Abstract
Summary: Extracting chemical features like Atom–Atom Mapping (AAM), Bond Changes (BCs) and Reaction Centres from biochemical reactions helps us understand the chemical composition of enzymatic reactions. Reaction Decoder is a robust command line tool, which performs this task with high accuracy. It supports standard chemical input/output exchange formats i.e. RXN/SMILES, computes AAM, highlights BCs and creates images of the mapped reaction. This aids in the analysis of metabolic pathways and the ability to perform comparative studies of chemical reactions based on these features.
Availability and implementation: This software is implemented in Java, supported on Windows, Linux and Mac OSX, and freely available at https://github.com/asad/ReactionDecoder
Contact: asad@ebi.ac.uk or s9asad@gmail.com
1 Introduction
Large-scale chemical reaction databases such as KEGG (Kanehisa et al., 2013), BRENDA (Chang et al., 2015), Rhea (Alcántara et al., 2012) and MetaCyc (Latendresse et al., 2012) link reactions to enzymes and provide data-mining opportunities for novel pathways (Hatzimanikatis et al., 2004; Rahman, 2007), and the discovery of drugs, natural products and green chemistry. One of the primary bottlenecks for automated analyses of these chemical reactions comes from the realizations of the imperfect quality of data, such as unmapped or unbalanced reactions. Accurate Atom–Atom Mapping (AAM)—the one-to-one correspondence between the substrate and product atoms (Gasteiger, 2003), will lead to correct prediction of bond changes (BCs) (Rahman et al., 2014) and the ability to locate the fate of interesting atoms or substructure across metabolic networks (Faulon and Bender, 2010) etc. Linking novel pathways or optimizing pathways of biological/commercial relevance demands better understanding of metabolic routes (Rahman, 2007) and pathway annotation (May et al., 2013).
We present Reaction Decoder Tool (RDT), a robust open source software for mining reaction features, i.e. BCs, reaction centres and to calculate similarity between reactions etc. The algorithm and competing tools have been published in our previous article (Rahman et al., 2014). Here we present the source code with relevant changes and below mentioned features.
2 Features
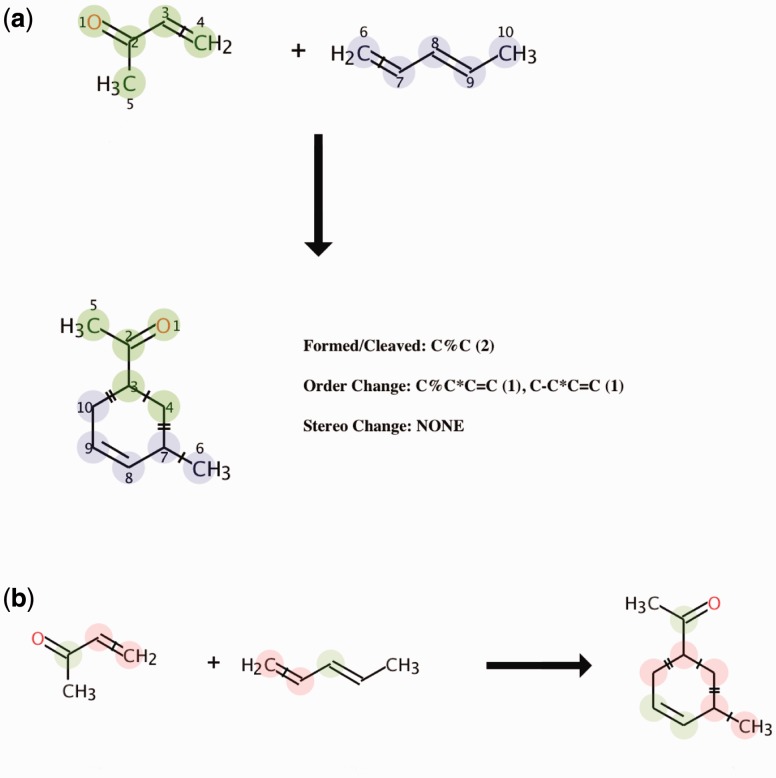
BCs in chemical reactions refers to the cleavage and formation of chemical bonds, changes in bond order and stereo changes, which are due to chemical processes such as chiral inversions or cis-trans isomerization(s). In the chemical reaction diagram and tables atoms are connected by a bond i.e. single ‘–‘, double ‘=’or ring ‘%’ etc. (Fig. 1a), for instance C–C means a single carbon–carbon bond that is cleaved (‘|’) or formed in the reaction (‘‖’). Bond order changes are represented by star ‘*’, e.g. C–C * C = C means a single carbon–carbon bond turning into double carbon-carbon bond or vice versa. Stereo changes are represented as atoms that change their absolute configuration, for instance C(R/S) means a carbon atom that changes from R to S configuration. A reaction centre is the collection of atoms and bonds that are changed during the reaction (Warr, 2014), also known as the local atomic environment around the atoms involved in BCs.
Fig. 1.
(a) AAM performed by RDT and the resulting BCs (i.e. formed/cleaved: C%C (Ring), Order Change: C%C*C = C, C-C*C = C) and (b) reaction centres are highlighted in pink colour in the image, where as neighbouring atoms are highlighted in green
The key features of this tool are:
Ability to perform AAM on chemical reactions catalyzed by enzymes (Fig. 1).
In a balanced reaction, the total number of atoms on the left side of the equation (Reactant), equals the total number of atoms on the right (Product). In unbalanced reactions a best ‘guess’ is made.
Generates images of the mapped reactions where matching substructures are highlighted.
Generates reaction patterns and BCs for input reactions.
The input format and resulting mapped reaction with AAM information can be SMILES or RXN file (Gasteiger, 2003).
The SMSD (Rahman et al., 2009) and CDK (Steinbeck et al., 2006) are used to process chemical information.
3 Usage and applications
Tools like EC-Blast (Rahman et al., 2014), FunTree (Sillitoe and Furnham, 2016), MACiE (Holliday et al., 2012) etc. use RDT in the background to mine and extract chemical information from thousands of enzyme reactions. The success rate of mapping is >99% when compared with manual AAM mappings (Rahman et al., 2014). Originally developed to explore enzyme reactions, the tool is also useful to explore any kind of organic chemical reaction (Martínez Cuesta et al., 2014).
4 Conclusion
Reaction Decoder is a robust tool to compute AAM and extract chemical features and calculate similarity between chemical reactions. This is coded in java and optimized to run as a computationally asynchronous process. It is distributed under GNU-GPL V3 license.
Acknowledgements
We would like to thank the CDK team, Sophie Therese Williams and anonymous users for their valuable feedback.
Funding
Authors would like to thank EMBL for funding this project. LB was partially funded by Marie Curie fellowship.
Conflict of Interest: none declared.
References
- Alcántara R. et al. (2012) Rhea—a manually curated resource of biochemical reactions. Nucleic Acids Res., 40, D754–D760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang A. et al. (2015) BRENDA in 2015: exciting developments in its 25th year of existence. Nucleic Acids Res., 43, D439–D446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faulon J.L, Bender A. (2010) Handbook of Chemoinformatics Algorithms. Chapman & Hall/CRC, Boca Raton, FL. [Google Scholar]
- Gasteiger J. (ed.) (2003) Applications, in Handbook of Chemoinformatics: From Data to Knowledge in 4 Volumes. Wiley-VCH, Verlag GmbH, Weinheim, Germany. [Google Scholar]
- Hatzimanikatis V. et al. (2004) Metabolic networks: enzyme function and metabolite structure. Curr. Opin. Struct. Biol., 14, 300–306. [DOI] [PubMed] [Google Scholar]
- Holliday G.L. et al. (2012) MACiE: exploring the diversity of biochemical reactions. Nucleic Acids Res., 40, D783–D789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M. et al. (2013) Data, information, knowledge and principle: back to metabolism in KEGG. Nucleic Acids Res., 42, D199–D205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latendresse M. et al. (2012) Accurate atom-mapping computation for biochemical reactions. J. Chem. Inf. Model., 52, 2970–2982. [DOI] [PubMed] [Google Scholar]
- Martínez Cuesta S. et al. (2014) The evolution of enzyme function in the isomerases. Curr. Opin. Struct. Biol., 26, 121–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- May J.W. et al. (2013) Metingear: a development environment for annotating genome-scale metabolic models. Bioinformatics, 29, 2213–2215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahman S.A. (2007) Pathway hunter tool (pht)- a platform for metabolic network analysis and potential drug targeting PhD Thesis, University of Cologne, Cologne, Germany. [Google Scholar]
- Rahman S.A. et al. (2014) EC-BLAST: a tool to automatically search and compare enzyme reactions. Nat. Methods, 11, 171–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahman S.A. et al. (2009) Small Molecule Subgraph Detector (SMSD) toolkit. J. Cheminf, 1, 12–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sillitoe I., Furnham N. (2016) FunTree: advances in a resource for exploring and contextualising protein function evolution. Nucleic Acids Res., 44, D317–D323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinbeck C. et al. (2006) Recent developments of the chemistry development kit (CDK) - an open-source java library for chemo- and bioinformatics. Curr. Pharm. Des., 12, 2111–2120. [DOI] [PubMed] [Google Scholar]
- Warr W.A. (2014) A Short Review of Chemical Reaction Database Systems, Computer-Aided Synthesis Design, Reaction Prediction and Synthetic Feasibility. Mol. Inf., 33, 469–476. [DOI] [PubMed] [Google Scholar]