Abstract
Purpose
MHC class I presentation of peptides allows T cells to survey the cytoplasmic protein milieu of host cells. During infection, presentation of self peptides is, in part, replaced by presentation of microbial peptides. However, little is known about the self peptides presented during infection, despite the fact that microbial infections alter host cell gene expression patterns and protein metabolism.
Experimental design
The self peptide repertoire presented by HLA-A*01;01, -A*02;01, -B*07;02, -B*35;01 and -B*45;01 was determined by mass spectrometry before and after vaccinia virus infection.
Results
We observed a profound alteration in the self peptide repertoire with hundreds of self peptides uniquely presented after infection for which we have coined the term ‘self peptidome shift’. The fraction of novel self peptides presented following infection varied for different HLA class I molecules. A large part (~40%) of the self peptidome shift was composed of peptides derived from type I interferon-inducible genes, consistent with cellular responses to viral infection. Interestingly, ~12% of self peptides presented after infection showed allelic variation when searched against ~300 human genomes.
Conclusion and clinical relevance
Self peptidome shift in a clinical transplant setting could result in alloreactivity by presenting new self peptides in context of infection-induced inflammation.
Keywords: infection, minor histocompatibility, peptidome, self peptides, transplantation
Introduction
MHC class I-restricted antigen processing and presentation inform T cells as to the internal state of the cell by binding cytoplasmic peptides and presenting them at the cell surface [1-3]. During homeostasis, these peptides are derived from self proteins and their presentation signifies normal cellular operations and as such is ignored by self-educated T cells. However, during infection, microbial peptides are processed and fed into this antigen presentation pathway alerting the immune system to the presence of a pathogen [4]. Activation of an innate immune response to the pathogen creates an inflammatory milieu that provides additional signals to the T cell, triggering its full activation.
Self peptide presentation at the immunological synapse contributes to T cell activation by lowering the activation threshold [5-7]. Since self peptides are continuously present at the immunological synapse, T cells strongly recognizing self peptide/MHC complexes (pMHC) must be deleted during thymic education to prevent the development of autoimmune disease [8]. However, T cell positive selection requires weak recognition of self pMHC complexes [9]. Peripheral self pMHC recognition in the absence of inflammation leads to tolerization [10]. Conversely, self peptide recognition in the context of the inflammatory stimulus emerging from a microbial infection can lead to activation of weakly self-reactive T cells and the development of autoimmune disease, e.g., diabetes, multiple sclerosis and polymyositis [11, 12]. Therefore, optimal peripheral T cell activation requires a combination of inflammatory signals, non-self pMHC recognition and low affinity self pMHC recognition to fully activate T cells.
Even though T cells strongly reactive to self pMHC are deleted during development, self peptides can act as minor histocompatibility antigens in the context of allograft transplantation if the genes encoding self peptides show allelic variation (i.e., when DNA sequences for the same gene differ between two or more variant alleles) within the human population [13-16]. Presentation of peptides containing allelic differences, termed allopeptides, by the HLA of donor transplanted tissue cells can activate recipient T cells leading to graft rejection. Alternatively, donor T cells may recognize recipient allopeptides leading to graft-versus-host disease (GVHD) even in a HLA-matched bone marrow transplant. Since immunosuppressive drugs given to the otherwise healthy transplant recipient suppress inflammation, allopeptide recognition by T cells should lead to tolerance. Nonetheless, once the graft has been accepted and the immunosuppressive drugs are withdrawn, subsequent infections would incite inflammatory conditions. Indeed, viral infections occur after transplantation in ~10-60% of immunosuppressed patients, leading to adverse effects on the host and/or transplanted organ [17, 18]. Recognition of new allopeptides that T cells have not been tolerized against, e.g., those that are not presented during homeostatic conditions, could result in T cell activation and immunopathology [19]. Hence, alterations in self peptides under inflammatory conditions can be detrimental to transplanted tissues/organs.
Despite their importance, little is known regarding the nature of the self peptide repertoire (peptidome) displayed during infection. Small-scale studies have reported little change in the self peptidome displayed by HLA-A*02;01 and HLA-B*07;02 after human immunodeficiency virus (HIV, [20]), influenza virus (INV, [21]), and measles virus (MeV, [22]) infections. Herein, we use a large-scale proteomics approach to study the dynamics of self peptides presented by five major HLA class I molecules, HLAA*01;01, -A*02;01, -B*07;02, -B*35;01, -B*45;01 before and after vaccinia virus (VACV) infection. In contrast to earlier studies (18-20), we observed a profound shift in the self peptidomes uniquely displayed by the five HLA class I molecules studied herein after VACV infection. The newly presented self peptides did not derive from any specific chromosomal region. A fraction (~40%) of them represented peptides derived from type I interferon-induced genes – consistent with the activation of cellular antiviral pathways – but also included other unrelated peptides, suggesting a global change in cellular protein metabolism in response to infection. Furthermore, population analyses of self peptides presented after infection revealed that a significant number of peptides were derived from proteins containing allelic variation(s). The frequency (~12%) of allelic variation was similar to the rate of complications reported for transplants between HLA-matched pairs [23, 24]. This changing repertoire may provide a possible mechanism for the initiation of allograft rejection or GVHD. Hence, sequencing of the transplant donor and recipient transcriptomes/proteomes could help uncover potential allopeptides that can complicate allograft outcomes [15, 16, 25-32].
Materials and Methods
Viruses
The Western Reserve strain of vaccinia virus (VACV; ATCC, VR-119) was grown in and titrated with BSC-40 cells as previously described [33].
Large-scale cell culture and VACV infection
Soluble HLA class I (sA1.1, sA2.1, sB7.2, sB35.1 and sB45.1) production and harvest were as described previously [34]. Briefly, ~1×109 viable cells were inoculated with VACV (MOI 0.1). Supernatants containing ~0.3—4.2mg/L sA1, sA2, sB7, sB35 or sB45 were collected at 24, 48, and 72 hrs post inoculation.
Isolation and fractionation of class I-associated peptides
sA1, sA2, sB7, sB35 and sB45 were affinity purified using W6/32-bound protein A Sepharose (GE Healthcare). Class I-associated peptide elution, separation and reversed-phase HPLC purification were all performed as previously described [35].
Mass spectrometry sequencing of eluted peptides
Lyophilized fractions were resuspended in 0.1% formic acid and subjected to reversed-phase microcapillary LC-nanoESI-MS/MS analysis using an Agilent 1100 binary HPLC pump and an LTQ linear ion trap mass spectrometer 2.2 (Thermofisher). A fritless, microcapillary column (100-μm inner diameter) was packed with 10 cm of 5-μm C18 reversed-phase material (Synergi 4u Hydro RP80a, Phenomenex) as previously described [36]. RPC fractionated peptides were loaded onto the column equilibrated in buffer A (0.1% formic acid, 5% acetonitrile) using a LCPacking's autosampler. Flow splitting was used to reduce the HPLC flow rate from 200μl/min to 0.3μl/min as previously described [36, 37]. Peptides from the microcapillary column were eluted directly into the linear ion LTQ mass spectrometer equipped with a microelectrospray source (James Hill Instrument Service). Peptides were eluted using a 60-min linear gradient from 0 to 60% buffer B (0.1% formic acid, 80% acetonitrile) at a flow rate of 0.3μl/min. During the gradient, the eluted ions were analyzed by one full precursor MS scan (400–2000 m/z) followed by five MS/MS scans of the five most abundant ions detected in the precursor MS scan while operating under dynamic exclusion. 65% of peaks were identified in replicate samples by this analysis. The program extractms2 was used to generate the ASCII peak list and identify +1 or multiply charged precursor ions from the native mass spectrometry data file [38]. Tandem spectra were searched with no protease specificity using SEQUEST-PVM [39] against a concatenated Human RefSeq protein database release May 2005 (28,818 entries), Vaccinia WR Copenhagen Uniprot protein database (760 entries) or a merged human and vaccinia FASTA database of protein sequences [40]. For multiply charged precursor ions (z ≥ +2), an independent search was performed on both the +2 and +3 mass of the parent ion. A weighted scoring matrix was used to select the most likely charge state of multiply charged precursor ions as previously described [41, 42]. Sequest search results were imported into Bioinformatic Graphical Comparative Analysis Tools (BIGCAT) and analyzed as previously described [33, 41]. Xcorr threshold of 1.5 for charge state 1, 2.0 for charge state 2, and 2.5 for charge state 3 and above was used to filter the peptides. This resulted in a false discovery rate (FDR) of 3.6%, which was calculated as the percentage of the number of peptide hits from the reversed database in the total number of peptides in the filtered list (FDR = number of reverse peptides/total number of peptides * 100).
Bioinformatics
Searches for publicly reported CD8 T cell epitopes and potential epitopes identified by algorithms were conducted through the Immune Epitope Database (http://www.iedb.org/). Peptide conservation amongst other Orthopoxviridae members was performed using BLAST search (NCBI). Self peptides were searched against the human proteome and nucleotide databases using the PAM30 matrix with the BLAST search program restricting the searches with the entrez criterion txid9606[orgn] to specify Homo sapiens. Genes encoding these peptide sequences were verified to be present in the HeLa genome by searching the translated HeLa Cell Genome Sequencing Studies (phs000640.v2.p1) database at the Database of Genotypes and Phenotypes (dbGaP), Bethesda (MD): National Center for Biotechnology Information, National Library of Medicine [43, 44]. Proteins were classified by functional class using the Panther Database version 9.0 [45, 46]. Innate immune responsive proteins were identified using the Interferome database [47]. So also, self peptides derived from known or potential oncogenes products were identified by BLAST search against the TSGene database [48]. Tissue expression of proteins was determined using the Tissue-specific Gene Expression and Regulation (TiGER) database [49].
Analysis
Microsoft Excel and PowerPoint, Prism GraphPad and Adobe Photoshop were used to analyze data and generate graphs and figures.
Results
VACV infection induces a profound self peptidome shift
To determine whether the presentation of self peptides changes after VACV infection, which is known to alter host protein metabolism [50-55], HeLa cells expressing secreted HLA A*01;01, A*02;01, B*07;02, B*35;01 and B*45;01, [34] molecules were infected with VACV. One, two and three days post infection, soluble class I molecules were affinity purified from culture supernatants of infected cells or uninfected controls. The associated peptides were acid eluted and fractionated by reversed-phase chromatography, as described previously [56]. Each of the resulting 150 fractions were individually subjected to mass analyses by 2D HPLC in-line with ESI-MS/MS. Peptide mass spectra so obtained were compared against both VACV and human proteomes to determine their origin and confirm sequence. Mass spectrometry data revealed the processing and presentation of numerous peptides derived from VACV, confirming infection of the HeLa cells (Figure 1, Table 1 and Supplemental Table 1; [33]).
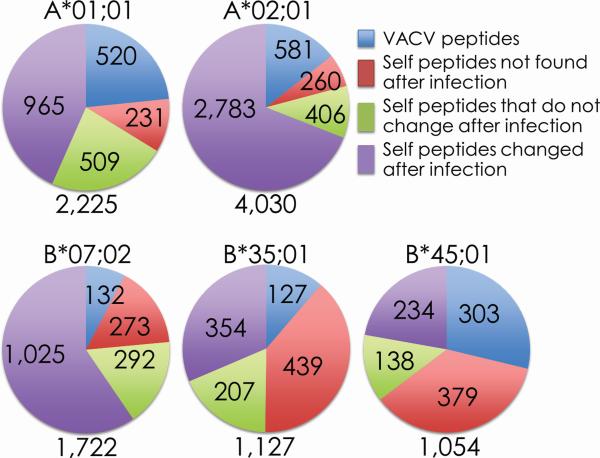
Figure 1. Numerous unique self and viral peptides are presented after VACV infection.
Mass spectrometry was used to sequence peptides eluted from the indicated soluble HLA class I molecules. Comparison of the pre- and post-infection samples identified a significant fraction of viral peptides (blue) presented after infection. In addition, large alterations in the self peptidome were observed with some peptides disappearing (red), unchanged (green), or newly presented (purple) after infection. The total number of peptides sequenced for each HLA class I molecule is annotated under each chart.
Table 1.
Characterization of HLA-B*35;01-restricted VACV-derived determinants presented during active infection
| VACCC ORFa | Sequence | Prior reportsb | Cnc | DPId | #Hitse | Functionf | Temporal Expg | VARVh | MONPV | ECTV |
|---|---|---|---|---|---|---|---|---|---|---|
| A4L83-91 | VPTATPAPI | [1] | 1.5598 | 1,3 | 10 | S | E/L | NSH | NSH | NSH |
| A8R25-34 | TPMIKENSGF | 1.717 | 1 | 3 | T | I/E | S8I | |||
| A10L111-119 | NPIINTHSF | [2] | 1.5417 | 3 | 1 | S | L | S8N | ||
| A10L457-465 | FPRKDKSIM | 1.5653 | 3 | 1 | S | L | ||||
| A10L853-862 | RPKILSMINY | 1.8534 | 1 | 7 | S | L | ||||
| A11R298-306 | SPVLNIVLF | 1.5163 | 1 | 7 | O | L | ||||
| A16L250-259 | YPKSNSGDKY | [2] | 1.8503 | 1 | 3 | S | L | |||
| A17L91-100 | LPLTSLVITY | [2] | 2.9936 | 1,3 | 8 | S | L | |||
| A18R52-61 | SPSVKTSLVF | 1.505 | 3 | 1 | T | E | S3C | |||
| A18R237-245 | TPRPANRIY | 1.6559 | 1,3 | 2 | T | E | A5S | |||
| A20R4-12 | LPVIFLPIF | 1.9299 | 3 | 2 | R | E | NSH | NSH | ||
| A20R162-170 | IPKYLEIEI | [1] | 1.8813 | 1 | 1 | R | E | K3N | K3N | |
| A21L99-107 | IPGFARSCY | 1.5227 | 3 | 1 | S | L | A5T | I1L;A5V | ||
| A24R663-671 | FPAEFRDGY | 1.9133 | 1,3 | 2 | T | E | ||||
| A24R1002-1010 | KPYASKVFF | [1] | 1.924 | 3 | 1 | T | E | A4E | ||
| A32L1103-1111 | IPISDYTGY | 2.0188 | 1,3 | 10 | T | E | ||||
| A37R129-138 | IPSKRLVTSF | 1.6865 | 1 | 1 | U | I/E | NSH | NSH | ||
| A37R240-248 | VPIKEQILY | 2.2542 | 1 | 1 | U | I/E | NSH | V1L | NSH | |
| A39R394-403 | MPQMKKILKM | 1.9143 | 1 | 1 | E/V | L | NSH | NSH | M1I;Q3R | |
| A44L326-335 | SPIFDVDVAF | 1.5898 | 1 | 1 | E/V | I/E | NSH | |||
| A51R94-102 | TPTGVYNYF | 1.6732 | 1 | 1 | U | E | ||||
| A55R213-221 | SPQVIKSLY | 1.838 | 1,3 | 4 | E/V | E | NSH | NSH | ||
| B3R39-48 | IPSTVKTNLY | [2] | 1.8943 | 1 | 2 | U | I/E | I1M | ||
| B8R70-78 | FPKNDFVSF | [1] ; [2] | 2.237 | 1 | 2 | E/V | E | K3N | ||
| B8R104-112 | PPTVTLTEY | 1.6996 | 3 | 1 | E/V | E | T5R | |||
| B8R158-167 | EPVTYDIDDY | [1] | 1.5432 | 3 | 1 | E/V | E | D6N | T4I | |
| B9R10-19 | FPSIIYSMSI | 1.5804 | 3 | 1 | U | E/L | NSH | NSH | ||
| B12R52-60 | KPLLSEIRF | 1.7458 | 1 | 1 | U | I/E | I4M;R8N | K1R | ||
| B16R4-12 | LPVIFLSIF | 1.5123 | 1 | 1 | E/V | L | NSH | S7P | NSH | |
| B16R52-61 | NPTQSDSGIY | 1.5403 | 1 | 1 | E/V | L | S5T | |||
| B16R76-84 | IPIDNGSNM | 1.6 | 1 | 4 | E/V | L | G6C;S7N | P2Q | S7N | |
| B17L181-190 | APLPGNVLVY | [1] | 1.8017 | 1,3 | 5 | U | E | L3Y | NSH | |
| C2L337-345 | LPNLITPRY | 2.4458 | 3 | 2 | E/V | E | ||||
| C9L130-138 | IPTCSNIQY | 1.5769 | 1,2,3 | 3 | U | E/L | NSH | NSH | ||
| D1R475-483 | VPIKFIAEF | 1.5121 | 1 | 1 | T | E | ||||
| D4Rl81-189 | HPAARDRQF | 1.6429 | 3 | 1 | R | E | R7H | R7H | ||
| D4R186-194 | SPVTTIVGY | 1.7325 | 1,3 | 6 | R | E | ||||
| D8L160-169 | LPSKLDYFTY | [1] | 1.8428 | 3 | 1 | S | L | T9K | K4T | K4T |
| D11L185-194 | TPIVNSVQEF | 1.6056 | 1,3 | 5 | T | L | I3V | |||
| D11L506-514 | MPTVDEDLF | 1.606 | 1 | 1 | T | L | ||||
| D12L34-43 | LPSLEYGANY | [2] | 1.6592 | 3 | 3 | T | E | |||
| D13L160-168 | TPFDVEDTF | [2] | 2.2957 | 1 | 8 | 0 | L | |||
| E1L10-18 | FPNITLKII | [2] | 1.9468 | 1,3 | 5 | T | E | F1L | F1L | |
| E3L117-125 | NPVTVINEY | 2.2336 | 1,2,3 | 21 | E/V | I/E | V5I | V5I | ||
| E8R233-241 | DPVLMFLLF | 1.736 | 3 | 2 | S | E/L | ||||
| E9L488-496 | LPQSMVFEY | 1.5988 | 3 | 2 | R | E | ||||
| E9L526-534 | FPYEGGKVF | [1] | 1.8446 | 3 | 2 | R | E | |||
| F1L162-170 | NPVKTIKMF | 1.7642 | 3 | 2 | U | E | K4E | K4E | K4E | |
| F2L26-35 | SPGAAGYDLY | [1] | 1.5367 | 1,2,3 | 4 | R | E | G3Y | NSH | |
| F3L435-443 | YPRDNPELI | 1.5905 | 3 | 1 | E/V | E | P2Q | |||
| G2R1-9 | MPFRDLILF | 2.2046 | 1 | 6 | T | E | ||||
| G9R69-77 | GPGGLSALL | [1] | 1.5564 | 3 | 1 | S | L | G4N | ||
| H1L133-142 | SPMLYFLYVY | 1.6313 | 3 | 2 | T | E/L | ||||
| H2R141-150 | DPSAQQFCQY | 1.585 | 3 | 2 | S | L | ||||
| H4L636-645 | EPTDASLKNF | 1.8267 | 3 | 1 | T | L | N9Q | E1K | ||
| H6R156-164 | SPDEIVIKF | 1.7386 | 3 | 3 | T | E/L | E4K | |||
| I1L53-62 | IPVDLVKSSF | [2] | 2.9981 | 1,3 | 3 | S | L | |||
| I4L670-678 | LPEDIKRVY | 2.1136 | 3 | 1 | R | E | ||||
| I6L159-167 | IPMSIISFF | [3] | 1.6008 | 1,3 | 5 | U | E/L | M3I | ||
| I7L153-161 | NPKVVKMKI | 1.5407 | 3 | 1 | S | L | ||||
| I12L15-23 | SPEDDLTDF | 1.6251 | 3 | 1 | U | N/A | P2Q | |||
| J6R177-185 | WPLLEIHQY | 2.5974 | 1 | 1 | T | E | ||||
| L3L291-299 | VPKEDYYFI | 2.2064 | 1 | 1 | T | L | ||||
| L4R37-45 | FPRSMLSIF | [1] ; [4] | 1.9615 | 1 | 1 | T | L | |||
| L5R10-18 | NPVFIEPTF | 1.6253 | 3 | 2 | S | L | ||||
| N2L147-155 | KPVYSYVLY | 1.5179 | 1,3 | 3 | 0 | I/E | V3I | V3I | ||
| O1L4-12 | YPEFARKAL | 1.543 | 1 | 2 | U | I/E | ||||
| O1L549-557 | IPITDSLSF | 1.6041 | 1,3 | 2 | U | I/E | D5E |
Open reading frames (ORF) and location of epitopes are defined based on Copenhagen reference strain (VACCC, ID 10249)
Prior reports according to immune epitope data base (IEDB; www.iedb.org); blank, this study
Correlation coefficient represents the number of peak identities determined between the theoretically and experimentally derived spectra for a given parent ion normalized to the charge state of the peptide
Days post infection of HeLa cultures with VACV at which the peptide was identified
Total number of times a given peptide sequence was identified by mass spectrometry
Protein function according to ([5]); S, structural (virion membrane and core); T, transcription; E/V, evasion/virulence; O, other; U, unknown; P, pseudogenes
Temporality of expression (shortened) according to ([6]): IE, immediate early; E; early; E/L, early/late; L, late; N/A, unidentified
Peptide homologies were identified using Netblast (blastcl3 at www.ncbi.nlm.nih.gov) using the following taxonomy id: VARV, variola virus, 10255; ECTV, ectromelia virus, 12643; MONPV, monkeypox virus, 10244
Amino acid changes for homologous poxviral epitopes; NSH, no significant homology; blank, conserved sequences with 100 % homology
Comparative proteome searches identified 520 A*01;01-, 581 A*02;01-, 132 B*07;02-, 127 B*35;01-, and 303 B*45;01-associated VACV-derived peptides that were presented only upon infection; of those, 34 A*01;01-, 109 A*02;01-, 65 B*07;02-, 68 B*35;01-, 49 B*45;01-restricted peptide sequences correlated with the MS/MS spectra's fragment ion data with high confidence (see Materials and Methods). Similar to prior reports, these naturally processed and presented peptides were derived from all functional and kinetic classes of VACV proteins (Table 1, Supplemental Table 1; [33, 57]).
To identify self peptides, MS/MS spectra of ligands eluted from the same class I preparation from which VACV peptides were identified were searched against a Human RefSeq protein database (release May 2005; 28,818 entries). These searches returned 1,705 A*01;01-, 3,449 A*02;01-, 1,590 B*07;02-, 1,000 B*35;01-, and 751 B*45;01-associated peptide sequences derived from the human proteome (Figure 1 and Figure 2). However, since these peptides were eluted from HeLa cells, we ascertained whether the peptides identified by MS/MS analysis were contained within proteins encoded by the HeLa genome. For this, the HeLa Cell Genome Sequencing Studies database (phs000640.v2.p1; Database of Genotypes and Phenotypes (dbGaP), National Center for Biotechnology Information, National Library of Medicine [43, 44]) was searched. Indeed, the peptide sequences reported herein were derived from proteins encoded by the HeLa genome (Table 2 and Supplementary Table 2).
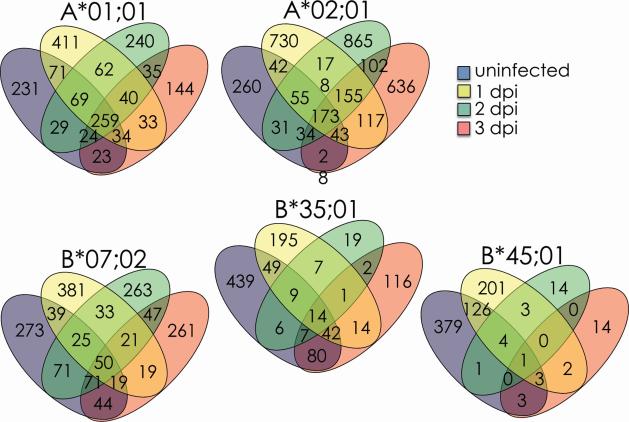
Figure 2. Self peptidome shift after infection with VACV WR strain.
The peptides presented by the indicated soluble HLA class I molecules were sequenced by mass spectrometry. The number of self peptides (Cn>1.5) detected 0, 1, 2 or 3 days after infection are reported in the single or overlapping regions for each HLA allele. The total number of peptides (Cn>1.5) sequenced is reported as n=# under each day.
Table 2.
Characterization of HLA-B*35;01-restricted self peptides presented during active infection
| Protein | Peptide sequencesa | Prior Reportsb | Interferon Responsivec | Cnd | DPIe | # Hitsf | Oncogene statusg | VACVh |
|---|---|---|---|---|---|---|---|---|
| AHSA1 | SPEELYRVF | 3.6857 | 1,3 | 4 | ||||
| AIM1 | LPDNSLKVF | 4 | 2.2947 | 3 | 2 | candidate | KVF7DGYi | |
| APEH | VPFKQGMEY | 1 | 2 | |||||
| ARHGEF18 | LPSGVGPEY | 1 | 2.3883 | 1 | 2 | |||
| ARPC4 | KPVEGYDISF | 3.4382 | 3 | 4 | ||||
| ATP5F1 | VPVPPLPEY | 1 | 2.4713 | 3 | 7 | |||
| ATP6V1B2 | HPIPDLTGY | 2 | 1 | 2 | ||||
| BLMH | KPLFNMEDKI | 2.1907 | 1 | 1 | ||||
| CANX | APPSSPKVTY | 1 | 2.5506 | 3 | 1 | |||
| CAPN1 | LPIKDGKLVF | 2.2217 | 3 | 1 | ||||
| CCT4 | HPTIISESF | [1] | 2.2973 | 1 | 2 | H1Y;I5F;S8T | ||
| CTNNA1 | NPVQALSEF | 2.6248 | 1 | 3 | ||||
| DDOST | FPDKPITQY | 1 | 2.8022 | 1,2 | 7 | |||
| DDX21 | SPPKDVESY | 2.4713 | 3 | 2 | ||||
| DDX50 | SPPQDVESY | 2.8877 | 2,3 | 5 | ||||
| DEK | FPFEKGSVQY | 2 | 2.6723 | 1,3 | 3 | known | ||
| DNAJC13 | LPVARFLKY | 1 | 2.0127 | 3 | 1 | |||
| EEF1G | FPAGKVPAF | 2.6279 | 1,3 | 5 | ||||
| EEF2 | LPSPVTAQKY | 3 | 3 | 4 | ||||
| LPVNESFGF | 2.275 | 1 | 3 | |||||
| EFHD2 | NPYTEFKEF | 2.7636 | 3 | 1 | ||||
| ERH | NPNSPSITY | [2] | 3.0901 | 1,2,3 | 15 | |||
| FH | MPTPVIKAF | 2 | 2.8021 | 1 | 6 | known* | ||
| FLNA | VPASLPVEF | 2 | 2.0951 | 3 | 2 | candidate | ||
| GLS | DPRLKECMDM | 16 | 2.1995 | 1 | 1 | |||
| GOT2 | LPIGGLAEF | 2.1101 | 1 | 5 | ||||
| HDGF | FPYEESKEKF | 1 | 3 | 5 | ||||
| HPRT1 | IPDKFVVGY | 3.1871 | 1,3 | 7 | ||||
| HSPA8 | IPTKQTQTF | 1 | 2.4455 | 3 | 2 | TQ6NF K4R;Q5K; Q7R |
||
| QPGVLIQVY | 2.2033 | 3 | 1 | |||||
| ILF2 | KPAPDETSF | 2.1768 | 1 | 1 | ||||
| ISOC1 | IPVIVTEQY | 1 | 2.4097 | 1 | 2 | |||
| LGALS3 | FPFESGKPF | 2.2508 | 1 | 1 | ||||
| LTA4H | VPYEKGFAL | 4 | 2.4068 | 3 | 3 | |||
| MPI | RPVEEIVTF | 1 | 2.4651 | 3 | 4 | candidate | ||
| MTHFD1 | TPVPGGVGPM | 1 | 2.5411 | 1 | 3 | |||
| MYO1C | APVGGHILSY | 2.3905 | 3 | 2 | ||||
| MYO1G | DPIGGHIHSY | 2.6728 | 3 | 4 | ||||
| NARG1 | TPLEEAIKF | 2.0495 | 1 | 1 | candidate | |||
| NDUFS2 | LPYFDRLDY | 2.1946 | 3 | 1 | ||||
| NIT2 | IPEEDAGKLY | 2.7275 | 1 | 1 | ||||
| NONO | RPSGKGIVEF | 1 | 2.4937 | 2,3 | 7 | |||
| NUP210 | FPAPAKAVVY | 7 | 2.0796 | 3 | 2 | known | ||
| PABPC1 | VPNPVINPY | 2 | 3.4778 | 1,2 | 2 | candidate | ||
| PDCD6IP | FPQPPQQSY | 2.1958 | 1 | 1 | candidate | |||
| PLEC1 | LPTEEQRVY | 2.171 | 3 | 2 | ||||
| PPA2 | EPMNPIKQY | 2.9655 | 3 | 1 | ||||
| PRPF8 | SPIPFPPLSY | 2 | 2.7818 | 1,3 | 17 | |||
| PSMD7 | LPINHQIIY | 2.6684 | 3 | 2 | candidate | |||
| RAD23A | FPVAGQKLIY | 2.7215 | 3 | 2 | ||||
| RAD23B | FPEGLVIQAY | 1 | 2.2462 | 1 | 1 | |||
| RPL15 | RPVPKGATY | 3 | 2.725 | 3 | 2 | |||
| RPN1 | APDELHYTY | 2.1773 | 1,3 | 2 | ||||
| SFRS2IP | LPADVQNYY | 2.6707 | 1 | 2 | known | |||
| SLC25A6 | IPKEQGVLSF | 7 | 2.2361 | 1 | 1 | |||
| SPTBN1 | YPNVNIHNF | 1 | 2.5082 | 3 | 2 | |||
| SRRM2 | SPRVPLSAY | 2.2579 | 3 | 2 | ||||
| STIP1 | NPFNMPNLY | 2.2604 | 1 | 1 | ||||
| SYNCRIP | DPYYGYEDF | 1 | 3.1554 | 3 | 3 | candidate | ||
| SYTL3 | RPDGTLNSF | 2.1761 | 1 | 1 | G4S;T5E; F9S | |||
| TMOD3 | IPIPTLKDF | 1 | 2.6011 | 1 | 3 | |||
| TMPO | TPFKGGTLF | 2 | 2.7071 | 1,3 | 8 | |||
| FPEISTRPPL | 2.6193 | 3 | 6 | |||||
| TOP2A | LPVKGFRSY | 2 | 2.2794 | 1 | 1 | |||
| TUBB3 | YPDRIMNTF | 1 | 2.6326 | 3 | 1 | |||
| UBE2L3 | YPFKPPKITF | 2.0431 | 1 | 1 | ||||
| VCP | YPVEHPDKF | 2.9714 | 1 | 4 |
Potential peptides were determined to be derived from proteins encoded by the HeLa genome.
Prior reports according to immune epitope data base (IEDB; www.iedb.org); blank, this study
Number of entries reporting type I interferon responsiveness for the protein
Correlation coefficient represents the number of peak identities determined between the theoretically and experimentally derived spectra for a given parent ion normalized to the charge state of the peptide
Days post infection of HeLa cultures with VACV at which the peptide was identified
Total number of times a given peptide sequence was identified by mass spectrometry
Peptides derived from proteins that represent known or potential oncogenes
indicates mutated self peptide
Amino acid substitutions compared with VACV proteome. Only amino acid substitutions from sequences >66% identical are annotated. Blank, no significant homology
Amino acid changes for homologous VACV epitopes; blank, no significant homology
In order to determine the reproducibility of peptide identification, peptides presented by HLA-A*02;01 were eluted from two independently infected cell samples and sequenced by MS/MS. Replicate analyses of the self peptides eluted from sA2.1 molecules expressed by VACV-infected cells revealed that ~60-80% of the HLA-bound peptides were presented at the same time points in both experiments (Supplemental Table 3).
Comparisons of the self peptides sequenced before and during infection revealed that all five HLA class I molecules investigated here presented numerous novel self peptide sequences after VACV infection. Ranging from ~30 to ~80% of the sequenced peptides, we identified 965 A*01;01- (~56%), 2,783 A*02;01- (~80%), 1,025 B*07;02- (~64%), 354 B*35;01- (~35%), and 234 B*45;01 (~30%)-associated human peptide sequences that were presented solely after infection. Some of the new self peptides were presented stably throughout the course of the infection while others (majority) were presented in a kinetic fashion: one, two or three days post infection (Figure 2), likely reflecting the kinetics of protein expression from which a given peptide was derived. These data, in conjunction with the high reproducibility of the self peptides between replicate experiments, indicate that VACV infection induced a self peptidome shift that was much more profound than those induced by INV, MeV and HIV infections [20-22].
Characterization of the self peptidome shift reveals that VACV infection up-regulates type I IFN-regulated and other cellular pathways
We previously reported that about a fifth of the VACV peptides presented by HLA class I molecules were recognized during a natural infection [56]. We reasoned that the nonantigenic VACV peptides may have close sequence homology to the self peptides that are uniquely presented by infected cells. To narrow the analysis of these potentially homologous peptides, focus was laid on 169 A*01;01-, 309 A*02;01-, 157 B*07;02-, 71 B*35;01-, and 107 B*45;01-associated peptide sequences that correlated with the MS/MS spectra's fragment ion data with high confidence (see Materials and Methods). These peptides were further interrogated to determine the mechanism(s) that induced the presentation of altered self peptidomes (Table 2 and Supplemental Table 2). In initial analyses, self peptides were compared against the VACV proteome to determine whether self peptides uniquely presented after infection had similarity to VACV peptides. On average only about 4.7% (range 2.5%—8.2%) of the self peptides were ≥66.6% identical, i.e., less than three amino acid changes, when compared to potential VACV peptides (Table 2 and Supplemental Table 2). Hence, there was very little sequence similarity between self and viral peptides presented after infection.
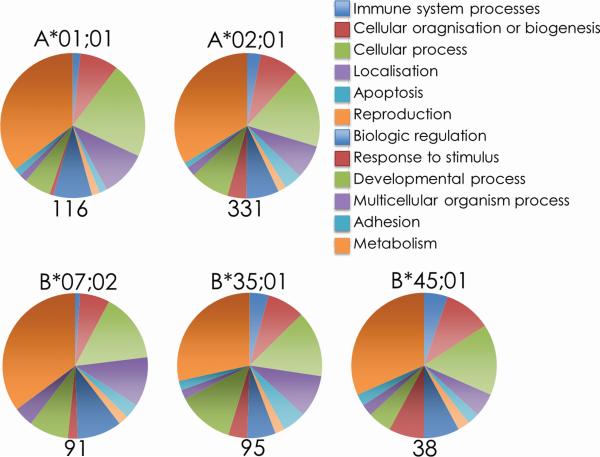
The presentation of unique self peptides after infection had been reported to result from the activation of multiple cellular pathways [20, 22]. Therefore, we classified the proteins from which these unique infection-induced self peptides were derived based upon their cellular functions by searching the Panther database [45, 46]. This analysis revealed that the proteins from which self peptides uniquely presented after infection were derived were distributed amongst multiple functional categories (Figure 3 and Supplemental Table 2). There was little alteration in the proportion of each functional protein family after VACV infection compared with uninfected samples (Supplemental Figure 1), suggesting that no particular functional family was induced in response to infection.
Figure 3. Proteins from which newly presented peptides are derived represent numerous functional families.
Panther GO Biological Process identification of the proteins from which self peptides uniquely presented during VACV infection of HeLa cells are derived. Proteins were searched using the Panther database and presented as the proportion of peptides allocated to each functional classifications.
HeLa cells respond to infection with a type I interferon response by up regulating internal innate sensors and mediators to defend against infection [58]. Therefore, we specifically sought to identify whether interferon responsive proteins were up regulated following VACV infection by searching the Interferome database for proteins known to be responsive to type I interferons [47]. Approximately, 40% of the self peptides uniquely presented after infection were attributed to type I interferon signaling (Table 2 and Supplemental Table 2) suggesting that the remaining 60% of the newly presented peptides were derived from host proteins other than those directly responsive to interferon signaling (Figure 3). These proteins are either uniquely expressed post infection or are differentially process by interferon-induced immunoproteasome in infected cells, — compared to the resting proteasome of uninfected controls. Therefore, alterations in host cell metabolism, possibly through interferon signaling, might play a significant role in the presentation of an infection-induced self peptidome.
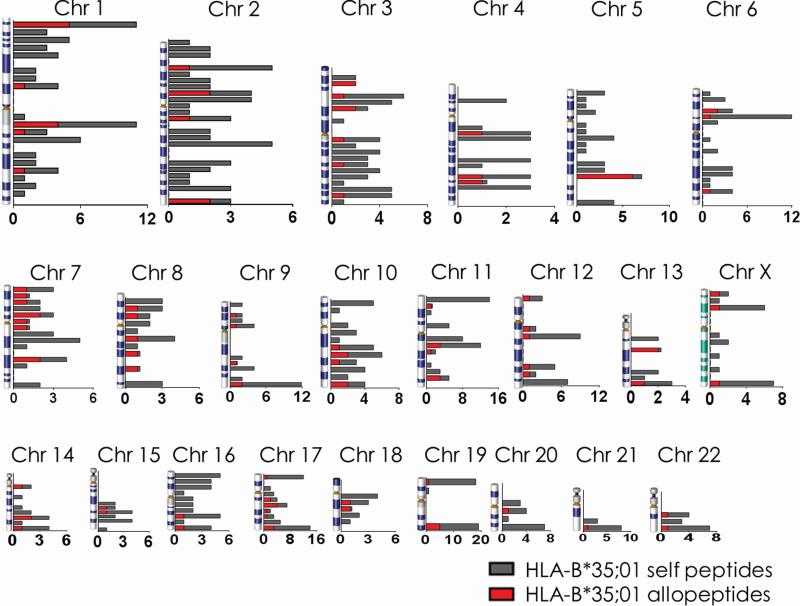
Recent drafts of the human proteome revealed that proteins routinely expressed by cells were encoded by genes distributed across all chromosomes, but rarely, if at all, from chromosome 21 and not at all from the Y chromosome [25, 26]. As well because our analyses so far indicated very little preference for protein families from which the infection-induced self peptides are derived, we mapped the chromosomal location of the genes that encode proteins from which newly presented peptides were derived in order to identify a particular chromosomal region containing the majority of newly presented peptides. The control of this region would perhaps point to a possible mechanism for this presentation. The infection-induced self peptides identified herein were derived from proteins encoded by genes distributed across all chromosomes with no concerted enrichment evident at any particular region of the human genome (grey bars, Figure 4 and Supplemental Figure 2). Similar to the reported proteome maps, few peptides were derived from proteins encoded from chromosome 21, but chromosomes 20 and 22 as well. No proteins were identified as encoded from the Y chromosome; this is consistent with the source HeLa cells being derived from a female subject [59]. Collectively, these data suggested that the shift in self peptide presentation by HLA class I molecules post VACV infection represented a global change in the overall protein metabolism of the cell and not a specific response to the infection.
Figure 4. Peptides presented after infection by HLA-B*35;01 are derived from proteins encoded by genes dispersed across the chromosomes.
The number of proteins from which peptides presented only after infection were derived is enumerated for HLA-B*35;01 (gray bars). Due to size limitations, the locations of each peptide were grouped according to major banding patterns for each chromosome (e.g., 1p36) along the vertical axis, as in reference [26]). Note the scale of the x-axis varies for each chromosome. The number of proteins containing allopeptides in the human population are enumerated for each location (red bars).
Several peptides uniquely presented after infection are derived from oncogenes
Proteomics and proteogenomics approaches have recently identified a few neo-epitopes derived from tumor specific antigens [56, 60-64]. The success of these approaches was predicated on the knowledge of the tumor ‘mutome’ —a collection of mutations within the tumor cell under study— encoded by non-synonymous single nucleotide polymorphisms revealed by exome and/or transcriptome analyses [57, 60-64]. As HeLa cells were originally isolated from a cervical tumor, we determined whether the collection of self peptides presented by the five HLA class I molecules under study here were derived from the HeLa ‘mutome’. Therefore, the proteins from which self peptides were derived were searched against the TSGene Database containing 184 tumor samples, including 28 cervical cancers [48]. On average, ~10% of self peptides were derived from proteins that are known or potential oncogenes (Table 2 and Supplemental Table 2). Self peptides derived from known oncogene products or candidate cancer proteins were then searched against the TSGene Database using BLAST to specifically identify self peptides that matched the mutated cancer protein sequence. We identified 16 HLA-A*01;01-, 34 HLA-A*02;01-, 13 HLA-B*07;02-, 22 HLAB*35;01-, and 3 HLA-B*45;01-restricted self peptide sequences that match known cancer associated mutations (* in Table 2 and Supplemental Table 2). Hence, a direct analysis of the five self peptidomes revealed that a fraction of the self peptides presented by class I molecules were derived from HeLa mutome.
Self peptidome presented after infection contains allopeptides
T cell activation by peptides induced for presentation under inflammatory conditions may be of great significance for HLA-matched transplant recipients responding to infections. In this regard, it is noteworthy that immunosuppressed transplant recipients are susceptible to cytomegalovirus infections [65-68]. This infection is known to induce acute allograft rejection [67, 68] and GVHD [65, 66]. These adverse outcomes would be further augmented if new peptides presented during infection contained allelic differences between the donor and recipient tissues as in allopeptides. Hence, recipient T cells would not be tolerized to such allopeptides. Therefore, in order to determine whether the self peptidomes presented after infection contains allopeptides, BLAST searches of the peptide sequences were performed against translated sequences from 297 human genomes. This analysis revealed variations of one to two amino acids in ~12% (range 9.6%—13.2%) of the peptides newly presented after infection (Table 3; Supplemental Table 4; red bars in Figure 4 and Supplemental Figure 2). This percentage is well above the false discovery rate calculated and presented in Materials and Methods suggesting the observed variability may be biologically relevant. In comparison, only about 2.7% (range 2.3%—3.4%) of the self peptides presented before infection displayed variation amongst the human population. This is less than the false discovery rate suggesting that the presentation of allopeptides in uninfected cells may be an artifact of incorrect peptide identification. The percentage of allopeptides presented after infection (12%) is similar to previous reports of variations in human MHC class I-associated peptides arising from genetic polymorphisms in the population [69].
Table 3.
Potential allopeptides presented by HLA-B*35;01 after infection with VACV
| Gene | Variants | VACVc | ||
|---|---|---|---|---|
| Aa | Bb | Cb | ||
| PABPC1 | VPNPVINPY | VPHPVINPY | ||
| CNOT8 | FPSIYDVKY | SPSIYDVKY | FPVIYDVKY | |
| UPP1 | FPALFGDVKF | FPALFGDVKV | LPAMFGDVKF | |
| LOC441837 | DPFIDLNYM | DPFIDLKYM | ||
| LOC387820 | SPEDIKKAY | SPEEIKKAY | ||
| ACTB | APEEHPVLL | APEEHPILL | ||
| CNOT7 | FPVIYDVKY | FPSIYDVKY | ||
| USP11 | TPARDYNNSY | TPARDYSNSS | ||
| UBE2D2 | YPFKPPKVAF | YPFKPPKVTF | ||
| C1D | YPVEIHEYL | YPVEIHDYL | ||
| ADAR | NPISGLLEY | NPVSGLLEY | ||
| DDX50 | SPPQDVESY | SPPQDIESY | ||
| ITM2B | DPANIVHDF | DPADIIHDF | ||
| DDOST | FPDKPITQY | FPDKPITQV | FPDKRITQY | |
| KIAA0828 | GPFKPNYYRY | GPFKPDHYRY | ||
| CCND1 | TPHDFIEHF | TPHDFIEHI | ||
| HNRPD | TPEEKIREYF | ATEEKIREYF | ||
| MLSTD2 | NPFHWGEVEY | NPFHWGEVGM | ||
| THRAP1 | KPINKSEHL | KPVNKSEHL | ||
| EFHD2 | NPYTEFKEF | NPYTEFPEF | ||
| CCT6A | HPRIITEGF | HPRIIAEGF | ||
| BACH1 | SPEPGQRTF | *PEPGQRTF | ||
| HSPA4L | APFSKVITF | APFSKVLTF | ||
| NIT2 | IPEEDAGKLY | IPEEDAGKLD | ||
| RPL15 | RPVPKGATY | HPVPKGATY | RPVPKGVTY | |
| OTUB1 | FPEGSEPKVY | FPEGSEPQVY | ||
| ROR2 | FPELGGGHAY | FPELNGGHSY | ||
| ARGBP2 | FPISYVEKL | FPISYVEKP | ||
| MYO1G | DPIGGHIHSY | DPIGGHINNY | ||
| DEK | FPFEKGSVQY | FPFEKGSAQY | ||
| UBE2E1 | YPFKPPKVTF | YPFKPPKITF | ||
| KIF1A | IPQLCEDLF | IPQLCEELF | ||
| TUBB3 | YPDRIMNTF | YPDRIINTF | ||
| CTNNA1 | NPVQALSEF | SPVQALSEF | ||
| SF3B4 | RPITVSYAF | HPITVSYAF | ||
| ATP6V1B2 | HPIPDLTGY | HPIPDLTGF | ||
| C20orf172 | HPIHQGITEL | HPIHQGITEV | ||
| ADAR | NPVGGLLEY | NPVSGLLEY | ||
| MTHFD1 | TPVPGGVGPM | TPVSGGVGPM | ||
| SMARCA2 | APSVVKISY | APSVVKVSY | ||
| KIAA1374 | TPYPAILHEY | PPYPAILHEY | ||
| SPTBN1 | YPNVNIHNF | YPNVNVHNF | ||
| KIAA1102 | SPLGGERPF | SPLGGQRPF | ||
| UBAP2 | NPYPGDVTKF | NPYSGDVTKF | ||
| SMARCA5 | APFHQLRISY | APFHQLRIQY | ||
| NT5C | FPEEPHVPL | SPEEPHVPL | ||
| DIAPH1 | NPVSWVQTF | NPVSWVESF | ||
| CTPS | RPIKPSPPY | RPMKPSPPY | ||
| HSPA8 | IPTKQTQTF | IPTKQTQIF | TQ6NF K4R;Q5K;Q7R |
|
| SUHW2 | NPIVLLSDF | NPIVLLSNF | ||
| CP | FPRTPGIWL | FPRTPGLWL | ||
| MYO1C | APVGGHILSY | IPVGGHIISY | ||
| UBP1 | SPWPDAPTAY | SPWPDASTAY | ||
| DHX38 | TPLPTPSYKY | TPLPAPSYKY | ||
| LPP | YPVTGPKKTY | CPVTSPKKTY | ||
| FLJ10706 | LPLWQHISF | SPLWQHIGF | ||
| FHL2 | KPITTGGVTY | MPITTGGVTY | ||
| TTF1 | FPFRDIFYY | FPSRDIFYY | ||
| UBE2D1 | YPFKPPKIAF | YPFKPPKITF | ||
| C6orf150 | VPRIQLEEY | VPRIQLEDF | ||
| HDHD4 | KPAPSIFYY | LPAPSIFYY | ||
| KIF21A | HPNNVVSVKY | HPNNVVSIKY | ||
| LOC441032 | HPGQISAGY | HPGQISSGY | ||
| AHCY | GPFKPDHYRY | CPFKPDHYRY | ||
| FLJ14827 | APHTNGPQDL | VPHTNGPQDL | ||
| BNIP2 | MPESSQPNY | TPESSQPNY | ||
| UBE2V2 | LPQPPEGQTY | LPQPPEGQCY | ||
| TUSC4 | HPTLGPKITY | HLTLGPKITY | ||
| NME2 | RPFFPGLVKY | RPFFAGLVKY | ||
| TCP1 | HPTSVISGY | HPTSVISSY | ||
| FLNB | IPYLPITNF | VPYLPITNF | ||
| LOC220717 | VPHSIINGY | VPHSIIDGY | ||
| PSMC4 | LPLTHFELY | LPVTHFELY | ||
| ZNF581 | SPCPQPLAF | SPCPQPLPF | ||
| TFCP2L2 | LPLNIQVDTY | LPLNIQIDTY | ||
| TOP2A | LPVKGFRSY | LPVNGFRSY | ||
| PHCA | YPWLRGLGY | YPWLRGLGI | ||
| ASCC3L1 | RPVPLEQTY | RPVPLERTY | ||
| SLC25A6 | IPKEQGVLS F | IPKDQGVLSF | ||
| LOC391387 | HPWKVMPDL | HPWEVMPDL | ||
| LOC339077 | LPKLEKAARL | LPKLERAARL | ||
| HSPA8 | QPGVLIQVY | QPGVFIQVY | ||
| SUHW1 | NPIVLLSNF | NPIVLLSDF | ||
| VIL2 | FPWSEIRNI | FPWNEIRNI | ||
| ING1 | LPIDPNEPTY | MPVDPNEPTY | ||
| FLJ14803 | HPKYPDGKTF | HPKYRDGKTF | ||
| LOC391387 | HPWKVMPDLY | HPWEVMPDLY | ||
| GAPD | APSADAPMF | TPSADAPMF | ||
| CARD8 | HPHPEDIKF | RPHPEDIKF | HRHPEDIKF | |
| NACA | SPASDTYIVF | SPASDTYVVF | ||
| TPMT | DPTKHPGPPF | DPTKHAGPPF | ||
| LOC391062 | RPNSNGSQFF | PNTNGSQFF | ||
| ANXA7 | YPQPPSQSY | YPQPPSQSI | ||
| RYR1 | SPHEQEIKFF | SPRDQEIKFF | ||
| ADIPOR2 | APLQEKVVF | PPLQEKVVF | ||
| FLJ10774 | IPWTVSEQF | IPWTVSEQV | ||
| ALB | VPQVSTPTL | VPEVSTPTL | ||
| TIP120A | GPLVSKVKEY | GPLVVKVKEY | ||
| PTPRS | WPDHGVPEY | WPDHGVPEH | ||
| GRK5 | KPENILLDDY | KPENILLDDH | E3D;D8F;Y10S | |
| ENO1 | SPDQLADLY | SPNQLADLY | ||
| ARHGEF1 | VPVPPNVAF | PPVPPNVAF | ||
| SORCS3 | SPVHCLLPF | SPVHCLLPQ | DPVHCLLPY | |
| MAGI 1 | KPGEGLGMY | KPSEGLGMY | ||
| ITGB4 | RPLQGYSVEY | RPLQGYSVAS | ||
| HLA-C | HPLSDHEATL | HPISDHEATL | ||
| SLC17A6 | MPLAGILVQY | MPLAGVLVQY | ||
| UBE2L3 | YPFKPPKITF | YPFKPPKVTF | ||
| PIK3C2B | LPQLVQALKY | LSQLVQALKY | ||
| CDC42 | FPSEYVPTVF | FPGEYVPTVF | ||
| ProSAPi P1 | DPGRDPLLAF | DPGKEPLLAF | ||
| AATF | LPQPDVFPLF | LPQPDVFPVF | ||
| RER1 | LPTKQNEEF | YPTKQNEEF | LPTIQNEEF | |
| GRK5 | SPDYWGLGCL | SPDWWGLGCL | ||
| PACS2 | LPIAEAMLTY | LPVAEAMLT | ||
| NDUFB9 | FPDSPGGTSY | FPDSPRGTSY | ||
| LIPC | QPGCHFLELY | QPGCHSLELY | ||
| HNRPK | FPNTETNGEF | FPNTETSGEF | ||
| DHRS4 | SPSPGFSPY | IPSPGFSPY | ||
| METAP2 | FPKGQECEY | FPKGQESEY | ||
| SMOC1 | RPLPGTSTRY | RPIPGTSTRY | ||
| DHX40 | MPDHVIPEI | MPDHVIPQF | ||
| PREI3 | TPKECPAIDY | TPKECRAIDY | ||
| PCDHB5 | APETVVAVF | SPETVVAVF | ||
| SMARCE1 | MPSTPGFVGY | MPSTPSFVGC | ||
| DSC3 | IPCSMQENSL | IPCSMLENSL | ||
| RHOC | FPEVYVPTVF | FPEEYVPTVF | ||
| NOTCH1 | CPPGFTGSY | CPPGFTGDY | ||
| GLCCI1 | CPDKNKVNF | CPDKNKVHF | ||
| BIRC5 | DPIGPGTVAY | DPIGPGTVA | ||
| SMURF2 | NPYYGLFQY | NPYYGLFEY | ||
| MEN1 | APDPPGGLTY | APDPHGGLTY | ||
| NUP210L | SPLTPGLAIY | KSLTPGLAIY | ||
| ING5 | MPVDPNEPTY | LPIDPNEPTY | ||
| JUP | SPVESVLFY | SPVDSVLFY | ||
| UBE2D2 | SPYQGGVFF | RPYQGGVFF | ||
| GPNMB | GPQLMEVTVY | GPQFMEVTVY | ||
| PURB | LPAQGLIEF | LPAQGMIEF | ||
| CPA4 | LPVANPDGY | LPVTNPDGY | ||
| LOC440059 | CPIMDLTLY | CPIMDLTL | ||
| CDADC1 | LPDANTDFY | LPDANTDLY | ||
| TINP1 | TPQGAVPAY | IPQGAVPAY | ||
| IFI16 | MPPSTPSSSF | MPPTTPSSSF | ||
| AATF | LPQPDVFPL | LPQPDVFPVF | ||
| FMNL3 | DPSVTRKKF | DPAVTRKKF | ||
| B4GALT6 | APGIANTYLF | APGIVNTYLF | ||
| GPR | VPVVVVFLFL | IPVVVIFLFL | ||
| ATXN10 | HPDKKIVAY | CPDKKIVAY | ||
Indicates corresponding amino acid is deleted in variant
Peptide sequence identified by mass spectrometry
Alternate peptide sequences identified by BLAST search
Amino acid substitutions compared with VACV proteome. Only amino acid substitutions from sequences >66% identical are annotated. Blank, no significant homology.
We noted that the frequency of allopeptides (12%) was similar to the reported rates of unsuccessful transplants (10%) suggesting a possible relationship between allopeptides and transplantation success. However, the presentation of allopeptides would only affect clinical outcomes if those peptides are presented by cells in transplanted tissues. Search of the TiGER tissue expression database revealed that 75% of the proteins from which the allopeptides were derived are expressed at variable levels in commonly transplanted tissues such as the liver, heart, lung, kidney and colon ([49]; Figure 5 and Supplemental Figure 3).
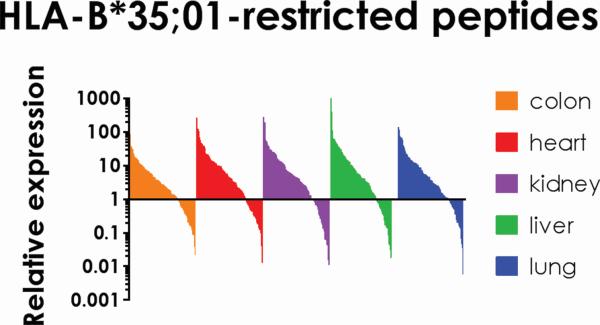
Figure 5. Proteins from which newly presented HLA-B*35;01-restricted allopeptides are derived are expressed in transplanted tissues.
Proteins expressed by the colon, heart, kidney, liver and lung were identified by searching the TiGER database of tissue expression. This database consists of expression data from microarray, real-time PCR, and proteomic. Studies and is reported as a fold change compared with housekeeping genes/proteins used in each respective assay. These proteins are variably expressed in the tissue yet all tissues express some proteins from which new peptides were derived after infection.
Discussion
Cumulatively, we have shown a profound shift in self peptides presented by MHC class I molecules after infection of HeLa cells with VACV. Depending on the HLA allele, the new self peptides represented between 30—80% of all peptides presented during infection. They were derived from proteins encoded by genes belonging to multiple cellular functional families and were broadly distributed across chromosomes. We did note that ~40% of the proteins from which peptides were derived were responsive to type I interferon signaling which may play at least some role in the generation of these peptides uniquely presented after infection. Critically, a subset of the altered peptides contained allelic variations within the human population and the proteins from which they were derived were expressed in commonly transplanted tissues suggesting the potential to negatively affect the outcome of clinical tissue/organ allografts.
This study significantly extends previous reports of self peptides that are uniquely presented by HLA-A*02;01 and HLA-B*07;02 during INV, MeV and HIV infections [20-22]. While these previous studies reported the presentation of only 20 self peptides uniquely presented during infection, here, we report over 1,000 peptides presented by five different HLA class I molecules solely after infection with VACV, the largest study of its kind to date. Herein, we identified 4 of the 20 peptides uniquely presented after infection with INV [21], 3 of the 15 peptides uniquely presented after infection with HIV [20], and neither of the two type I interferon-induced peptides presented after MeV infection [22]. Despite analysis of infected HeLa ,similar to previous studies, we observed two orders of magnitude greater number of peptides represented within the altered self peptidome than in the previous studies [20-22]. Perhaps the limited numbers of the total self peptides reported from past studies to be uniquely presented after infection with HIV, INV and measles was insufficient to detect a larger peptidome shift [20-22]. Critically however, alterations in the self peptidome have now been observed for five different HLA class I molecules and in response to multiple infections (HLA-A*02;01 and HLA-B*07;02): HIV, INV, MeV and VACV.
Despite a common core of host proteins responsive to infection [70], there is little overlap among other host proteins that are involved in cellular responses to VACV, MeV, HIV and INV infection [71-79]. This may, in part, explain the differences in the self peptidome shift reported here for VACV infection compared with HIV, MeV, and INV [20-22]. In addition, VACV encodes over 200 proteins [80, 81], including a number of proteins that alter cellular functions and immune response [79, 82-89]. In comparison, HIV genome encodes 15 proteins [90], INV genome encodes 17 proteins [91], and MeV genome encodes 8 proteins [92]. The large number of proteins encoded by VACV many result in substantially different effects on the host cell protein metabolism compared with the limited host range factors of HIV, INV and MeV.
In addition to virus-induced changes in host cell metabolism, 40% of peptides uniquely presented after infection were derived from proteins responsive to host interferon signaling. Although not as effective as type II interferon, type I interferons have also been reported to induce immunoproteasomes [93-100], which in turn alters the processing of self and viral peptides for presentation by MHC class I molecules [101]. In response to both type I and type II interferon signaling, the metabolism of HeLa cells is altered [102-109]. Although it remains unknown whether VACV infection leads to upregulation and function of immunoproteasomes, the presentation of newly processed peptides represents a change in intracellular protein turnover that will include antigen processing by the immunoproteasome. It is also possible that the extent of self peptidome shift might vary between different viral infections and, if so, the mechanism underlying this process could be of interest for future studies. For example, whether levels of cellular interferon response correlates with the magnitude of the self peptidome shift for different classes of viruses.
In recent years, much attention has been directed to the production of unique peptides through alternate translation pathways [110-117]. These cryptic peptides are produced from translation of alternate reading frames [118-122], read-through into the untranslated region [123], and frame shift mutations [124-126]. The peptides uniquely presented during VACV infection reported herein map to protein coding regions; however, it is possible that these proteins are generated through these alternate translation mechanisms. Determining whether such peptides are generated by translation of alternate reading frames or through mutations occurring during translation will require further analyses as infection [114, 115, 120] and perhaps interferon signaling may accentuate this process.
Infection of HeLa cells was confirmed by the identification of VACV-derived peptides presented by all HLA molecules studied. Those viral peptides presented by HLA-A*02;01 and -B*07;02 were shown to contain a subset of peptides that elicit protective immune responses in HLA class I-transgenic mice [33]. With similar characterization, a subset of the HLA-A*01;01-, -B*35;01-, and -B*45;01-restricted peptides reported here may also represent CD8 T cell epitopes useful for the development of next-generation vaccines.
Examination of self peptides presented by the five different HLA class I molecules revealed that each HLA molecule had a different propensity to present an altered self peptidome after infection. The greatest variety of newly presented peptides was observed with HLA-A*02;01 with 80.7% of the total peptides sequenced presented only after infection. HLA-B*07;02 and -A*01;01 had an intermediate shift in the peptidome with 64.4% and 56.6%, respectively, of total peptides presented only after infection. The presentation of unique self peptides after infection was less striking for HLA-B*35;01 (35.4%) and -B*45;01 (31.2%). It remains to be determined whether this variability among HLA class I molecules to present a shifted peptide repertoire after infection has a biological consequence, e.g., whether it correlates with the susceptibility to allograft rejection.
Biologically, the presentation of self peptides by MHC class I molecules annotates the internal state of the cell. Our data indicated that VACV infection profoundly impacted the cellular metabolism and, hence, the altered state of the cell. Significantly, a fraction of the self peptides presented by class I molecules were derived from HeLa mutome. This finding implies that the knowledge of the self peptidomes of non-cancer and cancer cells from the same individual can reveal neo-epitopes that can be targeted by tumor-specific T cells. Considering the finding that viral infections can alter the presentation of self peptides, cancer therapies based on oncolytic viruses can coax the tumor cell to display neo-epitopes that are coded by genes induced by viral infections [127, 128].
Of clinical concern is whether T cells recognize the peptides from the shifted self peptidome. Aire-regulated peptide expression in the thymus is thought to lead to the presentation of self peptides derived from proteins ordinarily not expressed by the thymus [129, 130]. However, if these peptides are presented only after infection and are not presented during T cell development, self-reactive T cells may persist in the periphery. Following infection, T cells may be activated under an inflammatory environment causing additional immunopathology. Recognition of cross-reactive peptides seems to be dependent upon only select amino acid residues that vary for each particular T cell receptor [131]. Similarities within five amino acids between self and foreign peptides are sufficient to induce cross-reactive T cells after infection potentially resulting in autoimmunity [132]. Yet, identification of these peptides proves to be difficult, as no single immunologic property is able to computationally predict immunogenic peptides [56].
Our results may help to explain the complication and ~10% failure rate of transplantation even amongst HLA-matched allograft recipients. Of the peptides that were identified to be uniquely presented after infection, ~12% displayed allelic variation amongst the human population. These potential allopeptides in transplanted tissues would remain innocuous until the recipient acquired an infection. The presentation of new peptides during the inflammatory conditions generated in response to infection may result in T cell activation and recognition of these potential allo-epitopes. Again, if certain HLA class I molecules are less likely to present novel peptides after infection, patients that express them would be less susceptible to GVHD or allograft rejection. Analysis of the proteomes of donor and recipient transplant patient may identify these allopeptides prior to committing to the organ transplant. Following the recent publication of drafts of the human proteome, these comparisons may become more commonplace, leading to better proteomic matches for organ transplants, preventing allopeptide presentation after infection, and ultimately more successful transplants [25-28].
Supplementary Material
Statement of clinical relevance.
MHC-encoded class I molecules present peptides derived from cellular proteins to CD8 informing them of a cell's milieu intérieur (cellular homeostasis). Cellular homeostasis is altered under a variety of stressed conditions, including during microbial infections. Whilst there were hints that infected cells altered the presentation of self peptide repertoire, this notion was not fully explored. Herein, through mass spectrometric analysis of thousands of self peptides isolated from HLA class I molecules (the largest study of its kind to date), we observed a dramatic shift in the self peptide repertoires presented after infection of HeLa cells with vaccinia virus. Of significant clinical import was the revelation that a fraction of the self peptides were derived from tumor-specific antigens. Furthermore, the self peptides uniquely presented after infection contained variants of such peptides —called allopeptides— within the human population. Approximately 12% of the self peptides uniquely presented after infection were potential allopeptides, the recognition of which can result in graft-versus-host-disease or transplant rejection. The proportion of allopeptides was very similar to the reported rate of transplant complication and failure (~10%). Our study suggests that deep sequencing and proteomics analyses of self peptides may enhance the success of clinical transplant outcome and tumor immunotherapies.
Acknowledgements
This work was supported by a VA Merit Award BX001444 as well as National Institutes of Health contract AI040079 and grants HL054977 and HL121139 to S.J. and by a National Institutes on Minority Health and Health Disparities grant 2G12MD007592 to support CTS.
Abbreviations
- BLAST
basic local alignment search tool
- Cn
Correlation coefficient
- GVHD
graft versus host disease
- HIV
human immunodeficiency virus
- HLA
human leukocyte antigen
- INV
influenza virus
- MeV
measles virus
- MPXV
monkeypox virus
- VACV
vaccinia virus
- VARV
variola virus
Footnotes
Conflicts of Interest
The authors have no conflicts of interest to declare.
References
- 1.Hickman HD, Luis AD, Buchli R, Few SR, et al. Toward a definition of self: proteomic evaluation of the class I peptide repertoire. J Immunol. 2004;172:2944–2952. doi: 10.4049/jimmunol.172.5.2944. [DOI] [PubMed] [Google Scholar]
- 2.Perreault C. The Origin and Role of MHC Class I-Associated Self-Peptides. Progress in Molecular Biology and Translational Science. 2010;92:41–60. doi: 10.1016/S1877-1173(10)92003-6. [DOI] [PubMed] [Google Scholar]
- 3.de Verteuil D, Granados DP, Thibault P, Perreault C. Origin and plasticity of MHC I-associated self peptides. Autoimmun Rev. 2012;11:627–635. doi: 10.1016/j.autrev.2011.11.003. [DOI] [PubMed] [Google Scholar]
- 4.Yewdell JW, Bennink JR, Hosaka Y. Cells process exogenous proteins for recognition by cytotoxic T lymphocytes. Science. 1988;239:637–640. doi: 10.1126/science.3257585. [DOI] [PubMed] [Google Scholar]
- 5.Yachi PP, Ampudia J, Gascoigne NR, Zal T. Nonstimulatory peptides contribute to antigen-induced CD8-T cell receptor interaction at the immunological synapse. Nat Immunol. 2005;6:785–792. doi: 10.1038/ni1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yachi PP, Lotz C, Ampudia J, Gascoigne NR. T cell activation enhancement by endogenous pMHC acts for both weak and strong agonists but varies with differentiation state. J Exp Med. 2007;204:2747–2757. doi: 10.1084/jem.20062610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Anikeeva N, Lebedeva T, Clapp AR, Goldman ER, et al. Quantum dot/peptide-MHC biosensors reveal strong CD8-dependent cooperation between self and viral antigens that augment the T cell response. Proc Natl Acad Sci U S A. 2006;103:16846–16851. doi: 10.1073/pnas.0607771103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zinkernagel RM, Callahan GN, Klein J, Dennert G. Cytotoxic T cells learn specificity for self H-2 during differentiation in the thymus. Nature. 1978;271:251–253. doi: 10.1038/271251a0. [DOI] [PubMed] [Google Scholar]
- 9.Adamopoulou E, Tenzer S, Hillen N, Klug P, et al. Exploring the MHC-peptide matrix of central tolerance in the human thymus. Nat Commun. 2013;4 doi: 10.1038/ncomms3039. [DOI] [PubMed] [Google Scholar]
- 10.Matzinger P, Zamoyska R, Waldmann H. Self tolerance is H-2-restricted. Nature. 1984;308:738–741. doi: 10.1038/308738a0. [DOI] [PubMed] [Google Scholar]
- 11.Fissolo N, Haag S, de Graaf KL, Drews O, et al. Naturally presented peptides on major histocompatibility complex I and II molecules eluted from central nervous system of multiple sclerosis patients. Mol Cell Proteomics. 2009;8:2090–2101. doi: 10.1074/mcp.M900001-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.van der Werf N, Kroese FG, Rozing J, Hillebrands JL. Viral infections as potential triggers of type 1 diabetes. Diabetes Metab Res Rev. 2007;23:169–183. doi: 10.1002/dmrr.695. [DOI] [PubMed] [Google Scholar]
- 13.Macdonald WA, Chen Z, Gras S, Archbold JK, et al. T cell allorecognition via molecular mimicry. Immunity. 2009;31:897–908. doi: 10.1016/j.immuni.2009.09.025. [DOI] [PubMed] [Google Scholar]
- 14.Bharat A, Mohanakumar T. Allopeptides and the alloimmune response. Cell Immunol. 2007;248:31–43. doi: 10.1016/j.cellimm.2007.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Spencer CT, Gilchuk P, Dragovic SM, Joyce S. Minor histocompatibility antigens: presentation principles, recognition logic and the potential for a healing hand. Curr Opin Organ Transplant. 2010;15:512–525. doi: 10.1097/MOT.0b013e32833c1552. [DOI] [PubMed] [Google Scholar]
- 16.Spencer CT, Joyce S. Know thyself: Variations in self peptidomes and their immunologic consequences. Am Soc Histocompatibility & Immunogenetics Quart. 2012;36:28–36. [Google Scholar]
- 17.Weikert BC, Blumberg EA. Viral Infection after Renal Transplantation: Surveillance and Management. Clinical Journal of the American Society of Nephrology. 2008;3:S76–S86. doi: 10.2215/CJN.02900707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Martin-Gandul C, Mueller NJ, Pascual M, Manuel O. The Impact of Infection on Chronic Allograft Dysfunction and Allograft Survival After Solid Organ Transplantation. Am J Transplant. 2015 doi: 10.1111/ajt.13486. [DOI] [PubMed] [Google Scholar]
- 19.Ohashi PS, Oehen S, Buerki K, Pircher H, et al. Ablation of “tolerance” and induction of diabetes by virus infection in viral antigen transgenic mice. Cell. 1991;65:305–317. doi: 10.1016/0092-8674(91)90164-t. [DOI] [PubMed] [Google Scholar]
- 20.Hickman HD, Luis AD, Bardet W, Buchli R, et al. Cutting edge: class I presentation of host peptides following HIV infection. J Immunol. 2003;171:22–26. doi: 10.4049/jimmunol.171.1.22. [DOI] [PubMed] [Google Scholar]
- 21.Wahl A, Schafer F, Bardet W, Hildebrand WH. HLA class I molecules reflect an altered host proteome after influenza virus infection. Hum Immunol. 2010;71:14–22. doi: 10.1016/j.humimm.2009.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Herberts CA, van Gaans-van den Brink J, van der Heeft E. d., van Wijk M, et al. Autoreactivity against induced or upregulated abundant self-peptides in HLA-A*0201 following measles virus infection. Hum Immunol. 2003;64:44–55. doi: 10.1016/s0198-8859(02)00707-3. [DOI] [PubMed] [Google Scholar]
- 23.Dierselhuis M, Goulmy E. The relevance of minor histocompatibility antigens in solid organ transplantation. Curr Opin Organ Transplant. 2009;14:419–425. doi: 10.1097/MOT.0b013e32832d399c. [DOI] [PubMed] [Google Scholar]
- 24.Gratwohl A, Dohler B, Stern M, Opelz G. H-Y as a minor histocompatibility antigen in kidney transplantation: a retrospective cohort study. Lancet. 2008;372:49–53. doi: 10.1016/S0140-6736(08)60992-7. [DOI] [PubMed] [Google Scholar]
- 25.Kim MS, Pinto SM, Getnet D, Nirujogi RS, et al. A draft map of the human proteome. Nature. 2014;509:575–581. doi: 10.1038/nature13302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wilhelm M, Schlegl J, Hahne H, Moghaddas Gholami A, et al. Mass-spectrometry-based draft of the human proteome. Nature. 2014;509:582–587. doi: 10.1038/nature13319. [DOI] [PubMed] [Google Scholar]
- 27.Spierings E, Kim YH, Hendriks M, Borst E, et al. Multicenter analyses demonstrate significant clinical effects of minor histocompatibility antigens on GvHD and GvL after HLA-matched related and unrelated hematopoietic stem cell transplantation. Biol Blood Marrow Transplant. 2013;19:1244–1253. doi: 10.1016/j.bbmt.2013.06.001. [DOI] [PubMed] [Google Scholar]
- 28.Spierings E, Goulmy E. Minor histocompatibility antigen typing by DNA sequencing for clinical practice in hematopoietic stem-cell transplantation. Methods Mol Biol. 2012;882:509–530. doi: 10.1007/978-1-61779-842-9_29. [DOI] [PubMed] [Google Scholar]
- 29.Dierselhuis MP, Spierings E, Drabbels J, Hendriks M, et al. Minor H antigen matches and mismatches are equally distributed among recipients with or without complications after HLA identical sibling renal transplantation. Tissue Antigens. 2013;82:312–316. doi: 10.1111/tan.12209. [DOI] [PubMed] [Google Scholar]
- 30.van der Torren CR, van Hensbergen Y, Luther S, Aghai Z, et al. Possible role of minor h antigens in the persistence of donor chimerism after stem cell transplantation; relevance for sustained leukemia remission. PLoS ONE. 2015;10:e0119595. doi: 10.1371/journal.pone.0119595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vogt MH, van den Muijsenberg JW, Goulmy E, Spierings E, et al. The DBY gene codes for an HLA-DQ5-restricted human male-specific minor histocompatibility antigen involved in graft-versus-host disease. Blood. 2002;99:3027–3032. doi: 10.1182/blood.v99.8.3027. [DOI] [PubMed] [Google Scholar]
- 32.Spierings E, Vermeulen CJ, Vogt MH, Doerner LE, et al. Identification of HLA class II-restricted H-Y-specific T-helper epitope evoking CD4+ T-helper cells in H-Y-mismatched transplantation. Lancet. 2003;362:610–615. doi: 10.1016/S0140-6736(03)14191-8. [DOI] [PubMed] [Google Scholar]
- 33.Gilchuk P, Spencer CT, Conant SB, Hill T, et al. Discovering naturally processed antigenic determinants that confer protective T cell immunity. J Clin Invest. 2013;123:1976–1987. doi: 10.1172/JCI67388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Prilliman K, Lindsey M, Zuo Y, Jackson KW, et al. Large-scale production of class I bound peptides: assigning a signature to HLA-B*1501. Immunogenetics. 1997;45:379–385. doi: 10.1007/s002510050219. [DOI] [PubMed] [Google Scholar]
- 35.Joyce S, Tabaczewski P, Angeletti RH, Nathenson SG, et al. A nonpolymorphic major histocompatibility complex class Ib molecule binds a large array of diverse self-peptides. J Exp Med. 1994;179:579–588. doi: 10.1084/jem.179.2.579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Link AJ, LaBaer J, Cold Spring Harbor Laboratory Proteomics : a Cold Spring Harbor Laboratory course manual. 2009;viii:228. [Google Scholar]
- 37.Link AJ, Jennings JL, Washburn MP. Analysis of protein composition using multidimensional chromatography and mass spectrometry. Curr Protoc Protein Sci. 2004 doi: 10.1002/0471140864.ps2301s34. Chapter 23, Unit 23 21. [DOI] [PubMed] [Google Scholar]
- 38.Gerbasi VR, Link AJ. The myotonic dystrophy type 2 protein ZNF9 is part of an ITAF complex that promotes cap-independent translation. Mol Cell Proteomics. 2007;6:1049–1058. doi: 10.1074/mcp.M600384-MCP200. [DOI] [PubMed] [Google Scholar]
- 39.Sadygov RG, Eng J, Durr E, Saraf A, et al. Code developments to improve the efficiency of automated MS/MS spectra interpretation. J Proteome Res. 2002;1:211–215. doi: 10.1021/pr015514r. [DOI] [PubMed] [Google Scholar]
- 40.Eng JK, Mccormack AL, Yates JR. An Approach to Correlate Tandem Mass-Spectral Data of Peptides with Amino-Acid-Sequences in a Protein Database. Journal of the American Society for Mass Spectrometry. 1994;5:976–989. doi: 10.1016/1044-0305(94)80016-2. [DOI] [PubMed] [Google Scholar]
- 41.McAfee KJ, Duncan DT, Assink M, Link AJ. Analyzing proteomes and protein function using graphical comparative analysis of tandem mass spectrometry results. Mol Cell Proteomics. 2006;5:1497–1513. doi: 10.1074/mcp.T500027-MCP200. [DOI] [PubMed] [Google Scholar]
- 42.Link AJ, Eng J, Schieltz DM, Carmack E, et al. Direct analysis of protein complexes using mass spectrometry. Nat Biotechnol. 1999;17:676–682. doi: 10.1038/10890. [DOI] [PubMed] [Google Scholar]
- 43.Mailman MD, Feolo M, Jin Y, Kimura M, et al. The NCBI dbGaP database of genotypes and phenotypes. Nat Genet. 2007;39:1181–1186. doi: 10.1038/ng1007-1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tryka KA, Hao L, Sturcke A, Jin Y, et al. NCBI's Database of Genotypes and Phenotypes: dbGaP. Nucleic Acids Res. 2014;42:D975–979. doi: 10.1093/nar/gkt1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mi H, Muruganujan A, Thomas PD. PANTHER in 2013: modeling the evolution of gene function, and other gene attributes, in the context of phylogenetic trees. Nucleic Acids Res. 2013;41:D377–386. doi: 10.1093/nar/gks1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mi H, Muruganujan A, Casagrande JT, Thomas PD. Large-scale gene function analysis with the PANTHER classification system. Nat. Protocols. 2013;8:1551–1566. doi: 10.1038/nprot.2013.092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rusinova I, Forster S, Yu S, Kannan A, et al. INTERFEROME v2.0: an updated database of annotated interferon-regulated genes. Nucleic Acids Research. 2013;41:D1040–D1046. doi: 10.1093/nar/gks1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhao M, Sun J, Zhao Z. TSGene: a web resource for tumor suppressor genes. Nucleic Acids Res. 2013;41:D970–976. doi: 10.1093/nar/gks937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Liu X, Yu X, Zack DJ, Zhu H, et al. TiGER: a database for tissue-specific gene expression and regulation. BMC Bioinformatics. 2008;9:271. doi: 10.1186/1471-2105-9-271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rice AP, Roberts BE. Vaccinia virus induces cellular mRNA degradation. Journal of Virology. 1983;47:529–539. doi: 10.1128/jvi.47.3.529-539.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pedley S, Cooper RJ. The Inhibition of HeLa Cell RNA Synthesis Following Infection with Vaccinia Virus. Journal of General Virology. 1984;65:1687–1697. doi: 10.1099/0022-1317-65-10-1687. [DOI] [PubMed] [Google Scholar]
- 52.Becker Y, Joklik WK. MESSENGER RNA IN CELLS INFECTED WITH VACCINIA VIRUS. Proceedings of the National Academy of Sciences. 1964;51:577–585. doi: 10.1073/pnas.51.4.577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bablanian R, Esteban M, Baxt B, Sonnabend JA. Studies on the Mechanisms of Vaccinia Virus Cytopathic Effects. Journal of General Virology. 1978;39:391–402. doi: 10.1099/0022-1317-39-3-391. [DOI] [PubMed] [Google Scholar]
- 54.Bablanian R, Coppola G, Scribani S, Esteban M. Inhibition of protein synthesis by vaccinia virus: IV. The role of low-molecular-weight viral RNA in the inhibition of protein synthesis. Virology. 1981;112:13–24. doi: 10.1016/0042-6822(81)90607-3. [DOI] [PubMed] [Google Scholar]
- 55.Guerra S, Lopez-Fernandez LA, Pascual-Montano A, Munoz M, et al. Cellular gene expression survey of vaccinia virus infection of human HeLa cells. J Virol. 2003;77:6493–6506. doi: 10.1128/JVI.77.11.6493-6506.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gilchuk P, Hill TM, Wilson JT, Joyce S. Discovering protective CD8 T cell epitopes--no single immunologic property predicts it! Curr Opin Immunol. 2015;34:43–51. doi: 10.1016/j.coi.2015.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Oseroff C, Kos F, Bui HH, Peters B, et al. HLA class I-restricted responses to vaccinia recognize a broad array of proteins mainly involved in virulence and viral gene regulation. Proc.Natl.Acad.Sci.U.S.A. 2005;102:13980–13985. doi: 10.1073/pnas.0506768102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kim O, Sun Y, Lai FW, Song C, et al. Modulation of type I interferon induction by porcine reproductive and respiratory syndrome virus and degradation of CREB-binding protein by non-structural protein 1 in MARC-145 and HeLa cells. Virology. 2010;402:315–326. doi: 10.1016/j.virol.2010.03.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Skloot R. The immortal life of Henrietta Lacks. 2011;xiv:381. [Google Scholar]
- 60.Gubin MM, Artyomov MN, Mardis ER, Schreiber RD. Tumor neoantigens: building a framework for personalized cancer immunotherapy. J Clin Invest. 2015:1–9. doi: 10.1172/JCI80008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Gubin MM, Zhang X, Schuster H, Caron E, et al. Checkpoint blockade cancer immunotherapy targets tumour-specific mutant antigens. Nature. 2014;515:577–581. doi: 10.1038/nature13988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yadav M, Jhunjhunwala S, Phung QT, Lupardus P, et al. Predicting immunogenic tumour mutations by combining mass spectrometry and exome sequencing. Nature. 2014;515:572–576. doi: 10.1038/nature14001. [DOI] [PubMed] [Google Scholar]
- 63.Kowalewski DJ, Schuster H, Backert L, Berlin C, et al. HLA ligandome analysis identifies the underlying specificities of spontaneous antileukemia immune responses in chronic lymphocytic leukemia (CLL). Proc Natl Acad Sci U S A. 2015;112:E166–175. doi: 10.1073/pnas.1416389112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Duan F, Duitama J, Al Seesi S, Ayres CM, et al. Genomic and bioinformatic profiling of mutational neoepitopes reveals new rules to predict anticancer immunogenicity. J Exp Med. 2014;211:2231–2248. doi: 10.1084/jem.20141308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Cantoni N, Hirsch HH, Khanna N, Gerull S, et al. Evidence for a Bidirectional Relationship between Cytomegalovirus Replication and acute Graft-versus-Host Disease. Biology of Blood and Marrow Transplantation. 2010;16:1309–1314. doi: 10.1016/j.bbmt.2010.03.020. [DOI] [PubMed] [Google Scholar]
- 66.McCarthy AL, Malik Peiris JS, Taylor CE, Green MA, et al. Increase in severity of graft versus host disease by cytomegalovirus. Journal of Clinical Pathology. 1992;45:542–544. doi: 10.1136/jcp.45.6.542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Beam E, Razonable R. Cytomegalovirus in Solid Organ Transplantation: Epidemiology, Prevention, and Treatment. Current Infectious Disease Reports. 2012;14:633–641. doi: 10.1007/s11908-012-0292-2. [DOI] [PubMed] [Google Scholar]
- 68.Linares L, Sanclemente G, Cervera C, Hoyo I, et al. Influence of Cytomegalovirus Disease in Outcome of Solid Organ Transplant Patients. Transplantation Proceedings. 2011;43:2145–2148. doi: 10.1016/j.transproceed.2011.05.007. [DOI] [PubMed] [Google Scholar]
- 69.Granados DP, Sriranganadane D, Daouda T, Zieger A, et al. Impact of genomic polymorphisms on the repertoire of human MHC class I-associated peptides. Nat Commun. 2014;5:3600. doi: 10.1038/ncomms4600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Jenner RG, Young RA. Insights into host responses against pathogens from transcriptional profiling. Nat Rev Micro. 2005;3:281–294. doi: 10.1038/nrmicro1126. [DOI] [PubMed] [Google Scholar]
- 71.Konig R, Stertz S, Zhou Y, Inoue A, et al. Human host factors required for influenza virus replication. Nature. 2010;463:813–817. doi: 10.1038/nature08699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Karlas A, Machuy N, Shin Y, Pleissner K-P, et al. Genome-wide RNAi screen identifies human host factors crucial for influenza virus replication. Nature. 2010;463:818–822. doi: 10.1038/nature08760. [DOI] [PubMed] [Google Scholar]
- 73.Brass AL, Huang IC, Benita Y, John SP, et al. The IFITM Proteins Mediate Cellular Resistance to Influenza A H1N1 Virus, West Nile Virus, and Dengue Virus. Cell. 2009;139:1243–1254. doi: 10.1016/j.cell.2009.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zhou H, Xu M, Huang Q, Gates AT, et al. Genome-Scale RNAi Screen for Host Factors Required for HIV Replication. Cell Host & Microbe. 2008;4:495–504. doi: 10.1016/j.chom.2008.10.004. [DOI] [PubMed] [Google Scholar]
- 75.König R, Zhou Y, Elleder D, Diamond TL, et al. Global Analysis of Host-Pathogen Interactions that Regulate Early-Stage HIV-1 Replication. Cell. 2008;135:49–60. doi: 10.1016/j.cell.2008.07.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Beard PM, Griffiths SJ, Gonzalez O, Haga IR, et al. A Loss of Function Analysis of Host Factors Influencing Vaccinia virus Replication by RNA Interference. PLoS ONE. 2014;9:e98431. doi: 10.1371/journal.pone.0098431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Brass AL, Dykxhoorn DM, Benita Y, Yan N, et al. Identification of Host Proteins Required for HIV Infection Through a Functional Genomic Screen. Science. 2008;319:921–926. doi: 10.1126/science.1152725. [DOI] [PubMed] [Google Scholar]
- 78.Guerra S, López-Fernández LA, Pascual-Montano A, Muñoz M, et al. Cellular Gene Expression Survey of Vaccinia Virus Infection of Human HeLa Cells. Journal of Virology. 2003;77:6493–6506. doi: 10.1128/JVI.77.11.6493-6506.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Assarsson E, Greenbaum JA, Sundström M, Schaffer L, et al. Kinetic analysis of a complete poxvirus transcriptome reveals an immediate-early class of genes. Proceedings of the National Academy of Sciences. 2008;105:2140–2145. doi: 10.1073/pnas.0711573105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Chou W, Ngo T, Gershon PD. An Overview of the Vaccinia Virus Infectome: a Survey of the Proteins of the Poxvirus-Infected Cell. Journal of Virology. 2012;86:1487–1499. doi: 10.1128/JVI.06084-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Chung C-S, Chen C-H, Ho M-Y, Huang C-Y, et al. Vaccinia Virus Proteome: Identification of Proteins in Vaccinia Virus Intracellular Mature Virion Particles. Journal of Virology. 2006;80:2127–2140. doi: 10.1128/JVI.80.5.2127-2140.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Barber GN. Host defense, viruses and apoptosis. Cell Death Differ. 2001;8:113–126. doi: 10.1038/sj.cdd.4400823. [DOI] [PubMed] [Google Scholar]
- 83.Durbin JE, Hackenmiller R, Simon MC, Levy DE. Targeted disruption of the mouse Stat1 gene results in compromised innate immunity to viral disease. Cell. 1996;84:443–450. doi: 10.1016/s0092-8674(00)81289-1. [DOI] [PubMed] [Google Scholar]
- 84.Meraz MA, White JM, Sheehan KC, Bach EA, et al. Targeted disruption of the Stat1 gene in mice reveals unexpected physiologic specificity in the JAK-STAT signaling pathway. Cell. 1996;84:431–442. doi: 10.1016/s0092-8674(00)81288-x. [DOI] [PubMed] [Google Scholar]
- 85.Symons JA, Alcamí A, Smith GL. Vaccinia virus encodes a soluble type I interferon receptor of novel structure and broad species soecificity. Cell. 1995;81:551–560. doi: 10.1016/0092-8674(95)90076-4. [DOI] [PubMed] [Google Scholar]
- 86.Smith GL, Symons JA, Khanna A, Vanderplasschen A, et al. Vaccinia virus immune evasion. Immunol Rev. 1997;159:137–154. doi: 10.1111/j.1600-065x.1997.tb01012.x. [DOI] [PubMed] [Google Scholar]
- 87.Kirwan S, Merriam D, Barsby N, McKinnon A, et al. Vaccinia virus modulation of natural killer cell function by direct infection. Virology. 2006;347:75–87. doi: 10.1016/j.virol.2005.11.037. [DOI] [PubMed] [Google Scholar]
- 88.Smith GL, Symons JA, Alcami A. Immune modulation by proteins secreted from cells infected by vaccinia virus. Arch Virol Suppl. 1999;15:111–129. doi: 10.1007/978-3-7091-6425-9_8. [DOI] [PubMed] [Google Scholar]
- 89.Moss B. Poxvirus Cell Entry: How Many Proteins Does it Take? Viruses. 2012;4:688–707. doi: 10.3390/v4050688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Watts JM, Dang KK, Gorelick RJ, Leonard CW, et al. Architecture and secondary structure of an entire HIV-1 RNA genome. Nature. 2009;460:711–716. doi: 10.1038/nature08237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Dubois J, Terrier O, Rosa-Calatrava M. Influenza Viruses and mRNA Splicing: Doing More with Less. mBio. 2014;5 doi: 10.1128/mBio.00070-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Bankamp B, Takeda M, Zhang Y, Xu W, et al. Genetic Characterization of Measles Vaccine Strains. Journal of Infectious Diseases. 2011;204:S533–S548. doi: 10.1093/infdis/jir097. [DOI] [PubMed] [Google Scholar]
- 93.McCarthy MK, Weinberg JB. The immunoproteasome and viral infection: a complex regulator of inflammation. Front Microbiol. 2015;6:21. doi: 10.3389/fmicb.2015.00021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Shin EC, Seifert U, Kato T, Rice CM, et al. Virus-induced type I IFN stimulates generation of immunoproteasomes at the site of infection. J Clin Invest. 2006;116:3006–3014. doi: 10.1172/JCI29832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Freudenburg W, Gautam M, Chakraborty P, James J, et al. Reduction in ATP levels triggers immunoproteasome activation by the 11S (PA28) regulator during early antiviral response mediated by IFNbeta in mouse pancreatic beta-cells. PLoS ONE. 2013;8:e52408. doi: 10.1371/journal.pone.0052408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Freudenburg W, Gautam M, Chakraborty P, James J, et al. Immunoproteasome Activation During Early Antiviral Response in Mouse Pancreatic beta-cells: New Insights into Auto-antigen Generation in Type I Diabetes? J Clin Cell Immunol. 2013;4 doi: 10.4172/2155-9899.1000141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Fabunmi RP, Wigley WC, Thomas PJ, DeMartino GN. Interferon gamma regulates accumulation of the proteasome activator PA28 and immunoproteasomes at nuclear PML bodies. J Cell Sci. 2001;114:29–36. doi: 10.1242/jcs.114.1.29. [DOI] [PubMed] [Google Scholar]
- 98.Nathan, James A, Spinnenhirn V, Schmidtke G, Basler M, et al. Immuno- and Constitutive Proteasomes Do Not Differ in Their Abilities to Degrade Ubiquitinated Proteins. Cell. 2013;152:1184–1194. doi: 10.1016/j.cell.2013.01.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Robek MD, Garcia ML, Boyd BS, Chisari FV. Role of Immunoproteasome Catalytic Subunits in the Immune Response to Hepatitis B Virus. Journal of Virology. 2007;81:483–491. doi: 10.1128/JVI.01779-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Kuzina ES, Chernolovskaya EL, Kudriaeva AA, Zenkova MA, et al. Immunoproteasome enhances intracellular proteolysis of myelin basic protein. Dokl Biochem Biophys. 2013;453:300–303. doi: 10.1134/S1607672913060070. [DOI] [PubMed] [Google Scholar]
- 101.Kincaid EZ, Che JW, York I, Escobar H, et al. Mice completely lacking immunoproteasomes show major changes in antigen presentation. Nat Immunol. 2012;13:129–135. doi: 10.1038/ni.2203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Shaw AC, Rossel Larsen M, Roepstorff P, Justesen J, et al. Mapping and identification of interferon gamma-regulated HeLa cell proteins separated by immobilized pH gradient two-dimensional gel electrophoresis. Electrophoresis. 1999;20:984–993. doi: 10.1002/(SICI)1522-2683(19990101)20:4/5<984::AID-ELPS984>3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- 103.Shirey KA, Jung JY, Maeder GS, Carlin JM. Upregulation of IFN-gamma receptor expression by proinflammatory cytokines influences IDO activation in epithelial cells. J Interferon Cytokine Res. 2006;26:53–62. doi: 10.1089/jir.2006.26.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Zhang HM, Yuan J, Cheung P, Chau D, et al. Gamma Interferon-Inducible Protein 10 Induces HeLa Cell Apoptosis through a p53-Dependent Pathway Initiated by Suppression of Human Papillomavirus Type 18 E6 and E7 Expression. Molecular and Cellular Biology. 2005;25:6247–6258. doi: 10.1128/MCB.25.14.6247-6258.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Yang W, Tan J, Liu R, Cui X, et al. Interferon-γ Upregulates Expression of IFP35 Gene in HeLa Cells via Interferon Regulatory Factor-1. PLoS ONE. 2012;7:e50932. doi: 10.1371/journal.pone.0050932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Lizard G, Chignol MC, Chardonnet Y, Schmitt D. Differences of reactivity to interferon gamma in HeLa and CaSki cells: a combined immunocytochemical and flow-cytometric study. J Cancer Res Clin Oncol. 1996;122:223–230. doi: 10.1007/BF01209650. [DOI] [PubMed] [Google Scholar]
- 107.Muller U, Steinhoff U, Reis LF, Hemmi S, et al. Functional role of type I and type II interferons in antiviral defense. Science. 1994;264:1918–1921. doi: 10.1126/science.8009221. [DOI] [PubMed] [Google Scholar]
- 108.Stark GR, Kerr IM, Williams BR, Silverman RH, et al. How cells respond to interferons. Annu Rev Biochem. 1998;67:227–264. doi: 10.1146/annurev.biochem.67.1.227. [DOI] [PubMed] [Google Scholar]
- 109.Zhou A, Hassel BA, Silverman RH. Expression cloning of 2-5A-dependent RNAase: a uniquely regulated mediator of interferon action. Cell. 1993;72:753–765. doi: 10.1016/0092-8674(93)90403-d. [DOI] [PubMed] [Google Scholar]
- 110.Schwab SR, Li KC, Kang C, Shastri N. Constitutive display of cryptic translation products by MHC class I molecules. Science. 2003;301:1367–1371. doi: 10.1126/science.1085650. [DOI] [PubMed] [Google Scholar]
- 111.Weinzierl AO, Maurer D, Altenberend F, Schneiderhan-Marra N, et al. A cryptic vascular endothelial growth factor T-cell epitope: identification and characterization by mass spectrometry and T-cell assays. Cancer Res. 2008;68:2447–2454. doi: 10.1158/0008-5472.CAN-07-2540. [DOI] [PubMed] [Google Scholar]
- 112.Andersen RS, Andersen SR, Hjortso MD, Lyngaa R, et al. High frequency of T cells specific for cryptic epitopes in melanoma patients. Oncoimmunology. 2013;2:e25374. doi: 10.4161/onci.25374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Li C, Goudy K, Hirsch M, Asokan A, et al. Cellular immune response to cryptic epitopes during therapeutic gene transfer. Proc Natl Acad Sci U S A. 2009;106:10770–10774. doi: 10.1073/pnas.0902269106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Rutkowski MR, Ho O, Green WR. Defining the mechanism(s) of protection by cytolytic CD8 T cells against a cryptic epitope derived from a retroviral alternative reading frame. Virology. 2009;390:228–238. doi: 10.1016/j.virol.2009.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Cardinaud S, Consiglieri G, Bouziat R, Urrutia A, et al. CTL escape mediated by proteasomal destruction of an HIV-1 cryptic epitope. PLoS Pathog. 2011;7:e1002049. doi: 10.1371/journal.ppat.1002049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Shastri N, Nguyen V, Gonzalez F. Major histocompatibility class I molecules can present cryptic translation products to T-cells. J Biol Chem. 1995;270:1088–1091. doi: 10.1074/jbc.270.3.1088. [DOI] [PubMed] [Google Scholar]
- 117.Malarkannan S, Afkarian M, Shastri N. A rare cryptic translation product is presented by Kb major histocompatibility complex class I molecule to alloreactive T cells. J Exp Med. 1995;182:1739–1750. doi: 10.1084/jem.182.6.1739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Schwab SR, Shugart JA, Horng T, Malarkannan S, et al. Unanticipated antigens: translation initiation at CUG with leucine. PLoS Biol. 2004;2:e366. doi: 10.1371/journal.pbio.0020366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Malarkannan S, Horng T, Shih PP, Schwab S, et al. Presentation of out-of-frame peptide/MHC class I complexes by a novel translation initiation mechanism. Immunity. 1999;10:681–690. doi: 10.1016/s1074-7613(00)80067-9. [DOI] [PubMed] [Google Scholar]
- 120.Cardinaud S, Moris A, Fevrier M, Rohrlich PS, et al. Identification of cryptic MHC I-restricted epitopes encoded by HIV-1 alternative reading frames. J Exp Med. 2004;199:1053–1063. doi: 10.1084/jem.20031869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Starck SR, Ow Y, Jiang V, Tokuyama M, et al. A distinct translation initiation mechanism generates cryptic peptides for immune surveillance. PLoS ONE. 2008;3:e3460. doi: 10.1371/journal.pone.0003460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Starck SR, Jiang V, Pavon-Eternod M, Prasad S, et al. Leucine-tRNA initiates at CUG start codons for protein synthesis and presentation by MHC class I. Science. 2012;336:1719–1723. doi: 10.1126/science.1220270. [DOI] [PubMed] [Google Scholar]
- 123.Goodenough E, Robinson TM, Zook MB, Flanigan KM, et al. Cryptic MHC class I-binding peptides are revealed by aminoglycoside-induced stop codon read-through into the 3' UTR. Proc Natl Acad Sci U S A. 2014;111:5670–5675. doi: 10.1073/pnas.1402670111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Garrison KE, Champiat S, York VA, Agrawal AT, et al. Transcriptional errors in human immunodeficiency virus type 1 generate targets for T-cell responses. Clin Vaccine Immunol. 2009;16:1369–1371. doi: 10.1128/CVI.00410-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Zook MB, Howard MT, Sinnathamby G, Atkins JF, et al. Epitopes derived by incidental translational frameshifting give rise to a protective CTL response. J Immunol. 2006;176:6928–6934. doi: 10.4049/jimmunol.176.11.6928. [DOI] [PubMed] [Google Scholar]
- 126.Garbe Y, Maletzki C, Linnebacher M. An MSI tumor specific frameshift mutation in a coding microsatellite of MSH3 encodes for HLA-A0201-restricted CD8+ cytotoxic T cell epitopes. PLoS ONE. 2011;6:e26517. doi: 10.1371/journal.pone.0026517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Wold WS, Toth K. Adenovirus vectors for gene therapy, vaccination and cancer gene therapy. Curr Gene Ther. 2013;13:421–433. doi: 10.2174/1566523213666131125095046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Knipe DM, Howley PM. Fields virology. 2013;2 [Google Scholar]
- 129.Coutinho A, Caramalho I, Seixas E, Demengeot J. Thymic commitment of regulatory T cells is a pathway of TCR-dependent selection that isolates repertoires undergoing positive or negative selection. Curr Top Microbiol Immunol. 2005;293:43–71. doi: 10.1007/3-540-27702-1_3. [DOI] [PubMed] [Google Scholar]
- 130.Park Y, Moon Y, Chung HY. AIRE-1 (autoimmune regulator type 1) as a regulator of the thymic induction of negative selection. Ann N Y Acad Sci. 2003;1005:431–435. doi: 10.1196/annals.1288.073. [DOI] [PubMed] [Google Scholar]
- 131.Birnbaum ME, Mendoza JL, Sethi DK, Dong S, et al. Deconstructing the peptide-MHC specificity of T cell recognition. Cell. 2014;157:1073–1087. doi: 10.1016/j.cell.2014.03.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Nelson, Ryan W, Beisang D, Tubo, Noah J, Dileepan T, et al. T Cell Receptor Cross-Reactivity between Similar Foreign and Self Peptides Influences Naive Cell Population Size and Autoimmunity. Immunity. 2015;42:95–107. doi: 10.1016/j.immuni.2014.12.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
References
- 1.Gilchuk P, Spencer CT, Conant SB, Hill T, et al. Discovering naturally processed antigenic determinants that confer protective T cell immunity. J Clin Invest. 2013;123:1976–1987. doi: 10.1172/JCI67388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Moutaftsi M, Peters B, Pasquetto V, Tscharke DC, et al. A consensus epitope prediction approach identifies the breadth of murine T(CD8+)-cell responses to vaccinia virus. Nat Biotechnol. 2006;24:817–819. doi: 10.1038/nbt1215. [DOI] [PubMed] [Google Scholar]
- 3.Oseroff C, Peters B, Pasquetto V, Moutaftsi M, et al. Dissociation between epitope hierarchy and immunoprevalence in CD8 responses to vaccinia virus western reserve. J Immunol. 2008;180:7193–7202. doi: 10.4049/jimmunol.180.11.7193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Moise L, McMurry JA, Buus S, Frey S, et al. In silico-accelerated identification of conserved and immunogenic variola/vaccinia T-cell epitopes. Vaccine. 2009;27:6471–6479. doi: 10.1016/j.vaccine.2009.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Assarsson E, Sidney J, Oseroff C, Pasquetto V, et al. A quantitative analysis of the variables affecting the repertoire of T cell specificities recognized after vaccinia virus infection. J Immunol. 2007;178:7890–7901. doi: 10.4049/jimmunol.178.12.7890. [DOI] [PubMed] [Google Scholar]
- 6.Assarsson E, Greenbaum JA, Sundstrom M, Schaffer L, et al. Kinetic analysis of a complete poxvirus transcriptome reveals an immediate-early class of genes. Proc Natl Acad Sci U S A. 2008;105:2140–2145. doi: 10.1073/pnas.0711573105. [DOI] [PMC free article] [PubMed] [Google Scholar]
References Cited
- 1.Schittenhelm RB, Dudek NL, Croft NP, Ramarathinam SH, et al. A comprehensive analysis of constitutive naturally processed and presented HLA-C*04:01 (Cw4)-specific peptides. Tissue Antigens. 2014;83:174–179. doi: 10.1111/tan.12282. [DOI] [PubMed] [Google Scholar]
- 2.Barber LD, Percival L, Arnett KL, Gumperz JE, et al. Polymorphism in the alpha 1 helix of the HLA-B heavy chain can have an overriding influence on peptide-binding specificity. J Immunol. 1997;158:1660–1669. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.