Abstract
Bromodomain containing proteins PB1, SMARCA4, and SMARCA2 are important components of SWI/SNF chromatin remodeling complexes. We identified bromodomain inhibitors that target these proteins and display unusual binding modes involving water displacement from the KAc binding site. The best compound binds the fifth bromodomain of PB1 with a KD of 124 nM, SMARCA2B and SMARCA4 with KD values of 262 and 417 nM, respectively, and displays excellent selectivity over bromodomains other than PB1, SMARCA2, and SMARCA4.
Introduction
The structure of chromatin, the complex of proteins and DNA through which DNA is packaged, is regulated by the post-translational modification of histone tails and the action of chromatin remodeling complexes such as the switch/sucrose nonfermenting (SWI/SNF) complex. The multisubunit SWI/SNF complex is based on the mutually exclusive helicase/ATPase proteins SMARCA2 and SMARCA4, with additional core subunits and regulatory or specificity proteins. Many components contain domains that recognize histone post-translational modifications, enabling cross-talk between the two mechanisms of regulation.
The SWI/SNF complex is subject to considerable research interest, since components are mutated in many cancers1−3 and are implicated in developmental disorders.4 In some tumor types, mutations within the SWI/SNF complex lead to context-specific vulnerabilities, such as dependence on SMARCA2/4 in SMARCB1 (Snf5) mutant rhabdoid tumors5 or the essential presence of SMARCA2 in SMARCA4 mutant lung cancer.6,7 The biological roles of the complexes have led to interest in generating chemical probes that target different domains within the multisubunit complex, to assist the elucidation of their function and therapeutic relevance.8 Bromodomain containing proteins (BCPs) are prevalent in SWI/SNF complexes and are present in SMARCA2/4 helicases, BRD7/9, and PB1. SMARCA2/4 and BRD7/9 contain one bromodomain each, while PB1 has six distinct bromodomains. In yeast SWI/SNF bromodomains are implicated in enhancing chromatin remodeling efficiency, loci targeting, and protein–protein interactions.9,10 In eukaryotes the SMARCA4 bromodomain is reported to act in the DNA damage response.11,12 Mutations in PB1’s bromodomains have been found in clear cell renal carcinoma and linked to genome instability and aneuploidy.13,14
Bromodomains are protein modules of ∼110 amino acids that recognize acetylated lysine (KAc) and share a common structure of four α helices linked by flexible loop regions and have been popular targets for chemical probe development.15,16 Sixty-one bromodomains are expressed in the human proteome, across 42 BCPs, which can be grouped into subfamilies based on amino acid sequence and structural alignment.17 Chemical probes have now been developed for multiple subfamilies and have helped identify new biology.18,19 In particular, (+)JQ1 and IBET-151 have demonstrated the therapeutic potential of bromodomain and extraterminal (BET) proteins in cancer, inflammation, and heart failure.20−22
The chemical probes I-BRD923 and LP99,24 targeting BRD9 and BRD7/BRD9, respectively, have been developed to study some bromodomains in the SWI/SNF complex. Recently a chemical probe, PFI-3, targeting the bromodomains in SMARCA2 and SMARCA4, as well as the fifth bromodomain of PB1 (PB1(5)), has been developed based on a salicyclic acid fragment hit and been used to demonstrate a role for SMARCA4 in embryonic stem cell maintenance.25 These results demonstrate that SWI/SNF BCPs are druggable, despite being predicted to be more challenging to target due to a featureless binding pocket lacking a clearly defined hydrophobic groove and ZA binding channel as in BRD4(1) (Supporting Information).17,26 A short ZA loop and a wider entrance than BET bromodomains have been identified as factors that may alter ligand binding mode in the structurally related bromodomains PB1(5) and SMARCA2/4, while PB1(2–4) are defined by an aromatic residue in the pocket that is proposed to act as a “lid”.26 Given the varied structural nature of the six bromodomains in PB1, one single probe is unlikely to be able to target all simultaneously. Several compounds with different chemotypes may be required.
In the course of library screening, 1 (Figure 1) and other related analogues were found to be weak PB1(5) inhibitors but with poor aqueous solubility. In this work we describe the optimization of 1, leading to a family of compounds with increased potency, improved physicochemical properties, and varied selectivity within the SWI/SNF bromodomains, including the potential to target PB1 selectively over SMARCA4 and SMARCA2 bromodomains.
Figure 1.
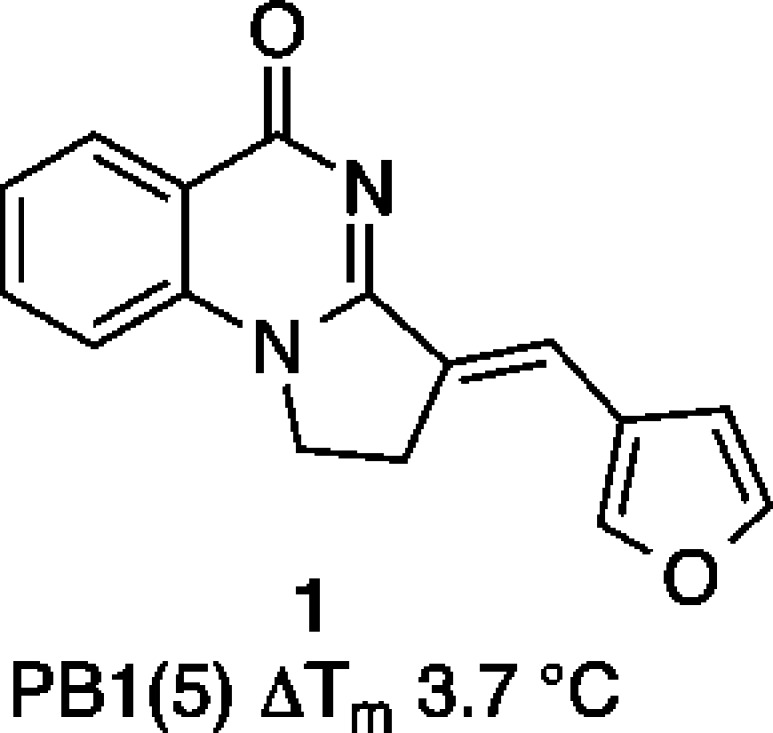
Initial hit from screening of a commercial library.
Results and Discussion
The library of commercial analogues of 1 showed varied response by differential scanning fluorimetry (DSF) against PB1(5). DSF has been demonstrated to be an operationally simple assay, suitable for rapid assessment of potency and selectivity for bromodomains.19,24,27 Limited SAR from the library indicated sensitivity to the size of the aromatic side chain and that the 2,3-dihydropyrrolo[1,2-a]quinazolin-5(1H)-one core bound within the KAc binding site (Supporting Information). We proposed that planarity of the compounds with conjugated aromatic ring substituents contributed to the poor solubility of the series and that analogues with greater sp3 character in their substituents, or the potential for ionization, would have improved aqueous solubility.28
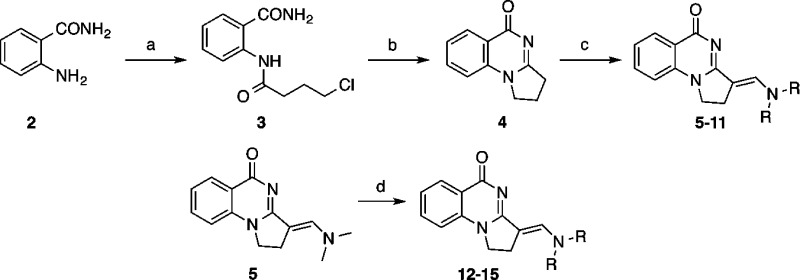
Compounds were synthesized by the routes shown in Scheme 1. Amidation of 2-aminobenzamide 2 with 4-chlorobutanoyl chloride29 gave amide 3, which could be cyclized to the 2,3-dihydropyrrolo[1,2-a]quinazolin-5(1H)-one core 4 using potassium tert-butoxide. The isomer was confirmed on the basis of NOE correlations and subsequent crystallography. The weak nucleophilicity of the core was then exploited by reacting compound 4 with substituted N,N-dialkylformamides using Vilsmeier–Haack conditions, giving analogues incorporating nitrogen-based aliphatic side chains solely as the E regioisomer. This chemistry was unsuitable for piperazine-based formamides. For these side chains, analogues 12–15 were synthesized as single regioisomers by displacement of the dimethylamino group in 5 using cyclic secondary amines and DMAP catalysis. Due to the potential for hydrolysis of the enamine-like moiety, the aqueous stability of 5 was assessed by 1H NMR in a D2O time course experiment and was stable for >24 h. All analogues appeared to have increased aqueous solubility compared to 1. Their binding to PB1(5) was assessed by the operationally simple DSF assay (Table 1). Although the simple scaffold 4 itself showed no interaction, aliphatic analogues 5–15 generally showed improved binding to PB1(5). The shape of the side chain was a key factor in determining activity; inclusion of propyl side chains in 7 led to moderate activity, but variation to isopropyl in 8 caused a loss of binding. Six-membered aliphatic rings (10–14) were better tolerated than analogues containing six-membered aromatic side chains (see Supporting Information), likely due to their greater flexibility. However, expansion to a seven-membered ring reduced activity.
Scheme 1. General Route to Core 2,3-Dihydropyrrolo[1,2-a]quinazolin-5(1H)-ones and Their Derivatives.
Reagents and conditions: (a) 4-chlorobutanoyl chloride, Et3N, THF, 0 °C to rt, 12 h, 92%; (b) tBuOK, THF, rt, 1 h, 77%; (c) N,N-dialkylformamide, POCl3, CH2Cl2, 70 °C, 16–74%; (d) H-NR2, DMAP, EtOH, 70 °C, 24–96 h, 39–81%. For R see Table 1.
Table 1. Effect of Aliphatic Side Chain Substitution on Binding to PB1(5) and SMARCA4 by DSF and AlphaScreen.
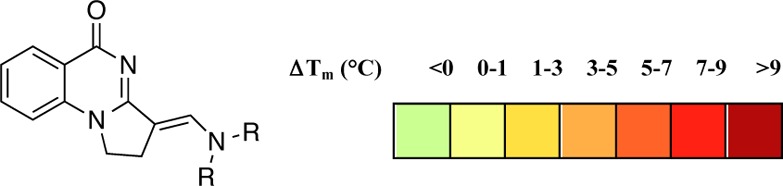
Values shown are the average of three replicates and standard deviation by DSF assay.
Values shown are the average of two replicates by AlphaScreen assay.
IC50 not determined.
It was anticipated that compounds targeting PB1(5) might interact with the SMARCA2 and SMARCA4 bromodomains due to the similarity of their binding pockets. We confirmed the binding of these analogues to the SMARCA4 bromodomain in an AlphaScreen assay (Table 1), which established that the compounds had affinities for this bromodomain ranging from 6.2 to 22.0 μM.
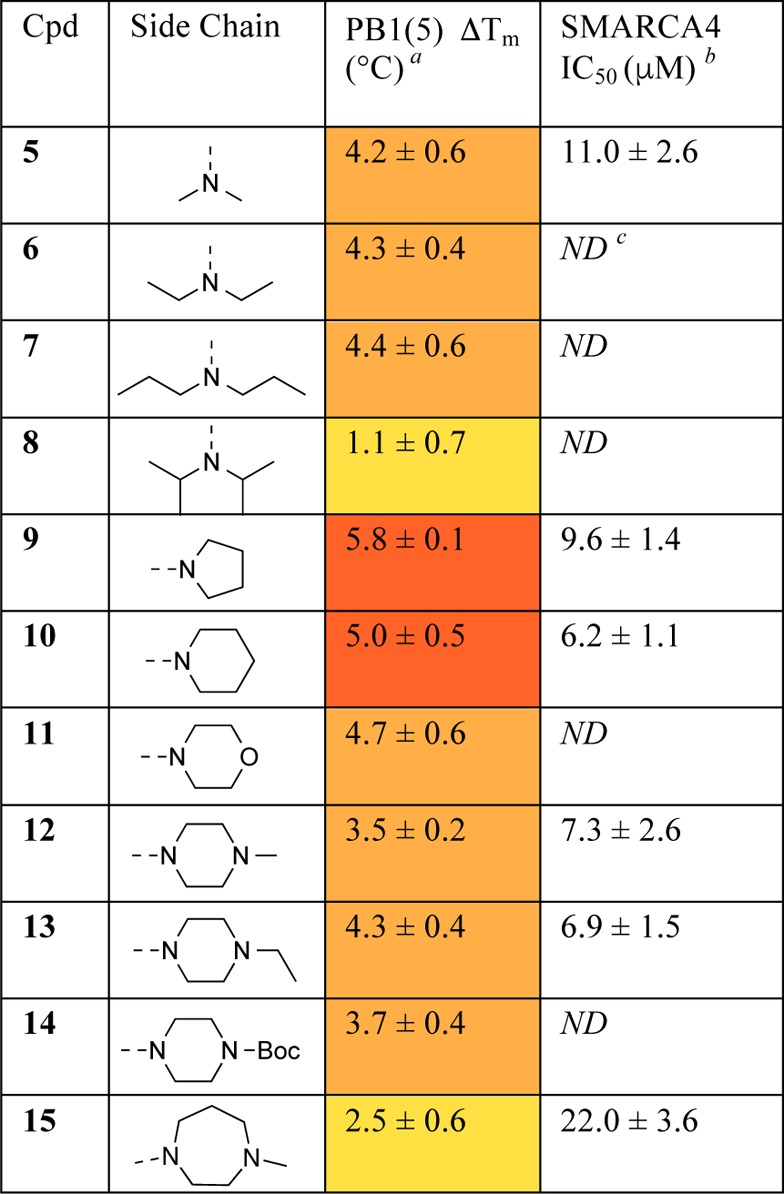
The improved solubility and activity of the compounds allowed us to conduct isothermal calorimetry (ITC) experiments with analogues 9 and 10. These had KD values of 2.3 and 1.2 μM, respectively, for the PB1(5) bromodomain. In both 9 and 10, the entropy change upon binding (TΔS) contributed favorably to binding (TΔS values of 4.7 and 5.3 kcal/mol, respectively). In addition, interaction of these two inhibitors with PB1(5) was driven by enthalpic contributions (ΔH of −2.9 and −2.7 kcal/mol, respectively). The underlying molecular mechanism for the observed thermodynamics was evident on analysis of the binding mode of 10, revealed by a cocrystal with PB1(5) bromodomain (Figure 2).
Figure 2.
Binding modes of bromodomain inhibitors. (a) Cocrystal structure of 10 with PB1(5) at 2.3 Å (PDB code 5FH6). Hydrogen bonds are shown by black dashed lines. (b) Apo-crystal structure of PB1(5) (PDB code 3G0J) showing conserved crystallographic waters (red spheres) which are displaced in (a).
This structure demonstrated that the rigid core of 10 sits deep in the hydrophobic KAc binding site of PB1(5), displacing the four waters observed in the apo-form and most BRD-ligand crystal structures. The carbonyl of the 5,6-dihydropyrimidin-4(1H)-one ring formed a direct hydrogen bond to Tyr696, while the amide hydrogen-bonds to Asn739. This binding mode is similar to that adopted by PFI-3 and its salicylic acid precursor, which also displace conserved waters and form direct hydrogen bonds to Tyr696 and Asn739.26 These interactions are in marked contrast to those in most bromodomain inhibitors, in which a KAc mimetic, directly hydrogen-bonding to the asparagine residue, forms a water-mediated hydrogen bond network to tyrosine using the structural conserved waters. The crystal structure highlighted the narrowness of the PB1(5) pocket, which the compound occupies well, and indicated that the side chain is key in orienting the core within the pocket. This helped explain the lack of activity of the unsubstituted core. The piperidine side chain projects out from the pocket forming a close interaction with the hydrophobic rim of the binding site, explaining the loss in activity in 8 and 15, which contain more sterically demanding side chains.
The carbonyl of Met731 was observed to be in an unsolvated cavity in the structure, and it was hypothesized that substitution on the aromatic ring ortho to the carbonyl group might enable an additional interaction in this region. Compounds 16–20 could be accessed by incorporating substituted 2-aminobenzamides into the synthetic methodology previously developed. Compound 21 featuring hydroxyl substitution was obtained by deprotection of the methoxy analogues using BBr3. Detailed procedures are in the Supporting Information.
Screening these analogues against PB1(5) by DSF showed that substitution of chlorine at R2 in 18 doubled the observed ΔTm in comparison to unsubstituted 5, while bromine substitution gave a smaller increase (Table 2). All other substitutions reduced binding compared with analogue 5. This SAR provides valuable compounds suitable as negative controls for the exploration of the biological activity of these compounds.
Table 2. Effect of Aromatic Substitution on Binding to the PB1(5) Bromodomain.
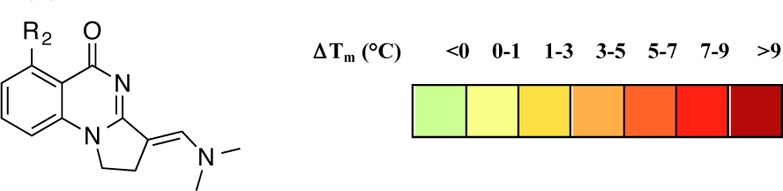
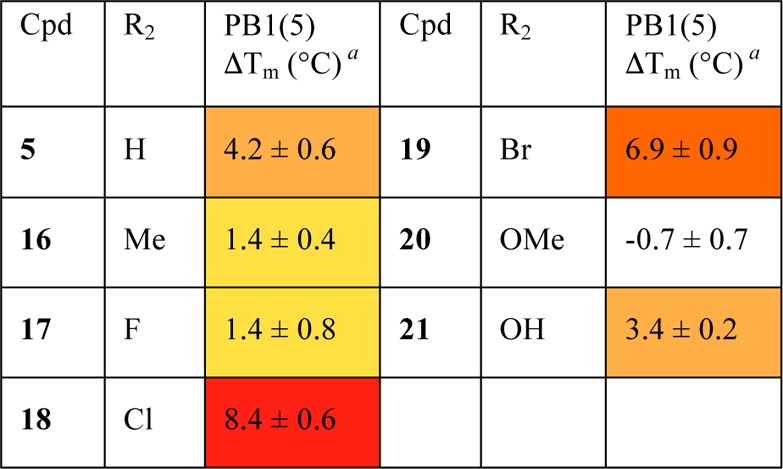
Values shown are the average of three replicates and standard deviation.
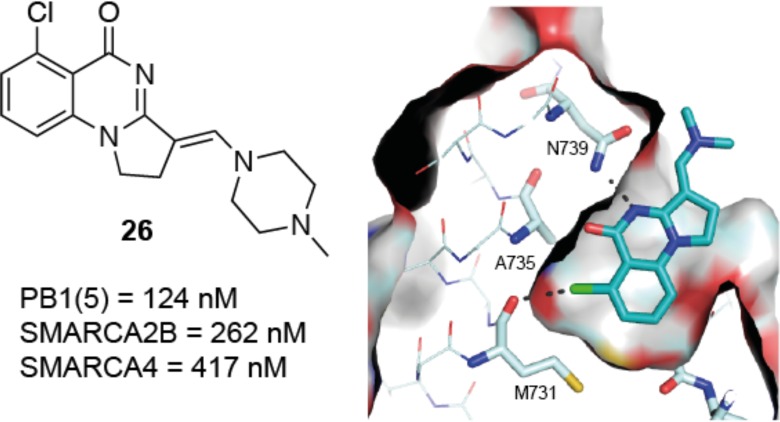
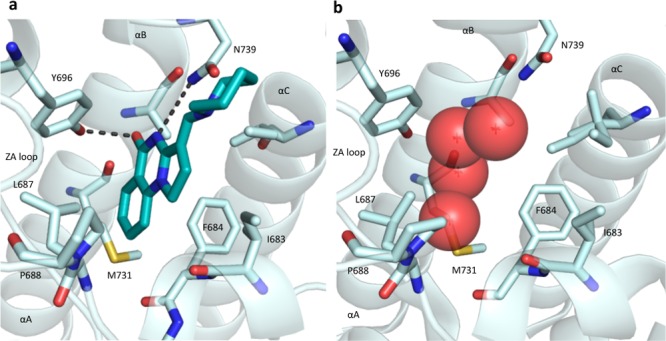
A KD of 137 nM for PB1(5) was obtained for 18 by ITC, with a significant increase in the enthalpic contribution to binding (ΔH = −6.0 kcal/mol). Since Cl/Br inclusion also increased binding of the core scaffold (ΔTm of 2.5 and 3.1 °C, respectively; see Supporting Information), we proposed that the improved interaction was due to formation of a halogen bond to the carbonyl of Met731. While bromination would be predicted to give a stronger halogen bond,30 the effect is countered by steric constraints within the cavity.
This interaction was confirmed by a cocrystal structure of 18 with PB1(5), in which the chlorine occupies the cavity previously observed with a separation of 3.2 Å from the carbonyl oxygen of Met731. This is consistent with a halogen bond (Figure 3).30 A slight twist in the scaffold, to maximize interactions with both Tyr696 and Asn739, can be observed. Comparison with PFI-3 shows that the interaction with Met731 has not been previously exploited (Supporting Information).
Figure 3.
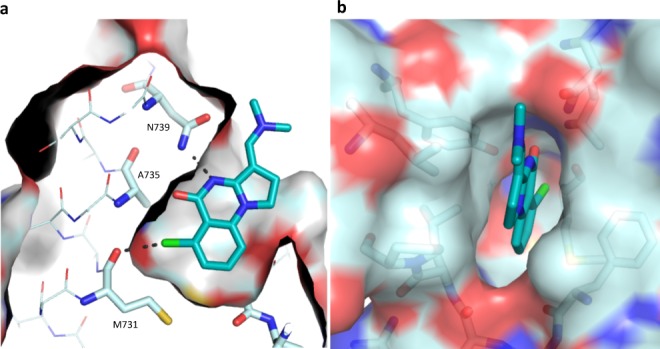
Cocrystal structure of 18 with PB1(5) at 1.5 Å (PDB code 5FH7): (a) cut-through illustrating positioning of chlorine in cavity; (b) surface view illustrating fit in PB1(5) pocket.
To improve the aqueous solubility of the chlorinated derivatives, we varied the enamine substituent using the existing methodology to access analogues 22–27, the majority of which retained their affinity for the target as assessed by DSF (Table 3). Compounds 24, 26, and 27 had greatly improved DMSO and aqueous solubility. The binding of these compounds to PB1(5) was assessed by ITC (Table 4), and all displayed similar KD values to 18.
Table 3. Structure–Activity Relationships of Chlorinated Derivatives against SWI/SNF Bromodomains.
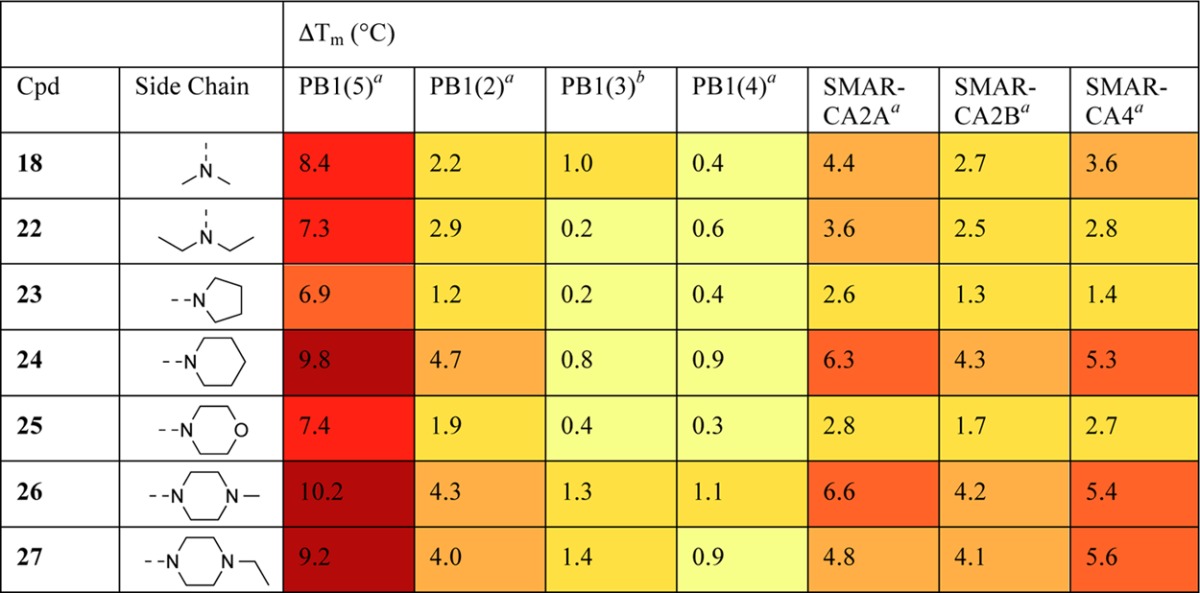
Values shown are the average of three replicates. Standard deviation is <0.6 °C.
Values are a single measurement.
Table 4. Binding of Key Compounds to PB1(5) Was Assessed by Isothermal Calorimetry.
| compd | K × 106 (M–1) | KD (nM) | ΔH (kcal/mol) | TΔS (kcal/mol) |
|---|---|---|---|---|
| 18 | 7.29 ± 0.91 | 137 (157–122) | –6.2 | 3.0 |
| 24 | 7.86 ± 0.89 | 127 (143–114) | –5.2 | 4.0 |
| 26 | 8.09 ± 0.66 | 124 (135–114) | –5.0 | 4.3 |
| 27 | 7.05 ± 0.68 | 142 (157–129) | –4.4 | 4.7 |
| 28 | 5.88 ± 0.60 | 170 (189–154) | –6.8 | 2.3 |
As shown in Table 3, the analogues were screened by DSF against other bromodomains within the SWI/SNF complex, including other structurally distinct bromodomains within protein PB1, to determine their selectivity. When interpreting this data, it should be noted that different proteins do not behave identically in DSF assays, and ΔTm values should not be viewed as an absolute scale. For example, probe PFI-3 has KD values by ITC of 89 and 48 nM for SMARCA4 and PB1(5), respectively, but ΔTm shifts of 5.1 and 7.5 °C.25
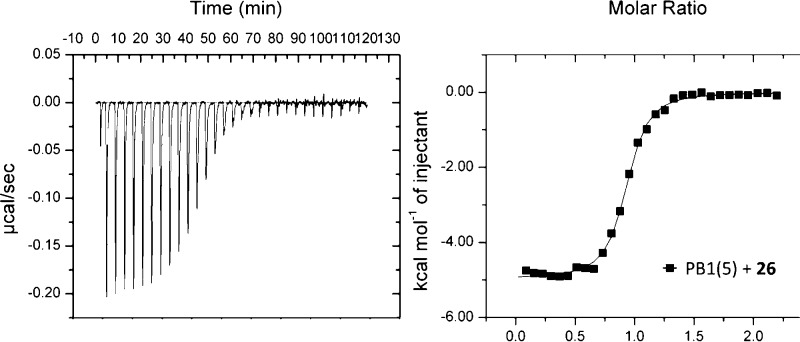
In our screening, compounds appeared to interact with the bromodomains in both SMARCA2 isoforms, A and B, and in SMARCA4 and PB1(5). ITC was used to further assess the binding of 26 (Figure 4) to SMARCA2B and SMARCA4, giving KD values of 262 and 417 nM, respectively. Binding to the SMARCA bromodomains had not been observed prior to inclusion of the chlorine substitution, suggesting that the proposed halogen bond is again important for binding.
Figure 4.
Representative ITC trace, measured using 26 and the bromodomain of PB1(5); 30 consecutive injections of PB1(5) into a solution of 26 in 20 mM HEPES, 150 mM NaCl, 0.05 mM TCEP. Raw heats (left) and normalized injection heats with a nonlinear least-squares fit for a single binding site model (right) are shown.
The bromodomain PB1(2) also demonstrated a moderate response, despite containing a broader KAc binding pocket than PB1(5). By contrast, minimal protein stabilization was observed with bromodomains PB1(3) and PB1(4). This may be due to the tyrosine residue in the ZA loop which acts as a “lid” to the pocket in these bromodomains.26
We were interested to establish whether further side chain variation could alter selectivity within SWI/SNF bromodomains (Table 5). Analogues 28–31 incorporating alkene side chains were synthesized by reaction of 6-chloro-2,3-dihydropyrrolo[1,2-a]quinazolin-5(1H)-one (18c) with the appropriate aliphatic aldehydes under basic conditions (see Supporting Information for full methods). These were screened by DSF, which suggested reduced binding to PB1(5) for the majority of ligands except 28. When assessed by ITC, 28 had a KD of 170 nM with PB1(5). DSF results suggested an intriguing change in selectivity among SWI/SNF bromodomains. In contrast to all previous compounds tested in this report, and the chemical probe PFI-3,2628 showed a stronger interaction with PB1(2) than with the SMARCA2/4 bromodomains. The weak binding to SMARCA4 was confirmed by ITC, with a KD of 2.03 μM for the interaction with 28.
Table 5. Structure–Activity Relationships of Compounds Bearing Alkene Based Side Chains against SWI/SNF Bromodomainsa.
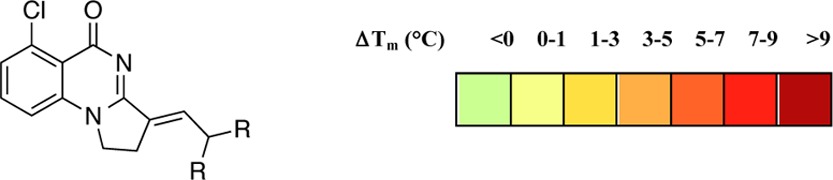
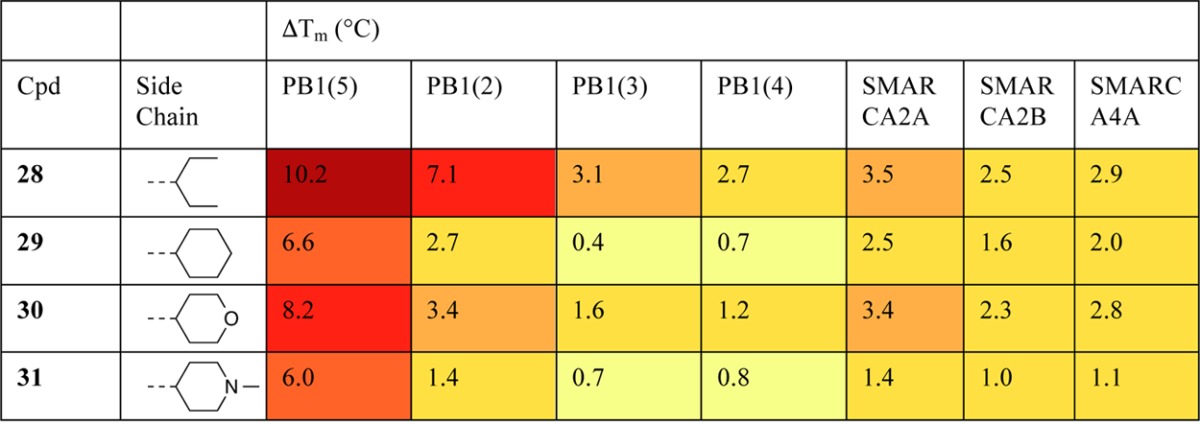
Values shown are the average of three replicates. Standard deviation is <0.6 °C. For PB1(3), values are a single measurement.
Understanding the reason for the change in affinity would help design of future chemical probes with selectivity for bromodomains not yet targeted such as PB1(2–4) and offers the intriguing possibility of targeting PB1 selectively over SMARCA2/4. We obtained a cocrystal structure of 28 with PB1(5) (Figure 5) to understand its unique features. This revealed a shift in the ZA loop region of PB1(5), making the KAc pocket wider and increasing its similarity to the PB1(2) KAc pocket. The ethyl side chains interact with the hydrophobic edges of the pocket and begin to occupy the ill-defined peptide-binding channel in PB1(5). Although no structure was obtained to confirm this, we propose that in SMARCA4, Ile1543 blocks this channel, preventing the ethyl side chains from being accommodated in the smaller pocket.
Figure 5.
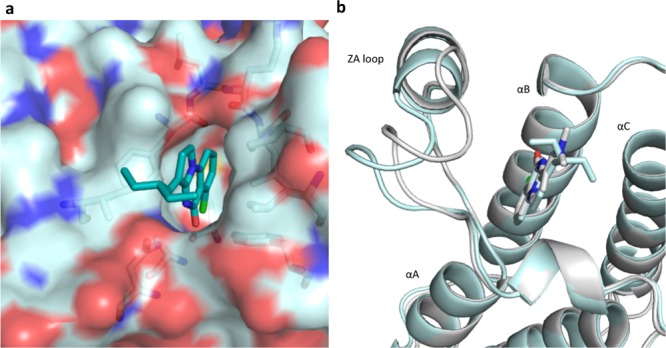
(a) Cocrystal structure of 28 with PB1(5) at 1.6 Å (PDB code 5FH8). (b) Overlay of PDB code 5FH7 (gray) of 18 and PDB code 5FH8 (cyan) of 28 showing the shift in the ZA loop of the PB1(5) domain on binding of 28.
We assessed the selectivity of the most potent pan-PB1, SMARCA2/4 analogue 26 against representative bromodomains (PCAF, BRD4(1), CREBBP, TRIM33B) from other subfamilies using DSF (Figure 6). No significant binding was observed.
Figure 6.
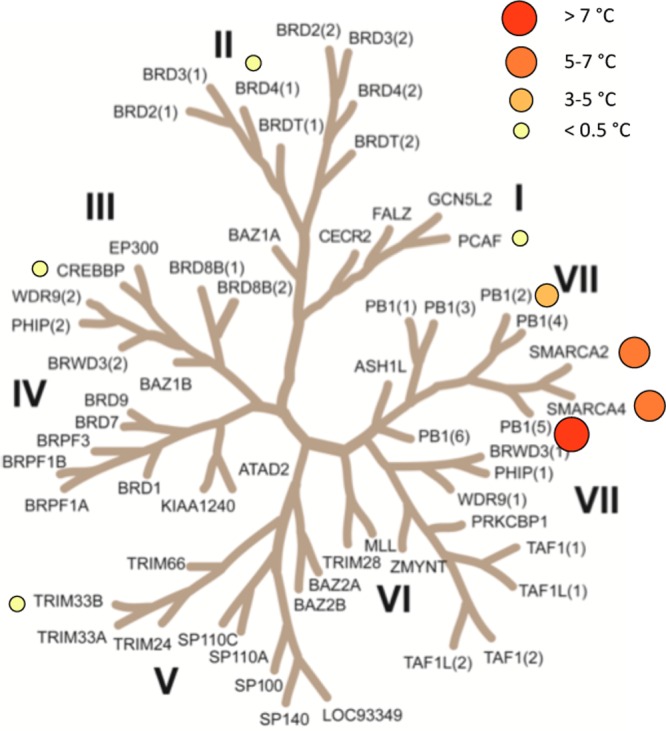
Selectivity of 26. Inhibitor 26 was screened at 10 μM against selected bromodomains by DSF assay. Temperature shifts for screened proteins are shown on the phylogenetic tree.
Finally, the ability of 26 to displace SMARCA2 from chromatin was assessed using a fluorescence recovery after photobleaching (FRAP) assay (Figure 7).31 U2OS cells were transfected with a GFP-linked SMARCA2 construct. To increase global histone acetylation and thereby the assay window the broad-spectrum histone-deacetylase (HDAC) inhibitor, suberoylanilide hydroxamic acid (SAHA) was used. Cells transfected with a construct carrying the N1464F mutation, unable to bind to chromatin via the bromodomain, were used as a positive control. Treatment of cells with 26 at 1 or 5 μM reduced FRAP recovery times back to that of the N1464F mutant and unstimulated levels, indicating that 26 was able to displace full length SMARCA2 from chromatin.
Figure 7.
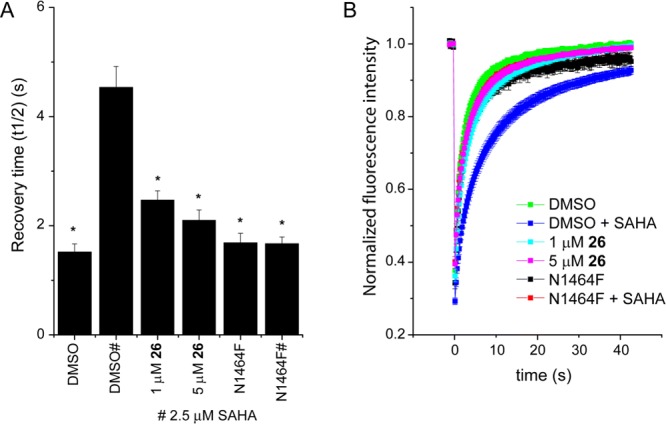
Compound 26 inhibits SMARCA2 association with chromatin in cells. (A) FRAP half recovery times of GFP-SMARCA2 are significantly decreased when treated with 26 at 1 or 5 μM as indicated. Cells expressing the SMARCA2 bromodomain-inactivating mutant (N1464) were analyzed as comparison. Significant differences to cells treated with SAHA of p < 0.0001 are shown by ∗. (B) Time dependence of fluorescence recovery in the bleached area of cells expressing wt or mutant GFP-SMARCA2 with the corresponding treatment.
Conclusions
We describe the optimization of an inhibitor series targeting bromodomains found within the SWI/SNF complex from a weakly potent hit with poor physicochemical properties. Improvement of solubility has allowed cocrystal structures to be obtained demonstrating the important role of water displacement in the binding of these inhibitors. Chlorination of the series has demonstrated the potential for exploitation of previously unexplored interactions deep within the PB1(5) KAc binding pocket through halogen bonding. Side chain variation in 28 shows that the second and fifth bromodomains of PB1 can be targeted selectively over the SMARCA2/4 helicases, in contrast to the selectivity shown by the chemical probe PFI-3. Lead inhibitor 26 displays good affinity for PB1(5), SMARCA4, and SMARCA2 as assessed by ITC, excellent selectivity within the bromodomain family, and the ability to displace SMARCA2 from chromatin in cells, making it suitable as a chemical probe with a distinct chemotype to PFI-3 and for further development of SWI/SNF bromodomain inhibitors.
Acknowledgments
C.L.S. was funded by the Cambridge Ph.D. Training Programme in Chemical Biology and Molecular Medicine. We gratefully acknowledge the EPSRC (SVL, Grants EP/K099494/1 and EP/K039520/1). The SGC is a registered charity (No. 1097737) that received funds from AbbVie, Bayer Pharma AG, Boehringer Ingelheim, the Canada Foundation for Innovation, Genome Canada, GlaxoSmithKline, Janssen, Lilly Canada, the Novartis Research Foundation, the Ontario Ministry of Economic Development, and Innovation, Pfizer, Takeda, and the Wellcome Trust (Grant 092809/Z/10/Z).
Glossary
Abbreviations Used
- ATP
adenosine triphosphate
- BCP
bromodomain containing protein
- BET
bromodomain and extraterminal domain
- BRD7
bromodomain containing protein 7
- BRD9
bromodomain containing protein 9
- DMAP
N,N-dimethyl-4-aminopyridine
- DSF
differential scanning fluorimetry
- FRAP
fluorescence recovery after photobleaching
- GFP
green fluorescent protein
- HDAC
histone deacetylase
- ITC
isothermal titration calorimetry
- KAc
acetyl-lysine
- NOE
nuclear Overhauser effect
- PB1
polybromo-1
- PB1(X)
Xth bromodomain of PB1
- PDB
Protein Data Bank
- SAHA
suberoylanilide hydroxamic acid
- SAR
structure–activity relationship
- SMARCA2/4
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily A, member 2/4
- SWI/SNF
switch/sucrose nonfermenting
Supporting Information Available
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acs.jmedchem.5b01997. In addition to the indicated Supporting Information PDF and CSV files, additional data related to this publication are available at https://www.repository.cam.ac.uk/handle/1810/254994.
The authors declare no competing financial interest.
Supplementary Material
References
- Shain A. H.; Pollack J. R. The Spectrum of SWI/SNF Mutations, Ubiquitous in Human Cancers. PLoS One 2013, 8, e55119. 10.1371/journal.pone.0055119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadoch C.; Crabtree G. R. Mammalian SWI/SNF Chromatin Remodeling Complexes and Cancer: Mechanistic Insights Gained from Human Genomics. Sci. Adv. 2015, 1, e1500447. 10.1126/sciadv.1500447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson B. G.; Roberts C. W. M. SWI/SNF Nucleosome Remodellers and Cancer. Nat. Rev. Cancer 2011, 11, 481–492. 10.1038/nrc3068. [DOI] [PubMed] [Google Scholar]
- Son E. Y.; Crabtree G. R. The Role of BAF (mSWI/SNF) Complexes in Mammalian Neural Development. Am. J. Med. Genet., Part C 2014, 166, 333–349. 10.1002/ajmg.c.31416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X.; Sansam C. G.; Thom C. S.; Metzger D.; Evans J. A.; Nguyen P. T. L.; Roberts C. W. M. Oncogenesis Caused by Loss of the SNF5 Tumor Suppressor Is Dependent on Activity of BRG1, the ATPase of the SWI/SNF Chromatin Remodeling Complex. Cancer Res. 2009, 69, 8094–8101. 10.1158/0008-5472.CAN-09-0733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oike T.; Ogiwara H.; Tominaga Y.; Ito K.; Ando O.; Tsuta K.; Mizukami T.; Shimada Y.; Isomura H.; Komachi M.; Furuta K.; Watanabe S.-I.; Nakano T.; Yokota J.; Kohno T. A Synthetic Lethality-Based Strategy to Treat Cancers Harboring a Genetic Deficiency in the Chromatin Remodeling Factor BRG1. Cancer Res. 2013, 73, 5508–5518. 10.1158/0008-5472.CAN-12-4593. [DOI] [PubMed] [Google Scholar]
- Hoffman G. R.; Rahal R.; Buxton F.; Xiang K.; McAllister G.; Frias E.; Bagdasarian L.; Huber J.; Lindeman A.; Chen D.; Romero R.; Ramadan N.; Phadke T.; Haas K.; Jaskelioff M.; Wilson B. G.; Meyer M. J.; Saenz-Vash V.; Zhai H.; Myer V. E.; Porter J. A.; Keen N.; McLaughlin M. E.; Mickanin C.; Roberts C. W. M.; Stegmeier F.; Jagani Z. Functional Epigenetics Approach Identifies BRM/SMARCA2 as a Critical Synthetic Lethal Target in BRG1-Deficient Cancers. Proc. Natl. Acad. Sci. U. S. A. 2014, 111, 3128–3133. 10.1073/pnas.1316793111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel M. N.; Halling-Brown M. D.; Tym J. E.; Workman P.; Al-Lazikani B. Objective Assessment of Cancer Genes for Drug Discovery. Nat. Rev. Drug Discovery 2013, 12, 35–50. 10.1038/nrd3913. [DOI] [PubMed] [Google Scholar]
- Hassan A. H.; Neely K. E.; Workman J. L. Histone Acetyltransferase Complexes Stabilize SWI/SNF Binding to Promoter Nucleosomes. Cell 2001, 104, 817–827. 10.1016/S0092-8674(01)00279-3. [DOI] [PubMed] [Google Scholar]
- Chatterjee N.; Sinha D.; Lemma-Dechassa M.; Tan S.; Shogren-Knaak M. A.; Bartholomew B. Histone H3 Tail Acetylation Modulates ATP-Dependent Remodeling through Multiple Mechanisms. Nucleic Acids Res. 2011, 39, 8378–8391. 10.1093/nar/gkr535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H.-S.; Park J.-H.; Kim S.-J.; Kwon S.-J.; Kwon J. A Cooperative Activation Loop among SWI/SNF, Gamma-H2AX and H3 Acetylation for DNA Double-Strand Break Repair. EMBO J. 2010, 29, 1434–1445. 10.1038/emboj.2010.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon S.-J.; Lee S. S.-A. S.-K.; Na J.; Lee S. S.-A. S.-K.; Lee H.; Park J.; Chung J.-K.; Youn H.; Kwon J. Targeting BRG1 Chromatin Remodeler via Its Bromodomain for Enhanced Tumor Cell Radiosensitivity in Vitro and in Vivo. Mol. Cancer Ther. 2015, 14, 597–607. 10.1158/1535-7163.MCT-14-0372. [DOI] [PubMed] [Google Scholar]
- Brownlee P. M.; Chambers A. L.; Oliver A. W.; Downs J. a. Cancer and the Bromodomains of BAF180. Biochem. Soc. Trans. 2012, 40, 364–369. 10.1042/BST20110754. [DOI] [PubMed] [Google Scholar]
- Brownlee P. M.; Chambers A. L.; Cloney R.; Bianchi A.; Downs J. A. BAF180 Promotes Cohesion and Prevents Genome Instability and Aneuploidy. Cell Rep. 2014, 6, 973–981. 10.1016/j.celrep.2014.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filippakopoulos P.; Knapp S. Targeting Bromodomains: Epigenetic Readers of Lysine Acetylation. Nat. Rev. Drug Discovery 2014, 13, 337–356. 10.1038/nrd4286. [DOI] [PubMed] [Google Scholar]
- Muller S.; Filippakopoulos P.; Knapp S. Bromodomains as Therapeutic Targets. Expert Rev. Mol. Med. 2011, 13, e29. 10.1017/S1462399411001992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filippakopoulos P.; Picaud S.; Mangos M.; Keates T.; Lambert J.-P.; Barsyte-Lovejoy D.; Felletar I.; Volkmer R.; Müller S.; Pawson T.; Gingras A.-C.; Arrowsmith C. H.; Knapp S. Histone Recognition and Large-Scale Structural Analysis of the Human Bromodomain Family. Cell 2012, 149, 214–231. 10.1016/j.cell.2012.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hay D. A.; Fedorov O.; Martin S.; Singleton D. C.; Tallant C.; Wells C.; Picaud S.; Philpott M.; Monteiro O. P.; Rogers C. M.; Conway S. J.; Rooney T. P. C.; Tumber A.; Yapp C.; Filippakopoulos P.; Bunnage M. E.; Müller S.; Knapp S.; Schofield C. J.; Brennan P. E. Discovery and Optimization of Small-Molecule Ligands for the CBP/p300 Bromodomains. J. Am. Chem. Soc. 2014, 136, 9308–9319. 10.1021/ja412434f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammitzsch A.; Tallant C.; Fedorov O.; O’Mahony A.; Brennan P. E.; Hay D. A.; Martinez F. O.; Al-Mossawi M. H.; de Wit J.; Vecellio M.; Wells C.; Wordsworth P.; Müller S.; Knapp S.; Bowness P. CBP30, a Selective CBP/p300 Bromodomain Inhibitor, Suppresses Human Th17 Responses. Proc. Natl. Acad. Sci. U. S. A. 2015, 112, 10768–10773. 10.1073/pnas.1501956112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delmore J. E.; Issa G. C.; Lemieux M. E.; Rahl P. B.; Shi J.; Jacobs H. M.; Kastritis E.; Gilpatrick T.; Paranal R. M.; Qi J.; Chesi M.; Schinzel A. C.; McKeown M. R.; Heffernan T. P.; Vakoc C. R.; Bergsagel P. L.; Ghobrial I. M.; Richardson P. G.; Young R. A.; Hahn W. C.; Anderson K. C.; Kung A. L.; Bradner J. E.; Mitsiades C. S. BET Bromodomain Inhibition as a Therapeutic Strategy to Target c-Myc. Cell 2011, 146, 904–917. 10.1016/j.cell.2011.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawson M. A.; Prinjha R. K.; Dittmann A.; Giotopoulos G.; Bantscheff M.; Chan W.-I.; Robson S. C.; Chung C.; Hopf C.; Savitski M. M.; Huthmacher C.; Gudgin E.; Lugo D.; Beinke S.; Chapman T. D.; Roberts E. J.; Soden P. E.; Auger K. R.; Mirguet O.; Doehner K.; Delwel R.; Burnett A. K.; Jeffrey P.; Drewes G.; Lee K.; Huntly B. J. P.; Kouzarides T. Inhibition of BET Recruitment to Chromatin as an Effective Treatment for MLL-Fusion Leukaemia. Nature 2011, 478, 529–533. 10.1038/nature10509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicodeme E.; Jeffrey K. L.; Schaefer U.; Beinke S.; Dewell S.; Chung C.-W.; Chandwani R.; Marazzi I.; Wilson P.; Coste H.; White J.; Kirilovsky J.; Rice C. M.; Lora J. M.; Prinjha R. K.; Lee K.; Tarakhovsky A. Suppression of Inflammation by a Synthetic Histone Mimic. Nature 2010, 468, 1119–1123. 10.1038/nature09589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theodoulou N. H.; Bamborough P.; Bannister A. J.; Becher I.; Bit R. A.; Che K. H.; Chung C.; Dittmann A.; Drewes G.; Drewry D. H.; Gordon L.; Grandi P.; Leveridge M.; Lindon M.; Michon A.; Molnar J.; Robson S. C.; Tomkinson N. C. O.; Kouzarides T.; Prinjha R. K.; Humphreys P. G. Discovery of I-BRD9, a Selective Cell Active Chemical Probe for Bromodomain Containing Protein 9 Inhibition. J. Med. Chem. 2016, 59, 1425–1439. 10.1021/acs.jmedchem.5b00256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark P. G. K.; Vieira L. C. C.; Tallant C.; Fedorov O.; Singleton D. C.; Rogers C. M.; Monteiro O. P.; Bennett J. M.; Baronio R.; Müller S.; Daniels D. L.; Méndez J.; Knapp S.; Brennan P. E.; Dixon D. J. LP99: Discovery and Synthesis of the First Selective BRD7/9 Bromodomain Inhibitor. Angew. Chem., Int. Ed. 2015, 54, 6217–6221. 10.1002/anie.201501394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fedorov O.; Castex J.; Tallant C.; Owen D. R.; Martin S.; Aldeghi M.; Monteiro O.; Filippakopoulos P.; Picaud S.; Trzupek J. D.; Gerstenberger B. S.; Bountra C.; Willmann D.; Wells C.; Philpott M.; Rogers C.; Biggin P. C.; Brennan P. E.; Bunnage M. E.; Schüle R.; Günther T.; Knapp S.; Müller S. Selective Targeting of the BRG/PB1 Bromodomains Impairs Embryonic and Trophoblast Stem Cell Maintenance. Sci. Adv. 2015, 1, e1500723. 10.1126/sciadv.1500723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidler L. R.; Brown N.; Knapp S.; Hoelder S. Druggability Analysis and Structural Classification of Bromodomain Acetyl-Lysine Binding Sites. J. Med. Chem. 2012, 55, 7346–7359. 10.1021/jm300346w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fedorov O.; Lingard H.; Wells C.; Monteiro O. P.; Picaud S.; Yapp C.; Philpott M.; Martin S. J.; Felletar I.; Marsden B.; Filippakopoulos P.; Muller-knapp S.; Knapp S.; Brennan P. E. [1,2,4]Triazolo[4,3-A]phthalazines: Inhibitors of Diverse Bromodomains. J. Med. Chem. 2014, 57, 462–476. 10.1021/jm401568s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovering F.; Bikker J.; Humblet C. Escape from Flatland: Increasing Saturation as an Approach to Improving Clinical Success. J. Med. Chem. 2009, 52, 6752–6756. 10.1021/jm901241e. [DOI] [PubMed] [Google Scholar]
- Dunn A. D.; Kinnear K. I. New Reactions of Deoxyvasicinone. Part 4. J. Heterocycl. Chem. 1986, 23, 53–57. 10.1002/jhet.5570230111. [DOI] [Google Scholar]
- Lu Y.; Shi T.; Wang Y.; Yang H.; Yan X.; Luo X.; Jiang H.; Zhu W. Halogen Bonding-a Novel Interaction for Rational Drug Design?. J. Med. Chem. 2009, 52, 2854–2862. 10.1021/jm9000133. [DOI] [PubMed] [Google Scholar]
- Philpott M.; Rogers C. M.; Yapp C.; Wells C.; Lambert J.-P.; Strain-Damerell C.; Burgess-Brown N. a; Gingras A.-C.; Knapp S.; Müller S. Assessing Cellular Efficacy of Bromodomain Inhibitors Using Fluorescence Recovery after Photobleaching. Epigenet. Chromatin 2014, 7, 14. 10.1186/1756-8935-7-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.