Abstract
Purpose
Disseminated tumor cells (DTCs) detected in the bone marrow of breast cancer patients identifies women at high risk of recurrence. DTCs are traditionally detected by immunocytochemical staining for cytokeratins or single gene expression measurements, which limit both specificity and sensitivity. We evaluated the Nanostring nCounter™ (NC) platform for multi-marker, gene expression-based detection and classification of DTCs in the bone marrow of breast cancer patients.
Experimental Design
Candidate genes exhibiting tumor cell specific expression were identified from microarray data sets and validated by qRT-PCR analysis in non-malignant human BM and identical samples spiked with predefined numbers of molecularly diverse breast tumor cell lines. Thirty-eight validated transcripts were designed for the nCounter™ platform and a subset of these transcripts was technically validated against qRT-PCR measurements using identical spiked bone marrow controls. Bilateral iliac crest bone marrow aspirates were collected and analyzed from twenty breast cancer patients, prior to neoadjuvant therapy, using the full 38 gene nCounter™ code set.
Results
Tumor cell specific gene expression by nCounter™ was detected with a sensitivity of one cancer cell per 1×106 nucleated bone marrow cells after optimization. Measurements were quantitative, log linear over a twenty-fold range, and correlated with qRT-PCR measurements. Using the nCounter™ 38-gene panel, 6 of 8 patients (75%) who developed metastatic disease had detectable expression of at least one transcript. Notably, three of these patients had detectable expression of ERBB2 in their bone marrow, despite the fact that their corresponding primary tumors were HER2/ERBB2 negative and therefore did not receive trastuzumab therapy. Four of these patients also expressed the PTCH1 receptor, a newly recognized therapeutic target based on hedgehog signaling pathway inhibition.
Conclusions
The presumptive detection and classification of DTCs in the bone marrow of breast cancer patients, based on sensitive and quantitative, multi-marker detection of gene expression using the nCounter™ platform provides an opportunity to both predict early distant recurrence and more importantly, identify opportunities for preventing the spread of disease based on the expression of unique, therapeutically actionable gene targets.
Translational relevance
This study demonstrates the application of a new technology for multiplexed gene expression-based detection of disseminated tumor cells in the bone marrow of breast cancer patients, and identifies at least two therapeutically targetable genes that are frequently expressed in BM of patients who develop metastatic disease.
Keywords: breast cancer, disseminated tumor cells, gene expression, Nanostring nCounter
Introduction
Approximately 40,000 women die from breast cancer each year as a consequence of metastatic disease development. Data suggests that only a small, unique subset of cells within a primary breast tumor (less than 1%) possess metastatic potential [1,2]. Furthermore, molecular profiles of primary, heterogeneous tumors may demonstrate limited resemblance to those tumor cells that eventually progress to metastatic foci [3]. Clinically, therapies that reduce primary tumor mass by simply eliminating proliferating cells often fail to cure patients, particularly with respect to metastatic disease [4]. To develop new therapeutic strategies to control and monitor distant disease development, it is essential to identify and target the intermediary cells in the metastatic process since these cells likely have biological behavior and therapeutic vulnerabilities which differ from the primary tumor [5].
Disseminated tumor cells (DTCs) can be detected in the bone marrow (BM) of 30-40% of early stage breast cancer patients[6]. Several large multi-institutional clinical trials have documented that the presence of BM DTCs at the time of diagnosis identifies women at high risk of recurrence. Those women with detectable DTC after completion of all adjuvant therapy are at higher risk of recurrent disease development than those women with no detectable DTC in their BM[7,8]. The proposed biological basis for this clinical observation is that BM serves as a reservoir that allows DTCs to adapt and disseminate to other organs[9]. Alternatively, it is possible that clinically detected BM DTCs may be a marker of body-wide dissemination of tumor cells rather than the body's only long-term reservoir[10-12]. Regardless, DTC detection allows the identification of women at high risk of recurrence and the ability to monitor response in those women with clinically undetectable disease.
Although the detection of DTCs in breast cancer is still investigational, its incorporation into clinical management of breast cancer patients is currently the focus of much research. Standardized guidelines have been published for the immnocytological (ICC) detection of DTCs[13], which is the current gold standard and has a reported sensitivity ranging from 1 DTC in 105 -106 leukocytes[14]. Antibodies against epithelial-specific antigens such as cytokeratins (CK) or EpCAM, surface adhesion molecules, or growth factor receptors have been used for the detection of carcinoma cells due to the absence of more tumor-specific target antigens[15]. Other studies have assessed DTC detection using assays that detect single epithelial- or organ-specific transcripts such as cytokeratin 19 (KRT19), MUC1[16], urokinase-type plasminogen activator receptor (uPAR) [17], EpCAM (TACSTD1), and mammaglobin (SCGB2A2)[18-23]. Expression of SCGB2A2, KRT19, and TWIST1 in the BM of breast cancer patients have individually demonstrated prognostic significance[24-27]. However, because individual gene expression is not always absolutely tumor-specific, false positive results can be observed due to low level transcription of epithelial or breast tissue-specific genes in normal BM cells [28,29]. Conversely, given the known molecular heterogeneity of DTCs, it is unlikely that a single gene marker will be suitable for the detection of DTCs in all breast cancer patients. For example, KRT19 is not expressed by all DTCs; its expression is often lost in breast cancer cells during tumor progression and epithelial to mesenchymal transition (EMT) [30,31].
A multi-marker assay approach has the potential to overcome some of the above concerns by allowing the simultaneous, composite detection of multiple genes in a single sample [32-34,26]. Analysis of DTC-associated molecular profiles could improve both sensitivity and specificity of DTC detection itself, and also provide prognostic or therapeutically predictive information for patients as well. The Nanostring nCounter™ (NC) assay platform allows for the simultaneous, quantitative detection of multiple (up to 600) nucleic acid targets in a single sample with a sensitivity and reproducibility comparable to qRT-PCR [35,36]. Accordingly, we have mined existing gene expression microarray data to identify and validate gene transcripts whose expression is specifically associated with breast tumor cells and absent in normal bone marrow cellular constituents. We have adopted a panel of 38 of these genes to the NC platform, and in this study, demonstrate how the assay can be used with sufficient sensitivity to profile clinical BM samples from breast cancer patients to detect and potentially classify rare BM-associated DTCs. In a cohort of twenty patients, eight of whom had distant metastatic recurrence within 48 months, composite patterns of gene expression identified patients more likely to develop metastatic disease and who might be candidates for targeted therapeutics directed to both ERBB2 and PTCH1 gene expression.
Methods
Study Population
Control bone marrow specimens were collected from healthy female volunteers after informed consent. DTC-positive bone marrow previously assessed by immunohistochemistry was obtained retrospectively from a biospecimen collection associated with a previously reported therapeutic trial[37]. Specimens for the test clinical cohort were also obtained retrospectively from an independent, ongoing IRB approved bone marrow and tissue banking breast cancer protocol at our institution. All patient bone marrow aspirates were collected prior to the initiation of any therapy. An overview of clinical samples used in this study is shown in Supplemental Table 1.
Bone Marrow Aspiration, Processing, and RNA Isolation
For all breast cancer cases, twenty ml of bone marrow was collected from the right and left anterior iliac crests, subjected to hypotonic RBC lysis, washing, and nucleated cell counting within two hours from the time of collection. For controls, bone marrow was harvested unilaterally. For whole bone marrow analysis, 5×106 nucleated cells were pelleted and immediately snap frozen for subsequent RNA isolation. For mononuclear cell enrichment, whole bone marrow was diluted five-fold in RPMI media and processed using a standard Ficoll-Hypaque gradient technique. The enriched mononuclear cell fraction was then counted and 5×106 mononuclear cells were pelleted and snap frozen for subsequent RNA isolation. SKBR3 (ER-/Her2+), MDA/MB231 (ER-/Her2-), and ZR75 (ER+/Her2-) human breast cancer cells were trypsinized from plates during log growth phase, washed, counted, diluted into the prepared nucleated cell fraction of normal human BM samples at incremental five-fold dilutions from 1:1000 to 1:500,000 tumor cells per nucleated cell count, pelleted, and snap frozen for subsequent RNA isolation. All RNA isolations were performed from snap frozen cell pellets using Trizol reagent (Invitrogen) as previously described[27]. RNA was quantified by A260 using a Nanodrop spectrophotometer and qualitatively assessed using an Agilent bioanalyzer. All RNAs used for qRT-PCR or NC analysis demonstrated a RIN score of > 7.0.
qRT-PCR Analysis
SyBr green or TaqMan Assay-on-Demand primer/probe sets for indicated transcripts were purchased from Applied Biosystems. Primer/probe sequences are shown in Supplemental Table 2. One microgram of total RNA was reverse-transcribed and 1/100th of the cDNA template was used in each replicate qPCR reactions. Each reaction consisted of 1ul of cDNA, TaqMan Master Mix (Applied Biosystems), and 5× primer/probe set in a total volume of 20ul, following the manufacturer's standard protocol. For each transcript/sample, duplicate or triplicate reactions were performed in an ABI 7500 FAST Sequence Detection System. Replicated sample assays with a cycle threshold (CT) value difference of greater than 1.5 (outliers) were excluded from analysis. Each run with a gene primer set was normalized to CT values of GAPDH primer set on the same cDNA sample. This value was in turn normalized across the entire sample set by using the average delta CT from an identical analysis using a set of four normal volunteer BM specimens, using the ddCT method [38]. In samples where a specific transcript was not detected (CT>40), the fold difference in expression was defined as 1.0 (equal to that of the normal bone BM average).
Nanostring nCounter Assay
The nCounter assay was performed with the 38 gene codeset as previously described8 with the exception that input RNA was increased from 0.5 ug to 5 ug to increase the sensitivity for detecting transcripts expressed only by rare DTC populations. Eleven positive and nine negative controls spikes were used to calibrate the assay. Mean hybridization counts of the negative controls were subtracted from all other transcript counts to correct for background[34]. Patient samples were run in duplicate or triplicate and the average value taken for analysis. Reference values for each gene were calculated from cohorts of 8 and 11 bone marrow samples from healthy female volunteers. All expression values were normalized based on the average expression of three internal control transcripts (TBP, RPR0, POL2RA). For binary analysis, a sample was considered positive for gene expression if hybridization counts were 2 standard deviations above the mean of the healthy control bone marrow population.
Statistical Methods
Spearman's rank correlation coefficient was calculated on PCR and NC data after removing values at the extreme of measurement. Coefficient of variation was calculated on normal bone marrow specimens to assess the variance within controls. Assessment of variance within patient samples was carried out treating differences between patient samples as signal and differences within patient samples as noise.
Results
Target Gene Selection
To identify gene transcripts that could potentially serve as specific markers for DTCs in the context of patient bone marrow, we analyzed gene expression microarray data from 80 primary breast tumors, 79 serial bone marrow samples with and without known DTC involvement (based on immunocytochemistry) from 25 breast cancer patients, 5 bone marrow samples from healthy volunteers, as well as additional breast tumor data sets in literature[39,27,40]. Comparative expression analysis of these data sets identified genes that had detectable microarray-based expression in either primary tumors and/or ICC-positive BM specimens, but were not detectable in similar data sets from healthy volunteer bone marrow or in ICC-negative breast cancer patient bone marrow. A total of sixty-nine genes met these criteria (Supplemental Table 3). To further validate their tumor-specific expression, each gene was quantified by qRT-PCR using a set of healthy volunteer bone marrow samples and three exemplar breast tumor cell lines of differing molecular subtypes: SKBR3 (HER2+), ZR75 (ER+), or MDAMB231 (HER2-/ER-). Of the 69 genes chosen, 27 genes met the criteria of undetectable or nearly undetectable expression (defined as Ct > 37) in healthy volunteer BM and a minimum of twenty-fold (range 20-18,000) higher expression in at least one of three breast cancer cell lines relative to the healthy control population. Eleven genes (CCND1, ESR1, FGFR4, Gli2, Gli3, GSC, IGFBP4, PDGFRB, PTCH1, SMO, SNAI1) were expressed at less than twenty fold over that in healthy BM but were included in the panel due to their biological and/or clinical relevance (Supplemental Table 4). Figure 1 lists the genes that comprise the full 38-gene panel, along with their biological process associations.
Figure 1.
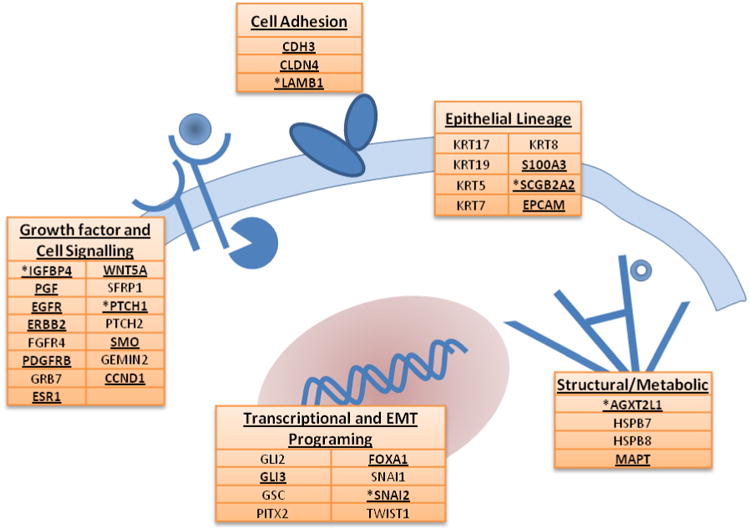
Genes represented in the 38-gene DTC panel and their biological pathway associations. Transcripts detected in the twenty-patient pilot cohort are highlighted in bold underline and those associated with early metastatic recurrence are denoted by asterisks.
nCounter Performance Comparison
Transcript expression analysis by qualitative or semi-quantitative RT-PCR remains the most documented approach for molecular detection of breast cancer DTCs, primarily because of the requisite sensitivity that amplification methods can provide. To determine whether the non-amplification based nCounter platform could have sufficient sensitivity to detect gene expression from rare DTCs amidst a high background of normal bone marrow constituents, we examined the expression of a subset of nine genes (Twist1[27], SNAI2[41], EPCAM[42,43], SCGB2A2[44], EGFR[45], PITX2[46], S100A3[27], KRT17, and KRT19[47]) from the 38 gene panel in the three exemplar breast cancer cell lines serially diluted into normal human BM. As shown in Table 1 and Figure 2, expression measurements were quantitative and varied over a 20-fold linear dynamic range. Consistent with previous reports [48,49], not all of the nine genes were detected in each of the cell lines, demonstrating the necessity of a multi-gene marker panel for detecting DTC expression from cells of varying molecular phenotypes. Although KRT19 transcript was detected in BM samples spiked with each of the cell lines, each cell line demonstrated a unique pattern of expression of one or more genes.
Table 1.
Nanostring nCounter detection of nine DTC-associated transcripts in three different breast tumor cell lines spiked into healthy donor bone marrow at defined cell number dilutions. Numbers indicate total hybridization counts for each transcript in each sample, after subtracting transcript counts from those obtained from healthy bone marrow controls. Transcripts demonstrating detectable expression proportional with dilution of each cell line are indicated in bold. Low hybridization counts that are constant despite changing tumor cell dilutions, even after background subtraction, are attributed to signal from non-malignant bone marrow cellular constituents.
| Cell Line | SKBR3 | MDA-MB231 | ZR75 | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Dilution | 1×10-5 | 2×10-4 | 5×10-3 | 1×10-5 | 2×10-4 | 5×10-3 | 1×10-3 | 1×10-5 | 2×10-4 | 5×10-3 | 1×10-3 |
| KRT17 | - | - | - | - | - | - | - | - | - | - | - |
| KRT19 | 51 | 424 | 1,408 | - | 42 | 202 | 1,588 | 90 | 400 | 1,252 | 5,580 |
| PITX2 | - | - | - | - | - | - | - | - | - | - | - |
| S100A3 | 13 | 5 | 13 | 8 | 14 | 25 | 122 | 9 | 3 | 7 | 5 |
| SCGB2A2 | 0 | 2 | 5 | - | - | - | - | - | 7 | 23 | 112 |
| SNAI2 | 4 | 4 | 7 | 8 | 20 | 49 | 350 | 11 | 3 | 3 | 12 |
| EPCAM | 38 | 20 | 61 | 9 | 19 | 6 | 32 | 3 | 37 | 169 | 761 |
| TWIST1 | 10 | - | 2 | - | 8 | - | 3 | 4 | 7 | 11 | 2 |
| EGFR | 7 | 14 | - | 2 | 26 | 108 | 783 | 14 | 25 | 15 | 49 |
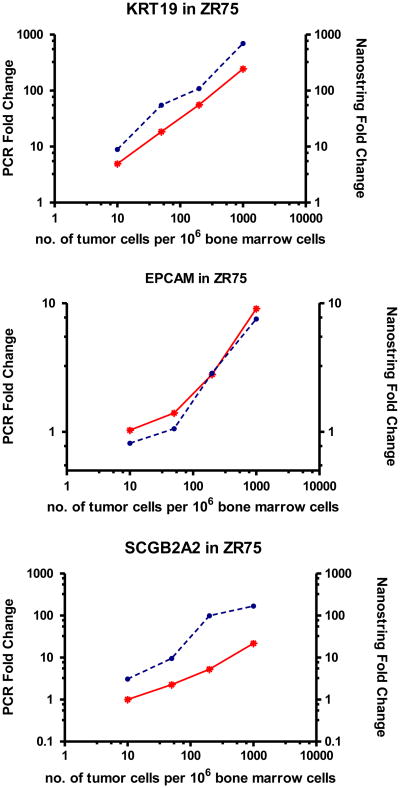
Figure 2.
nCounter and qRT-PCR detection of gene expression in control bone marrow spiked with increasing numbers of ZR75 breast tumor cells. Units on the right axis show the fold-difference of nCounter signal counts (red line) relative to normal bone marrow background. Units on left axis show fold-difference of expression (calculated by ddCT method) relative to normal bone marrow using qRT-PCR (blue dotted line). Axis are plotted in log scale.
In parallel, the same sample dilution series were used for single gene qRT-PCR expression analysis. For those genes with detectable expression by nCounter assay, expression levels measured by qRT-PCR and nCounter demonstrated good correlation over the full dynamic range (Figure 2). Spearman's rank correlation coefficient between qRT-PCR (expressed as fold increase over expression in non-malignant bone marrow) and nCounter (expressed as hybridization counts over that from non-malignant bone marrow) for KRT19, S100A3, SCGB2A2, TACSTD1, and TWIST1 ranged from .80 to .99 in those cell lines that expressed the individual genes.
Using 0.5 ug of input RNA, KRT19 expression could be detected at a level of one cancer cell per 100,000 nucleated BM cells. However, the limits of reported detection using conventional immunocytochemistry have been reported at one cancer cell per 1 million nucleated bone marrow cells, theoretically ten-times more sensitive[50]. Therefore, to increase the sensitivity of the assay, we modified the standard protocol to perform both mononuclear cell enrichment (a standard procedure employed for ICC analysis) and increased input RNA to five micrograms, a quantity of analyte that is still readily obtainable from a routine bone marrow aspiration. Mononuclear cell enrichment increased expression signal levels by approximately two fold while, as expected, a ten fold increase in RNA input increased expression signals proportionally by ten fold as well (Supplemental Table 5). Not surprisingly, some transcripts that previously demonstrated nominal expression in normal bone marrow cell constituents (SNAI2, TACSTD1, S100A3, KRT17) had higher background expression using the modified protocol, while background expression of several other transcripts (KRT19, EGFR, TWIST1, SCGB2A2, PITX2) was unaltered by the protocol modification. Importantly, the protocol modification allowed us to achieve a theoretical sensitivity of one DTC per one million nucleated BM cells, the generally accepted limit of sensitivity observed with the ‘gold-standard’ ICC detection.
Analysis of Bone Marrow from Breast Cancer Patients
To demonstrate the clinical applicability of this method, the nCounter nine-gene panel was first tested in five bone marrow samples collected from patients with stage II/III breast cancer prior to any treatment, who had previously documented DTCs in their bone marrow by ICC (Table 2). A ‘normal range’ for bone marrow expression of each gene was established by analyzing bone marrow from eight female healthy volunteers. Intra-class correlation coefficient which compares within patient variance to total noise variance was calculated for the nine genes and this ranged from .622 to .909. At least one gene transcript was detected above background bone marrow signal in four of the five breast cancer bone marrow samples. Although, from this small sample set, a composite pattern of expression based on primary tumor molecular phenotype was not obvious, two ER-/Her2- breast cancer cases had detectable bone marrow expression of several genes known to be expressed in this subtype of primary breast cancer (EPCAM, SNAI2, KRT17). Interestingly, KRT19 gene expression, a molecular marker often used for detection of DTCs, was detected in only one of the five patient specimens.
Table 2.
nCounter signal detection of DTC-associated transcripts in BM specimens from five breast cancer patients with previously documented DTCs by ICC, who also developed metastatic disease. Molecular phenotype and clinical characteristics of the five tumors are shown, including the number of DTCs bone marrow cells detected by immunocytochemistry (ICC) per two million mononucleated cells. Mean signal counts from triplicate assays for each gene transcript in each BM sample are shown with the standard deviation from triplicate samples; values which exceed two standard deviations above the mean of eight bone marrow samples from healthy individuals are in bold. Note that EPCAM and SCGB2A2 expression in sample 7547 approached two standard deviations above the reference range, but did not exceed this threshold.
| Pt ID | Control BM | 2904 | 9645 | 9290 | 7547 | 6335 |
|---|---|---|---|---|---|---|
| ER | -- | neg | neg | pos | pos | neg |
| PR | -- | neg | neg | pos | pos | neg |
| Her-2 | -- | neg | neg | neg | neg | pos |
| DTCs by ICC | -- | 2 | 4 | 3 | 4 | 10 |
| Months to recurrence | -- | 15 | 12 | 48 | 15 | 22 |
| Metastatic site | -- | Liver | Lung | Bone | Ovary | Liver, Bone |
| TWIST1 | 13 ± 8 | 8 ± 9 | 6 ± 2 | 10 ± 4 | 2 ± 1 | 68 ± 13 |
| SNAI2 | 32 ± 40 | 18 ± 19 | 315 ± 10 | 66 ± 11 | 55 ± 9 | 31 ± 3 |
| EPCAM | 293 ± 134 | 694 ± 283 | 213 ±13 | 446 ± 70 | 397 ± 12 | 178 ± 57 |
| SCGB2A2 | 5 ± 15 | 1 ± 1 | 1 ± 1 | 1 ± 1 | 15 ± 5 | 1 ± 1 |
| EGFR | 21 ± 12 | 11 ± 14 | 27 ± 3 | 28 ± 6 | 17 ± 4 | 100 ± 21 |
| S100A3 | 32 ± 13 | 57 ± 18 | 28 ± 1 | 78 ± 20 | 37 ± 10 | 38 ± 11 |
| PITX2 | 10 ± 8 | 4 ± 2 | 5 ± 0 | 10 ± 6 | 1 ± 2 | 105 ± 13 |
| KRT17 | 91 ± 48 | 48 ± 40 | 258 ± 25 | 101 ± 15 | 27 ± 7 | 118 ± 29 |
| KRT19 | 34 ± 19 | 22 ± 14 | 19 ± 5 | 42 ± 5 | 9 ± 2 | 139 ± 18 |
Having documented performance characteristics of the assay and applicability to primary bone marrow samples from breast cancer patients with known DTCs, we analyzed retrospectively collected bilateral bone marrow from an independent cohort of twenty clinical stage II/III breast cancer patients and an additional reference set of 11 healthy female volunteers, using the entire 38 gene panel. Eight of twenty patients developed metastatic disease within 2-48 months of primary diagnosis (mean 23 months), while twelve had no evidence of metastatic disease at 3-5 years of follow-up. As shown in Figure 3, 20 transcripts of the 38 gene panel were expressed above background in at least one bone marrow sample, while at least one transcript was expressed above background in 17 patients (85%). None of the bone marrow specimens in this cohort had detectable expression of KRT19 and only four patients (20%) had detectable expression of EPCAM, two genes commonly used for the molecular detection or selection of DTCs. Most patients demonstrated unique patterns of gene expression in bone marrow. However, with only a few notable exceptions (Figure 3, patient 3709) patterns of gene expression were highly concordant between right and left bone marrow samples from the same patient in 11 of 20 cases.
Figure 3. Unsupervised hierarchical clustering based on a 38-gene expression profile in bone marrow samples from breast cancer patients.
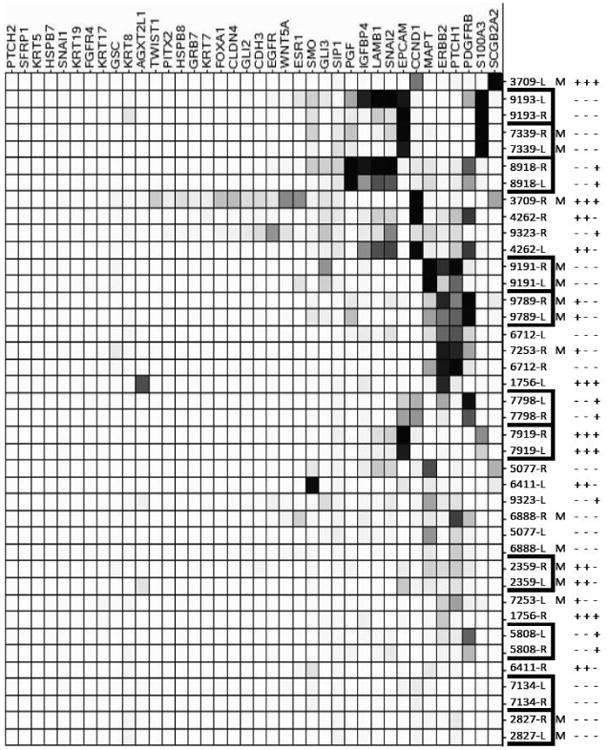
Each row represents an individual sample (either left- L or right-R sampling) from 20 individual patients with a known breast tumor molecular phenotype (ER, PR, Her2 indicated with +/-). Eight patients developed metastatic disease (denoted by M) within 5 years of diagnosis. Each column represents relative expression of each gene in each sample. Expression is scaled based on number of SD above the mean of a set of 11 health control bone marrow samples analyzed in the same assay. Gene transcripts and samples are ordered and grouped based upon similar patterns of expression. Corresponding values for normalized hybridization counts for differentially expressed genes are shown in Supplemental Table 7.
Unsupervised, hierarchical clustering of expression data demonstrated no obvious subclassification of cases, particularly with regard to molecular phenotype of the primary tumor or the occurrence of eventual metastases. Although 6 of the 8 patients (75%) who developed metastatic disease had detectable expression of at least one of the 38 genes in their bone marrow, patients who remained disease-free had a mean number of detectable transcripts that was not significantly different than that of patients with metastatic relapse. However, at the level of individual gene analysis, two transcripts (SNAIL2 and LAMB1) demonstrated a significantly different level of expression in bone marrow from patients who relapsed versus those that remained disease-free (p = .0013 and .0014 respectively with correction for multiple comparisons). One additional transcript from the 38-gene signature (IGFBP4) also demonstrated expression differences that were close to significance between disease free and metastatic patient populations (p=.058). Notably, several genes that are known targets for therapeutic intervention: ERBB2, PTCH1, and PDGFRB, were also expressed in the bone marrow of patients who eventually developed metastatic disease. Of the eight patients with metastatic relapse, one or more of these therapeutic targets were expressed in four patients (50%). Four of six patients with PTCH1 positive marrow eventually developed metastatic disease and therefore, could have potentially benefited from adjuvant therapy directed to gene in the Hedgehog pathway. Three of four patients (Figure 4) with HER2 negative tumors and ERBB2 positive DTC eventually developed metastasis, while none of the patients with HER2 positive tumors and ERBB2 positive bone marrow who received therapy with traztuzumab developed metastasis. For this set of patients, there was good concordance between detection of selected genes by PCR and NS (Supplemental Table 7)
Figure 4.
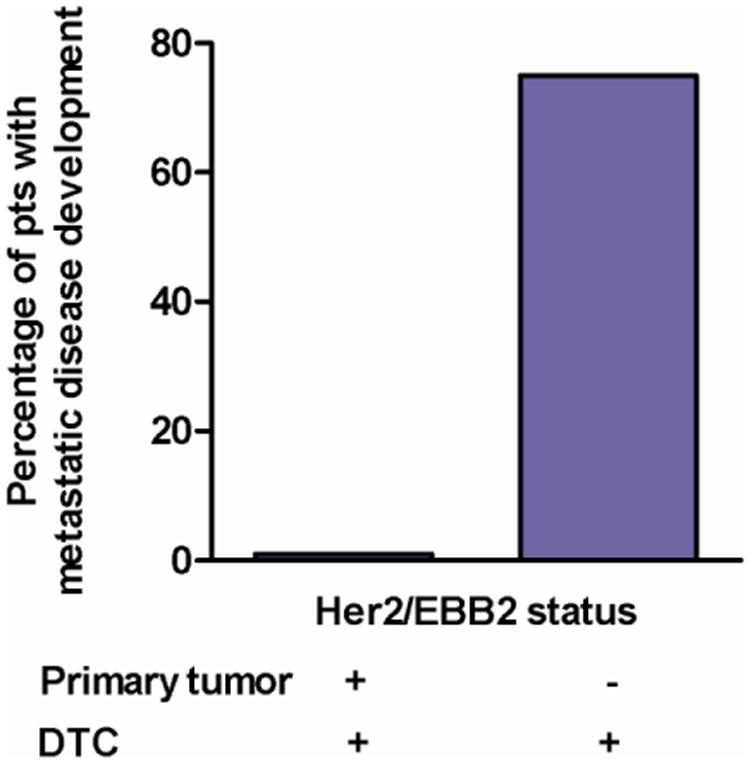
Recurrent disease development in patients with ERBB2-positive DTCs. Patients with Her2 positive tumors received chemotherapy with trastuzumab. Patients with Her2-negative tumors received cytotoxic chemotherapy alone. All patients had ERBB2-positive DTCs. All recurrences detected within 24 months (mean=19) of diagnosis.
Discussion
The presence of DTCs in the bone marrow of breast cancer patient has important prognostic and therapeutic implications. Adjuvant therapy administration is generally based on the primary tumor characteristics and lymph node status. However, those women with no detectable disease in their lymph nodes or bone marrow remain at very low risk of recurrence and may not benefit from adjuvant treatment[51]. Conversely, patients who have completed adjuvant therapy, but retain residual micro-metastatic disease could benefit from further therapy. DTCs are heterogeneous [52,53], can persist for years and remain a predictor for disease recurrence[54], may exhibit a stem cell-like chemotherapy resistant phenotype, [55] and likely have varying metastatic capabilities. Patients with detectable DTCs in their BM after chemotherapy treatment have a particularly poor prognosis indicating that chemotherapy does not eliminate all DTCs and that those subpopulations of DTCs which survive cytotoxic chemotherapy have a high metastatic potential[56]. Persistence of DTCs in BM years after diagnosis and initial therapy has been shown to be an indicator of subsequent systemic treatment failure [7,8,32]. Persistence or disappearance of DTCs after systemic treatment could therefore be used as a surrogate marker of treatment response [57]. Assays that detect and molecularly classify residual micro-metastatic disease could greatly benefit and guide the treatment of patients.
Currently the ‘gold ‘standard’ for DTC detection is ICC which is laborious and often limited in sensitivity and specificity given the heterogeneity of DTCs. Molecular assays based on detection of single, ‘epithelial specific’ transcripts such as EPCAM or KRT19 often perform no better than ICC based methods[58]. Given the known heterogeneity of DTCs, multiplexed biomarkers could provide improved sensitivity and specificity for detection and classification, but the ability to effectively multiplex multiple markers in a single reaction while maintaining high specificity and sensitivity for each marker in clinical specimens has been limited and few studies have been published regarding the genomic profiling of DTCs[14]. A major challenge lies in identifying a panel of genes that are biologically relevant, representative of all possible breast cancer molecular subtypes, and has very absent or minimal expression in non-malignant bone marrow constituents. Thus, a gene such as CD44, whose expression is related specifically to the breast cancer stem cell phenotype and metastases, is not useful in this assay since it is expressed in multiple hemapoietic cells[58]
The Nanostring nCounter platform provides an attractive approach for performing multiplexed gene expression assays in a single reaction and can quantitatively detect expression of up to 600 gene transcripts in a single reaction with similar sensitivity and reproducibility as amplification based methods34. Most importantly, the assay does not require high quality RNA and the direct readout of transcript copy number provides a higher precision for detecting gene expression originating from minor cell populations. In fact, we were surprised to find in our validation studies that the assay could detect minimally to moderately abundant transcripts specifically emanating from as few as one tumor cell in a background of one million bone marrow cells, comparable to current ICC detection methods. This sensitivity was facilitated by enrichment of the mononuclear cell fraction of bone marrow, increasing assay RNA input by ten-fold without signal saturation, and careful selection of transcripts which demonstrated absent or minimal expression in bone marrow cell populations.
Despite selecting target transcripts which were expressed in one or more of three archetypical breast tumor cell lines of varying molecular phenotype, several of the genes in our 38-gene panel were not detected in the twenty cases of primary bone marrow from breast cancer patients analyzed in this study. It is possible that the expression of many of these genes is restricted to tumor cell lines or that our patient population in this study was too small to fully sample the diversity of DTC molecular phenotypes. For example we found that 1 of 5 samples in our first validation cohort was TWIST1 expression positive, but none of the twenty samples in the second cohort had detectable levels of expression above background. In this regard, the flexibility of easily introducing or removing transcript probes in an iterative design approach for the nCounter assay is proved particularly useful for the current study and for future studies will allow the easy introduction of new targetable genes.
Conversely, we found that 65% of patients in our patient cohort demonstrated expression of at least one transcript in at least one of their bilateral bone marrow specimens. This far exceeds previously reported positivity based upon ICC detection of DTCs and clearly, many patients with a positive gene signature remained relapse free for more than five years. There are several possible explanations for this result. First, although gene expression levels were referenced to a population of healthy female controls, it is possible that the reference population size was not sufficiently large to fully capture inter-individual variability that may exist in healthy bone marrow. Second, since it is not possible to specifically equate expression from the 38-gene panel with the presence of tumor cells, gene expression in some patients could originate from rare bone marrow cell elements specifically present in the cancer patient population. Such a cancer-specific, ‘stromal response’ could still be biologically and clinically significant, even if it is not indicative of actual DTCs residing in the bone marrow. Finally, it is possible that 65% of our population do actually harbor DTCs in their bone marrow. Detection by cytokeratin ICC may underestimate the percentage of DTC positive patients, particularly for those DTCs that have undergone epithelial to mesenchymal transition and concomitant loss of cytokeratin expression [60]. Since only 6 of 17 patients (30%) with positive gene expression developed distant metastases within five years, it is possible that some bone marrow DTCs may have attenuated metastatic potential that can be directly related to their individual expression profiles.
In this regard, it was striking to see that patterns of DTC expression demonstrated such variability between patients. This cannot likely be attributed to biological noise since bilateral bone marrow aspirates demonstrated, except in a few cases, high concordance in expression patterns. In fact, it might be expected that in some patients, due to sampling bias, bilateral samplings may not be completely concordant. The molecular diversity of gene expression observed in this small subset of patients as well the biological processes that cancer cells undergo during their transition in the hematological system underscores the need for a multiplexed gene expression panel. Furthermore, patterns of DTC gene expression did not appear to be directly related to the molecular phenotype of the primary tumor. In fact, several tumors that were Her2 negative corresponded to patient bone marrow samples with ERBB2 expression, while two of the six patients with Her2 positive tumors had detectable expression of ERBB2 in their bone marrow, and were also positive for other transcripts. Although the composite 38-gene expression pattern did not immediately classify patients based upon disease relapse and in fact, more than half of patients with a transcript positive bone marrow did not relapse, we identified several individual genes within the 38-gene signature that did reach a significant correlation with disease relapse- LAMB1, and SNAI2. In fact, SNAI2 expression was inversely correlated with disease relapse suggesting that expression patterns may be related to ‘favorable’ as well as ‘unfavorable’ DTC molecular phenotypes.
Beyond providing prognostic information, several genes in the 38-gene panel are also relevant therapeutic targets- ERBB2, EGFR, ESR1, PDGFB, PTCH1- and were expressed in patient bone marrow specimens, often in non-overlapping patterns. This finding, together with the fact that expression of individual target genes such as ERBB2 are often expressed discordantly in primary tumors and corresponding DTCs, suggests a utility for using DTC molecular profiles to ‘personalize’ adjuvant therapy in a prospective clinical trial setting. Although all specimens analyzed in the current study were collected only prior to therapy, this assay could also be applied to monitoring therapeutic efficacy in the adjuvant setting for those patients without radiographic or clinical evidence of disease. Bone marrow aspirations for DTCs performed before and following therapy and at times points during follow-up could lead to real-time assessments of therapeutic efficacy, and identify those patients in need of continued therapy. Subsets of DTCs that persist or are eliminated by therapy could be monitored and targeted. This type of assay platform for clinical samples could facilitate translation of multi-gene biomarkers into the clinic, identify women at high risk of breast cancer recurrence and provide guidance on tailored therapies based on the molecular profile of micro-metastatic breast tumor cells.
Supplementary Material
Supplemental Table 1. Clinical characteristics of the specimens analyzed
Supplemental Table 2. PCR Primer Probes for 38-gene panel
Supplemental Table 3. Genes screened for the nCounter assay panel which are expressed in breast cancer cells and demonstrate absent or low expression in normal bone marrow.
Supplemental Table 4. Realtive Gene expression* in Breast cancer cell lines compared to normal bone marrow
Supplemental Table 5. Enhanced nCounter signal detection of DTC-associated transcripts using increased input RNA and Ficoll mononuclear cell enrichment. A 1:1 mixture of MDA-MB231 and ZR75 breast tumor cells were spiked into normal volunteer control BM at a dilution of 1×10-4 tumor cells per nucleated BM cells. Cells were either used for direct RNA isolation or first enriched for the mononuclear cell fraction prior to RNA isolation. The indicated quantity of resulting RNA was used for the 9 gene nCounter assay. Numbers indicate total hybridization counts from each sample after subtraction of background signal from normal bone marrow controls and absolute hybridization counts of normal bone marrow controls themselves.
Supplemental Table 6. Comparison of sample analysis by NS and PCR
Supplemental Table 7. Normalized Nanostring hybridization counts* for patient Bone marrow specimens
Acknowledgments
We thank Philippa Webster, Jeannette Nussbaum, Paul Rasmussen and Malini Maysuria at Nanostring for their assistance with this project. This study was supported by a grant from the Komen Foundation.
Footnotes
Conflict of Interest: None of the authors have any conflict of interest to report including related honoraria, investments, or research funding to report.
Disclosure: None of the authors have any disclosures or conflicts of interest.
References
- 1.Sahai E. Illuminating the metastatic process. Nat Rev Cancer. 2007;7(10):737–749. doi: 10.1038/nrc2229. [DOI] [PubMed] [Google Scholar]
- 2.Weigelt B, Peterse JL, van't Veer LJ. Breast cancer metastasis: markers and models. Nat Rev Cancer. 2005;5(8):591–602. doi: 10.1038/nrc1670. [DOI] [PubMed] [Google Scholar]
- 3.Prat A, Perou CM. Deconstructing the molecular portraits of breast cancer. Mol Oncol. 2011;5(1):5–23. doi: 10.1016/j.molonc.2010.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sorlie T, Perou CM, Tibshirani R, Aas T, Geisler S, Johnsen H, Hastie T, Eisen MB, van de Rijn M, Jeffrey SS, Thorsen T, Quist H, Matese JC, Brown PO, Botstein D, Lonning PE, Borresen-Dale AL. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc Natl Acad Sci U S A. 2001;98(19):10869–10874. doi: 10.1073/pnas.191367098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lin H, Balic M, Zheng S, Datar R, Cote RJ. Disseminated and circulating tumor cells: Role in effective cancer management. Crit Rev Oncol Hematol. 2011;77(1):1–11. doi: 10.1016/j.critrevonc.2010.04.008. [DOI] [PubMed] [Google Scholar]
- 6.Braun S, Pantel K, Muller P, Janni W, Hepp F, Kentenich CR, Gastroph S, Wischnik A, Dimpfl T, Kindermann G, Riethmuller G, Schlimok G. Cytokeratin-positive cells in the bone marrow and survival of patients with stage I, II, or III breast cancer. N Engl J Med. 2000;342(8):525–533. doi: 10.1056/NEJM200002243420801. doi:MJBA-420801[pii] [DOI] [PubMed] [Google Scholar]
- 7.Janni W, Rack B, Schindlbeck C, Strobl B, Rjosk D, Braun S, Sommer H, Pantel K, Gerber B, Friese K. The persistence of isolated tumor cells in bone marrow from patients with breast carcinoma predicts an increased risk for recurrence. Cancer. 2005;103(5):884–891. doi: 10.1002/cncr.20834. [DOI] [PubMed] [Google Scholar]
- 8.Wiedswang G, Borgen E, Karesen R, Qvist H, Janbu J, Kvalheim G, Nesland JM, Naume B. Isolated tumor cells in bone marrow three years after diagnosis in disease-free breast cancer patients predict unfavorable clinical outcome. Clin Cancer Res. 2004;10(16):5342–5348. doi: 10.1158/1078-0432.CCR-04-024510/16/5342[pii]. [DOI] [PubMed] [Google Scholar]
- 9.Pantel K, Brakenhoff RH, Brandt B. Detection, clinical relevance and specific biological properties of disseminating tumour cells. Nat Rev Cancer. 2008;8(5):329–340. doi: 10.1038/nrc2375. doi:nrc2375[pii]10.1038/nrc2375. [DOI] [PubMed] [Google Scholar]
- 10.Jenkins DE, Hornig YS, Oei Y, Dusich J, Purchio T. Bioluminescent human breast cancer cell lines that permit rapid and sensitive in vivo detection of mammary tumors and multiple metastases in immune deficient mice. Breast Cancer Res. 2005;7(4):R444–454. doi: 10.1186/bcr1026. doi:bcr1026[pii]10.1186/bcr1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kurebayashi J, McLeskey SW, Johnson MD, Lippman ME, Dickson RB, Kern FG. Quantitative demonstration of spontaneous metastasis by MCF-7 human breast cancer cells cotransfected with fibroblast growth factor 4 and LacZ. Cancer Res. 1993;53(9):2178–2187. [PubMed] [Google Scholar]
- 12.Naumov GN, MacDonald IC, Weinmeister PM, Kerkvliet N, Nadkarni KV, Wilson SM, Morris VL, Groom AC, Chambers AF. Persistence of solitary mammary carcinoma cells in a secondary site: a possible contributor to dormancy. Cancer Res. 2002;62(7):2162–2168. [PubMed] [Google Scholar]
- 13.Fehm T, Braun S, Muller A, Janni W, Gebauer G, Marth C, Schindlbeck C, Wallwiener D, Borgen E, Naume B, Pantel K, Solomayer EF. A concept for the standardized detection of disseminated tumor cells in bone marrow from patients with primary breast cancer and its clinical implementation. Cancer. 2006;107(5):885–892. doi: 10.1002/cncr.22076. [DOI] [PubMed] [Google Scholar]
- 14.Vincent-Salomon A, Bidard FC, Pierga JY. Bone marrow micrometastasis in breast cancer: review of detection methods, prognostic impact and biological issues. J Clin Pathol. 2008;61(5):570–576. doi: 10.1136/jcp.2007.046649. doi:jcp.2007.046649[pii]10.1136/jcp.2007.046649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pantel K, Alix-Panabieres C, Riethdorf S. Cancer micrometastases. Nat Rev Clin Oncol. 2009;6(6):339–351. doi: 10.1038/nrclinonc.2009.44. [DOI] [PubMed] [Google Scholar]
- 16.de Cremoux P, Extra JM, Denis MG, Pierga JY, Bourstyn E, Nos C, Clough KB, Boudou E, Martin EC, Muller A, Pouillart P, Magdelenat H. Detection of MUC1-expressing mammary carcinoma cells in the peripheral blood of breast cancer patients by real-time polymerase chain reaction. Clin Cancer Res. 2000;6(8):3117–3122. [PubMed] [Google Scholar]
- 17.Pierga JY, Bonneton C, Magdelenat H, Vincent-Salomon A, Nos C, Boudou E, Pouillart P, Thiery JP, de Cremoux P. Real-time quantitative PCR determination of urokinase-type plasminogen activator receptor (uPAR) expression of isolated micrometastatic cells from bone marrow of breast cancer patients. Int J Cancer. 2005;114(2):291–298. doi: 10.1002/ijc.20698. [DOI] [PubMed] [Google Scholar]
- 18.Ferrucci PF, Rabascio C, Gigli F, Corsini C, Giordano G, Bertolini F, Martinelli G. A new comprehensive gene expression panel to study tumor micrometastasis in patients with high-risk breast cancer. Int J Oncol. 2007;30(4):955–962. doi: 10.3892/ijo.30.4.955. [DOI] [PubMed] [Google Scholar]
- 19.Schoenfeld A, Kruger KH, Gomm J, Sinnett HD, Gazet JC, Sacks N, Bender HG, Luqmani Y, Coombes RC. The detection of micrometastases in the peripheral blood and bone marrow of patients with breast cancer using immunohistochemistry and reverse transcriptase polymerase chain reaction for keratin 19. Eur J Cancer. 1997;33(6):854–861. doi: 10.1016/s0959-8049(97)00014-2. doi:S0959804997000142[pii] [DOI] [PubMed] [Google Scholar]
- 20.Smith BM, Slade MJ, English J, Graham H, Luchtenborg M, Sinnett HD, Cross NC, Coombes RC. Response of circulating tumor cells to systemic therapy in patients with metastatic breast cancer: comparison of quantitative polymerase chain reaction and immunocytochemical techniques. J Clin Oncol. 2000;18(7):1432–1439. doi: 10.1200/JCO.2000.18.7.1432. [DOI] [PubMed] [Google Scholar]
- 21.Berois N, Varangot M, Aizen B, Estrugo R, Zarantonelli L, Fernandez P, Krygier G, Simonet F, Barrios E, Muse I, Osinaga E. Molecular detection of cancer cells in bone marrow and peripheral blood of patients with operable breast cancer. Comparison of CK19, MUC1 and CEA using RT-PCR. Eur J Cancer. 2000;36(6):717–723. doi: 10.1016/s0959-8049(99)00338-x. doi:S0959-8049(99)00338-X[pii] [DOI] [PubMed] [Google Scholar]
- 22.Bossolasco P, Ricci C, Farina G, Soligo D, Pedretti D, Scanni A, Deliliers GL. Detection of micrometastatic cells in breast cancer by RT-pCR for the mammaglobin gene. Cancer Detect Prev. 2002;26(1):60–63. doi: 10.1016/s0361-090x(02)00009-0. [DOI] [PubMed] [Google Scholar]
- 23.Becker S, Becker-Pergola G, Fehm T, Wallwiener D, Solomayer EF. Time is an important factor when processing samples for the detection of disseminated tumor cells in blood/bone marrow by reverse transcription-PCR. Clin Chem. 2004;50(4):785–786. doi: 10.1373/clinchem.2003.02551050/4/785[pii]. [DOI] [PubMed] [Google Scholar]
- 24.Benoy IH, Elst H, Philips M, Wuyts H, Van Dam P, Scharpe S, Van Marck E, Vermeulen PB, Dirix LY. Real-time RT-PCR detection of disseminated tumour cells in bone marrow has superior prognostic significance in comparison with circulating tumour cells in patients with breast cancer. Br J Cancer. 2006;94(5):672–680. doi: 10.1038/sj.bjc.6602985. doi:6602985[pii]10.1038/sj.bjc.6602985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Farmen RK, Nordgard O, Gilje B, Shammas FV, Kvaloy JT, Oltedal S, Heikkila R. Bone marrow cytokeratin 19 mRNA level is an independent predictor of relapse-free survival in operable breast cancer patients. Breast Cancer Res Treat. 2008;108(2):251–258. doi: 10.1007/s10549-007-9592-x. [DOI] [PubMed] [Google Scholar]
- 26.Benoy IH, Elst H, Philips M, Wuyts H, Van Dam P, Scharpe S, Van Marck E, Vermeulen PB, Dirix LY. Prognostic significance of disseminated tumor cells as detected by quantitative real-time reverse-transcriptase polymerase chain reaction in patients with breast cancer. Clin Breast Cancer. 2006;7(2):146–152. doi: 10.3816/CBC.2006.n.024. [DOI] [PubMed] [Google Scholar]
- 27.Watson MA, Ylagan LR, Trinkaus KM, Gillanders WE, Naughton MJ, Weilbaecher KN, Fleming TP, Aft RL. Isolation and molecular profiling of bone marrow micrometastases identifies TWIST1 as a marker of early tumor relapse in breast cancer patients. Clin Cancer Res. 2007;13(17):5001–5009. doi: 10.1158/1078-0432.CCR-07-0024. doi:13/17/5001[pii]10.1158/1078-0432.CCR-07-0024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ring A, Smith IE, Dowsett M. Circulating tumour cells in breast cancer. Lancet Oncol. 2004;5(2):79–88. doi: 10.1016/S1470-2045(04)01381-6S1470204504013816[pii]. [DOI] [PubMed] [Google Scholar]
- 29.Ballestrero A, Garuti A, Bertolotto M, Rocco I, Boy D, Nencioni A, Ottonello L, Patrone F. Effect of different cytokines on mammaglobin and maspin gene expression in normal leukocytes: possible relevance to the assays for the detection of micrometastatic breast cancer. Br J Cancer. 2005;92(10):1948–1952. doi: 10.1038/sj.bjc.6602563. doi:6602563[pii]10.1038/sj.bjc.6602563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Woelfle U, Sauter G, Santjer S, Brakenhoff R, Pantel K. Down-regulated expression of cytokeratin 18 promotes progression of human breast cancer. Clin Cancer Res. 2004;10(8):2670–2674. doi: 10.1158/1078-0432.ccr-03-0114. [DOI] [PubMed] [Google Scholar]
- 31.Willipinski-Stapelfeldt B, Riethdorf S, Assmann V, Woelfle U, Rau T, Sauter G, Heukeshoven J, Pantel K. Changes in cytoskeletal protein composition indicative of an epithelial-mesenchymal transition in human micrometastatic and primary breast carcinoma cells. Clin Cancer Res. 2005;11(22):8006–8014. doi: 10.1158/1078-0432.CCR-05-0632. doi:11/22/8006[pii]10.1158/1078-0432.CCR-05-0632. [DOI] [PubMed] [Google Scholar]
- 32.Slade MJ, Singh A, Smith BM, Tripuraneni G, Hall E, Peckitt C, Fox S, Graham H, Luchtenborg M, Sinnett HD, Cross NC, Coombes RC. Persistence of bone marrow micrometastases in patients receiving adjuvant therapy for breast cancer: results at 4 years. Int J Cancer. 2005;114(1):94–100. doi: 10.1002/ijc.20655. [DOI] [PubMed] [Google Scholar]
- 33.Ring AE, Zabaglo L, Ormerod MG, Smith IE, Dowsett M. Detection of circulating epithelial cells in the blood of patients with breast cancer: comparison of three techniques. Br J Cancer. 2005;92(5):906–912. doi: 10.1038/sj.bjc.6602418. doi:6602418[pii]10.1038/sj.bjc.6602418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Balducci E, Azzarello G, Valori L, Toffolatti L, Bolgan L, Valenti MT, Bari M, Pappagallo GL, Ausoni S, Vinante O. A new nested primer pair improves the specificity of CK-19 mRNA detection by RT-PCR in occult breast cancer cells. Int J Biol Markers. 2005;20(1):28–33. doi: 10.1177/172460080502000105. [DOI] [PubMed] [Google Scholar]
- 35.Geiss GK, Bumgarner RE, Birditt B, Dahl T, Dowidar N, Dunaway DL, Fell HP, Ferree S, George RD, Grogan T, James JJ, Maysuria M, Mitton JD, Oliveri P, Osborn JL, Peng T, Ratcliffe AL, Webster PJ, Davidson EH, Hood L, Dimitrov K. Direct multiplexed measurement of gene expression with color-coded probe pairs. Nat Biotechnol. 2008;26(3):317–325. doi: 10.1038/nbt1385. doi:nbt1385[pii]10.1038/nbt1385. [DOI] [PubMed] [Google Scholar]
- 36.Malkov VA, Serikawa KA, Balantac N, Watters J, Geiss G, Mashadi-Hossein A, Fare T. Multiplexed measurements of gene signatures in different analytes using the Nanostring nCounter Assay System. BMC Res Notes. 2009;2:80. doi: 10.1186/1756-0500-2-80. doi:1756-0500-2-80[pii]10.1186/1756-0500-2-80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Aft R, Naughton M, Trinkaus K, Watson M, Ylagan L, Chavez-MacGregor M, Zhai J, Kuo S, Shannon W, Diemer K, Herrmann V, Dietz J, Ali A, Ellis M, Weiss P, Eberlein T, Ma C, Fracasso PM, Zoberi I, Taylor M, Gillanders W, Pluard T, Mortimer J, Weilbaecher K. Effect of zoledronic acid on disseminated tumour cells in women with locally advanced breast cancer: an open label, randomised, phase 2 trial. The lancet oncology. 2010;11(5):421–428. doi: 10.1016/S1470-2045(10)70054-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25(4):402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 39.Lin Y, Lin S, Watson M, Trinkaus KM, Kuo S, Naughton MJ, Weilbaecher K, Fleming TP, Aft RL. A gene expression signature that predicts the therapeutic response of the basal-like breast cancer to neoadjuvant chemotherapy. Breast Cancer Res Treat. 2009 doi: 10.1007/s10549-009-0664-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Perou CM, Sorlie T, Eisen MB, van de Rijn M, Jeffrey SS, Rees CA, Pollack JR, Ross DT, Johnsen H, Akslen LA, Fluge O, Pergamenschikov A, Williams C, Zhu SX, Lonning PE, Borresen-Dale AL, Brown PO, Botstein D. Molecular portraits of human breast tumours. Nature. 2000;406(6797):747–752. doi: 10.1038/35021093. [DOI] [PubMed] [Google Scholar]
- 41.Olmeda D, Moreno-Bueno G, Flores JM, Fabra A, Portillo F, Cano A. SNAI1 is required for tumor growth and lymph node metastasis of human breast carcinoma MDA-MB-231 cells. Cancer Res. 2007;67(24):11721–11731. doi: 10.1158/0008-5472.CAN-07-2318. doi:67/24/11721[pii]10.1158/0008-5472.CAN-07-2318. [DOI] [PubMed] [Google Scholar]
- 42.Cimino A, Halushka M, Illei P, Wu X, Sukumar S, Argani P. Epithelial cell adhesion molecule (EpCAM) is overexpressed in breast cancer metastases. Breast Cancer Res Treat. 2010;123(3):701–708. doi: 10.1007/s10549-009-0671-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fehm T, Hoffmann O, Aktas B, Becker S, Solomayer EF, Wallwiener D, Kimmig R, Kasimir-Bauer S. Detection and characterization of circulating tumor cells in blood of primary breast cancer patients by RT-PCR and comparison to status of bone marrow disseminated cells. Breast Cancer Res. 2009;11(4):R59. doi: 10.1186/bcr2349. doi:bcr2349[pii]10.1186/bcr2349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tjensvoll K, Oltedal S, Farmen RK, Shammas FV, Heikkila R, Kvaloy JT, Gilje B, Smaaland R, Nordgard O. Disseminated tumor cells in bone marrow assessed by TWIST1, cytokeratin 19, and mammaglobin A mRNA predict clinical outcome in operable breast cancer patients. Clin Breast Cancer. 2010;10(5):378–384. doi: 10.3816/CBC.2010.n.050. doi:0221385427253021[pii]10.3816/CBC.2010.n.050. [DOI] [PubMed] [Google Scholar]
- 45.Liu Z, Fusi A, Schmittel A, Tinhofer I, Schneider A, Keilholz U. Eradication of EGFR-positive circulating tumor cells and objective tumor response with lapatinib and capecitabine. Cancer Biol Ther. 2010;10 doi: 10.4161/cbt.10.9.13323. doi:13323[pii]10.4161/cbt.10.9.13323. [DOI] [PubMed] [Google Scholar]
- 46.Dasgupta N, Watson M, Fleming T, Trinkaus K, Ferguson A, Aft R. TWIST1 and PITX2 expression in bone marrow of women with clinical stage II/III breast cancer identifies patients at risk for the development of early metastatic disease. San Antonio Breast Cancer Symposium. 2008 abstract #108. [Google Scholar]
- 47.Chen Y, Zou TN, Wu ZP, Zhou YC, Gu YL, Liu X, Jin CG, Wang XC. Detection of cytokeratin 19, human mammaglobin, and carcinoembryonic antigen-positive circulating tumor cells by three-marker reverse transcription-PCR assay and its relation to clinical outcome in early breast cancer. Int J Biol Markers. 2010;25(2):59–68. doi: 10.1177/172460081002500201. doi:60BC8313-C7D7-4AC1-8E66-68B5E0D9714A[pii] [DOI] [PubMed] [Google Scholar]
- 48.Obermayr E, Sanchez-Cabo F, Tea MK, Singer CF, Krainer M, Fischer MB, Sehouli J, Reinthaller A, Horvat R, Heinze G, Tong D, Zeillinger R. Assessment of a six gene panel for the molecular detection of circulating tumor cells in the blood of female cancer patients. BMC Cancer. 2010;10:666. doi: 10.1186/1471-2407-10-666. doi:1471-2407-10-666[pii]10.1186/1471-2407-10-666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Neve RM, Chin K, Fridlyand J, Yeh J, Baehner FL, Fevr T, Clark L, Bayani N, Coppe JP, Tong F, Speed T, Spellman PT, DeVries S, Lapuk A, Wang NJ, Kuo WL, Stilwell JL, Pinkel D, Albertson DG, Waldman FM, McCormick F, Dickson RB, Johnson MD, Lippman M, Ethier S, Gazdar A, Gray JW. A collection of breast cancer cell lines for the study of functionally distinct cancer subtypes. Cancer Cell. 2006;10(6):515–527. doi: 10.1016/j.ccr.2006.10.008. doi:S1535-6108(06)00314-X[pii]10.1016/j.ccr.2006.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tanja F, Stephan B, Volkmar M, Wolfgang J, Gerhard G, Christian M, Christian S, Diethelm W, Elin B, Bjorn N, Klaus P, Erich S. A concept for the standardized detection of disseminated tumor cells in bone marrow from patients with primary breast cancer and its clinical implementation. Cancer. 2006;107(5):885–892. doi: 10.1002/cncr.22076. [DOI] [PubMed] [Google Scholar]
- 51.Braun S, Vogl FD, Naume B, Janni W, Osborne MP, Coombes RC, Schlimok G, Diel IJ, Gerber B, Gebauer G, Pierga JY, Marth C, Oruzio D, Wiedswang G, Solomayer EF, Kundt G, Strobl B, Fehm T, Wong GY, Bliss J, Vincent-Salomon A, Pantel K. A pooled analysis of bone marrow micrometastasis in breast cancer. N Engl J Med. 2005;353(8):793–802. doi: 10.1056/NEJMoa050434. doi:353/8/793[pii]10.1056/NEJMoa050434. [DOI] [PubMed] [Google Scholar]
- 52.Klein CA, Blankenstein TJ, Schmidt-Kittler O, Petronio M, Polzer B, Stoecklein NH, Riethmuller G. Genetic heterogeneity of single disseminated tumour cells in minimal residual cancer. Lancet. 2002;360(9334):683–689. doi: 10.1016/S0140-6736(02)09838-0. [DOI] [PubMed] [Google Scholar]
- 53.Thurm H, Ebel S, Kentenich C, Hemsen A, Riethdorf S, Coith C, Wallwiener D, Braun S, Oberhoff C, Janicke F, Pantel K. Rare expression of epithelial cell adhesion molecule on residual micrometastatic breast cancer cells after adjuvant chemotherapy. Clin Cancer Res. 2003;9(7):2598–2604. [PubMed] [Google Scholar]
- 54.Janni W, Hepp F, Rjosk D, Kentenich C, Strobl B, Schindlbeck C, Hantschmann P, Sommer H, Pantel K, Braun S. The fate and prognostic value of occult metastatic cells in the bone marrow of patients with breast carcinoma between primary treatment and recurrence. Cancer. 2001;92(1):46–53. doi: 10.1002/1097-0142(20010701)92:1<46::aid-cncr1290>3.0.co;2-6. [DOI] [PubMed] [Google Scholar]
- 55.Balic M, Lin H, Young L, Hawes D, Giuliano A, McNamara G, Datar RH, Cote RJ. Most early disseminated cancer cells detected in bone marrow of breast cancer patients have a putative breast cancer stem cell phenotype. Clin Cancer Res. 2006;12(19):5615–5621. doi: 10.1158/1078-0432.CCR-06-0169. doi:12/19/5615[pii]10.1158/1078-0432.CCR-06-0169. [DOI] [PubMed] [Google Scholar]
- 56.Braun S, Kentenich C, Janni W, Hepp F, de Waal J, Willgeroth F, Sommer H, Pantel K. Lack of effect of adjuvant chemotherapy on the elimination of single dormant tumor cells in bone marrow of high-risk breast cancer patients. J Clin Oncol. 2000;18(1):80–86. doi: 10.1200/JCO.2000.18.1.80. [DOI] [PubMed] [Google Scholar]
- 57.Lacroix M. Significance, detection and markers of disseminated breast cancer cells. Endocr Relat Cancer. 2006;13(4):1033–1067. doi: 10.1677/ERC-06-0001. doi:13/4/1033[pii]10.1677/ERC-06-0001. [DOI] [PubMed] [Google Scholar]
- 58.Tjensvoll K, Gilje B, Oltedal S, Shammas VF, Kvaloy JT, Heikkila R, Nordgard O. A small subgroup of operable breast cancer patients with poor prognosis identified by quantitative real-time RT-PCR detection of mammaglobin A and trefoil factor 1 mRNA expression in bone marrow. Breast Cancer Res Treat. 2009;116(2):329–338. doi: 10.1007/s10549-008-0204-1. [DOI] [PubMed] [Google Scholar]
- 59.Al-Hajj M, Wicha MS, Benito-Hernandez A, Morrison SJ, Clarke MF. Prospective identification of tumorigenic breast cancer cells. Proc Natl Acad Sci U S A. 2003;100(7):3983–3988. doi: 10.1073/pnas.05302911000530291100[pii]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kasimir-Bauer S, Hoffmann O, Wallwiener D, Kimmig R, Fehm T. Expression of stem cell and epithelial-mesenchymal transition markers in primary breast cancer patients with circulating tumor cells. Breast cancer research : BCR. 2012;14(1):R15. doi: 10.1186/bcr3099. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Table 1. Clinical characteristics of the specimens analyzed
Supplemental Table 2. PCR Primer Probes for 38-gene panel
Supplemental Table 3. Genes screened for the nCounter assay panel which are expressed in breast cancer cells and demonstrate absent or low expression in normal bone marrow.
Supplemental Table 4. Realtive Gene expression* in Breast cancer cell lines compared to normal bone marrow
Supplemental Table 5. Enhanced nCounter signal detection of DTC-associated transcripts using increased input RNA and Ficoll mononuclear cell enrichment. A 1:1 mixture of MDA-MB231 and ZR75 breast tumor cells were spiked into normal volunteer control BM at a dilution of 1×10-4 tumor cells per nucleated BM cells. Cells were either used for direct RNA isolation or first enriched for the mononuclear cell fraction prior to RNA isolation. The indicated quantity of resulting RNA was used for the 9 gene nCounter assay. Numbers indicate total hybridization counts from each sample after subtraction of background signal from normal bone marrow controls and absolute hybridization counts of normal bone marrow controls themselves.
Supplemental Table 6. Comparison of sample analysis by NS and PCR
Supplemental Table 7. Normalized Nanostring hybridization counts* for patient Bone marrow specimens