1) and human cytomegalovirus (HCMV).
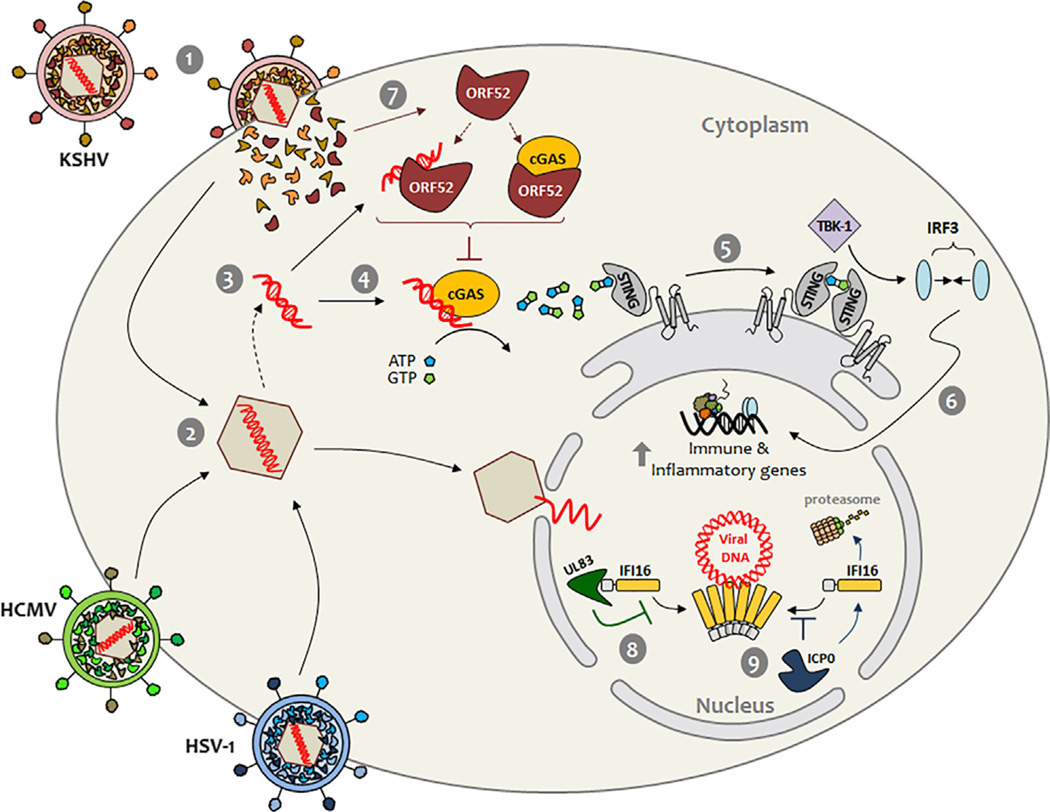
Figure 1. Herpesvirus Strategies for Abating Host DNA Sensing.
Fusion of the viral lipid envelope with the plasma membrane of host cells releases viral tegument proteins and the nucleocapsid containing the virus double-stranded DNA genome (1–2). During its transit to the nucleus, the nucleocapsid may be disrupted, releasing viral DNA into the cytosol (3). Here cGAS binds to the viral DNA, stimulating cGAMP production from ATP and GTP (4). Subsequently, cGAMP triggers STING to activate protein kinase TBK-1 (5), in turn activating transcription factor IRF3. Upon dimerization, IRF3 enters the nucleus and stimulates antiviral gene expression (6). As shown by Wu et al. (2015), during KSHV infection, the tegument protein ORF52 obstructs cGAS function through the sequestration of viral DNA substrate and/or an interaction, which directly alters cGAS enzymatic activity (7). In contrast, the HSV-1 E3 ubiquitin ligase ICP0 promotes degradation of the nuclear DNA sensor IFI16 (8), whereas the HCMV tegument protein UL83 inhibits IFI16 by blocking its oligomerization (9).
Although the discovery of DNA sensors is a major step forward in understanding the barriers to pathogen replication, it represents only one side of the host-pathogen interaction. On the opposing side are the diverse viral immune evasion strategies, which have remained less characterized. Progress has been made in recent years, in which a few virus factors that inhibit DNA sensors during herpesvirus infections have been identified. During HSV-1 infection, the viral E3 ubiquitin ligase ICP0 was shown to promote the proteasome-dependent degradation of IFI16 (Orzalli et al., 2012) (Figure 1, step 8). In contrast, during HCMV infection, the viral tegument protein pUL83 was shown to bind IFI16, preventing its DNA-dependent oligomerization (Li et al., 2013) (Figure 1, step 9). Both of these viral strategies effectively abate IFI16- and STING-dependent IFN expression. Surprisingly, given the enormously expanded interest in DNA sensing, no immunoevasion mechanism targeting cGAS has yet been described.
Here, Wu et al. (2015) address this important gap in knowledge by identifying a viral strategy for inhibiting cGAS. The study is a true tour de force with respect to the diversity of cellular, biochemical, and molecular techniques employed to reveal a virus immunoevasion mechanism during infection with Kaposi sarcoma-associated herpesvirus (KSHV). Specifically, the authors define the poorly characterized tegument protein ORF52 as a potent inhibitor of the central cGAS-STING signaling axis (Figure 1, step 7). For this, each KSHV open-reading frame (>80) was individually assayed for its ability to attenuate an IFN reporter driven by cGAS activity. Of the KSHV ORFs that reduced IFN reporter stimulation, only ORF52 displayed both DNA-binding activity and cytoplasmic localization. ORF52 also inhibited the stimulation of IRF3 in human monocytes upon challenge with either DNA substrates or Vaccinia virus, a DNA virus that replicates in the cytoplasm. Interestingly, cells stimulated with cGAMP still initiated immune signaling independent of ORF52 expression, suggesting that ORF52 may directly inhibit cGAS function, rather than affecting a downstream pathway component. To substantiate this model, the authors demonstrated in vitro that purified ORF52 drastically reduces cGAS production of cGAMP in the presence of DNA. Thus, ORF52 may function directly by modulating cGAS (i.e., its DNA binding or catalysis of cGAMP production) or indirectly by sequestering its DNA substrate. The authors provide substantial evidence for both ORF52 functions. Using DNA competitive binding assays, ORF52 was shown to affect the ability of cGAS to bind its DNA substrate. Furthermore, using an impressive mutagenesis screen, the authors demonstrate that upon loss of binding to DNA, ORF52 can no longer inhibit cGAMP production in vitro. The authors go on to establish by immunoaffinity purification that ORF52 and cGAS can also interact in a DNA-independent manner, mapping the domains that mediate this interaction. Thus, although ORF52 may to some extent compete with cGAS for its DNA substrate, it seemingly has a second mechanism for directly targeting cGAS. Furthermore, this inhibitory mechanism seems to be specific for cGAS, as ORF52 expression did not affect the functions of AIM2, another cytoplasmic DNA sensor. This virus immune evasion mechanism was validated in both human monocytes and epithelial cells, as the authors show that KSHV-induced immune responses through the cGAS-STING signaling axis are significantly elevated in the absence of ORF52. Mechanism aside, the positive impact of ORF52 on KSHV fitness is underscored by its conserved function across the gammaherpesvirus family. Similar to their KSHV counterpart, ORF52 homologs from human Epstein-Barr virus, Rhesus monkey rhadinovirus, and Murid gammaherpesvirus 68 all bind to both cGAS and DNA, inhibit cGAMP production in vitro, and attenuate IRF3 activation upon DNA challenge. Altogether, Wu et al. (2015) provide considerable evidence that ORF52 is a critical KSHV factor targeting a central component of the mammalian immune system— the cGAS-STING signaling axis.
Considering all the recent findings in DNA sensing, this is clearly an exciting and evolving research area. So, it is not surprising that many questions still remain. For instance, like all herpesviruses, KSHV deposits its DNA genome into the nucleus, where it is replicated by the viral DNA polymerase. It is therefore challenging to reconcile how cGAS, thought to be predominantly cytoplasmic, has the opportunity to sense the viral genome. The disruption of the viral capsid within the cytosol may explain this phenomenon, but this remains to be further explored. Also, herpesviruses can establish a latent infection characterized by a stably integrated, but transcriptionally silent, viral genome within certain cell types. This raises further questions about how cGAS sensing would be affected during productive versus latent viral infection. Additionally, cGAS has been observed to interact with the DNA sensor IFI16, promoting IFI16-dependent responses to other herpesviruses. Therefore, there are likely additional components and crosstalk pathways that require further characterization. Reinforcing the immunomodulatory role of the cGAS-STING axis, recent reports showed that cGAMP produced in response to HIV-1 and mouse CMV infections is packaged into progeny virions and transferred to naive cells, stimulating their immune responses (Bridgeman et al., 2015; Gentili et al., 2015). This may serve to rapidly disseminate IFN signaling locally or to slow the progress of subsequent de novo infections. Thus, further understanding the interaction between pathogens and the cGAS-STING axis, as well as continued research for new mechanisms of viral immune evasion, is a high priority in the race toward rational drug design. For example, additional clarification of the mechanism by which ORF52 specifically inhibits cGAS has the promise to be an important contributor to drug design efforts and to expand the current understanding of the antagonistic interplay between pathogens and host immunity.
REFERENCES
- Ablasser A, Goldeck M, Cavlar T, Deimling T, Witte G, Röhl I, Hopfner KP, Ludwig J, Hornung V. Nature. 2013;498:380–384. doi: 10.1038/nature12306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bridgeman A, Maelfait J, Davenne T, Partridge T, Peng Y, Mayer A, Dong T, Kaever V, Borrow P, Rehwinkel J. Science. 2015:aab3632. doi: 10.1126/science.aab3632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gentili M, Kowal J, Tkach M, Satoh T, Lahaye X, Conrad C, Boyron M, Lombard B, Durand S, Kroemer G, et al. Science. 2015:aab3628. doi: 10.1126/science.aab3628. [DOI] [PubMed] [Google Scholar]
- Li T, Chen J, Cristea IM. Cell Host Microbe. 2013;14:591–599. doi: 10.1016/j.chom.2013.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orzalli MH, Broekema NM, Diner BA, Hancks DC, Elde NC, Cristea IM, Knipe DM. Proc. Natl. Acad. Sci. USA. 2015;112:E1773–E1781. doi: 10.1073/pnas.1424637112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orzalli MH, DeLuca NA, Knipe DM. Proc. Natl. Acad. Sci. USA. 2012;109:E3008–E3017. doi: 10.1073/pnas.1211302109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoggins JW, Wilson SJ, Panis M, Murphy MY, Jones CT, Bieniasz P, Rice CM. Nature. 2011;472:481–485. doi: 10.1038/nature09907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun L, Wu J, Du F, Chen X, Chen ZJ. Science. 2013;339:786–791. doi: 10.1126/science.1232458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J, Sun L, Chen X, Du F, Shi H, Chen C, Chen ZJ. Science. 2013;339:826–830. doi: 10.1126/science.1229963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J-j, Li W, Shao Y, Avey D, Fu B, Gillen J, Hand T, Ma S, Liu X, Miley W, Konrad A, et al. Cell Host Microbe. 2015;18:333–344. doi: 10.1016/j.chom.2015.07.015. this issue. [DOI] [PMC free article] [PubMed] [Google Scholar]