Abstract
Motivation: CRISPR-derived RNA guided endonucleases (RGENs) have been widely used for both gene knockout and knock-in at the level of single or multiple genes. RGENs are now available for forward genetic screens at genome scale, but single guide RNA (sgRNA) selection at this scale is difficult.
Results: We develop an online tool, Cas-Database, a genome-wide gRNA library design tool for Cas9 nucleases from Streptococcus pyogenes (SpCas9). With an easy-to-use web interface, Cas-Database allows users to select optimal target sequences simply by changing the filtering conditions. Furthermore, it provides a powerful way to select multiple optimal target sequences from thousands of genes at once for the creation of a genome-wide library. Cas-Database also provides a web application programming interface (web API) for advanced bioinformatics users.
Availability and implementation: Free access at http://www.rgenome.net/cas-database/.
Contact: sangsubae@hanyang.ac.kr or jskim01@snu.ac.kr
Supplementary information: Supplementary data are available at Bioinformatics online.
1 Introduction
RNA guided endonucleases (RGENs) derived from the Type II clustered regularly interspaced short palindromic repeats (CRISPR)-Cas (CRISPR associated) system, an adaptive immune response in bacteria and archaea, have been usefully harnessed in many genome engineering applications such as gene knockout and knock-in in various organisms (Doudna and Charpentier, 2014; Kim and Kim, 2014; Sander and Joung, 2014; Shalem et al., 2015). Recently, a few groups have undertaken genome-wide Cas9-mediated genetic screens (Chen et al., 2015; Gilbert et al., 2014; Hart et al., 2015; Koike-Yusa et al., 2014; Konermann et al., 2014; Shalem et al., 2014; Wang et al., 2014; Zhou et al., 2014) in human and other mammalian cells. The selection of target sequences is an initial, rate-limiting step in RGEN applications. We and other groups have developed a number of web-based online tools or command-line programs for single-guide RNA (sgRNA) design or off-target site identification (Aach et al., 2014; Bae et al., 2014a; Cradick et al., 2014; Doench et al., 2014; Heigwer et al., 2014; Hsu et al., 2013; Lei et al., 2014; Montague et al., 2014; Naito et al., 2015; O’Brien and Bailey, 2014; Park et al., 2015; Sander et al., 2010; Upadhyay and Sharma, 2014; Xiao et al., 2014; Xie et al., 2014). Although they are very useful for choosing sgRNAs that target one gene, selection of sgRNAs at the genome scale is still challenging.
Previously, two genome-wide databases for SpCas9 design (Hodgkins et al., 2015; MacPherson and Scherf, 2015) have been built conceptually. However, although they are useful for selecting sgRNAs that target a small number of genes, neither offers an easy way to select optimal target sequences at once from thousands of genes for genome-wide library construction in a variety of organisms. Here we describe Cas-Database, a web-based genome-wide gRNA design tool for SpCas9 nucleases for genome-scale screening experiments. Cas-Database contains all available targets of SpCas9 nucleases that recognizes 5′-NGG-3′ protospacer-adjacent motif (PAM) sequences in all coding sequence (CDS) regions throughout the whole genome of a selected organism, based on the Ensembl database (Cunningham et al., 2014). Each target site has the following information: GC content, relative cleavage position in the CDS, constitutive exon coverage, a microhomolgy-associated out-of-frame score (Bae et al., 2014b), and potential off-target sequences with up to 2-nt mismatches. In addition, JBrowse (Skinner et al., 2009) is used to display all available target sites graphically in a zoomable interface with genomic location information for the user’s convenience.
Cas-Database basically provides a fast and easy way to select optimal target sequences in genes of interest from a variety of organisms. Additionally, it offers a powerful way to select optimal target sequences in many genes simultaneously. Selecting sgRNA sequences that target each gene is as easy as online shopping because of the use of a ‘cart’ system, similar to what is used in online shopping malls, which was implemented using cutting-edge web development techniques such as AJAX (asynchronous JavaScript and XML).
Currently, Cas-Database supports sgRNA design in 12 different organisms, including five vertebrates (human, rat, mouse, pig and zebrafish), one insect (Drosophila melanogaster), one nematode (Caenorhabditis elegans) and five plants (Arabidopsis thaliana, tomato, banana, grapes and soybean). All processes required to generate the genome-wide database have now been automated using a scripts pipeline so that we can easily update information about the existing organisms or add new organisms to the database. We are planning to continue to add and support more organisms in the future.
2 Methods
2.1 Target selection for SpCas9 nucleases
The latest whole genome sequence and associated annotation data from each organism in the Ensembl database were automatically retrieved and saved to our server using a homemade program written in Python language. To allow easy access to the annotation database, we used the Biomart protocol (Kasprzyk, 2011; Kasprzyk et al., 2004) for communicating with the Ensembl server. After retrieving the genome sequence and annotation data associated with each organism, we first searched for all possible targets that contain 5′-NGG-3′ PAM sequences in CDS regions and then calculated several characteristics of each target, e.g. sgRNA GC content, relative cleavage position in the CDS, common exon coverage throughout the various transcripts of the gene, and a microhomology-associated out-of-frame score.
2.2 Searching for potential off-target sites
After identifying all possible targets for SpCas9 nucleases in CDS regions and calculating the characteristics of each target, we next used the Cas-OFFinder program (Bae et al., 2014a) to search for potential off-target sites that differ from each on-target site by up to 2nt and that contain 5′-NGG-3′ or 5′-NAG-3′ PAM sequences. Because this step is very time-consuming, we ran Cas-OFFinder on an OpenCL-enabled cluster computer such as Chundoong (http://chundoong.snu.ac.kr/) for all sites in parallel. During this process, we periodically validated that each searching node had finished correctly. After validation, all Cas-OFFinder output files were retrieved from the Chundoong server and moved to our local storage site. Then, for each selected target the total number of potential off-targets was counted; information about the genomic location of these potential off-targets, such as whether they reside in CDS, UTR, intron or intergenic regions, was also evaluated.
2.3 Inserting information into the database
We rearranged all of the resulting data and constructed a SQL database that contains all possible SpCas9 nuclease targets in CDS regions and related characteristics of each target. We chose PostgreSQL (http://www.postgresql.org/) as the database server, which showed the best performance in our case.
2.4 Web interface
We built a web interface for Cas-Database using the Python web framework Django (https://www.djangoproject.com/). Because of its modern and easily-implemented database application program interface (database API), the algorithm for storing and retrieving data from the PostgreSQL database is simplified and web site maintenance is easy. For creating the HTML part of the interface, we used the web development framework Bootstrap (http://getbootstrap.com/) and the JavaScript framework JQuery (https://jquery.com/). We also implemented asynchronous data upload and download to achieve a fast response time for each user using the asynchronous JavaScript and XML (AJAX) technique. Because the results are retrieved from the server asynchronously, user requests will be run in parallel rather than in sequence. In conclusion, searching and filtering functions will be operated and displayed immediately after a user changes the input data or filtering condition.
3 Results and discussion
3.1 Cas-Database overview
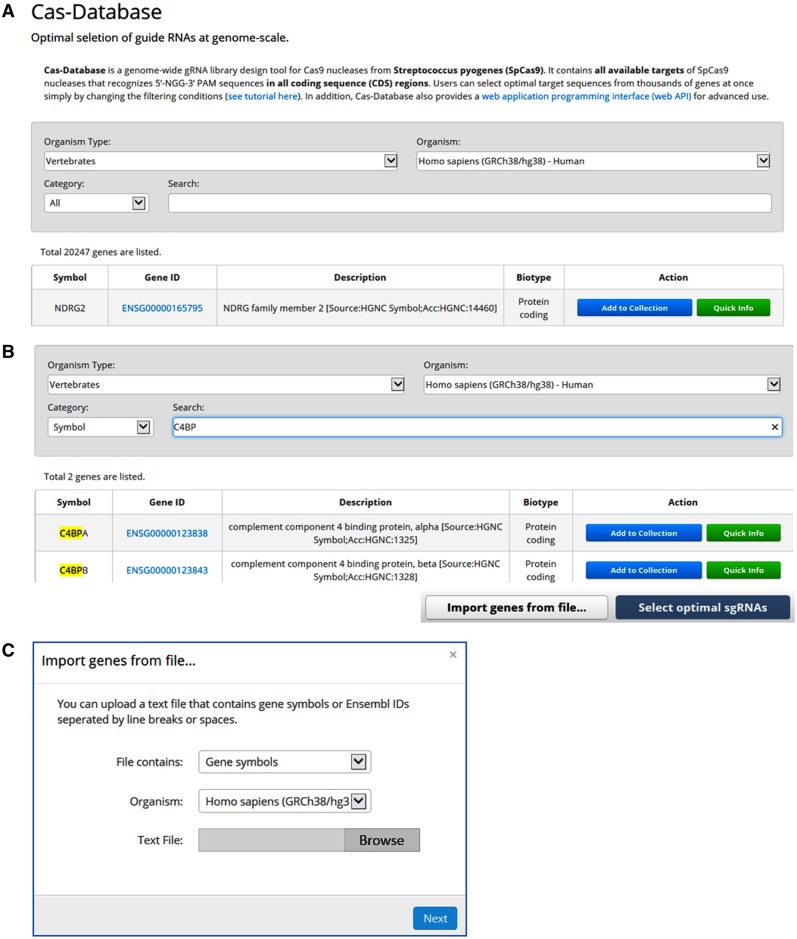
Cas-Database provides a simple and easy way to design optimal sgRNAs. All available genes from a selected organism are listed on the main page of the Cas-Database website, as shown in Figure 1A. Users can easily search for desired genes by querying with the gene symbol, Ensembl ID or gene description. The search results are instantly displayed on the screen after every keystroke through the use of the AJAX web technique (Fig. 1B).
Fig. 1.
Cas-Database overview. (A) All available genes from selected organisms are listed with their Ensembl ID, description and biotype information on the main page of the Cas-Database web site. Users can preview gene information and manually select optimal sgRNAs for each gene using the ‘Quick Info’ function, or select sgRNAs for many genes automatically using the ‘Add to Collection’ function. (B) Cas-Database’s top search panel provides a powerful searching function. Users can easily search for a desired gene by querying the gene symbol, Ensembl ID or gene description. The results are updated instantly on the screen via the AJAX web technique. (C) When working with hundreds or thousands of genes, users can upload text files that contain Ensembl IDs or gene symbols separated by line breaks or spaces
For a desired gene, users can use the ‘Quick Info’ function or the ‘Add to collection’ function. Basically, the ‘Quick Info’ function is useful for selecting sgRNAs manually for each gene; one can easily preview all available targets in a specific gene by clicking on the ‘Quick info’ button. Moreover, users can add additional desired genes—just as items are added to a shopping cart online—and select sgRNAs for many genes simultaneously using the ‘Add to collection’ function. In addition, hundreds or thousands of genes can be imported from text files that contain gene symbols or Ensembl ID lists, with individual entries separated by line breaks or spaces (Fig. 1C).
3.2 Use of the ‘Quick Info’ function
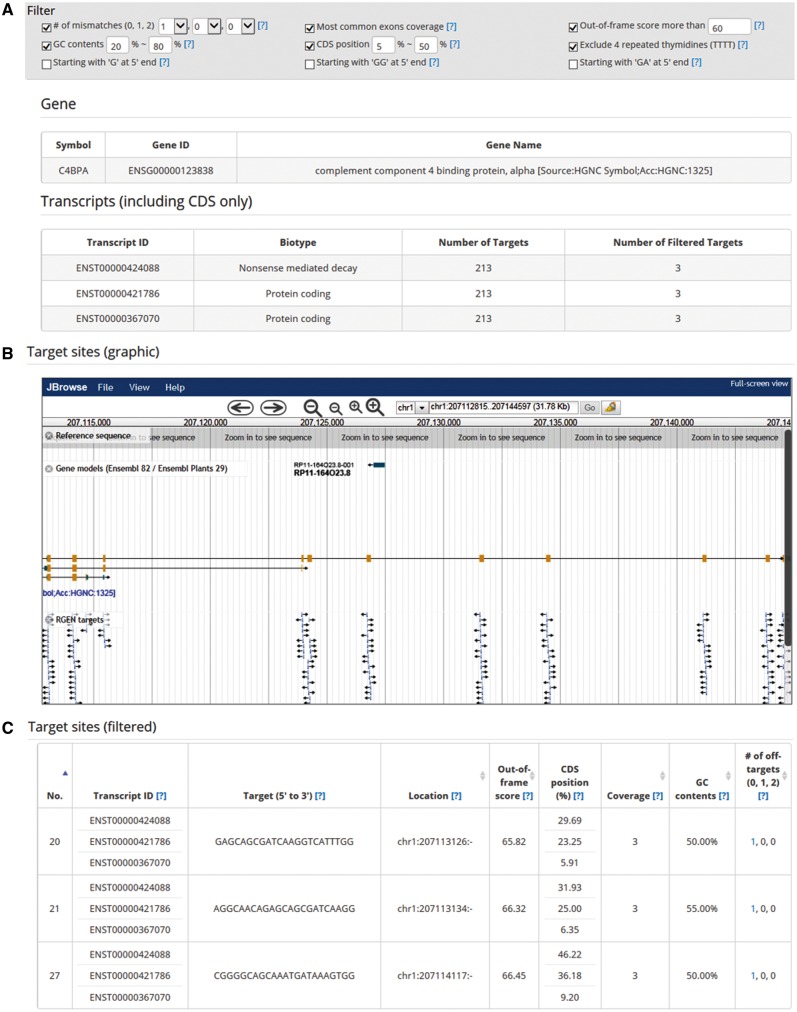
Cas-Database provides an easy way to preview sgRNAs that target a specific gene. Clicking on the ‘Quick info’ button immediately provides detailed information about the gene, transcripts and targets in a new dialog box (Fig. 2A–C). Furthermore, the genomic locus of each target is displayed on the graphical genome browser, JBrowse (Skinner et al., 2009) (Fig. 2B). If one clicks on a target in the browser, the corresponding target (Fig. 2C) will be shown on the Cas-Database web page for added convenience.
Fig. 2.
The ‘Quick Info’ function of Cas-Database. Clicking on the ‘Quick Info’ button as described for Figure 1A results in the rapid display of useful information about the selected gene: (A) gene description, transcript variant information that includes the CDS region, (B) location of sgRNAs in the gene visualized by JBrowse and (C) all filtered sgRNAs with useful descriptions. Additionally, all target sites are listed below, with transcript ID, GC content, genomic location, relative position in the CDS, exon coverage throughout all transcript variants of the gene, a microhomology-associated out-of-frame score and the number of mismatched nucleotides. Users can alter filter conditions at the top of the dialog
Cas-Database also offers a powerful filtering function. Using the filtering feature at the top of the web page, as shown in Figure 2A, users can change the filtering conditions and rapidly preview the new results.
Filtering criteria include GC content, relative position in the CDS, exon coverage throughout all transcript variants of the gene, a microhomology-associated out-of-frame score (Bae et al., 2014b), excluding four thymidine nucleotides in tandem (Braglia et al., 2005) and the number of mismatched nucleotides.
3.3 Use of the ‘Add to Collection’ function
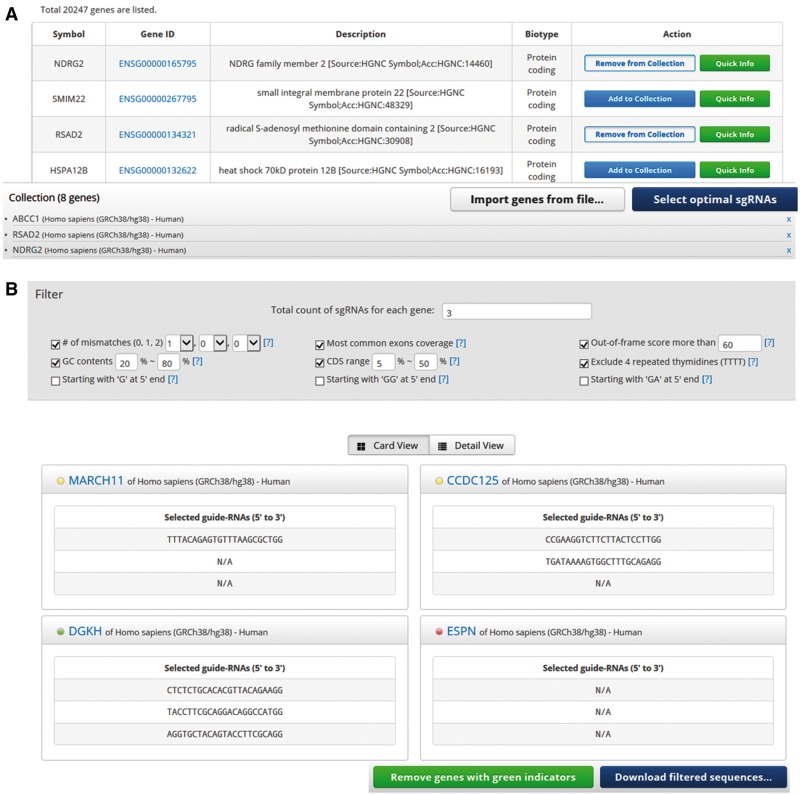
Cas-Database also provides a novel function that allows the selection of sgRNAs from hundreds or thousands of genes at once through the use of a ‘cart’ system. Users can either collect desired genes on the main web page by clicking on the ‘Add to Collection’ button (Fig. 3A) or by uploading a text file as discussed above and shown in Figure 1C. After all desired genes are collected, clicking on the ‘Select optimal sgRNAs’ button will open the results page, which will list all available sgRNAs filtered by the default conditions as shown in Figure 3B. Users can easily change the filtering conditions, including the total count of sgRNAs for each gene, in the filter section that appears at the top of the resulting page (Fig. 3B). Because the AJAX technique is used, the retrieving processes run independently of each other, resulting in fast loading speeds; e.g. the loading time for 1000 genes is about 2 min in the default conditions.
Fig. 3.
The ‘Quick Info’ function of Cas-Database. The ‘Add to Collection’ function of Cas-Database. (A) Cas-Database provides a unique and easy way to select many genes at once by the implementation of a ‘cart’ system. Users can collect desired genes by clicking on the ‘Add to Collection’ button or by uploading a text file as described in the Figure 1C legend. Note that one can also select genes from different organisms. By clicking on the ‘Select optimal gRNAs’ button, users will proceed to the next step. (B) The results page will list all available sgRNAs filtered by the default conditions on the top panel. A colored indicator represents the selection status for each gene, e.g. green (selected completely), yellow (selected partially) or red (not selected at all). A user can download only the genes for which sgRNAs were successfully designed, and repeat the process for the remaining genes by changing the filter criteria and clicking on the ‘Remove “green” genes from list’ button
Whether enough sgRNAs have been selected for each gene after filtering is indicated by colored indicators—green (selected completely), yellow (selected partially) or red (not selected at all), as shown in Figure 3B. In this step, users can download either sequences for all selected sgRNAs targeting each gene or for some sgRNAs selected completely from each gene with a green indicator. In other words, users can download sequences only for genes for which sgRNAs were selected completely under the initial filtering conditions. After that, users can eliminate genes with green indicators from the list by clicking the ‘Remove green genes from list’ button, and then can alter the filtering conditions again. After resetting the filtering conditions, users can download the genes with green indicators again and repeat this process until sgRNAs have been selected completely in the most remained genes. Finally, if a few genes are still left, users can manually select targets for those genes using the ‘Quick info’ function on the main page. As a result, users can select optimal sgRNAs from hundreds or thousands of genes and download a list of sgRNAs for every gene with the target specific information.
3.4 Web API
For users familiar with bioinformatics, Cas-Database also provides a web application programming interface (web API). If users send queries through hypertext transfer protocol (HTTP) requests to our database server, the results will be returned in the JavaScript oriented notation (JSON) format. Thus, researchers can easily create their own simple homemade scripts for automated data retrieval. Details about the web interface are described in the Supplementary data.
3.5 Update of Cas-Database
The entire process of database creation, from retrieving an organism’s genome sequence and its associated annotation data from the ENSEMBL server to creating a new database, is totally automated by our homemade scripts. The time to build a new database depends on the organism’s genome size, e.g. the creation of a new database for zebrafish (1.32 GB) took about three days.
Currently, Cas-Database supports 12 different model organisms, including five vertebrates (human, rat, mouse, pig and zebrafish), one insect (D.melanogaster), one nematode (C.elegans) and five plants (A.thaliana, tomato, banana, grapes and soybean), and we are planning to update Cas-Database continuously to obtain the most recent versions of genomic sequences for the existing organisms. We also plan to add data from other organisms in the ENSEMBL database. In addition, we will update the database to allow for alternative CRISPR/Cas nucleases such as Cprf1 (Zetsche et al., 2015).
4 Conclusion
Cas-Database is an easy-to-use web-based tool for designing sgRNAs for SpCas9 nucleases on a genome scale. It can be applied to construct optimal sgRNA libraries that target thousands of coding sequences in 12 different organisms for genome-wide knockout screening experiments. Because Cas-Database contains all available targets in CDS regions as well as target-related information, including data about potential off-target sites, users can easily access the data through the interactive web interface or web API. The web interface was made using cutting-edge web development techniques such as AJAX, so the website is highly responsive to user input and the output results load quickly.
Supplementary Material
Funding
This work was supported by the Plant Molecular Breeding Center of Next Generation BioGreen 21 Program (PJ01119201) and the research fund of Hanyang University (HY-2015) to S.B. and Institute for Basic Science (IBS-R021-D1) to J.-S.K.
Conflict of Interest: none declared.
References
- Aach J. et al. (2014) CasFinder: flexible algorithm for identifying specific Cas9 targets in genomes. bioRxiv, 005074. [Google Scholar]
- Bae S. et al. (2014a) Cas-OFFinder: a fast and versatile algorithm that searches for potential off-target sites of Cas9 RNA-guided endonucleases. Bioinformatics, 30, 1473–1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bae S. et al. (2014b) Microhomology-based choice of Cas9 nuclease target sites. Nat. Methods, 11, 705–706. [DOI] [PubMed] [Google Scholar]
- Braglia P. et al. (2005) Sequence context effects on oligo(dT) termination signal recognition by Saccharomyces cerevisiae RNA polymerase III. J. Biol. Chem., 280, 19551–19562. [DOI] [PubMed] [Google Scholar]
- Chen S. et al. (2015) Genome-wide CRISPR screen in a mouse model of tumor growth and metastasis. Cell, 160, 1246–1260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cradick T.J. et al. (2014) COSMID: a web-based tool for identifying and validating CRISPR/Cas Off-target sites. Mol. Ther. Nucleic Acids, 3, e214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunningham F. et al. (2014) Ensembl 2015. Nucleic Acids Res., 43, D662–D669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doench J.G. et al. (2014) Rational design of highly active sgRNAs for CRISPR-Cas9-mediated gene inactivation. Nat. Biotechnol., 32, 1262–1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doudna J.A., Charpentier E. (2014) The new frontier of genome engineering with CRISPR-Cas9. Science, 346, 1258096.. [DOI] [PubMed] [Google Scholar]
- Gilbert L.A. et al. (2014) Genome-scale CRISPR-mediated control of gene repression and activation. Cell, 159, 647–661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hart T. et al. (2015) High-resolution CRISPR screens reveal fitness genes and genotype-specific cancer liabilities. Cell, 163, 1515–1526. [DOI] [PubMed] [Google Scholar]
- Heigwer F. et al. (2014) E-CRISP: fast CRISPR target site identification. Nat. Methods, 11, 122–123. [DOI] [PubMed] [Google Scholar]
- Hodgkins A. et al. (2015) WGE: A CRISPR database for genome engineering. Bioinformatics, 31, 3078–3080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu P.D. et al. (2013) DNA targeting specificity of RNA-guided Cas9 nucleases. Nat. Biotechnol., 31, 827–832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasprzyk A. (2011) BioMart: driving a paradigm change in biological data management. Database, doi:10.1093/database/bar049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasprzyk A. et al. (2004) EnsMart: a generic system for fast and flexible access to biological data. Genome Res., 14, 160–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H., Kim J.S. (2014) A guide to genome engineering with programmable nucleases. Nat. Rev. Genet., 15, 321–334. [DOI] [PubMed] [Google Scholar]
- Koike-Yusa H. et al. (2014) Genome-wide recessive genetic screening in mammalian cells with a lentiviral CRISPR-guide RNA library. Nat. Biotechnol., 32, 267–273. [DOI] [PubMed] [Google Scholar]
- Konermann S. et al. (2014) Genome-scale transcriptional activation by an engineered CRISPR-Cas9 complex. Nature, 517, 583–588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei Y. et al. (2014) CRISPR-P: a web tool for synthetic single-guide RNA design of CRISPR-system in plants. Mol. Plant, 7, 1494–1496. [DOI] [PubMed] [Google Scholar]
- MacPherson C.R., Scherf A. (2015) Flexible guide-RNA design for CRISPR applications using Protospacer Workbench. Nat. Biotechnol., 33, 805–806. [DOI] [PubMed] [Google Scholar]
- Montague T.G. et al. (2014) CHOPCHOP: a CRISPR/Cas9 and TALEN web tool for genome editing. Nucleic Acids Res., 42, W401–W407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naito Y. et al. (2015) CRISPRdirect: software for designing CRISPR/Cas guide RNA with reduced off-target sites. Bioinformatics, 31, 1120–1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Brien A., Bailey T.L. (2014) GT-Scan: identifying unique genomic targets. Bioinformatics, 30, 2673–2675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J. et al. (2015) Cas-Designer: a web-based tool for choice of CRISPR-Cas9 target sites. Bioinformatics, 31, 4014–4016. [DOI] [PubMed] [Google Scholar]
- Sander J.D. et al. (2010) ZiFiT (Zinc Finger Targeter): an updated zinc finger engineering tool. Nucleic Acids Res., 38, W462–W468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sander J.D., Joung J.K. (2014) CRISPR-Cas systems for editing, regulating and targeting genomes. Nat. Biotechnol., 32, 347–355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shalem O. et al. (2014) Genome-scale CRISPR-Cas9 knockout screening in human cells. Science, 343, 84–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shalem O. et al. (2015) High-throughput functional genomics using CRISPR–Cas9. Nat. Rev. Genet., 16, 299–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skinner M.E. et al. (2009) JBrowse: a next-generation genome browser. Genome Res., 19, 1630–1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Upadhyay S.K., Sharma S. (2014) SSFinder: High throughput CRISPR-Cas target sites prediction tool. Biomed Res. Int., 2014, 742482.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T. et al. (2014) Genetic screens in human cells using the CRISPR-Cas9 system. Science, 343, 80–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao A. et al. (2014) CasOT: a genome-wide Cas9/gRNA off-target searching tool. Bioinformatics, 30, 1180–1182. [DOI] [PubMed] [Google Scholar]
- Xie S. et al. (2014) sgRNAcas9: a software package for designing CRISPR sgRNA and evaluating potential off-target cleavage sites. PLoS One, 9, e100448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zetsche B. et al. (2015) Cpf1 is a single RNA-guided endonuclease of a class 2 CRISPR-Cas system. Cell, 163, 759–771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y. et al. (2014) High-throughput screening of a CRISPR/Cas9 library for functional genomics in human cells. Nature, 509, 487–491. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.