ABSTRACT
Muscle contraction brings about movement and locomotion in animals. However, muscles have also been implicated in several atypical physiological processes including immune response. The role of muscles in immunity and the mechanism involved has not yet been deciphered. In this paper, using Drosophila indirect flight muscles (IFMs) as a model, we show that muscles are immune-responsive tissues. Flies with defective IFMs are incapable of mounting a potent humoral immune response. Upon immune challenge, the IFMs produce anti-microbial peptides (AMPs) through the activation of canonical signaling pathways, and these IFM-synthesized AMPs are essential for survival upon infection. The trunk muscles of zebrafish, a vertebrate model system, also possess the capacity to mount an immune response against bacterial infections, thus establishing that immune responsiveness of muscles is evolutionarily conserved. Our results suggest that physiologically fit muscles might boost the innate immune response of an individual.
KEY WORDS: Muscle, Drosophila, Zebrafish, Infection, Immunity, Anti-microbial peptides
Summary: Using fruit fly and zebrafish models, we show that skeletal muscles are immune responsive tissues; they mount innate immune responses during bacterial infection – an evolutionarily conserved defense mechanism.
INTRODUCTION
Host defense mechanisms against invading pathogens have evolved over time, resulting in higher vertebrates with extremely complex, multi-layered immune systems. Despite decades of research, our understanding of the immune system is not yet complete. With its powerful genetic and genomic tools and absence of typical adaptive immunity, Drosophila melanogaster has proved to be an excellent model for studying the intricacies of innate immunity (Brennan and Anderson, 2004; Kounatidis and Ligoxygakis, 2012; Igboin et al., 2012).
The Drosophila immune system is fairly well studied and is conventionally categorized into a cellular arm and a humoral arm. The cellular arm mainly comprises phagocytosis by plasmatocytes (predominant blood cells) (Lemaitre and Hoffmann, 2007; Kounatidis and Ligoxygakis, 2012). Anti-microbial peptides (AMPs), short cationic peptides that limit the growth of the invading microbes by forming pores or by other unknown mechanisms, are primarily produced by the fat body and are the hallmark of humoral immunity (Imler and Bulet, 2005; Brogden, 2005). Before systemic infection by a microbe, the boundary defense, comprising the cuticle and localized production of AMPs by the epithelium, takes care of several intrusions (Brey et al., 1993; Davis and Engstrom, 2012). Tissues endowed with this defense include the reproductive tract, tracheae, gastrointestinal tract and malpighian tubules (Tzou et al., 2000; Verma and Tapadia, 2012; Davis and Engstrom, 2012). Once a pathogen breaches these defenses, the fat body-mediated systemic AMP response in the hemocoel takes over.
Besides the fat body, Drosophila may also utilize other tissues for immune response. Several studies in Drosophila suggest that muscles affect multiple physiological processes apart from their contractile function. Although muscle-specific signaling pathways regulate the organism's oxidative stress resistance and lifespan (Tohyama and Yamaguchi, 2010; Demontis and Perrimon, 2010; Vrailas-Mortimer et al., 2011), whether muscles play a role in surviving an infection is not known. Interestingly, expression of muscle structural genes, like actin, flightin, troponin I (also known as wings up A), troponin C etc., are induced early on as a defense response of Drosophila to Pseudomonas aeruginosa pathogenesis (Apidianakis et al., 2005). These genes are speculated to have a role in tissue reconstruction upon trauma that acts in concert with immune responses (Apidianakis et al., 2007). However, these genes are not induced upon sterile injury where, intuitively, expression of tissue-reconstruction genes should be increased (Apidianakis et al., 2007). This suggests that muscles might have a more direct role in immunity.
To decipher the function of muscles in immunity we chose the indirect flight muscles (IFMs) of Drosophila as a model system because several of the muscle genes induced upon infection are specific to IFM (Apidianakis et al., 2005). Moreover, presence of several IFM-specific isoforms of many structural genes allows the manipulation of the IFMs without affecting other body muscles (Nongthomba et al., 2004, 2007). IFMs are structurally similar to vertebrate skeletal muscles and physiologically similar to cardiac muscles (Peckham et al., 1990; Moore, 2006), allowing the possibility of extrapolating the findings to higher organisms.
Here, we report that IFMs of Drosophila are capable of producing AMPs, and that this immune response mounted by IFMs is essential for flies to survive bacterial infection. We further establish that vertebrate skeletal muscles also respond to an immune challenge.
RESULTS
IFMs are required for surviving microbial infections
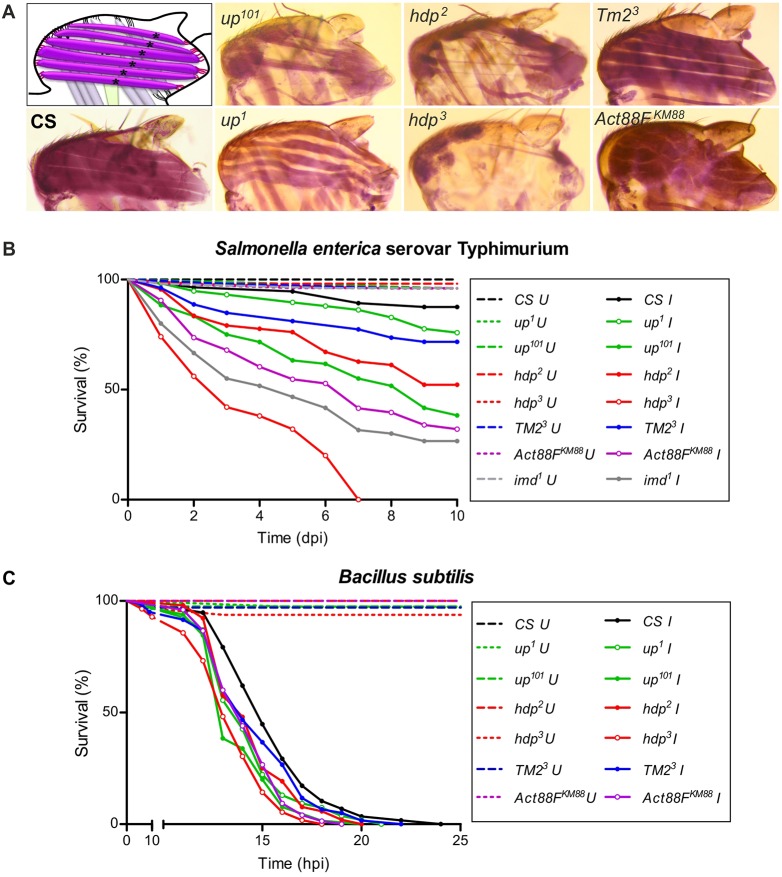
Mutants of Drosophila muscle structural genes were selected for the study. Being flightless, these mutants could not be distinguished at the functional level (Fig. S1D); however, at the structural level, they showed defects of the IFMs that varied through a spectrum amongst the mutants (Fig. 1A). Most mutants showed hypercontraction (up1, up101, wupAhdp−2, wupAhdp−3) (Nongthomba et al., 2003, 2004, 2007), hypomorph for Tropomyosin Tm23 resembled the wild type in IFM morphology despite being flightless, IFM-specific actin-null Act88FKM88 had a wavy appearance of the IFMs, and IFM-specific Troponin I-null wupAhdp−3 had the most severely affected IFMs with no muscles (Fig. 1A).
Fig. 1.
IFM mutants are susceptible to bacterial infection. (A) Hematoxylin-stained hemithoraces of the fly strains used in the study, alongside a representative schematic, show a spectrum of defects in the IFMs. CS: the wild-type fly strain Canton-S. (B,C) Survival curves of the muscle mutants post-infection with (B) Salmonella and (C) Bacillus subtilis by septic injury. A log-rank test was done for estimating the significance of the survival curves. (B) Salmonella infected versus uninfected: Canton-S, P=0.0094; up1, P=0.0034; up101, P<0.0001; hdp2, P<0.0001; hdp3, P<0.0001; TM23, P=0.0006; Act88FKM88, P<0.0001. Mutant versus wild type: up1, P=0.1171; up101, P<0.0001; hdp2, P<0.0001; hdp3, P<0.0001; TM23, P=0.0364; Act88FKM88, P<0.0001. (C) Bacillus infected versus uninfected: All fly strains, P<0.0001. Mutant versus wild type: up1, P=0.0139; up101, P=0.0002; hdp2, P=0.0346; hdp3, <0.0001; TM23, P=0.1646; Act88FKM88, P=0.0009. n>80. Open circles, IFM-specific mutants; dashed lines, uninfected controls; U, uninfected; I, infected; dpi, days post-infection; hpi, hours post-infection.
As compared with wild-type strain Canton-S, all the muscle structural gene mutants (‘muscle mutants’ hereafter) showed reduced survival upon infection by septic injury, both with Gram-negative Salmonella infection (Fig. 1B) and Gram-positive Bacillus infection (Fig. 1C), or by continuous feeding with Gram-negative Escherichia coli (Fig. S1E). Interestingly, the susceptibility to infection was not dependent on whether the mutation was manifested throughout the body or IFM-specific. Rather, the order of susceptibility was proportional to the increasing order of defects in IFMs.
The data was evaluated statistically to assess the degree of correlation between IFM defects and susceptibility to infection. Analysis of sarcomere length and IFM area (Fig. S1A-C) allowed us to rank the mutants in terms of overall defects (Fig. S2B). Mutants were also ranked for their survival after infection (Fig. S2A). Upon correlating all the susceptibility ranks with the gross IFM defect ranks, a positive correlation coefficient of r=0.8199 (P=0.0003) was obtained, where the number of pairs compared was 14 (Fig. S2C,D). A statistically significant correlation between increased IFM defects and reduced survival of flies after bacterial infection suggests an important role of IFMs in surviving infection challenges.
Optimal infection-induced AMP production requires intact IFMs
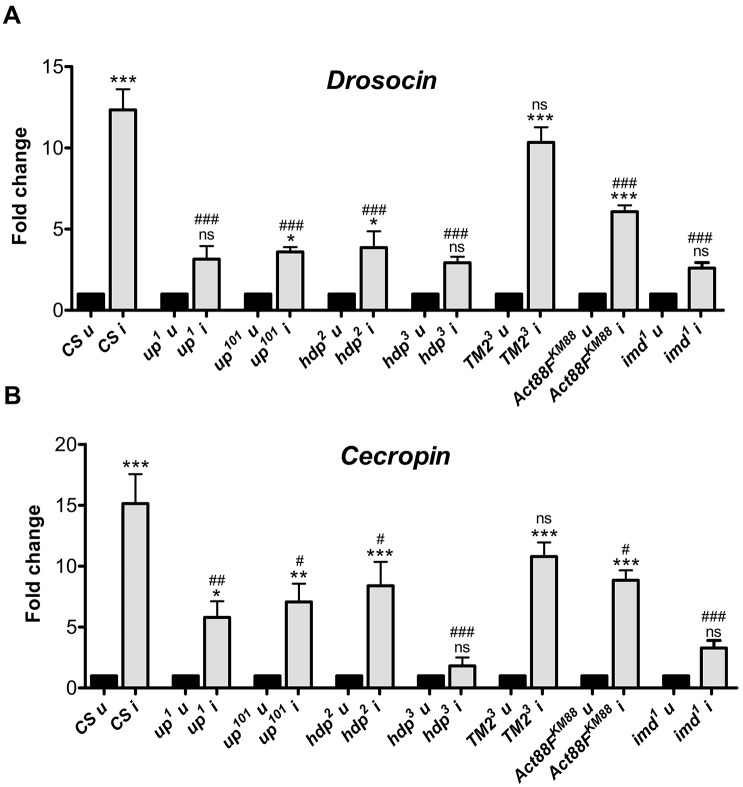
Upon bacterial infection, the Drosophila innate immune system induces the expression of a specific set of AMPs directed against the infecting bacteria by primarily activating either the Toll or the immune deficiency (Imd) pathway (Lemaitre et al., 1997). Cecropin and Drosocin, two AMPs downstream of the Imd pathway, show a substantial increase in their expression six hours post-infection with Salmonella (Fig. 2A,B). In most muscle mutants, AMPs induction was diminished compared with wild-type flies post-infection (Fig. 2A,B); although basal expression of AMPs was comparable to that of naïve wild-type flies (Fig. S3A). wupAhdp−3 flies demonstrated the weakest response to infection with no induction of either AMP. In Tm23 flies, increases in expression of both the AMPs were comparable to wild type (Fig. 2A,B), and AMP induction in the other mutants was moderate. As would be expected, reduced AMP induction correlated well with both reduced infection survival (Figs 1, 2; n=28, Spearman r=0.607, P=0.0006) and the degree of IFM defect (Fig. 2; Fig. S2; n=14, Spearman r=0.847, P=0.0001).
Fig. 2.
Induction of AMPs in muscle mutants upon infection is sub-optimal. (A,B) qRT-PCR analyses for estimation of induction of gene expression for anti-microbial peptides (A) Drosocin and (B) Cecropin upon infection. The mRNA was extracted from whole flies 6 h after infection with Salmonella. The values represented are mean±s.e.m. of three independent replicates. *, significant difference in expression of infected versus uninfected; #, significant difference in induction levels of infected mutant versus infected wild type. ns, P>0.05; */#, P<0.05; **/##, P<0.001; ***/###, P<0.0001. CS, the wild-type fly strain Canton-S; u, uninfected; i, infected.
IFMs produce AMPs upon infection
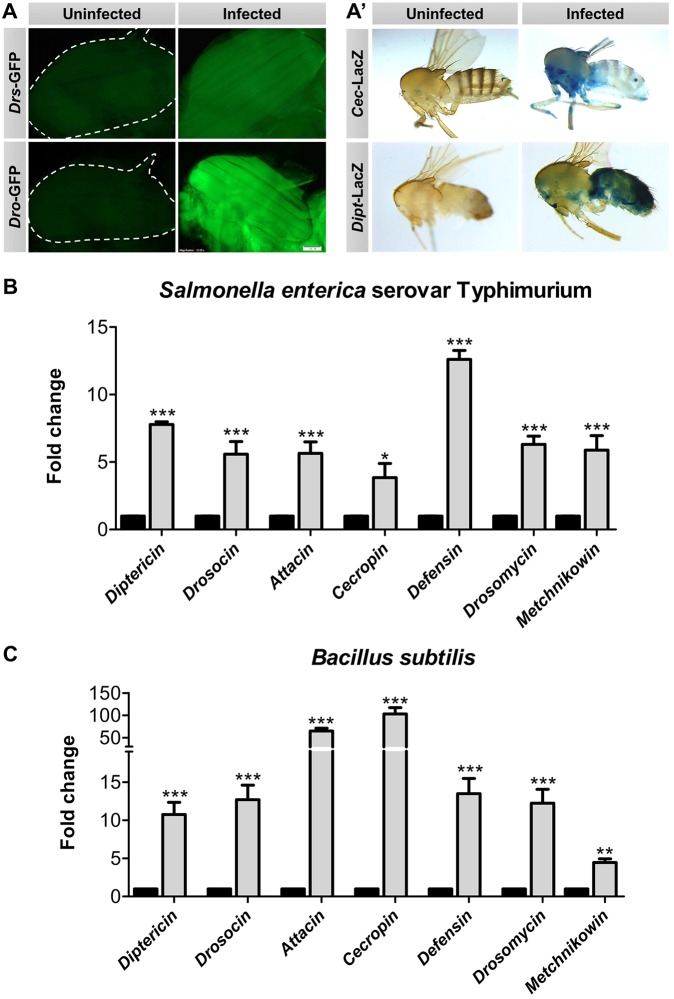
The loss of induction of AMPs in muscle mutants indicated that IFM might be an immunologically active tissue that produces AMPs upon infection. To test this hypothesis, we took two approaches. Firstly, flies with AMP reporter constructs were infected to visualize if any expression could be identified in IFM. Expression of the tested AMPs – Drosocin, Drosomycin, Cecropin and Diptericin – were elevated in the IFMs of infected flies, and also in other known body organs viz. fat and epithelia (Fig. 3A,A′).
Fig. 3.
IFMs induce AMPs upon infection. (A,A′) Infected AMP reporter fly strains show increased expression of AMPs in their IFMs compared with naïve IFMs. Drs-GFP (Drosomycin) flies were infected with Gram-positive Staphylococcus aureus. Dro-GFP (Drosocin), Dipt-LacZ (Diptericin) and Cec-LacZ (Cecropin) flies were infected with Gram-negative Enterobacter cloacae. Flies were visualized 12 h post-infection. Scale bar: 100 µm. (B,C) Change in expression of the seven Drosophila AMPs in the IFMs of wild-type flies when infected with (B) Salmonella or (C) Bacillus subtilis. The mRNA was isolated from IFMs 6 hpi. Solid columns represent uninfected controls for each of the genes tested. The values represented are mean±s.e.m. of three independent replicates. *, significant difference in expression of infected versus uninfected. *P<0.05; **P<0.001; ***P<0.0001.
Secondly, the induction of AMPs in the IFMs of infected wild-type flies was assessed six hours post-infection by quantitative reverse transcriptase PCR (qRT-PCR). All seven AMPs of Drosophila, viz. Attacin, Cecropin, Diptericin, Drosocin, Defensin, Metchnikowin and Drosomycin, were induced in IFMs upon Salmonella infection (Fig. 3B). Interestingly, the levels of AMPs responsive to Gram-positive bacteria and/or fungi (Defensin, Drosomycin and Metchnikowin) were also elevated in IFMs after a Gram-negative bacterial infection (Fig. 3B). Similar induction of all the AMPs, though at a higher level, was observed when the wild-type flies were infected with a Gram-positive bacterial species, Bacillus subtilis (Fig. 3C). Bacillus subtilis has DAP-type peptidoglycans (ligands of Gram-negative bacteria used to activate specific immune response) and thus, infection also caused induction of Gram-negative bacteria-responsive AMPs. This showed that IFMs are indeed an immune responsive tissue and can produce AMPs against both Gram-negative and Gram-positive bacteria.
IFMs produce AMP upon infection using canonical signaling pathways
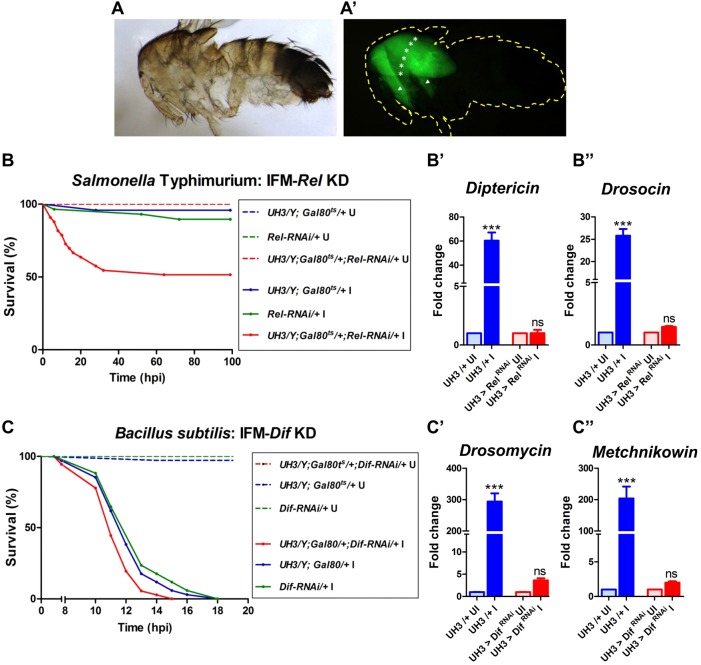
To test if the AMP-production response of IFMs upon infection is via canonical signaling pathways (Toll and Imd), we took a genetic approach. The transcription factors downstream of the Toll and Imd pathways, Dorsal-related immunity factor (Dif) and Relish (Rel), respectively, were knocked down individually under spatio-temporal control (i.e. only in adult IFMs) using the Gal4-UAS system (Brand and Perrimon, 1993). We used UH3-Gal4, whose expression is restricted to IFMs in the adult flies (Fig. 4A,A′; Fig. S4) (Singh et al., 2014). The knockdown was specifically done in the adults as the Toll and Imd pathways are important during development. We predicted that the IFMs of infected test flies (with either Dif or Rel knocked down in IFMs) will fail to induce the effector molecules or AMPs, though AMPs from other immune organs will be induced unhindered, and thus survival post-infection might be reduced as compared with controls. Alternatively, if IFMs activate Toll- or Imd-independent pathways for inducing AMPs, there would be no effect of Dif or Rel knockdown on survival post-infection.
Fig. 4.
IFM-mediated immune response is essential for survival upon bacterial infection. (A,A′) Expression of UH3-Gal4 is restricted to IFMs in 2-day-old adults. Pattern of expression was visualized with UAS-GFP as a reporter. Yellow dotted line represents the body outline of the fly. White asterisks represent the dorsal longitudinal muscles and arrowheads indicate the dorso-ventral muscles, which together constitute the IFM. Also see Fig. S4. (B,C) Survival of male flies (n>50) infected with (B) Salmonella and (C) Bacillus subtilis. A log-rank test was done for estimating the significance of the survival curves. (B) Salmonella infected versus uninfected: UH3-Gal4/+ (Gal4 control), P=0.2207; Rel-RNAi/+ (UAS control), P=0.0640; UH3>Rel-RNAi (test), P<0.0001. Test versus infected Gal4 control, P=0.0003. Test versus infected UAS control, P=0.0009. (C) Bacillus infected versus uninfected: All fly strains, P<0.0001. Test (UH3>Dif-RNAi) versus infected Gal4 control (UH3-Gal4/+), P=0.0381. Test versus infected UAS control (Dif-RNAi/+), P=0.0046. Solid lines, infected flies; dashed lines, uninfected controls; KD, knockdown; U, uninfected; I, infected, (B′,B″,C′,C″) AMP gene induction in the IFMs upon infection as tested by qRT-PCR of tissues isolated at 6 hpi. Induction of (B′) Diptericin and (B″) Drosocin was entirely lost in flies with altered IFM-Imd signaling. Similarly increased expression of (C′) Drosomycin and (C″) Metchnikowin was entirely lost in flies with altered IFM-Toll signaling. The values represented are mean±s.e.m. of three replicates. *, significant difference in expression of infected versus uninfected. ns, P>0.05; ***P<0.0001.
Dif knockdown in IFMs significantly reduced the survival of flies upon infection with Bacillus subtilis (Fig. 4C). In agreement with the reduced survival, IFMs of these flies were unable to induce the Toll pathway-specific AMP genes, Drosomycin and Metchnikowin, post infection (Fig. 4C′,C″). Elevation in transcript levels of Defensin, an AMP downstream of both the Toll and Imd pathways, was also moderate (Fig. S5A). Similarly, Rel knockdown in IFM drastically reduced the survival of flies upon infection with Salmonella (Fig. 4B). Confirming the survival data, the IFMs of the flies with Rel knockdown were unable to induce AMP genes specific to the Imd pathway (Diptericin and Drosocin) (Fig. 4B′,B″). Cecropin, encoding an AMP downstream of both the Toll and Imd pathways, was induced, albeit to a reduced level (Fig. S5B).
Thus, AMPs are produced by IFMs through activating canonical immune signaling pathways and these IFM-produced AMPs are essential for the survival of flies upon infection.
Induction of TLR/NFκB signaling in muscles upon infection is conserved in vertebrates
IFMs of Drosophila are structurally similar to vertebrate skeletal muscles. This led to the possibility that active immune signaling networks might be conserved in vertebrate skeletal muscles. To test the same, adult Danio rerio (zebrafish) trunk muscles were taken as a representation of vertebrate skeletal muscles. Unlike Drosophila, NFκB (homologous to Toll signaling) and TNFα (homologous to Imd signaling) signaling in vertebrates leads to expression of cytokines and not AMPs (Pahl, 1999; Zhou et al., 2003). Therefore, we assessed the induction of cytokines (both pro- and anti-inflammatory) in trunk muscles of zebrafish with Salmonella (Fig. S7).
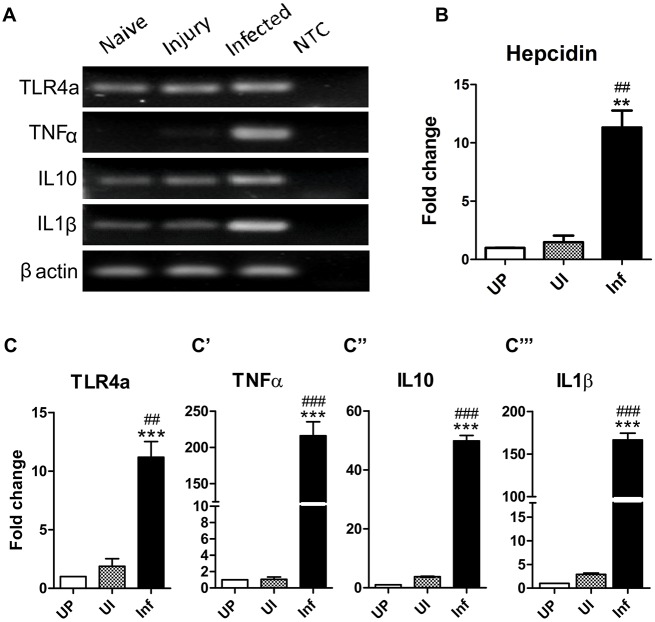
Trunk muscles of infected zebrafish showed massive inductions in expression of several cytokines (Fig. 5A). Substantial inductions (∼200-fold) were evident in the expression of pro-inflammatory cytokines IL1β (Fig. 5C‴) and TNFα (Fig. 5C′), indicating an acute phase inflammatory response. Interestingly, the anti-inflammatory cytokine IL10 was also significantly induced (Fig. 5C″), albeit to a lesser extent than the pro-inflammatory cytokines. Expression of TLR4a, a Toll-like receptor 4 paralog, was also induced upon infection (Fig. 5C). Unlike Drosophila, where injury alone can lead to induction in AMP expression, sterile injury in zebrafish did not induce cytokines [see unpricked control (UP) versus uninfected with sterile injury (UI) in Fig. 5].
Fig. 5.
Vertebrate skeletal muscles are immune responsive. (A,C-C‴) Adult zebrafish myotomes induce cytokine expression upon infection. (A) Agarose gel images of expression level of cytokine genes assessed by RT-PCR. qRT-PCR analysis was performed to estimate the induction in gene expression of the zebrafish cytokine: TLR4a (C), TNFα (C′), IL10α (C″) and IL1β (C‴). (B) Induction of Hepcidin expression, a vertebrate AMP, upon infection with Salmonella. NTC, no template control; UP, unpricked; UI, uninfected with sterile injury; Inf, infected. The values represented are mean±s.e.m. of three independent replicate experiments. *, significant difference in expression of infected versus unpricked; #, infected versus uninfected. **/##, P<0.001; ***/###, P<0.0001.
Increased expression of Hepcidin (a vertebrate AMP) has been reported following lipopolysaccharide challenge in mice (Pigeon et al., 2001) and bacterial challenge in fishes (Shike et al., 2002; Douglas et al., 2003). Trunk muscles of zebrafish also showed significant induction of expression of Hepcidin after bacterial challenge (Fig. 5B). This induction was specific to infection as a sterile injury did not show any change in expression.
DISCUSSION
The results reported in this study establish the IFMs of Drosophila as an immune responsive tissue. IFMs produce AMPs through canonical Imd and Toll pathways in response to bacterial infections. Mutant flies with defective IFMs produce suboptimal AMPs during infection and hence are more susceptible to bacterial infections. Further, using a zebrafish infection model we establish that the immune responsiveness of skeletal muscles is evolutionarily conserved.
The susceptibility of Drosophila mutants to infection mirrors the degree of overall defect in their IFMs. The overall physiology of IFMs is important for an optimal immune response. This can be elaborated by two results; firstly, wupAhdp−3, an IFM-specific Troponin-I-null mutant (Nongthomba et al., 2004), displayed the highest susceptibility to infection of all the mutants examined. Another Troponin-I mutant, wupAhdp−2, had higher survival rates following infection than wupAhdp−3. This was surprising as wupAhdp−2 has a missense mutation in a constitutive exon of the Troponin-I gene (wupA), resulting in the abnormal function of most muscles including the heart (Nongthomba et al., 2003; Wolf et al., 2006). Yet, wupAhdp−3 mutants, with only the flight and the jump muscles affected, are more susceptible to infection. This probably results from more severely affected IFMs (that are almost absent) in wupAhdp−3 compared with those in wupAhdp−2 mutants , indicating that IFM, compared with other muscles in the fly body, is important for survival upon infection.
Secondly, Act88FKM88 flies, which lack Actin in the IFMs (Okamoto et al., 1986), showed below-average survival upon infection. If it were the IFM mass alone that governed an efficient IFM-mediated immune response, then Act88FKM88, with the highest IFM mass, should have been more stress-resistant than the wild-type flies. If it were only IFM function, then the lowest survival rates would have been expected from Act88FKM88 flies, which lack the entire thin filament structure and have disorganized thick filaments (Beall et al., 1989). An average response from Act88FKM88 flies and the poorest response from hdp3 flies to bacterial infection confirm that both overall bulk and physiology of the IFMs are important for survival of flies in response to infection.
Our data support an effect of muscle defects on immune function, but indirect effects of these mutations on other physiological functions, and thereby a reduced immune response, might exist. Some facts support the former. Though all mutants used in the study were flightless, flighlessness alone (flies with clipped wings and intact IFMs) does not lead to an increase in susceptibility to infection (Fig. S6). Apart from flight, motor activity of IFM mutants (walking, climbing and food uptake) is comparable to wild type (data not shown). Naïve mutant flies do not have activated stress response pathways (Fig. S3B), suggesting that they are not stressed. A reduced ability to survive infections could also be an effect of the genetic backgrounds of the fly strains. We attempted to disprove this hypothesis by utilizing mutant fly lines from different genetic backgrounds; all yielded similar results. Thus, a deficiency of IFM-mediated immune response in mutant flies might be responsible for the reduction in surviving infections.
Infections lead to release of reactive oxygen species (ROS) early on (reviewed in Rada and Leto, 2008; Dickinson and Chang, 2011). Being highly diffusible, these reactive species can act as messengers and spread the alarm to distant cells (Dickinson and Chang, 2011). Muscles have a high mitochondrial load and thus produce large amounts of ROS themselves; levels of ROS sensors and quenchers are also substantially higher in muscles (Freyssenet, 2007; Barbieri and Sestili, 2012). Moreover, muscles are more sensitive to ROS than other tissues (Godenschwege et al., 2009). This raises the possibility of muscles utilizing ROS (generated during pathogen invasion) as indicators of stress, along with other pathogen associated molecular patterns as has been reported for fat bodies during gut infections (Wu et al., 2012). Possessing such sensitive machinery makes muscles extremely perceptive to infections, therefore making it an ideal tissue to evoke immune response.
Vertebrate homologs of the Drosophila Imd and Toll signaling pathways are functionally active in zebrafish skeletal muscles. After 6 h of bacterial infection, zebrafish muscles show high induction (∼200-fold) of pro-inflammatory cytokines IL1β and TNFα. A similar observation in zebrafish was reported earlier (Lin et al., 2009), but the role of muscles in immune response was not suggested. It is possible that the cytokine induction we found is from muscle-resident macrophages. Murine myoblast cell lines and skeletal muscles have been reported to produce pro-inflammatory cytokines when stimulated with Gram-negative bacterial lipopolysaccharide (Frost et al., 2002). Thus, the increase in cytokine expression, at least in part, must be through muscles. Along with pro-inflammatory cytokines, zebrafish muscles also induced expression of the anti-inflammatory cytokine IL10, though to a lesser extent. The production of anti-inflammatory cytokines clearly shows that zebrafish muscles also retain the ability to inhibit an immune response when not required.
Mouse skeletal muscles produce β-defensin-6, an AMP (Yamaguchi et al., 2001). Hepcidin is another AMP that is conserved among vertebrates (Shike et al., 2004). Although expression of Hepcidin is highest in the liver, Hepcidin mRNA has also been detected in other organs such as spleen, heart and stomach (Douglas et al., 2003; Ilyin et al., 2003; Shike et al., 2004). In this study, we not only show that Hepcidin is expressed in zebrafish trunk muscles, but also that its expression increases upon infection. Pathways of Hepcidin induction are presently not known; nonetheless, our result reiterates that skeletal muscles can sense and effectively respond to bacterial infection.
Unlike other immune organs (liver, thymus, neurons etc.), muscles possess the unique characteristic of muscular hypertrophy (the ability to build muscles). Exercise (strength and anaerobic training) leads to both sarcoplasmic as well as myofibrillar hypertrophy, thereby increasing muscle mass and strength (Rasch and Morehouse, 1957; Schoenfeld et al., 2013; Rai and Nongthomba, 2013). Moreover, studies in rodents show that a bout of vigorous physical activity before bacterial inoculation dramatically increases their resistance to infection (Reis Gonçalves et al., 2012; Wada et al., 1990; Cannon and Kluger, 1984). Moderate physical activity enhances natural killer cell activity and immune function in general (Pedersen and Ullum, 1994; Nielsen, 2013), but whether active hypertrophic muscles are more efficient in the immune response they mount is an unanswered question. If so, exercise might boost an individual's immunity and/or fitness indirectly by affecting overall physiology as well as directly by having more ammunition against infections and stressors.
MATERIALS AND METHODS
Fly strains
Canton-S was used as the wild-type strain. The IFM mutants used in the study were up1 (Nongthomba et al., 2007), up101 (Nongthomba et al., 2003), wupAhdp−2 (Nongthomba et al., 2003), wupAhdp−3 (Nongthomba et al., 2004), Tm23 (Mogami and Hotta, 1981), Act88FKM88 (Nongthomba et al., 2001). UH3-Gal4 (Singh et al., 2014), UAS-RelRNAi (BS#33661), UAS-DifRNAi (BS#30513) and Diptericin-LacZ (BS#55707) were procured from the Bloomington Drosophila Stock Center, Indiana. Drosomycin-GFP was a gift from Prof. Pradip Sinha (Indian Institute of Technology, Kanpur, India) Drosocin-GFP and Cecropin-LacZ were gifts from Dr M. Tapadia (Banaras Hindu University, Varanasi, India). Unless mentioned otherwise sample populations represented both sexes in equal proportion.
Microbial stocks
Salmonella enterica subsp. enterica, serovar Typhimurium (strain 12023) was a gift from Prof. D. Chakravortty (Indian Institute of Science, Bangalore, India). Escherichia coli was a gift from Prof. S. Mahadevan (Indian Institute of Science, Bangalore, India). Enterobacter cloacae (MTCC #7097), Bacillus subtilis (MTCC #441) and Staphylococcus aureus (MTCC #3160) were procured from the Microbial Type Culture Collection and Gene Bank (MTCC; Imtech, Chandigarh, India).
Infection studies
Infections were performed using the septic injury method (Apidianakis and Rahme, 2009). Salmonella was used for septic Gram-negative bacterial infections. Groups of at least 100 adults, aged one to two days, were randomized in two equal sets for the uninfected and infected groups (each containing equal number of males and females). Flies were pricked in the thorax using a sharpened tungsten needle dipped in the concentrated bacterial suspension (∼1012 colony forming units) for infection or sterile water for controls. Flies were maintained at 25°C and transferred to a fresh vial every three days. Dead flies were counted every 24 h until all the infected flies were dead.
For studies with Gram-positive infection an overnight culture of Bacillus subtilis or Staphylococcus aureus was used. The rest of the protocol was similar to the Gram-negative infection test except that dead flies were counted every hour from eight hours post-infection.
Kanamycin-resistant Escherichia coli was used for infections via feeding. Bacteria cultured overnight were diluted 1:2 with 5% sucrose-kanamycin solution. The diluted culture was inoculated onto LB-agar vials and incubated at 37°C for 12 h. Control vials were inoculated with 5% sucrose-kanamycin and incubated as for experimental vials. One- to two-day-old flies were introduced to these vials. Flies were transferred to fresh vials with a 12-h bacterial lawn every eight hours. Dead flies were counted every two hours until all the infected flies were dead.
Groups of five wild-type, germ-free fishes [five months old, treated with Chloramphenicol (40 mg/l for 24 h; HiMedia, Bangalore, India)] were anesthetized with Tricaine (1.68 mg/ml; Sigma-Aldrich, Bangalore, India) and infected by pricking the trunk with a sharp tungsten needle dipped in concentrated culture (∼1012 colony forming units) of Salmonella. Fish were let to recover from anesthetization and then assessed for GFP expression and survival for the next 24 h.
Imaging
Drosophila hemithoraces were fixed in 4% paraformaldehyde (Sigma-Aldrich) for 30 min. The tissues were then either: (a) stained with Mayer's Hematoxylin (that stains nuclei blue-black) for 15 min, de-stained in 0.3% acid-alcohol for two minutes, serially dehydrated in 50%, 70%, 90% and 100% alcohol for one hour each and finally cleared in methyl salicylate overnight; (b) incubated in LacZ staining solution (150 mM NaCl, 1 mM MgCl2, 3 mM K3[Fe(CN)6], 3 mM K4[Fe(CN)6], 0.3% Triton X-100 plus 0.25% X-gal) for 10 h at 37°C; or (c) washed with phosphate-buffered saline and directly visualized after mounting. Mounting was done using DPX Mountant (Qualigens) for bright-field imaging and in Vectashield (Vector Laboratories) for fluorescence imaging. Bright-field imaging was done using an Olympus SZX12 microscope (DSS Imagetech, Bangalore, India) and the images were photographed using an Olympus C-5060 camera (DSS Imagetech). Fluorescence imaging was done using Olympus IX81 fluorescence microscope.
qRT-PCR
Total RNA was extracted from the tissue of interest, either whole flies or muscles (according to experimental requirement), using TRIzol reagent (Sigma-Aldrich) by following manufacturer's instructions. Complementary DNA (cDNA) was prepared by using a first strand cDNA synthesis kit (RevertAid First Strand cDNA Synthesis Kit, Thermo-Scientific from Microphil, Bangalore, India) with 2 µg of RNA. The primers were used for qRT-PCRs for Drosophila and zebrafish genes are detailed in Table S1. qRT-PCR was run using Dynamo SYBR Green qPCR kit (Thermo-Scientific from Microphil) in a StepOnePlus™ Real Time PCR system (ABI). Experiments were repeated three times with independent biological repeats. Results were normalized to co-amplified house-keeping genes (rp49 for Drosophila and β-actin for zebrafish). Alterations in gene expression were calculated on the basis of the relative comparative threshold cycle (Ct) value, using the ΔΔCt method. Two-tailed Mann–Whitney tests were performed for estimating statistical significance.
Acknowledgements
We thank Prof. Dipshikha Chakravortty (Indian Institute of Science, Bangalore, India) for giving us the Salmonella strain. We thank Dr Madhu Tapadia (Benaras Hindu University, Varanasi, India) and Prof. Pradip Sinha (Indian Institute of Technology, Kanpur, India) for giving us the AMP-reporter fly strains. We also thank Dr Subhash Kairamkonda for his ideas and critical comments.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Author contributions
Conceptualization, U.N.; methodology, A.C.; investigation, A.C., D.R. and E.P.; writing – original draft, A.C.; writing – review and editing, A.C. and U.N. The final draft was approved by all the authors for submission.
Funding
We thank the Indian Institute of Science and the Department of Biotechnology, Ministry of Science and Technology, India, for funding this research. A.C. was supported by a research fellowship from the Council of Scientific and Industrial Research, Ministry of Human Resource Development, India. Financial assistance provided by the Indian University Grants Commission Departmental Research Support and Department of Science and Technology Fund for Improvement of S&T Infrastructure in Higher Educational Institutions, is also acknowledged.
Supplementary information
Supplementary information available online at http://dmm.biologists.org/lookup/suppl/doi:10.1242/dmm.022665/-/DC1
References
- Apidianakis Y. and Rahme L. G. (2009). Drosophila melanogaster as a model host for studying Pseudomonas aeruginosa infection. Nat. Protoc. 4, 1285-1294. 10.1038/nprot.2009.124 [DOI] [PubMed] [Google Scholar]
- Apidianakis Y., Mindrinos M. N., Xiao W., Lau G. W., Baldini R. L., Davis R. W. and Rahme L. G. (2005). Profiling early infection responses: pseudomonas aeruginosa eludes host defenses by suppressing antimicrobial peptide gene expression. Proc. Natl. Acad. Sci. USA 102, 2573-2578. 10.1073/pnas.0409588102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apidianakis Y., Mindrinos M. N., Xiao W., Tegos G. P., Papisov M. I., Hamblin M. R., Davis R. W., Tompkins R. G. and Rahme L. G. (2007). Involvement of skeletal muscle gene regulatory network in susceptibility to wound infection following trauma. PLoS ONE 2, e1356 10.1371/journal.pone.0001356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbieri E. and Sestili P. (2012). Reactive oxygen species in skeletal muscle signaling. J. Signal Transduct. 2012, 982794 10.1155/2012/982794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beall C. J., Sepanski M. A. and Fyrberg E. A. (1989). Genetic dissection of Drosophila myofibril formation: effects of actin and myosin heavy chain null alleles. Genes Dev. 3, 131-140. 10.1101/gad.3.2.131 [DOI] [PubMed] [Google Scholar]
- Brand A. H. and Perrimon N. (1993). Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development 118, 401-415. [DOI] [PubMed] [Google Scholar]
- Brennan C. A. and Anderson K. V. (2004). Drosophila: the genetics of innate immune recognition and response. Annu. Rev. Immunol. 22, 457-483. 10.1146/annurev.immunol.22.012703.104626 [DOI] [PubMed] [Google Scholar]
- Brey P. T., Lee W. J., Yamakawa M., Koizumi Y., Perrot S., Francois M. and Ashida M. (1993). Role of the integument in insect immunity: epicuticular abrasion and induction of cecropin synthesis in cuticular epithelial cells. Proc. Natl. Acad. Sci. USA 90, 6275-6279. 10.1073/pnas.90.13.6275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brogden K. A. (2005). Antimicrobial peptides: pore formers or metabolic inhibitors in bacteria? Nat. Rev. Microbiol. 3, 238-250. 10.1038/nrmicro1098 [DOI] [PubMed] [Google Scholar]
- Cannon J. G. and Kluger M. J. (1984). Exercise enhances survival rate in mice infected with Salmonella typhimurium. Proc. Soc. Exp. Biol. Med. 175, 518-521. 10.3181/00379727-175-41830 [DOI] [PubMed] [Google Scholar]
- Davis M. M. and Engström Y. (2012). Immune response in the barrier epithelia: lessons from the fruit fly Drosophila melanogaster. J. Innate. Immun. 4, 273-283. 10.1159/000332947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demontis F. and Perrimon N. (2010). FOXO/4E-BP signaling in Drosophila muscles regulates organism-wide proteostasis during aging. Cell 143, 813-825. 10.1016/j.cell.2010.10.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickinson B. C. and Chang C. J. (2011). Chemistry and biology of reactive oxygen species in signaling or stress responses. Nat. Chem. Biol. 7, 504-511. 10.1038/nchembio.607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douglas S. E., Gallant J. W., Liebscher R. S., Dacanay A. and Tsoi S. C. M. (2003). Identification and expression analysis of hepcidin-like antimicrobial peptides in bony fish. Dev. Comp. Immunol. 27, 589-601. 10.1016/S0145-305X(03)00036-3 [DOI] [PubMed] [Google Scholar]
- Freyssenet D. (2007). Energy sensing and regulation of gene expression in skeletal muscle. J. Appl. Physiol. 102, 529-540. 10.1152/japplphysiol.01126.2005 [DOI] [PubMed] [Google Scholar]
- Frost R. A., Nystrom G. J. and Lang C. H. (2002). Lipopolysaccharide regulates proinflammatory cytokine expression in mouse myoblasts and skeletal muscle. Am. J. Physiol. Regul. Integr. Comp. Physiol. 283, R698-R709. 10.1152/ajpregu.00039.2002 [DOI] [PubMed] [Google Scholar]
- Godenschwege T., Forde R., Davis C. P., Paul A., Beckwith K. and Duttaroy A. (2009). Mitochondrial superoxide radicals differentially affect muscle activity and neural function. Genetics 183, 175-184. 10.1534/genetics.109.103515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Igboin C. O., Griffen A. L. and Leys E. J. (2012). The Drosophila melanogaster host model. J. Oral. Microbiol. 4, 10368 10.3402/jom.v4i0.10368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ilyin G., Courselaud B., Troadec M.-B., Pigeon C., Alizadeh M., Leroyer P., Brissot P. and Loréal O. (2003). Comparative analysis of mouse hepcidin 1 and 2 genes: evidence for different patterns of expression and co-inducibility during iron overload. FEBS Lett. 542, 22-26. 10.1016/S0014-5793(03)00329-6 [DOI] [PubMed] [Google Scholar]
- Imler J.-L. and Bulet P. (2005). Antimicrobial peptides in Drosophila: structures, activities and gene regulation. Chem. Immunol. Allergy 86, 1-21. 10.1159/000086648 [DOI] [PubMed] [Google Scholar]
- Kounatidis I. and Ligoxygakis P. (2012). Drosophila as a model system to unravel the layers of innate immunity to infection. Open Biol. 2, 120075 10.1098/rsob.120075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemaitre B. and Hoffmann J. (2007). The host defense of Drosophila melanogaster. Annu. Rev. Immunol. 25, 697-743. 10.1146/annurev.immunol.25.022106.141615 [DOI] [PubMed] [Google Scholar]
- Lemaitre B., Reichhart J.-M. and Hoffmann J. A. (1997). Drosophila host defense: differential induction of antimicrobial peptide genes after infection by various classes of microorganisms. Proc. Natl. Acad. Sci. USA 94, 14614-14619. 10.1073/pnas.94.26.14614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin S.-B., Fan T.-W., Wu J.-L., Hui C.-F. and Chen J.-Y. (2009). Immune response and inhibition of bacterial growth by electrotransfer of plasmid DNA containing the antimicrobial peptide, epinecidin-1, into zebrafish muscle. Fish Shellfish Immunol. 26, 451-458. 10.1016/j.fsi.2009.01.008 [DOI] [PubMed] [Google Scholar]
- Mogami K. and Hotta Y. (1981). Isolation of Drosophila flightless mutants which affect myofibrillar proteins of indirect flight muscle. Mol. Gen. Genet. 183, 409-417. 10.1007/BF00268758 [DOI] [PubMed] [Google Scholar]
- Moore J. R. (2006). Stretch activation: towards a molecular mechanism. In Nature’s Versatile Engine: Insect Flight Muscle Inside and Out (ed. Vigoreaux J. O.), pp. 44-60. Texas, USA: Landes Bioscience/Eurekah.com. [Google Scholar]
- Nielsen H. G. (2013). Exercise and Immunity. In Current Issues in Sports and Exercise Medicine (ed. Hamlin M.), pp. 121-140. Rijeka, Croatia: InTech. 10.5772/54681 [DOI] [Google Scholar]
- Nongthomba U., Pasalodos-Sanchez S., Clark S., Clayton J. D. and Sparrow J. C. (2001). Expression and function of the Drosophila ACT88F actin isoform is not restricted to the indirect flight muscles. J. Muscle Res. Cell Motil. 22, 111-119. 10.1023/A:1010308326890 [DOI] [PubMed] [Google Scholar]
- Nongthomba U., Cummins M., Clark S., Vigoreaux J. O. and Sparrow J. C. (2003). Suppression of muscle hypercontraction by mutations in the myosin heavy chain gene of Drosophila melanogaster. Genetics 164, 209-222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nongthomba U., Clark S., Cummins M., Ansari M., Stark M. and Sparrow J. C. (2004). Troponin I is required for myofibrillogenesis and sarcomere formation in Drosophila flight muscle. J. Cell Sci. 117, 1795-1805. 10.1242/jcs.01024 [DOI] [PubMed] [Google Scholar]
- Nongthomba U., Ansari M., Thimmaiya D., Stark M. and Sparrow J. (2007). Aberrant splicing of an alternative exon in the Drosophila troponin-T gene affects flight muscle development. Genetics 177, 295-306. 10.1534/genetics.106.056812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamoto H., Hiromi Y., Ishikawa E., Yamada T., Isoda K., Maekawa H. and Hotta Y. (1986). Molecular characterization of mutant actin genes which induce heat-shock proteins in Drosophila flight muscles. EMBO J. 5, 589-596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pahl H. L. (1999). Activators and target genes of Rel/NF-kappaB transcription factors. Oncogene 18, 6853-6866. 10.1038/sj.onc.1203239 [DOI] [PubMed] [Google Scholar]
- Peckham M., Molloy J. E., Sparrow J. C. and White D. C. (1990). Physiological properties of the dorsal longitudinal flight muscle and the tergal depressor of the trochanter muscle of Drosophila melanogaster. J. Muscle Res. Cell Motil. 11, 203-215. 10.1007/BF01843574 [DOI] [PubMed] [Google Scholar]
- Pedersen B. K. and Ullum H. (1994). NK cell response to physical activity: possible mechanisms of action. Med. Sci. Sports Exerc. 26, 140-146. 10.1249/00005768-199402000-00003 [DOI] [PubMed] [Google Scholar]
- Pigeon C., Ilyin G., Courselaud B., Leroyer P., Turlin B., Brissot P. and Loreal O. (2001). A new mouse liver-specific gene, encoding a protein homologous to human antimicrobial peptide hepcidin, is overexpressed during iron overload. J. Biol. Chem. 276, 7811-7819. 10.1074/jbc.M008923200 [DOI] [PubMed] [Google Scholar]
- Rada B. and Leto T. L. (2008). Oxidative innate immune defenses by Nox/Duox family NADPH oxidases. Contrib. Microbiol. 15, 164-187. 10.1159/000136357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rai M. and Nongthomba U. (2013). Effect of myonuclear number and mitochondrial fusion on Drosophila indirect flight muscle organization and size. Exp. Cell Res. 319, 2566-2577. 10.1016/j.yexcr.2013.06.021 [DOI] [PubMed] [Google Scholar]
- Rasch P. J. and Morehouse L. E. (1957). Effect of static and dynamic exercises on muscular strength and hypertrophy. J. Appl. Physiol. 11, 29-34. [DOI] [PubMed] [Google Scholar]
- Reis Gonçalves C. T., Reis Gonçalves C. G., de Almeida F. M., dos Santos Lopes F. D. T. Q., dos Santos Durão A. C. C., dos Santos F. A., da Silva L. F. F., Marcourakis T., Castro-Faria-Neto H. C., de PaulaVieira R . et al. (2012). Protective effects of aerobic exercise on acute lung injury induced by LPS in mice. Crit. Care 16, R199 10.1186/cc11807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoenfeld B. J., Aragon A. A. and Krieger J. W. (2013). The effect of protein timing on muscle strength and hypertrophy: a meta-analysis. J. Int. Soc. Sports Nutr. 10, 53 10.1186/1550-2783-10-53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shike H., Lauth X., Westerman M. E., Ostland V. E., Carlberg J. M., Van Olst J. C., Shimizu C., Bulet P. and Burns J. C. (2002). Bass hepcidin is a novel antimicrobial peptide induced by bacterial challenge. Eur. J. Biochem. 269, 2232-2237. 10.1046/j.1432-1033.2002.02881.x [DOI] [PubMed] [Google Scholar]
- Shike H., Shimizu C., Lauth X. and Burns J. C. (2004). Organization and expression analysis of the zebrafish hepcidin gene, an antimicrobial peptide gene conserved among vertebrates. Dev. Comp. Immunol. 28, 747-754. 10.1016/j.dci.2003.11.009 [DOI] [PubMed] [Google Scholar]
- Singh S. H., Kumar P., Ramachandra N. B. and Nongthomba U. (2014). Roles of the troponin isoforms during indirect flight muscle development in Drosophila. J. Genet. 93, 379-388. 10.1007/s12041-014-0386-8 [DOI] [PubMed] [Google Scholar]
- Tohyama D. and Yamaguchi A. (2010). A critical role of SNF1A/dAMPKalpha (Drosophila AMP-activated protein kinase alpha) in muscle on longevity and stress resistance in Drosophila melanogaster. Biochem. Biophys. Res. Commun. 394, 112-118. 10.1016/j.bbrc.2010.02.126 [DOI] [PubMed] [Google Scholar]
- Tzou P., Ohresser S., Ferrandon D., Capovilla M., Reichhart J.-M., Lemaitre B., Hoffmann J. A. and Imler J.-L. (2000). Tissue-specific inducible expression of antimicrobial peptide genes in Drosophila surface epithelia. Immunity 13, 737-748. 10.1016/S1074-7613(00)00072-8 [DOI] [PubMed] [Google Scholar]
- Verma P. and Tapadia M. G. (2012). Immune response and anti-microbial peptides expression in Malpighian tubules of Drosophila melanogaster is under developmental regulation. PLoS ONE 7, e40714 10.1371/annotation/4b02305d-dcb8-40db-8f1f-1f7f0da51544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vrailas-Mortimer A., del Rivero T., Mukherjee S., Nag S., Gaitanidis A., Kadas D., Consoulas C., Duttaroy A. and Sanyal S. (2011). A muscle-specific p38 MAPK/Mef2/MnSOD pathway regulates stress, motor function, and life span in Drosophila. Dev. Cell 21, 783-795. 10.1016/j.devcel.2011.09.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wada M., Morimoto A., Watanabe T., Sakata Y. and Murakami N. (1990). Effects of physical training on febrile and acute-phase responses induced in rats by bacterial endotoxin or interleukin-1. J. Physiol. 430, 595-603. 10.1113/jphysiol.1990.sp018309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf M. J., Amrein H., Izatt J. A., Choma M. A., Reedy M. C. and Rockman H. A. (2006). Drosophila as a model for the identification of genes causing adult human heart disease. Proc. Natl. Acad. Sci. USA 103, 1394-1399. 10.1073/pnas.0507359103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu S.-C., Liao C.-W., Pan R.-L. and Juang J.-L. (2012). Infection-induced intestinal oxidative stress triggers organ-to-organ immunological communication in Drosophila. Cell Host Microbe 11, 410-417. 10.1016/j.chom.2012.03.004 [DOI] [PubMed] [Google Scholar]
- Yamaguchi Y., Fukuhara S., Nagase T., Tomita T., Hitomi S., Kimura S., Kurihara H. and Ouchi Y. (2001). A novel mouse β-Defensin, mBD-6, predominantly expressed in skeletal muscle. J. Biol. Chem. 276, 31510-31514. 10.1074/jbc.M104149200 [DOI] [PubMed] [Google Scholar]
- Zhou A., Scoggin S., Gaynor R. B. and Williams N. S. (2003). Identification of NF-kappaB-regulated genes induced by TNFalpha utilizing expression profiling and RNA interference. Oncogene 22, 2034-2044. 10.1038/sj.onc.1206262 [DOI] [PubMed] [Google Scholar]