Abstract
Spermatogonial stem cells (SSCs) maintain spermatogenesis throughout adulthood through balanced self-renewal and differentiation, yet the regulatory logic of these fate decisions is poorly understood. The transcription factors Sal-like 4 (SALL4) and promyelocytic leukemia zinc finger (PLZF; also known as ZBTB16) are known to be required for normal SSC function, but their targets are largely unknown. ChIP-seq in mouse THY1+ spermatogonia identified 4176 PLZF-bound and 2696 SALL4-bound genes, including 1149 and 515 that were unique to each factor, respectively, and 1295 that were bound by both factors. PLZF and SALL4 preferentially bound gene promoters and introns, respectively. Motif analyses identified putative PLZF and SALL4 binding sequences, but rarely both at shared sites, indicating significant non-autonomous binding in any given cell. Indeed, the majority of PLZF/SALL4 shared sites contained only PLZF motifs. SALL4 also bound gene introns at sites containing motifs for the differentiation factor DMRT1. Moreover, mRNA levels for both unique and shared target genes involved in both SSC self-renewal and differentiation were suppressed following SALL4 or PLZF knockdown. Together, these data reveal the full profile of PLZF and SALL4 regulatory targets in undifferentiated spermatogonia, including SSCs, which will help elucidate mechanisms controlling the earliest cell fate decisions in spermatogenesis.
KEY WORDS: Spermatogonial stem cells, PLZF, SALL4, DMRT1, ChIP-seq
Summary: ChIP-seq in undifferentiated mouse spermatogonia identifies both unique and overlapping targets of PLZF and SALL4, and reveals preferential binding to gene promoters and introns, respectively.
INTRODUCTION
Spermatogonial stem cells (SSCs) are undifferentiated male germ cells that sustain the spermatogenic lineage throughout adult life through balanced self-renewal and differentiation at the population level. In rodents, SSCs can be identified in whole-mount preparations of testicular seminiferous tubules [initially described by Clermont and Bustos-Obregon (1968)] as isolated Asingle spermatogonia. These Asingle SSCs are present on the basement membrane of seminiferous tubules and can be distinguished from committed, transit-amplifying progenitor spermatogonia that remain connected by intercellular cytoplasmic bridges (some Apaired and Aaligned chains of 4-16 cells). Progenitor spermatogonia are defined as undifferentiated spermatogonia that are committed to differentiate and thereafter undergo a finite number of self-renewing divisions. Each division of an Asingle SSC proceeds through an Apaired state, and thus every clone of Apaired spermatogonia has two potential developmental fates: (1) complete cytokinesis and self-renew to produce two new Asingles; or (2) remain as a clone and produce an Aaligned-4 chain of progenitors at the next mitosis (de Rooij and Grootegoed, 1998; Tegelenbosch and de Rooij, 1993). Disruption to the balance between these fate choices, leading to excessive SSC differentiation or self-renewal, impedes spermatogenesis by either depleting the stem cell pool or failing to produce differentiating germ cells to support spermatogenesis, respectively (de Rooij and Grootegoed, 1998).
Although the mechanisms that underlie and control SSC fate decisions are poorly understood, insights into the players could potentially be gleaned from knowledge of their molecular signature and distinguishing features. However, this has proven to be a difficult undertaking because SSCs are extremely rare [∼3000 per adult testis based on transplantation (Nagano, 2003)] and cannot be recognized prospectively in any species (Valli et al., 2015). SSCs, however, can be identified retrospectively by their ability to produce and maintain spermatogenesis in a functional transplantation assay that was initially described by Brinster and colleagues (Brinster and Avarbock, 1994; Brinster and Zimmermann, 1994). Indeed, SSC transplantation combined with cell sorting, in vitro gene knockdown, and gene knockout approaches have enabled systematic characterization of the phenotype of mouse SSCs (Aloisio et al., 2014; Buaas et al., 2004; Buageaw et al., 2005; Chan et al., 2014; Costoya et al., 2004; Kanatsu-Shinohara et al., 2014, 2004; Kubota et al., 2003; Oatley et al., 2011; Shinohara et al., 1999, 2000; Tokuda et al., 2007; Yang et al., 2013a,b). Moreover, gene knockout studies have been used extensively to demonstrate loss of spermatogenesis following loss of specific gene products and have often been coupled with lineage tracing to demonstrate expression of gene products in SSCs in vivo (Agbor et al., 2013; Aloisio et al., 2014; Ballow et al., 2006; Buaas et al., 2004; Costoya et al., 2004; Falender et al., 2005; Goertz et al., 2011; Greenbaum et al., 2006; Hobbs et al., 2012; Hu et al., 2013; Kanatsu-Shinohara et al., 2014; Lovasco et al., 2015; Meng et al., 2000; Nakagawa et al., 2007; Oatley et al., 2011; Raverot et al., 2005; Schlesser et al., 2008; Suzuki et al., 2012; Yang et al., 2013a; Yoshida et al., 2007, 2004). A majority of the gene products examined that are required for undifferentiated spermatogonial function are involved in transcriptional regulation [e.g. PLZF (ZBTB16) and SALL4], and thus might execute important regulatory circuits that control SSC fate.
Knockout studies for both PLZF and SALL4 have confirmed that they are essential for the proper control of SSC fate. Loss of PLZF results in progressive depletion of SSCs after the first wave of spermatogenesis leading to eventual spermatogenic arrest, apparently the result of a shift in the balance in SSC fate away from self-renewal and towards differentiation (Buaas et al., 2004; Costoya et al., 2004). Knockout of Sall4 causes embryonic death close to the time of implantation (Sakaki-Yumoto et al., 2006), but conditional Sall4 deletion with Vasa-Cre led to spermatogenic deficiencies consistent with defects in both the maintenance and differentiation in SSCs (Hobbs et al., 2012). Thus, PLZF and SALL4, which have expression patterns primarily restricted to SSCs and progenitor spermatogonia and which are required for ongoing spermatogenesis, are likely to execute transcriptional programs that are crucial for SSC self-renewal and/or differentiation. Identifying the repertoire of target genes regulated by the PLZF and SALL4 transcription factors in spermatogonia will help elucidate their mechanistic relationship with the control of SSC fate. In the present study, we identified the genomic targets of mouse PLZF and SALL4 in undifferentiated spermatogonia, which provides an important resource for uncovering the mechanisms regulating SSC and progenitor spermatogonia fate and function.
RESULTS
PLZF and SALL4 bind thousands of genomic sites in undifferentiated spermatogonia
In order to identify the target gene repertoires of PLZF and SALL4 in mouse spermatogonia we performed ChIP-seq using primary cultures of THY1+ spermatogonia, which are enriched for SSCs but also contain daughter progenitor spermatogonia (Kanatsu-Shinohara et al., 2005; Kubota et al., 2004; Schmidt and Brinster, 2011). THY1+ spermatogonia lie atop STO feeder cells and nearly all spermatogonia in these cultures expressed endogenous PLZF and SALL4 (Fig. S1). ChIP-qPCR for elongation phase-specific RNA polymerase II (CTD phospho-Ser2) demonstrated enrichment of the first introns of Actb and Gapdh compared with a non-expressed genomic region (Untr6) (Fig. S2A), confirming the suitability of chromatin from these cells for ChIP-seq.
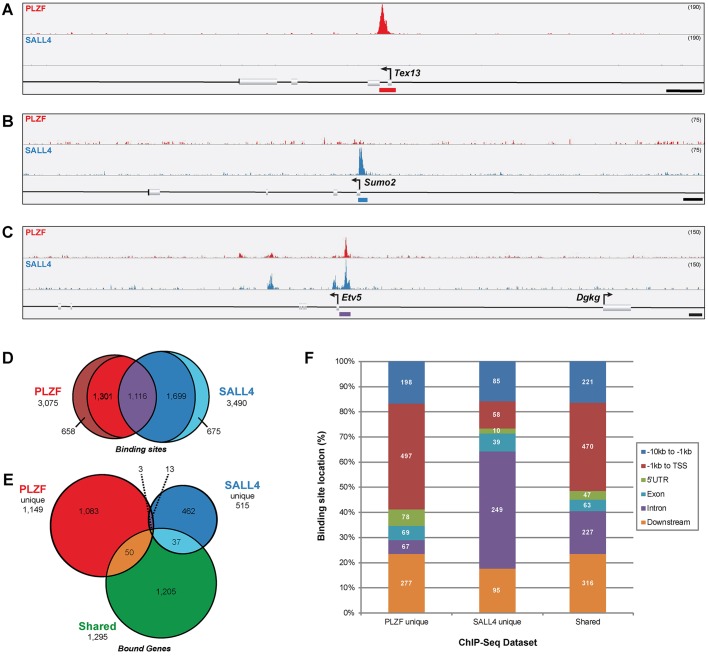
ChIP-seq for SALL4 and PLZF revealed thousands of putative binding sites, and to focus on the most biologically informative sites downstream analyses were limited to only those sites observed in all three biological replicates (Tables S1 and S2). Pairwise comparisons of peak heights demonstrated strong correlation among PLZF and SALL4 samples, respectively, but much lower correlation between PLZF and SALL4 samples (Fig. S3), demonstrating factor specificity to the binding site profiles. The correlation was somewhat lower between SALL4 sample 3 and the other two SALL4 samples (SALL4 samples 1 and 2), but excluding this replicate did not significantly change the results of subsequent analyses (data not shown). PLZF bound 3075 sites and SALL4 bound 3490 sites, including subsets that were unique to each transcription factor (658 PLZF and 675 SALL4) and 1116 that were shared (Fig. 1, Table S2), consistent with previous observations that PLZF and SALL4 physically interact (Hobbs et al., 2012). For example, binding to the Tex13 promoter was unique to PLZF (Fig. 1A, Fig. S4A), whereas Sumo2 promoter binding was unique to SALL4 (Fig. 1B, Fig. S4A), and yet both PLZF and SALL4 bound the Etv5 promoter (Fig. 1C, Fig. S4A). Some PLZF (1301) and SALL4 (1699) binding sites exhibited partial overlap with the other factor (that is, a peak was observed in one or two of the biological replicates for the other factor, but not in zero or three) and thus were not considered either unique or shared (see lists of all PLZF and all SALL4 binding sites in Table S2). Pairwise comparison of ChIP peak height between PLZF and SALL4 demonstrated the specificity of PLZF unique and SALL4 unique binding sites (Fig. S5). ChIP-qPCR was used to evaluate enrichment of four randomly selected binding sites, namely Katnb1-129, 17000065J11Rik-1056 and Igf2bp3-5969, which were all bound by both PLZF and SALL4, and Ptcd3-312, which was bound by only PLZF (Fig. S2B,C). As expected, we observed significant enrichment of genomic DNA (gDNA) for all four putative PLZF binding sites with PLZF ChIP (Fig. S2B), compared with the negative control Untr6, and significant enrichment of Katnb1-129, 17000065J11Rik-1056 and Igf2bp3-5969 gDNA with SALL4 ChIP (Fig. S2C), validating the ChIP-seq results.
Fig. 1.
PLZF and SALL4 bind unique and shared genomic sites in undifferentiated spermatogonia. (A-C) Histogram tracks show the abundance of PLZF and SALL4 ChIP DNA in mouse undifferentiated spermatogonia with binding site peaks indicated by colored bars below the reference sequence annotation. Some binding sites were unique to (A) PLZF (red curve, Tex13) or (B) SALL4 (blue curve, Sumo2), whereas others (C) were bound by both PLZF and SALL4 (shared, Etv5). Scale bars: 1 kb. Track height is indicated in parentheses at the top right. See Fig. S3 for all three biological replicates for each example ChIP peak. (D) Overlap between all PLZF (red) or SALL4 (blue) binding sites, including shared sites (purple) and unique sites (outer crescents, no overlap with the other factor). Non-unique and shared sites exhibiting partial overlap are also noted (1301 PLZF and 1699 SALL4). (E) Annotated genes within 10 kb of PLZF unique (red), SALL4 unique (blue) or shared (green) binding sites. Overlap between groups is from some genes that are located within 10 kb of more than one binding site. (F) Binding site positions relative to annotated genes were categorized as: −10 kb to −1 kb 5′ to the TSS (distal promoter); −1 kb to TSS (proximal promoter); 5′UTR; exon; intron; or downstream (transcription termination site +10 kb). The number of peaks (binding sites) falling into each category is indicated within the bars. For binding sites associated with multiple genes (Table 1), all relative positions are reported.
To understand the relationship between putative binding sites and annotated genes, and thus predict PLZF and SALL4 regulatory targets, binding sites were assigned to 4176 and 2696 annotated mRNA, miRNA and lncRNA genes located within 10 kb (Fig. S6, Table S2). We subsequently focused on genes that mapped to PLZF unique, SALL4 unique and shared sites (Fig. 1E). Most binding sites mapped to within 10 kb of at least one annotated gene (PLZF unique, 92.1%, 1149 genes; SALL4 unique, 64.3%, 515 genes; shared, 77.7%, 1295 genes; Table 1, Fig. 1E). However, a much larger proportion of SALL4 unique sites (35.7%) and shared binding sites (22.3%) were located >10 kb from any gene than were PLZF unique sites (7.9%) (Table 1). Conversely, more PLZF unique (365 sites, 55.5%) and shared (373 sites, 33.4%) binding sites were mapped to two or more annotated genes than SALL4 unique sites (88 sites, 13.0%) (Table 1). A small number of unique or shared binding sites (n=24) did not map to annotated mRNA or miRNA genes, but were within 10 kb of long intergenic non-coding RNA (lincRNA) genes (Table 1).
Table 1.
Numbers of binding sites mapped to annotated genes and lincRNAs, arranged by the number of genes
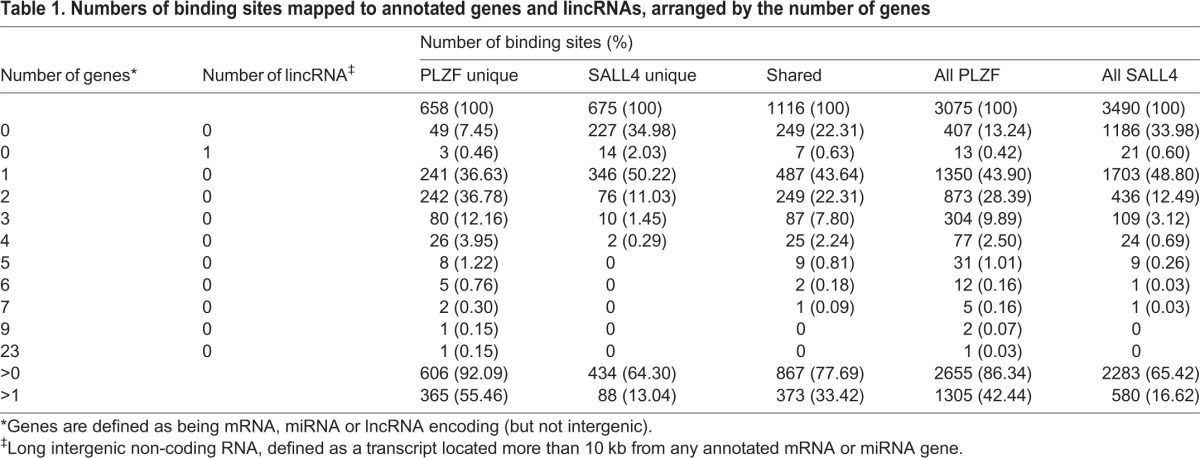
PLZF and SALL4 binding sites were also positioned differently relative to nearby annotated genes. More than half (58.6%) of PLZF unique binding sites were located in the 5′ flanking regions of nearby genes [−10 kb to transcription start site (TSS)] and 41.9% were within the first 1 kb upstream of the TSS (Fig. 1F). Similarly, more than half of shared binding sites (51.4%) occurred upstream of annotated genes and more than one-third were within the first 1 kb of 5′ flanking sequence (Fig. 1F). Surprisingly, nearly half (46.5%) of SALL4 unique binding sites were located within introns of annotated genes and only one-quarter (26.7%) were upstream of TSSs (Fig. 1F). Intron binding sites were more abundant among shared binding sites (16.9%) than among PLZF unique binding sites (5.6%).
PLZF and SALL4 binding motifs in undifferentiated spermatogonia
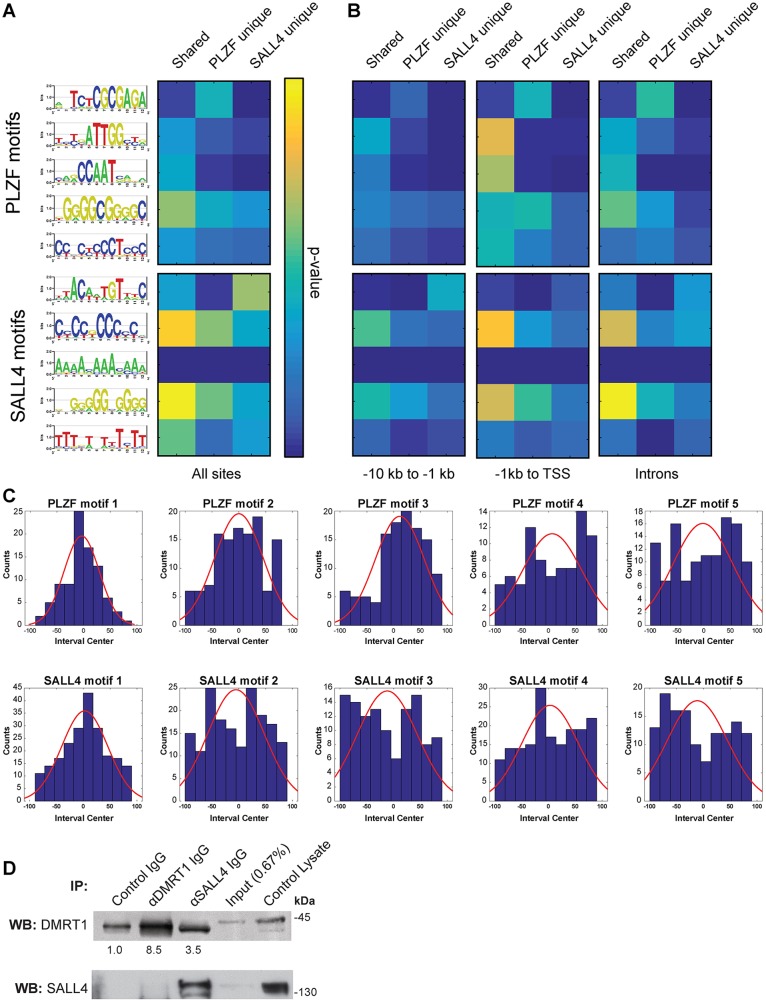
Motif analysis was subsequently performed to elaborate on which factor directs DNA binding at shared sites. MEME using the PLZF and SALL4 ChIP sequences defined motifs (Machanick and Bailey, 2011), and an in silico enrichment test was performed to determine the relative abundance of each motif among PLZF unique, SALL4 unique and shared binding sites (Fig. 2). As expected, the top five PLZF motifs were more significantly represented among PLZF unique sites than SALL4 unique sites, demonstrating specificity to PLZF (Fig. 2A, Table S3). Among these, the top PLZF motif (5′-TCTCGCGAGA-3′) was similar to a motif identified previously for another ZBTB family member (Mikula et al., 2010) and was more significantly represented among PLZF unique and shared sites (P≤2.64×10−24 and P≤1.87×10−3, respectively) than among SALL4 unique sites (P≤0.134). In comparison, a previously reported in vitro-selected PLZF motif [5′-A(T/G)(G/C)T(A/C)(A/C)AGT-3′ (Li et al., 1997)] was not significantly represented among PLZF unique sites (P≤0.403; Table S3). The second and third PLZF motifs represent opposite strands of a consensus CCAAT-box element and were probably identified because PLZF binds promoter regions (Maity and de Crombrugghe, 1998; Mantovani, 1998).
Fig. 2.
SALL4 and PLZF binding motifs in undifferentiated spermatogonia. (A) MEME-derived binding motifs for PLZF (top) and SALL4 (bottom) are shown as DNA base stack logos at each motif position, with their relative size indicative of frequency and total letter stack height reflecting information content at each position (bits). Motif scanning analysis (MAST) was performed with PLZF unique, SALL4 unique and shared binding sites and resulting hypergeometric P-values (Tables S3, S4) are presented as a heat map (yellow indicates lower P-value, see scale). (B) MAST using promoter and intron subsets of unique and shared binding sites (see Fig. 1F). (C) Histograms showing the position of the top five motifs in PLZF (top) and SALL4 (bottom) intervals normalized to the interval center (red curve shows regression). (D) Western blots for DMRT1 (top) and SALL4 (bottom) using control THY1+ spermatogonial lysate, IP input lysate, and eluates from control IgG, DMRT1 IgG or SALL4 IgG IPs. Average band intensities from three replicates are shown beneath the DMRT1 blots.
The top SALL4 motif (5′-ACAATGT-3′) was significantly enriched among SALL4 unique and shared sites (P≤1.20×10−55 and P≤3.27×10−8, respectively; Fig. 2A, Table S4). This binding motif is identical to the consensus binding site for Drosophila Doublesex protein and very similar to a motif derived from ChIP-on-chip for the mouse ortholog DMRT1 (Erdman et al., 1996; Murphy et al., 2010), suggesting DNA binding to these regions is not SALL4 autonomous. Four additional previously published SALL4 motifs identified in embryonic stem cells (ESCs) (Kim et al., 2008; Rao et al., 2010) were much less significantly represented among SALL4 unique binding sites (P≤1 to P≤6.48×10−3) as compared with the top spermatogonia SALL4 motif (Table S4). This suggested that SALL4 largely does not bind independently via these previously identified ESC motifs in undifferentiated spermatogonia. The top PLZF motif (5′-TCTCGCGAGA-3′) was also enriched in PLZF unique binding sites found in promoter regions (−10 kb to −1 kb, P≤9.88×10−11; −1 kb to TSS, P≤1.04×10−29) and the top SALL4 motif (5′-ACAATGT-5′) was enriched in both SALL4 unique and shared binding sites located in introns (P≤6.25×10−35 and P≤2.46×10−8) (Fig. 2B, Tables S5 and S6), following their broader binding preferences (Fig. 1F). Importantly, though, these two binding motifs rarely appeared in the same shared sites (P≤0.16 to P≤0.99, depending on sequence context; Table S6).
PLZF motifs 4 and 5 and SALL4 motifs 2-5 were low-complexity sequences composed largely of polynucleotide tracts (Fig. 2A, Table S4). PLZF-bound regions were relatively CG rich (∼55%), whereas SALL4-bound regions were more AT rich (∼65%) (Fig. S7). Analysis of the top 20 most frequent nucleotide k-mers (k≤4 bases) revealed that SALL4-bound regions exhibited a much higher frequency of poly(A) than PLZF, which is likely to explain the discovery of the A-rich motif for SALL4 (Fig. 2A, Fig. S7). To further explore the origin of these low-complexity motifs, we plotted the position of each motif relative to the center of the binding site (interval) sequences containing the motif. As expected, the top motif for PLZF and for SALL4 were most abundant at the center of the ChIP intervals, but the low-complexity motifs were widely distributed across the intervals (Fig. 2C). These data are consistent with the interpretation that the low-complexity motifs represent local sequence features around the position of PLZF and/or SALL4 genome interaction.
SALL4 may physically interact with DMRT1 in undifferentiated spermatogonia
Co-immunoprecipitation (IP) studies were performed with THY1+ spermatogonial lysates to determine whether SALL4 and DMRT1 physically interact in undifferentiated spermatogonia (Fig. 2D). IPs for SALL4 or DMRT1 demonstrated that each could be individually selected, but SALL4 IP eluates also contained protein that cross-reacted with DMRT1 antibody by western blot at an average of 3.5-fold above IgG (Fig. 2D, P≤0.046) (Agbor et al., 2013). The DMRT1 antibody recognized two bands of ∼41 kDa and ∼43 kDa, and the DMRT1 IP eluate was highly enriched for the slower migrating protein (mean 8.5-fold above IgG), as was the SALL4 IP eluate (Fig. 2D). Whereas the SALL4 antibody precipitated both SALL4A (upper band) and SALL4B, neither isoform was detectable in DMRT1 IPs (Fig. 2D). Together, these data suggest that SALL4 and DMRT1 may interact in undifferentiated spermatogonia and that this might account for DMRT1 binding sites observed with SALL4 ChIP.
PLZF/SALL4 binding is relevant to target gene expression
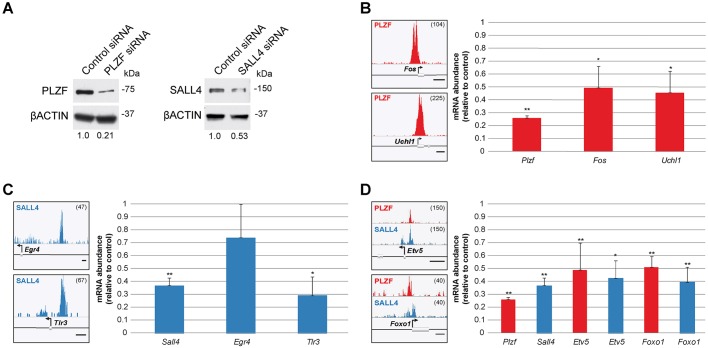
To validate the relevance of PLZF/SALL4 binding to putative target gene expression, we performed a series of siRNA-mediated knockdown studies to suppress PLZF or SALL4 in cultures of THY1+ spermatogonia (Oatley et al., 2009). This approach reduced PLZF and SALL4 protein levels to 21% and 53% of control levels, respectively (Fig. 3A). Knockdown of PLZF led to significant reductions in mRNA levels for Fos, Uchl1 and Bcl6b, which are target genes bound robustly by PLZF in their promoter regions (Fig. 3B, Fig. S8). Likewise, knockdown of SALL4 led to significant reduction in Tlr3 mRNA levels, a target bound by SALL4 in its promoter region (Fig. 3C), whereas mRNA levels for Egr4 were not significantly changed. Separate knockdowns for PLZF or SALL4 significantly suppressed mRNA levels of the shared target genes Etv5 and Foxo1 (Fig. 3C), but no significant changes were observed for shared targets Lhx1 or Pou5f1 (Fig. S8). These data confirm that PLZF and SALL4 binding is relevant to the expression of these putative target genes as revealed by ChIP.
Fig. 3.
PLZF and SALL4 are required to maintain expression levels of putative target genes in THY1+ spermatogonia. Pools of siRNAs targeting PLZF or SALL4 (or non-targeting control siRNA pools) were transfected into THY1+ spermatogonia. (A) PLZF and SALL4 protein levels were evaluated by western blot, normalized to β-ACTIN; the relative densitometry quantification is shown. (B-D) Histogram tracks demonstrate the abundance of ChIP DNA for example target genes for (B) PLZF (red curves, Fos and Uchl1), (C) SALL4 (blue curves, Egr4 and Tlr3), or (D) both PLZF and SALL4 (shared, Etv5 and Foxo1). Scale bars: 500 bp. Track height is indicated top right. Bar charts show mRNA abundance (2−ΔΔCt; mean±s.e.) for the indicated genes following knockdown of PLZF (red bars) or SALL4 (blue bars) relative to mRNA levels of non-targeting negative control siRNAs, including (B) Plzf and target genes Fos and Uchl1, (C) Sall4 and target genes Egr4 and Tlr3, and (D) shared target genes Etv5 and Lhx1. Data are from at least three replicate transfections. *P<0.05, **P<0.01 (Student's t-test). Plots of mRNA abundance for all genes examined after knockdown are shown in Fig. S8.
Pathways regulated by PLZF and SALL4 in undifferentiated spermatogonia
To better understand the biological roles for PLZF and SALL4 in undifferentiated spermatogonia, we performed gene ontology analysis on their bound gene repertoires. The top canonical pathway among genes bound only by PLZF was ‘hereditary breast cancer signaling', while the top pathway for genes bound only by SALL4 was ‘calcium-induced T lymphocyte apoptosis', and for shared genes it was the ‘unfolded protein response' (Table S7). Notably, genes involved in cell proliferation and differentiation were strongly represented among PLZF-bound genes, including those involved in oxidative phosphorylation and mitochondrial function, as well as EIF2 signaling (translational regulation), suggesting a role of PLZF in the control of spermatogonial differentiation (Table S7). Gene expression was among the top cell and molecular functions for both PLZF unique and shared genes (Table S7), suggesting that PLZF initiates biological cascades by regulating the expression of transcriptional regulators.
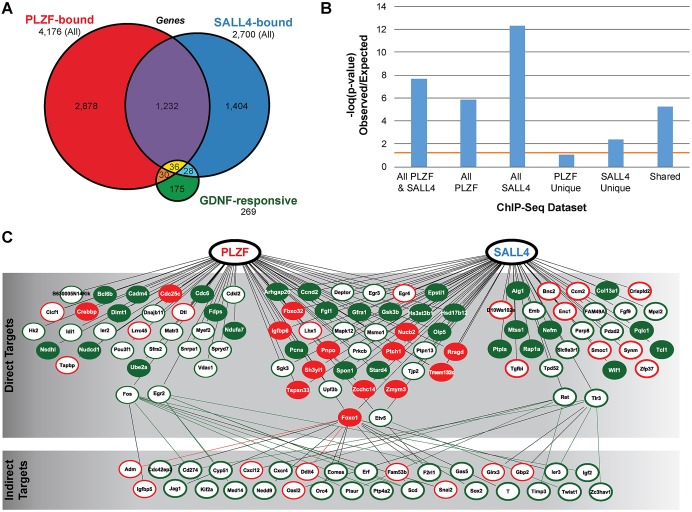
Inspection of the PLZF-bound and SALL4-bound gene lists revealed an abundance of genes involved in SSC self-renewal and differentiation, including Bcl6b, Etv5, Foxo1 and Lhx1 (Goertz et al., 2011; Oatley et al., 2006, 2007). Given that these genes were previously shown to be regulated by glial cell line-derived neurotrophic factor (GDNF), we compared the PLZF and SALL4 target gene lists with the 269 genes known to be GDNF responsive (Oatley et al., 2006). Among these GDNF-responsive genes, 30 were bound by PLZF, 28 by SALL4 and 36 by both PLZF and SALL4 (Fig. 4A), accounting for 35% of GDNF-responsive genes in THY1+ spermatogonia and a significant over-representation among the ChIP datasets (Fig. 4B). To illustrate this regulatory network, we plotted the direct PLZF-bound and SALL4-bound GDNF-responsive genes with color coding indicating the direction of GDNF regulation (Fig. 4C). Among the GDNF-responsive genes bound by PLZF and/or SALL4, six encode proteins (FOS, EGR2, FOXO1, ETV5, RET, TLR3) known to regulate another 32 GDNF-responsive genes (indirect targets, Fig. 4C). PLZF and SALL4 bound to genes that were both upregulated and downregulated by GDNF signaling, suggesting that both factors might act as both transcriptional activators and repressors in a gene-specific manner, consistent with previous reports (Filipponi et al., 2007; Hobbs et al., 2010; Yang et al., 2012).
Fig. 4.
SALL4 and PLZF bind a significant proportion of GDNF-responsive genes in undifferentiated spermatogonia. (A) Overlap between genes that are PLZF bound, SALL4 bound and GDNF responsive (Oatley et al., 2006). (B) Fisher's exact test results showing the representation of GDNF-responsive genes among those bound by PLZF and/or SALL4. Bars represent the −log(P-value) and those above the orange line are significant (P<0.05). (C) A hierarchy diagram of the 94 GDNF-responsive genes bound directly by PLZF and/or SALL4 (direct targets). The protein products of six PLZF/SALL4 direct target genes (FOS, EGR2, FOXO1, ETV5, RET and TLR3) are known to regulate 32 additional GDNF-responsive genes that are not directly bound by PLZF or SALL4 (indirect targets). Ellipse color reflects whether the GDNF-responsive genes are stimulated by GDNF (green) or inhibited by GDNF (red) and filled ellipses indicate genes that contain sites that are uniquely bound by PLZF or SALL4 or shared.
PLZF-bound and SALL4-bound genes are differentially expressed between spermatogenic cell types
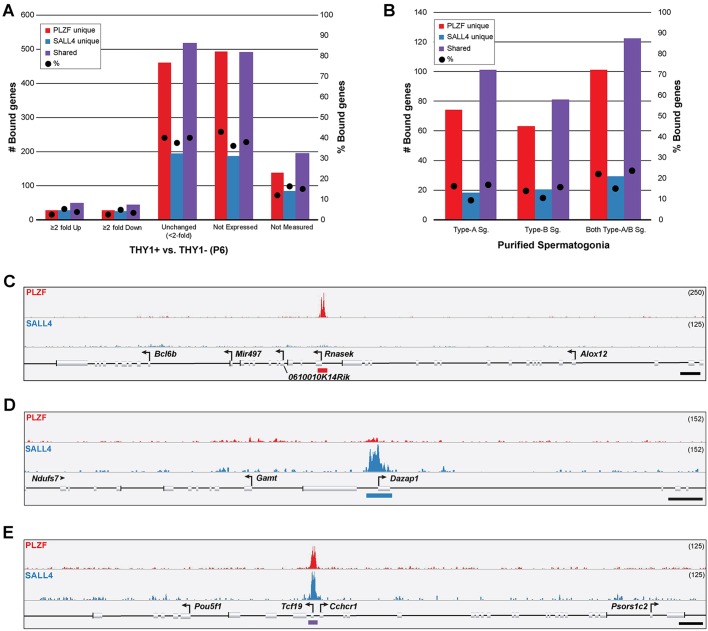
To elucidate the likely direction of gene regulation by PLZF and SALL4 for the majority of target genes in undifferentiated spermatogonia, we examined existing gene expression microarray databases from THY1-selected postnatal day (P) 6 mouse testis cells (Oatley et al., 2009). In these datasets, undifferentiated spermatogonia expressing PLZF and SALL4 from P6 testes are highly enriched in the THY1+ fraction and depleted from the THY1− fraction (Gassei and Orwig, 2013; Oatley et al., 2009). Among bound genes, 29 PLZF unique (2.5%), 27 SALL4 unique (5.2%) and 49 shared (3.8%) were expressed at ≥2-fold higher levels in THY1+ cells, correlating with positive regulation by PLZF/SALL4 (Fig. 5A, Table S8). Likewise, 29 PLZF unique (2.5%), 25 SALL4 unique (4.9%) and 44 shared (3.4%) genes were expressed at ≥2-fold higher levels in THY1− cells, correlating with negative regulation by PLZF/SALL4 (Fig. 5A, Table S8). A much larger proportion of bound genes were not differentially expressed between THY1+ and THY1− cells (<2-fold change; 37.5-40%), suggesting that these genes are not robustly regulated by either PLZF or SALL4 (Fig. 5A, Table S8).
Fig. 5.
Expression of PLZF/SALL4-bound genes in spermatogenic cells. (A) Number (left axis, bars) and percentage (right axis, dots) of genes uniquely bound by PLZF or SALL4 or shared according to expression in THY1+ or THY1− mouse pup testis cells (Oatley et al., 2009). (B) Number (left axis, bars) and percentage (right axis, dots) of genes uniquely bound by PLZF or SALL4 or shared that were not differentially expressed between THY1+ and THY1−, but which were differentially expressed between THY1+ and differentiating type A and type B spermatogonia (≥2-fold). (C-E) ChIP-seq tracks show example (C) PLZF unique, (D) SALL4 unique and (E) shared binding sites that were within 10 kb of multiple genes. Binding sites are indicated by colored bars below the reference sequence annotation. Scale bars: 1 kb. Track height is indicated top right.
One possible explanation for this observation is that PLZF/SALL4 might regulate genes that are differentially expressed later in spermatogenesis. To explore this possibility, we compared the list of PLZF/SALL4-bound genes that were not differentially expressed in THY1+ or THY1− cells with the transcriptomes of isolated differentiating type A and type B spermatogonia (Shima et al., 2004). We identified 101 PLZF unique (22%), 29 SALL4 unique (15%) and 122 shared (23.6%) genes that were differentially upregulated or downregulated (≥2-fold) in differentiating spermatogonia compared with THY1+ cells (Fig. 5B, Table S9). Thus, it is possible that PLZF and SALL4 in undifferentiated spermatogonia participate in repression/activation of a considerable cohort of genes that will become expressed/repressed later in spermatogenesis.
Additionally, given that the assignment of binding peaks to genes was based solely on proximity (within 10 kb), it was possible that some of the binding site gene assignments do not reflect a bona fide regulatory relationship. Indeed, 826 binding sites (365 PLZF unique, 88 SALL4 unique, and 373 shared) were assigned to two or more nearby annotated genes (Table 1, Fig. 5C-E). For example, PLZF binds a region of chromosome 11qB3 near five annotated genes (Bcl6b, Mir497, 0610010K14Rik, Rnasek, Alox12; Fig. 5C), yet among these five genes, only Bcl6b was ≥2-fold upregulated in the THY1+ population (Table S8) and Rnasek was upregulated in type B spermatogonia (Table S9, Fig. 5C). A SALL4 unique peak near the Ndufs7, Gamt and Dazap1 genes on chromosome 10qC1 is likely to target only Gamt, since this gene is differentially downregulated 5.2-fold in THY1− cells (Fig. 5D, Table S8), and neither Ndufs7 nor Dazap1 is differentially expressed between THY1+ and other spermatogonia (Tables S8, S9). Further, a shared binding site located on chromosome 17qB1 near the Pou5f1, Tcf19, Cchcr1 and Psors1c2 genes might be relevant to regulation of Pou5f1 (upregulated in THY1+), Tcf19 and Cchcr1 (upregulated in type A and type B spermatogonia, respectively; Fig. 5E, Table S8). Thus, binding sites found among several genes might only regulate the expression of a subset. Additionally, individual binding sites might serve multiple functions (activation or repression) dependent on the target gene in question.
DISCUSSION
PLZF and SALL4 are two zinc-finger-containing transcription factors expressed by Asingle SSCs and Apaired-Aaligned chains of progenitor spermatogonia and are known to be required for normal SSC function and spermatogenesis (Buaas et al., 2004; Costoya et al., 2004; Gassei and Orwig, 2013; Hobbs et al., 2012). Yet, relatively little is known about how PLZF and SALL4 exert their regulatory influence in the testis. In this study we identified thousands of PLZF and SALL4 binding sites and hypothesized that this information would reveal likely gene regulatory programs crucial for controlling SSC function and spermatogenesis. The results support this hypothesis and reveal considerable numbers of unique and shared regulatory targets in undifferentiated spermatogonia.
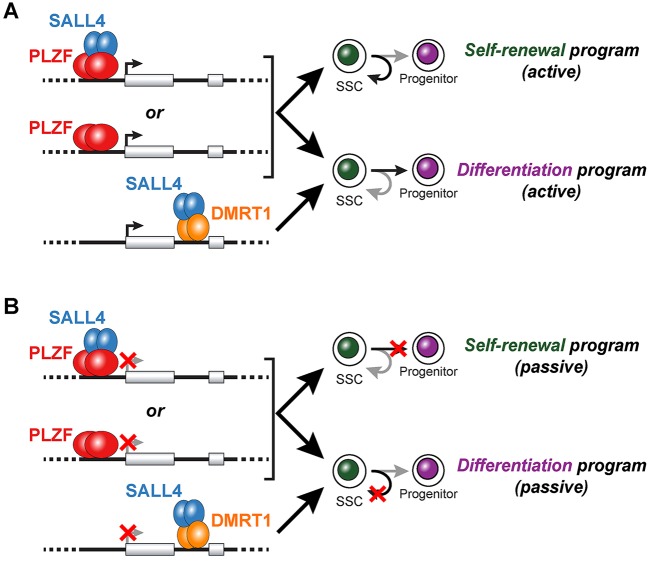
It is tempting to conclude that PLZF and SALL4 bind shared sites in the same cells at the same time, since they are largely co-expressed in undifferentiated spermatogonia. Results of our motif analyses support this conclusion. We typically observed either PLZF binding motifs or SALL4 binding motifs in shared binding sites, but a virtual absence of shared binding sites containing both motifs. This suggests that PLZF targets SALL4 binding (and vice versa) to a subset of shared binding sites in the same cells at the same time. These data are also consistent with previously characterized PLZF-SALL4 protein-protein interactions (Hobbs et al., 2012). However, the concept that SALL4 recruits PLZF away from cognate PLZF binding sites (and vice versa) in order to relieve transcriptional repression of target genes does not match our observations that knockdown of either PLZF or SALL4 suppressed shared target gene mRNA levels (Etv5, Foxo1). Rather, our data support cooperative gene activation by PLZF and SALL4 of shared target genes (Fig. 6A). Additional in-depth analyses of PLZF-SALL4 interaction at such genomic loci with temporal and single-cell resolution might help resolve these apparent discrepancies (Hermann et al., 2015).
Fig. 6.
Models of PLZF-SALL4 regulation of spermatogonial fate. Binding activities and putative regulatory roles of PLZF and SALL4 in undifferentiated spermatogonia. (A) PLZF dimers (with and without SALL4 interaction) bind to promoters and drive positive gene regulation to initiate gene expression programs associated with self-renewal (curved arrow, top) and differentiation (straight arrow, bottom). Prototypical targets are those involved in SSC self-renewal (Bcl6b, Etv5, Fos) and differentiation (Foxo1). SALL4 binding to introns might also be targeted by DMRT1 and promote the expression of genes involved in spermatogonial differentiation. (B) Passively, PLZF and SALL4 might also drive self-renewal and differentiation gene expression programs by repressing genes involved in the reciprocal cell fate. That is, as above, PLZF dimers (with and without SALL4) interact with gene promoters and SALL4 is targeted to introns by DMRT1. But in this case, the transcriptional effect is repression (red X) of both differentiation (top) and self-renewal (bottom) genes, blocking these fate pathways and leading to reciprocal passive increases in the opposing fate outcome (self-renewal, top; differentiation, bottom). Repression of later spermatogenic genes to prevent differentiation and passively enhance self-renewal would fit into this category. The extent to which these active and passive regulatory mechanisms, via PLZF alone, PLZF-SALL4 or SALL4-DMRT1 complexes occur simultaneously or separately is not known. Dimers are shown throughout, but the precise stoichiometry of DNA binding remains an open question.
Prior to the present study, a surprising paucity of regulatory targets for PLZF and SALL4 had been reported. Among the best characterized putative PLZF targets is Kit, which encodes a type-III receptor tyrosine kinase required for spermatogonial differentiation (Besmer et al., 1993; Dym et al., 1995; Koshimizu et al., 1992; Manova and Bachvarova, 1991; Sorrentino et al., 1991; Yoshinaga et al., 1991; Zhang et al., 2011). Previous studies demonstrated weak PLZF interaction with the Kit promoter by ChIP in enriched populations of mouse spermatogonia and the activation of Kit mRNA in Plzf mutants (Filipponi et al., 2007; Hobbs et al., 2012; Puszyk et al., 2013), which led to the conclusion that PLZF directly represses Kit transcription. We detected a shared binding site in the first intron of Kit (active region 17206, Table S2), yet we did not find any evidence of PLZF binding to the Kit promoter in THY1+ spermatogonia. Another putative PLZF target, DNA-damage-inducible transcript 4 (Ddit4; also known as Redd1) was identified in a study examining PLZF regulation of spermatogonial differentiation via mTORC1 (Hobbs et al., 2010). In our study, PLZF bound the Ddit4 promoter at −175 bp in two of three samples of THY1+ spermatogonia, and thus failed to reach the threshold for repeatability. Similarly few putative SALL4 target genes have been defined previously in spermatogonia. One report suggested that SALL4 represses Sall1 transcription in undifferentiated spermatogonia (Hobbs et al., 2012), but we observed no repeatable interaction between SALL4 and the Sall1 promoter in THY1+ spermatogonia. A potential explanation for these apparently disparate results between the current study and previous reports could relate to differences in the cell populations examined (e.g. cultured versus freshly isolated). While caution may be warranted in extending observations from cultured spermatogonia to their biology in vivo, use of cultured spermatogonia in this study enabled the production of sufficient cells for ChIP-seq and the use of a highly purified cell population. Indeed, spermatogonia obtained directly from the testis for analysis typically include cell population impurities [see analysis in Lambrot et al. (2015)].
The palindromic PLZF spermatogonial motif identified here (5′-TCTCGCGAGA-3′) is also bound by the three zinc-finger-containing transcription factor KAISO (also known as ZBTB33). This sequence was previously observed in human heterogeneous nuclear ribonucleoprotein K (HNRNPK) and ADP-ribosylation factor 3 (ARF3) gene promoters and is known to be bound by KAISO (Haun et al., 1993; Mikula et al., 2010). We found this same motif in a shared PLZF/SALL4 binding site in THY1+ spermatogonia in intron 1 of the mouse Hnrnpk gene and in a PLZF binding site in the promoter region of the mouse Arf3 gene (Table S2). KAISO binds the central two CpG dinucleotides (underlined above) in a methylation-dependent manner to mediate both target gene repression and activation (Prokhortchouk et al., 2001; Ruzov et al., 2004; Yoon et al., 2003). Although it seems likely that this palindromic motif is a bona fide spermatogonial PLZF binding sequence, additional studies will be required to expand on the potential for CpG methylation to influence PLZF binding and target gene expression in spermatogonia.
A surprising result of our SALL4 ChIP is the general lack of evidence to support any considerable autonomous SALL4 genomic binding in undifferentiated spermatogonia. That is, we observed a paucity of known SALL4 motifs in SALL4-bound genomic regions in undifferentiated spermatogonia. Rather, SALL4-bound sites were predominantly either co-occupied by PLZF and contained PLZF binding motifs (5′-TCTCGCGAGA-3′) or contained the DMRT1 motif (5′-ACAATGT-3′) with and without PLZF co-occupancy. Since all three factors, DMRT1, PLZF and SALL4, are co-expressed by undifferentiated spermatogonia (Murphy et al., 2010) (data not shown), these results suggest that SALL4 interaction with the mouse spermatogonial genome is not autonomous but facilitated by other DNA-binding proteins. Co-immunoprecipitation of SALL4 and DMRT1 with SALL4 antibodies supports the possibility that these proteins interact, either directly or indirectly, at the protein level. However, this result must be interpreted with caution given that reciprocal IPs with a DMRT1 antibody did not pull down SALL4. Although this negative result could be due to steric inhibition, changes to DMRT1 structure, or direct competition for the site of interaction, it is also possible that there is no stable direct or indirect interaction between SALL4 and DMRT1. Future experiments will be necessary to confirm the potential SALL4-DMRT1 interaction and determine whether such interactions participate in activating genes involved in spermatogonial differentiation (Fig. 6A) or in passively promoting differentiation by repressing self-renewal genes (Fig. 6B) (Lei et al., 2007; Matson et al., 2010). Independently, PLZF might regulate cell fate by actively promoting the expression of renewal or differentiation genes (Fig. 6A) or, conversely, might act more passively to promote cell fate by suppressing the expression of fate-related genes (Fig. 6B). So, PLZF may promote self-renewal by activating the expression of renewal genes or suppressing the transcription of differentiation genes. Similarly, PLZF-SALL4 interactions might promote self-renewal in two ways: by targeting SALL4 to PLZF-bound genes to promote the activation of genes involved in spermatogonial renewal (Fig. 6A) (Buaas et al., 2004; Costoya et al., 2004) or the suppression of differentiation genes (passive, Fig. 6B). For example, we identified a cohort of GDNF-responsive genes that might be regulatory targets of PLZF and/or SALL4. Since neither Plzf nor Sall4 is itself transcriptionally responsive to GDNF (Oatley et al., 2006), these data raise the intriguing possibility that PLZF and SALL4 are direct signaling mediators of GDNF signaling in undifferentiated spermatogonia.
In summary, this study identified the complete binding repertoires for two key transcription factors, PLZF and SALL4, in undifferentiated spermatogonia. These data provide fundamental insights into the potential modes by which each factor regulates target genes and the biological functions of their regulatory networks that lead to SSC self-renewal or differentiation, and are ultimately required for normal spermatogenesis and male fertility. These data also underscore the apparent complexity of PLZF and SALL4 interactions (Fig. 6), which might tailor their regulatory control over the fate of undifferentiated spermatogonia, including SSCs. These results will serve as a resource to the field to elucidate the molecular mechanisms controlling the earliest cell fate decisions in spermatogenesis.
MATERIALS AND METHODS
Animals and THY1+ spermatogonia cultures
All animal procedures were approved by the University of Texas at San Antonio Institutional Animal Care and Use Committee and were performed according to the NIH Guide for the Care and Use of Laboratory Animals. DBA/2 mice from The Jackson Laboratory were bred to produce male pups. Testes from 6- to 8-day-old pups were used to produce testis cell suspensions that were enriched for SSCs by THY1 selection, and cultured on mitomycin C-treated SNL76/7 feeders in a defined serum-free medium containing GDNF and FGF2, as described (Fig. S1) (Benavides-Garcia et al., 2015; Kanatsu-Shinohara et al., 2003; Nagano et al., 2003; Oatley and Brinster, 2006; Ogawa et al., 1997). There are no apparent differences in THY1+ spermatogonial cultures over the first 6 months [i.e. before passage 26 (Schmidt et al., 2011; Schmidt and Brinster, 2011)], and thus cultures between passages 5 and 20 (considered equivalent) were used for the various experiments in this study.
Indirect immunofluorescence
Cultured THY1+ spermatogonia were stained for PLZF and SALL4 as described (Hermann et al., 2009). PLZF (goat anti-PLZF, AF2944, R&D Systems; 1 µg/ml) or SALL4 (rabbit anti-SALL4, ab29112, Abcam; 2 µg/ml) antibodies were detected by indirect immunofluorescence with donkey anti-goat Alexa Fluor 488 and donkey anti-rabbit Alexa Fluor 488 secondary antibodies (A-11055 and A-21206, Life Technologies; 10 µg/ml) and counterstained with 1 μg/ml Hoechst 33342. Validation of positive immunoreactivity was confirmed by omission of the primary antibody. Images were acquired on an AxioVert CFL microscope (Zeiss) using an AxioCam MRc5 (Zeiss).
ChIP-seq
Chromatin from three independent biological preparations of THY1+ spermatogonia (each line was generated independently from multiple pups from one or more P6-P8 mouse litters) was used for ChIP essentially as described previously (Hermann et al., 2008; Hermann and Heckert, 2005). Briefly, THY1+ spermatogonia (without further purification) were fixed by addition of 1% (v/v) formaldehyde to the culture medium at room temperature for 15 min and fixation was quenched by addition of excess glycine (to 125 mM). Cells were scraped, pelleted and washed twice with Dulbecco's phosphate-buffered saline (DPBS) plus 0.5% IGEPAL. Cell pellets were suspended in lysis buffer (10 mM EDTA, 50 mM Tris pH 8.1, 1% SDS, 1× protease inhibitor cocktail) and disrupted with a Dounce homogenizer. Chromatin was sheared by sonication (average 300-500 bp) and quantified by spectrophotometry. An aliquot of chromatin (input) was treated with RNase A, proteinase K and heated to reverse the crosslinks, and then isolated by ethanol precipitation. Aliquots of chromatin (30 µg) were precleared with Protein A- or Protein G-agarose beads (Invitrogen) and then incubated with 5 μg of antibodies for PLZF (goat anti-PLZF, sc-11146, Santa Cruz Biotechnology), RNA polymerase II (mouse anti-Pol II CTD phospho-Ser2, 61083, Active Motif) or SALL4 (rabbit anti-SALL4, ab29112, Abcam) at 4°C overnight. Protein A- or Protein G-agarose beads were then used to isolate immune complexes from SALL4 and PLZF/Pol II IPs, respectively, and then washed, eluted with SDS buffer, and treated with both RNase A and proteinase K. Following overnight incubation at 65°C to reverse crosslinks, ChIP DNA was purified by phenol-chloroform extraction and ethanol precipitation. To confirm chromatin quality prior to next-generation sequencing, ChIP qPCR was performed for RNA Pol II CTD phospho-Ser2 in triplicate reactions with primers against intronic regions of Actb (+1831) and Gapdh (+2089) using SYBR Green Supermix (Bio-Rad) (Fig. S2A). qPCR signals were normalized to those obtained from input DNA and calculated as copies of DNA detected per 1000 genome equivalents of input DNA (1000 cells). A genomic region located in a gene desert on chromosome 6 (Untr6) served as a negative control, as described (Franco et al., 2012).
For next-generation sequencing, ChIP and input samples were prepared for amplification by converting overhangs into phosphorylated blunt ends, 3′ adenylation, ligation of Illumina adaptors and library size selection (175-225 bp) on an agarose gel. Adaptor-ligated libraries were then amplified for 18 cycles and the resulting DNAs were purified, quantified, and tested by qPCR to assess the quality of amplification reactions. Amplified DNA libraries were subjected to Illumina sequencing with Genome Analyzer II or HiSeq2000 instruments. Resulting reads were aligned to mouse genome (mm9) using ELAND (Illumina), sequence alignments (35 bases) were then 3′ extended in silico to a length of 110 bp and assigned to 32 bp bins along the genome, and peaks were called using the model-based analysis of ChIP-seq (MACS) algorithm (Zhang et al., 2008) with default parameters (P≤10−10), except that the genome length was specified with the ‘−g mm’ option for mouse (1.87e9).
PLZF and SALL4 binding sites were further analyzed using Genpathway proprietary software (Active Motif) that provides comprehensive information on genomic annotation, peak metrics, and sample comparisons for all binding regions. To compare peak metrics between two or more samples, overlapping binding sites were grouped into ‘active regions', which were defined as the union of overlapping binding sites from multiple biological replicates. All downstream analyses of the Chip-seq data were focused on active regions that were present in all three replicates, except where noted otherwise. Mapping genome features was performed with bedtools (Quinlan and Hall, 2010) using the mm9 annotation from the UCSC genome browser. Raw and processed data were submitted to NIH Gene Expression Omnibus (GEO) and Sequence Read Archive (SRA) databases under accession number GSE73390.
To validate the ChIP-seq results, four peaks (Katnb-129, 17000065J11Rik-1056, Igf2bp3-5969 and Ptcd3-312) were evaluated using ChIP qPCR using primers specific for putative PLZF-bound and SALL4-bound genomic regions (Table S10) and compared with enrichment at the negative control Untr6 genomic region as described above for Pol II ChIP (Fig. S2B,C).
Gene ontology analysis
Lists of genes bound by PLZF, SALL4 or both, as well as those unique to PLZF or SALL4 were analyzed by Ingenuity pathway analysis (Qiagen, build 366632M, content version 26127183, 1/2016). For comparison with GDNF-responsive genes, we used the list of genes previously shown to be significantly upregulated or downregulated following GDNF withdrawal and add-back to THY1+ spermatogonia culture medium (Oatley et al., 2006).
PLZF and SALL4 binding motif identification
Binding motifs for PLZF and SALL4 among the ChIP-seq datasets were determined using MEME (Machanick and Bailey, 2011). To ensure high-quality motif results, the binding sites in the replicated samples were pooled together, subjected to Repeat Masker, and all remaining sequences were evaluated (numbers of binding sequences for each MAST comparison can be found in Tables S3-S6). Genomic segments (200 bp) spanning the peak summit (the nucleotide position with the highest fragment density) of each putative binding site were used as input sequences for MEME. Command-line options of MEME included: minimum motif length, 6; maximum length, 12; minimum number of bindings, 10. To verify motif quality and perform motif enrichment analysis, the top motifs identified from PLZF and SALL4 binding sites by MEME were used to scan various genomic regions of interest (e.g. PLZF-bound regions that overlap with proximal promoters) and the corresponding random control sequences using MAST (Machanick and Bailey, 2011). Random control sequences (negative set) for a particular group of sequences (positive set) were generated by a first-order Markov model with the same dinucleotide frequency and sequence length distribution as the positive sequences. Statistical significance (P-value) of motif over-representation in a positive sequence group was calculated using Fisher's exact test:
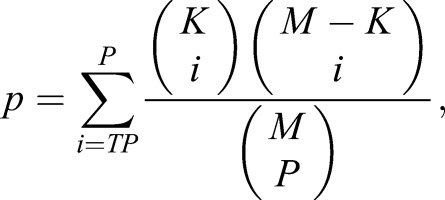 |
(1) |
where M is the total number of positive and negative sequences, K the number of positive sequences, P the number of positive and negative sequences containing the motif, and TP the number of positive sequences containing the motif. To perform the MAST validation using previously published motifs reported as position-specific weight matrices (PSWMs), the PSWMs were converted to position-specific scoring matrices (PSSMs) using the equation:
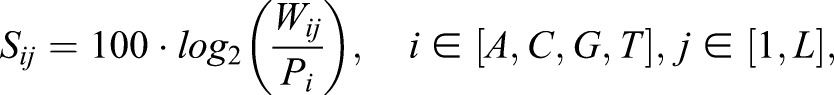 |
(2) |
where Sij (Wij) represents the score (probability) for observing nucleotide i in the j-th position of a putative binding site, Pi the background frequency of nucleotide i in the genome, and L the length of the motif.
SALL4 and DMRT1 co-immunoprecipitation
Lysates from THY1+ spermatogonia were used for IP experiments essentially as described (Elion, 2006). Briefly, 500 μg fresh lysate was incubated with 1 μg SALL4 IgG (rabbit anti-SALL4, ab29112, Abcam), DMRT1 IgG [rabbit anti-DMRT1 (Agbor et al., 2013)] or control rabbit IgG (ab37415, Abcam) at 4°C, followed by precipitation with Protein A-agarose beads (Millipore), and resolution of eluates by PAGE (10%) under denaturing SDS (DMRT1) or denaturing SDS and reducing (SALL4) conditions. Western blots were performed on nitrocellulose, and SALL4 or DMRT1 were detected with the same antibodies used for IP and horseradish peroxidase-conjugated secondary antibodies (goat anti-rabbit HRP, 0.04 µg/ml, sc-2054, Santa Cruz Biotechnology) and chemiluminescence (SuperSignal West Pico, Pierce). Molecular weight standards were the PageRuler Plus Prestained Protein Ladder (Life Technologies). Band intensities from three independent IP experiments were determined by densitometry using NIH ImageJ and the significance of differences was determined by Student's t-test.
PLZF and SALL4 knockdown by siRNA transfection
Knockdown of PLZF and SALL4 in cultured THY1+ spermatogonia was performed using siRNA transfection, as described with minor modifications (Oatley et al., 2006). Briefly, On-TARGETplus siRNA SMARTpools (GE Life Sciences Dharmacon) against mouse Sall4 (L-051702-01-0005), mouse Plzf (Zbtb16; L-040219-01-0005) or On-TARGETplus non-targeting siRNA pool (D-001810-10-05) were gently mixed with OptiMEM (Invitrogen) at 300 pmol siRNA per 106 cells transfected. RNAiMax transfection reagent (Invitrogen) was combined at 2% (v/v) with OptiMEM and equal volumes of the siRNA. Transfection reagent mixtures were then combined and incubated for 15 min at room temperature to form transfection complexes, added drop-wise to mechanically disrupted THY1+ spermatogonia on the day of passage, and plated in growth medium (OptiMEM base) on feeders. Medium was replaced after 18 h and THY1+ spermatogonia were subsequently collected by mechanical disruption 48 h post-transfection.
Target gene expression analysis by qRT-PCR
Cell pellets from 2.5×105 siRNA-treated THY1+ spermatogonia were washed with DPBS and total RNA was isolated using Trizol reagent (Invitrogen) following the manufacturer's recommendations and Phase Lock Gel tubes (5 Prime). RNA was DNase treated using the Turbo DNA-Free Kit (Ambion) and quantified by nanodrop spectrophotometry. Complementary DNA was synthesized from 1 μg RNA as described (Hermann et al., 2007) using SuperScript III reverse transcriptase (Life Technologies) and oligo(dT)18 priming, and resulting cDNAs were used in qPCR reactions. Primers for qPCR were validated for 90-100% efficiency (Table S10) and reactions were carried out in triplicate for each sample and primer set using Power SYBR Green PCR Master Mix (Applied Biosystems), 12.5 ng cDNA per reaction, and 250 nM primers on a 7300 Real-Time PCR System (Applied Biosystems). For Uchl1, a TaqMan gene expression assay (Mm00495900_m1) was used with TaqMan Universal PCR Master Mix. The relative mRNA abundance for each gene of interest was calculated using the ΔΔCt method, where Actb cDNA amplification was used for normalization to determine the fold-change value (2−ΔΔCt), and the significance of differences between control and knockdown samples was determined using Student's t-tests.
Confirmation of PLZF and SALL4 knockdown by western blot
Cell lysates were generated from 8×105 siRNA-treated THY1+ spermatogonia using RIPA buffer. Lysates (30 μg) were resolved by SDS-PAGE, transferred to nitrocellulose membrane, blocked in 5% Blotto (Santa Cruz Biotechnology), probed with primary antibodies for ACTB (mouse anti-ACTB, A1978, Sigma-Aldrich) and either PLZF (goat anti-PLZF, AF2944, R&D Systems) or SALL4 (same as IP), and detected with horseradish peroxidase-conjugated secondary antibodies (for PLZF, donkey anti-goat HRP; for SALL4, goat anti-rabbit HRP; for β-actin, goat anti-mouse-HRP; all from Santa Cruz Biotechnology) and chemiluminescence as noted above.
ChIP-seq dataset comparisons with transcriptomes
Existing gene expression microarray data were obtained from the NIH GEO database and used for comparison with the PLZF and SALL4 ChIP-seq datasets: SSC-enriched THY1+ and SSC-depleted THY1− fractions of P6 mouse testes [GSE14222 (Oatley et al., 2009)] and STAPUT-isolated differentiating type A and type B spermatogonia [GSE4193 (Shima et al., 2004)]. To enable intra- and inter-dataset comparison, data from both studies were evaluated using GeneSpring GX 13.0 (Agilent, build 211261). Raw intensity values were cross-normalized using quantile normalization without baseline transformation and using the robust multichip averaging (RMA) probe summarization algorithm (see Fig. S9). Gene-level expression analysis was then performed by merging probe signals according to the Entrez gene ID, genes with a normalized expression value above the 20th percentile in any one sample were retained and values from biological replicates were averaged. To determine differential gene expression, we performed one-way ANOVA with a corrected P-value cutoff of 0.05 (asymptotic computation) using a Benjamini and Hochberg false discovery rate multiple testing correction and Tukey's honestly significant difference test to identify genes with significant differential expression, as described previously (Cooper et al., 2014; Roy et al., 2010). Genes were considered differentially expressed when the fold-change was ≥2 and were then grouped into the following categories: ≥2-fold upregulated in THY1+; ≥2-fold downregulated in THY1+; no change in expression (≤2-fold change between THY1+ and THY1−); or not expressed. The resulting lists were compared with the ChIP-seq datasets. Putative PLZF and SALL4 target genes that were not differentially expressed in the THY1+ and THY1− transcriptomes were then compared with the differentiating type A and type B spermatogonia transcriptomes.
Acknowledgements
We thank Leslie Heckert (University of Kansas Medical Center) for graciously providing the DMRT1 antibody; the staff of the UTSA LARC for animal care; and Dr Ahmad Khalil (Case Western Reserve University) for assistance with lncRNA annotations.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Author contributions
D.L.L.: conception and design, collection and assembly of data, data analysis and interpretation, manuscript writing. Z.G.: collection and assembly of data, data analysis and interpretation, manuscript writing. K.M.: collection and assembly of data. Y.C.S.: collection and assembly of data, data analysis and interpretation. J.R.: conception and design, financial support, data analysis and interpretation, manuscript writing. B.P.H.: conception and design, financial support, administrative support, provision of study material, data analysis and interpretation, manuscript writing, final approval of manuscript.
Funding
This work was supported by the National Science Foundation [IIS-1218201 to J.R.]; the National Institutes of Health K99/R00 HD062687 to B.P.H.; P30 GM092334 to John McCarrey]; the Max and Minnie Tomerlin Voelcker Fund; the Helen Freeborn Kerr Charitable Foundation; and The University of Texas at San Antonio. This work also received bioinformatics support from Computational System Biology Core, funded by the National Institute on Minority Health and Health Disparities [G12MD007591] from the National Institutes of Health. Deposited in PMC for release after 12 months.
Data availability
Raw and processed ChIP-seq data are available at the NIH Gene Expression Omnibus (GEO) and Sequence Read Archive (SRA) databases under accession number GSE73390.
Supplementary information
Supplementary information available online at http://dev.biologists.org/lookup/suppl/doi:10.1242/dev.132761/-/DC1
References
- Agbor V. A., Tao S., Lei N. and Heckert L. L. (2013). A Wt1-Dmrt1 transgene restores DMRT1 to sertoli cells of Dmrt1(−/−) testes: a novel model of DMRT1-deficient germ cells. Biol. Reprod. 88, 51 10.1095/biolreprod.112.103135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aloisio G. M., Nakada Y., Saatcioglu H. D., Pena C. G., Baker M. D., Tarnawa E. D., Mukherjee J., Manjunath H., Bugde A., Sengupta A. L. et al. (2014). PAX7 expression defines germline stem cells in the adult testis. J. Clin. Invest. 124, 3929-3944. 10.1172/JCI75943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballow D., Meistrich M. L., Matzuk M. and Rajkovic A. (2006). Sohlh1 is essential for spermatogonial differentiation. Dev. Biol. 294, 161-167. 10.1016/j.ydbio.2006.02.027 [DOI] [PubMed] [Google Scholar]
- Benavides-Garcia R., Joachim R., Pina N. A., Mutoji K. N., Reilly M. A. and Hermann B. P. (2015). Granulocyte colony-stimulating factor prevents loss of spermatogenesis after sterilizing busulfan chemotherapy. Fertil. Steril. 103, 270-280.e8. 10.1016/j.fertnstert.2014.09.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Besmer P., Manova K., Duttlinger R., Huang E. J., Packer A., Gyssler C. and Bachvarova R. F. (1993). The kit-ligand (steel factor) and its receptor c-kit/W: pleiotropic roles in gametogenesis and melanogenesis. Dev. Suppl. 125-137. [PubMed] [Google Scholar]
- Brinster R. L. and Avarbock M. R. (1994). Germline transmission of donor haplotype following spermatogonial transplantation. Proc. Natl. Acad. Sci. USA 91, 11303-11307. 10.1073/pnas.91.24.11303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brinster R. L. and Zimmermann J. W. (1994). Spermatogenesis following male germ-cell transplantation. Proc. Natl. Acad. Sci. USA 91, 11298-11302. 10.1073/pnas.91.24.11298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buaas F. W., Kirsh A. L., Sharma M., McLean D. J., Morris J. L., Griswold M. D., de Rooij D. G. and Braun R. E. (2004). Plzf is required in adult male germ cells for stem cell self-renewal. Nat. Genet. 36, 647-652. 10.1038/ng1366 [DOI] [PubMed] [Google Scholar]
- Buageaw A., Sukhwani M., Ben-Yehudah A., Ehmcke J., Rawe V. Y., Pholpramool C., Orwig K. E. and Schlatt S. (2005). GDNF family receptor alpha1 phenotype of spermatogonial stem cells in immature mouse testes. Biol. Reprod. 73, 1011-1016. 10.1095/biolreprod.105.043810 [DOI] [PubMed] [Google Scholar]
- Chan F., Oatley M. J., Kaucher A. V., Yang Q.-E., Bieberich C. J., Shashikant C. S. and Oatley J. M. (2014). Functional and molecular features of the Id4+ germline stem cell population in mouse testes. Genes Dev. 28, 1351-1362. 10.1101/gad.240465.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clermont Y. and Bustos-Obregon E. (1968). Re-examination of spermatogonial renewal in the rat by means of seminiferous tubules mounted “in toto”. Am. J. Anat. 122, 237-247. 10.1002/aja.1001220205 [DOI] [PubMed] [Google Scholar]
- Cooper D. J., Walter C. A. and McCarrey J. R. (2014). Co-regulation of pluripotency and genetic integrity at the genomic level. Stem Cell Res. 13, 508-519. 10.1016/j.scr.2014.09.006 [DOI] [PubMed] [Google Scholar]
- Costoya J. A., Hobbs R. M., Barna M., Cattoretti G., Manova K., Sukhwani M., Orwig K. E., Wolgemuth D. J. and Pandolfi P. P. (2004). Essential role of Plzf in maintenance of spermatogonial stem cells. Nat. Genet. 36, 653-659. 10.1038/ng1367 [DOI] [PubMed] [Google Scholar]
- de Rooij D. G. and Grootegoed J. A. (1998). Spermatogonial stem cells. Curr. Opin. Cell Biol. 10, 694-701. 10.1016/S0955-0674(98)80109-9 [DOI] [PubMed] [Google Scholar]
- Dym M., Jia M. C., Dirami G., Price J. M., Rabin S. J., Mocchetti I. and Ravindranath N. (1995). Expression of c-kit receptor and its autophosphorylation in immature rat type A spermatogonia. Biol. Reprod. 52, 8-19. 10.1095/biolreprod52.1.8 [DOI] [PubMed] [Google Scholar]
- Elion E. A. (2006). Detection of protein-protein interactions by coprecipitation. Curr. Protoc. Mol. Biol. Chapter 20, Unit 20 10.1002/0471142727.mb2005s76 [DOI] [PubMed] [Google Scholar]
- Erdman S. E., Chen H. J. and Burtis K. C. (1996). Functional and genetic characterization of the oligomerization and DNA binding properties of the Drosophila doublesex proteins. Genetics 144, 1639-1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falender A. E., Freiman R. N., Geles K. G., Lo K. C., Hwang K., Lamb D. J., Morris P. L., Tjian R. and Richards J. S. (2005). Maintenance of spermatogenesis requires TAF4b, a gonad-specific subunit of TFIID. Genes Dev. 19, 794-803. 10.1101/gad.1290105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filipponi D., Hobbs R. M., Ottolenghi S., Rossi P., Jannini E. A., Pandolfi P. P. and Dolci S. (2007). Repression of kit expression by Plzf in germ cells. Mol. Cell. Biol. 27, 6770-6781. 10.1128/MCB.00479-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franco H. L., Rubel C. A., Large M. J., Wetendorf M., Fernandez-Valdivia R., Jeong J.-W., Spencer T. E., Behringer R. R., Lydon J. P. and DeMayo F. J. (2012). Epithelial progesterone receptor exhibits pleiotropic roles in uterine development and function. FASEB J. 26, 1218-1227. 10.1096/fj.11-193334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gassei K. and Orwig K. E. (2013). SALL4 expression in gonocytes and spermatogonial clones of postnatal mouse testes. PLoS ONE 8, e53976 10.1371/journal.pone.0053976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goertz M. J., Wu Z., Gallardo T. D., Hamra F. K. and Castrillon D. H. (2011). Foxo1 is required in mouse spermatogonial stem cells for their maintenance and the initiation of spermatogenesis. J. Clin. Invest. 121, 3456-3466. 10.1172/JCI57984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenbaum M. P., Yan W., Wu M.-H., Lin Y.-N., Agno J. E., Sharma M., Braun R. E., Rajkovic A. and Matzuk M. M. (2006). TEX14 is essential for intercellular bridges and fertility in male mice. Proc. Natl. Acad. Sci. USA 103, 4982-4987. 10.1073/pnas.0505123103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haun R. S., Moss J. and Vaughan M. (1993). Characterization of the human ADP-ribosylation factor 3 promoter. Transcriptional regulation of a TATA-less promoter. J. Biol. Chem. 268, 8793-8800. [PubMed] [Google Scholar]
- Hermann B. P. and Heckert L. L. (2005). Silencing of Fshr occurs through a conserved, hypersensitive site in the first intron. Mol. Endocrinol. 19, 2112-2131. 10.1210/me.2004-0244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hermann B. P., Hornbaker K. I., Maran R. R. M. and Heckert L. L. (2007). Distal regulatory elements are required for Fshr expression, in vivo. Mol. Cell. Endocrinol. 260-262, 49-58. 10.1016/j.mce.2006.01.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hermann B. P., Hornbaker K., Rice D. A., Sawadogo M. and Heckert L. L. (2008). In vivo regulation of follicle-stimulating hormone receptor by the transcription factors upstream stimulatory factor 1 and upstream stimulatory factor 2 is cell specific. Endocrinology 149, 5297-5306. 10.1210/en.2007-1199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hermann B. P., Sukhwani M., Simorangkir D. R., Chu T., Plant T. M. and Orwig K. E. (2009). Molecular dissection of the male germ cell lineage identifies putative spermatogonial stem cells in rhesus macaques. Hum. Reprod. 24, 1704-1716. 10.1093/humrep/dep073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hermann B. P., Mutoji K. N., Velte E. K., Ko D., Oatley J. M., Geyer C. B. and McCarrey J. R. (2015). Transcriptional and translational heterogeneity among neonatal mouse spermatogonia. Biol. Reprod. 92, 54 10.1095/biolreprod.114.125757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hobbs R. M., Seandel M., Falciatori I., Rafii S. and Pandolfi P. P. (2010). Plzf regulates germline progenitor self-renewal by opposing mTORC1. Cell 142, 468-479. 10.1016/j.cell.2010.06.041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hobbs R. M., Fagoonee S., Papa A., Webster K., Altruda F., Nishinakamura R., Chai L. and Pandolfi P. P. (2012). Functional antagonism between Sall4 and Plzf defines germline progenitors. Cell Stem Cell 10, 284-298. 10.1016/j.stem.2012.02.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y.-C., de Rooij D. G. and Page D. C. (2013). Tumor suppressor gene Rb is required for self-renewal of spermatogonial stem cells in mice. Proc. Natl. Acad. Sci. USA 110, 12685-12690. 10.1073/pnas.1311548110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanatsu-Shinohara M., Ogonuki N., Inoue K., Miki H., Ogura A., Toyokuni S. and Shinohara T. (2003). Long-term proliferation in culture and germline transmission of mouse male germline stem cells. Biol. Reprod. 69, 612-616. 10.1095/biolreprod.103.017012 [DOI] [PubMed] [Google Scholar]
- Kanatsu-Shinohara M., Toyokuni S. and Shinohara T. (2004). CD9 is a surface marker on mouse and rat male germline stem cells. Biol. Reprod. 70, 70-75. 10.1095/biolreprod.103.020867 [DOI] [PubMed] [Google Scholar]
- Kanatsu-Shinohara M., Ogonuki N., Iwano T., Lee J., Kazuki Y., Inoue K., Miki H., Takehashi M., Toyokuni S., Shinkai Y. et al. (2005). Genetic and epigenetic properties of mouse male germline stem cells during long-term culture. Development 132, 4155-4163. 10.1242/dev.02004 [DOI] [PubMed] [Google Scholar]
- Kanatsu-Shinohara M., Onoyama I., Nakayama K. I. and Shinohara T. (2014). Skp1-Cullin-F-box (SCF)-type ubiquitin ligase FBXW7 negatively regulates spermatogonial stem cell self-renewal. Proc. Natl. Acad. Sci. USA 111, 8826-8831. 10.1073/pnas.1401837111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J., Chu J., Shen X., Wang J. and Orkin S. H. (2008). An extended transcriptional network for pluripotency of embryonic stem cells. Cell 132, 1049-1061. 10.1016/j.cell.2008.02.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koshimizu U., Watanabe D., Tajima Y. and Nishimune Y. (1992). Effects of W (c-kit) gene mutation on gametogenesis in male mice: agametic tubular segments in Wf/Wf testes. Development 114, 861-867. [DOI] [PubMed] [Google Scholar]
- Kubota H., Avarbock M. R. and Brinster R. L. (2003). Spermatogonial stem cells share some, but not all, phenotypic and functional characteristics with other stem cells. Proc. Natl. Acad. Sci. USA 100, 6487-6492. 10.1073/pnas.0631767100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubota H., Avarbock M. R. and Brinster R. L. (2004). Culture conditions and single growth factors affect fate determination of mouse spermatogonial stem cells. Biol. Reprod. 71, 722-731. 10.1095/biolreprod.104.029207 [DOI] [PubMed] [Google Scholar]
- Lambrot R., Lafleur C. and Kimmins S. (2015). The histone demethylase KDM1A is essential for the maintenance and differentiation of spermatogonial stem cells and progenitors. FASEB J. 29, 4402-4416. 10.1096/fj.14-267328 [DOI] [PubMed] [Google Scholar]
- Lei N., Hornbaker K. I., Rice D. A., Karpova T., Agbor V. A. and Heckert L. L. (2007). Sex-specific differences in mouse DMRT1 expression are both cell type- and stage-dependent during gonad development. Biol. Reprod. 77, 466-475. 10.1095/biolreprod.106.058784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J.-Y., English M. A., Ball H. J., Yeyati P. L., Waxman S. and Licht J. D. (1997). Sequence-specific DNA binding and transcriptional regulation by the promyelocytic leukemia zinc finger protein. J. Biol. Chem. 272, 22447-22455. 10.1074/jbc.272.36.22447 [DOI] [PubMed] [Google Scholar]
- Lovasco L. A., Gustafson E. A., Seymour K. A., de Rooij D. G. and Freiman R. N. (2015). TAF4b is required for mouse spermatogonial stem cell development. Stem Cells 33, 1267-1276. 10.1002/stem.1914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machanick P. and Bailey T. L. (2011). MEME-ChIP: motif analysis of large DNA datasets. Bioinformatics 27, 1696-1697. 10.1093/bioinformatics/btr189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maity S. N. and de Crombrugghe B. (1998). Role of the CCAAT-binding protein CBF/NF-Y in transcription. Trends Biochem. Sci. 23, 174-178. 10.1016/S0968-0004(98)01201-8 [DOI] [PubMed] [Google Scholar]
- Manova K. and Bachvarova R. F. (1991). Expression of c-kit encoded at the W locus of mice in developing embryonic germ cells and presumptive melanoblasts. Dev. Biol. 146, 312-324. 10.1016/0012-1606(91)90233-S [DOI] [PubMed] [Google Scholar]
- Mantovani R. (1998). A survey of 178 NF-Y binding CCAAT boxes. Nucleic Acids Res. 26, 1135-1143. 10.1093/nar/26.5.1135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matson C. K., Murphy M. W., Griswold M. D., Yoshida S., Bardwell V. J. and Zarkower D. (2010). The mammalian doublesex homolog DMRT1 is a transcriptional gatekeeper that controls the mitosis versus meiosis decision in male germ cells. Dev. Cell 19, 612-624. 10.1016/j.devcel.2010.09.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng X., Lindahl M., Hyvonen M. E., Parvinen M., de Rooij D. G., Hess M. W., Raatikainen-Ahokas A., Sainio K., Rauvala H., Lakso M. et al. (2000). Regulation of cell fate decision of undifferentiated spermatogonia by GDNF. Science 287, 1489-1493. 10.1126/science.287.5457.1489 [DOI] [PubMed] [Google Scholar]
- Mikula M., Gaj P., Dzwonek K., Rubel T., Karczmarski J., Paziewska A., Dzwonek A., Bragoszewski P., Dadlez M. and Ostrowski J. (2010). Comprehensive analysis of the palindromic motif TCTCGCGAGA: a regulatory element of the HNRNPK promoter. DNA Res. 17, 245-260. 10.1093/dnares/dsq016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy M. W., Sarver A. L., Rice D., Hatzi K., Ye K., Melnick A., Heckert L. L., Zarkower D. and Bardwell V. J. (2010). Genome-wide analysis of DNA binding and transcriptional regulation by the mammalian Doublesex homolog DMRT1 in the juvenile testis. Proc. Natl. Acad. Sci. USA 107, 13360-13365. 10.1073/pnas.1006243107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagano M. C. (2003). Homing efficiency and proliferation kinetics of male germ line stem cells following transplantation in mice. Biol. Reprod. 69, 701-707. 10.1095/biolreprod.103.016352 [DOI] [PubMed] [Google Scholar]
- Nagano M., Ryu B.-Y., Brinster C. J., Avarbock M. R. and Brinster R. L. (2003). Maintenance of mouse male germ line stem cells in vitro. Biol. Reprod. 68, 2207-2214. 10.1095/biolreprod.102.014050 [DOI] [PubMed] [Google Scholar]
- Nakagawa T., Nabeshima Y.-I. and Yoshida S. (2007). Functional identification of the actual and potential stem cell compartments in mouse spermatogenesis. Dev. Cell 12, 195-206. 10.1016/j.devcel.2007.01.002 [DOI] [PubMed] [Google Scholar]
- Oatley J. M. and Brinster R. L. (2006). Spermatogonial stem cells. Methods Enzymol. 419, 259-282. 10.1016/S0076-6879(06)19011-4 [DOI] [PubMed] [Google Scholar]
- Oatley J. M., Avarbock M. R., Telaranta A. I., Fearon D. T. and Brinster R. L. (2006). Identifying genes important for spermatogonial stem cell self-renewal and survival. Proc. Natl. Acad. Sci. USA 103, 9524-9529. 10.1073/pnas.0603332103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oatley J. M., Avarbock M. R. and Brinster R. L. (2007). Glial cell line-derived neurotrophic factor regulation of genes essential for self-renewal of mouse spermatogonial stem cells is dependent on Src family kinase signaling. J. Biol. Chem. 282, 25842-25851. 10.1074/jbc.M703474200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oatley J. M., Oatley M. J., Avarbock M. R., Tobias J. W. and Brinster R. L. (2009). Colony stimulating factor 1 is an extrinsic stimulator of mouse spermatogonial stem cell self-renewal. Development 136, 1191-1199. 10.1242/dev.032243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oatley M. J., Kaucher A. V., Racicot K. E. and Oatley J. M. (2011). Inhibitor of DNA binding 4 is expressed selectively by single spermatogonia in the male germline and regulates the self-renewal of spermatogonial stem cells in mice. Biol. Reprod. 85, 347-356. 10.1095/biolreprod.111.091330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa T., Aréchaga J. M., Avarbock M. R. and Brinster R. L. (1997). Transplantation of testis germinal cells into mouse seminiferous tubules. Int. J. Dev. Biol. 41, 111-122. [PubMed] [Google Scholar]
- Prokhortchouk A., Hendrich B., Jorgensen H., Ruzov A., Wilm M., Georgiev G., Bird A. and Prokhortchouk E. (2001). The p120 catenin partner Kaiso is a DNA methylation-dependent transcriptional repressor. Genes Dev. 15, 1613-1618. 10.1101/gad.198501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puszyk W., Down T., Grimwade D., Chomienne C., Oakey R. J., Solomon E. and Guidez F. (2013). The epigenetic regulator PLZF represses L1 retrotransposition in germ and progenitor cells. EMBO J. 32, 1941-1952. 10.1038/emboj.2013.118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan A. R. and Hall I. M. (2010). BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26, 841-842. 10.1093/bioinformatics/btq033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao S., Zhen S., Roumiantsev S., McDonald L. T., Yuan G.-C. and Orkin S. H. (2010). Differential roles of Sall4 isoforms in embryonic stem cell pluripotency. Mol. Cell. Biol. 30, 5364-5380. 10.1128/MCB.00419-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raverot G., Weiss J., Park S. Y., Hurley L. and Jameson J. L. (2005). Sox3 expression in undifferentiated spermatogonia is required for the progression of spermatogenesis. Dev. Biol. 283, 215-225. 10.1016/j.ydbio.2005.04.013 [DOI] [PubMed] [Google Scholar]
- Roy C. D., Small C., Wang Y., Mueller P. R., Rebel V. I., Griswold M. D. and McCarrey J. R. (2010). Microarray-based analysis of cell-cycle gene expression during spermatogenesis in the mouse. Biol. Reprod. 83, 663-675. 10.1095/biolreprod.110.084889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruzov A., Dunican D. S., Prokhortchouk A., Pennings S., Stancheva I., Prokhortchouk E. and Meehan R. R. (2004). Kaiso is a genome-wide repressor of transcription that is essential for amphibian development. Development 131, 6185-6194. 10.1242/dev.01549 [DOI] [PubMed] [Google Scholar]
- Sakaki-Yumoto M., Kobayashi C., Sato A., Fujimura S., Matsumoto Y., Takasato M., Kodama T., Aburatani H., Asashima M., Yoshida N. et al. (2006). The murine homolog of SALL4, a causative gene in Okihiro syndrome, is essential for embryonic stem cell proliferation, and cooperates with Sall1 in anorectal, heart, brain and kidney development. Development 133, 3005-3013. 10.1242/dev.02457 [DOI] [PubMed] [Google Scholar]
- Schlesser H. N., Simon L., Hofmann M.-C., Murphy K. M., Murphy T., Hess R. A. and Cooke P. S. (2008). Effects of ETV5 (ets variant gene 5) on testis and body growth, time course of spermatogonial stem cell loss, and fertility in mice. Biol. Reprod. 78, 483-489. 10.1095/biolreprod.107.062935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt J. A. and Brinster R. L. (2011). Spermatogonial stem cell transplantation and culture. In Male Germline Stem Cells: Developmental and Regenerative Potential (ed. Orwig K. E. and Hermann B. P.), pp. 107-123. New York, NY: Humana Press. [Google Scholar]
- Schmidt J. A., Abramowitz L. K., Kubota H., Wu X., Niu Z., Avarbock M. R., Tobias J. W., Bartolomei M. S. and Brinster R. L. (2011). In vivo and in vitro aging is detrimental to mouse spermatogonial stem cell function. Biol. Reprod. 84, 698-706. 10.1095/biolreprod.110.088229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shima J. E., McLean D. J., McCarrey J. R. and Griswold M. D. (2004). The murine testicular transcriptome: characterizing gene expression in the testis during the progression of spermatogenesis. Biol. Reprod. 71, 319-330. 10.1095/biolreprod.103.026880 [DOI] [PubMed] [Google Scholar]
- Shinohara T., Avarbock M. R. and Brinster R. L. (1999). β1- and α6-integrin are surface markers on mouse spermatogonial stem cells. Proc. Natl. Acad. Sci. USA 96, 5504-5509. 10.1073/pnas.96.10.5504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinohara T., Orwig K. E., Avarbock M. R. and Brinster R. L. (2000). Spermatogonial stem cell enrichment by multiparameter selection of mouse testis cells. Proc. Natl. Acad. Sci. USA 97, 8346-8351. 10.1073/pnas.97.15.8346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorrentino V., Giorgi M., Geremia R., Besmer P. and Rossi P. (1991). Expression of the c-kit proto-oncogene in the murine male germ cells. Oncogene 6, 149-151. [PubMed] [Google Scholar]
- Suzuki H., Ahn H. W., Chu T., Bowden W., Gassei K., Orwig K. and Rajkovic A. (2012). SOHLH1 and SOHLH2 coordinate spermatogonial differentiation. Dev. Biol. 361, 301-312. 10.1016/j.ydbio.2011.10.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tegelenbosch R. A. J. and de Rooij D. G. (1993). A quantitative study of spermatogonial multiplication and stem cell renewal in the C3H/101 F1 hybrid mouse. Mutat. Res. 290, 193-200. 10.1016/0027-5107(93)90159-D [DOI] [PubMed] [Google Scholar]
- Tokuda M., Kadokawa Y., Kurahashi H. and Marunouchi T. (2007). CDH1 is a specific marker for undifferentiated spermatogonia in mouse testes. Biol. Reprod. 76, 130-141. 10.1095/biolreprod.106.053181 [DOI] [PubMed] [Google Scholar]
- Valli H., Phillips B. T., Orwig K. E., Gassei K. and Nagano M. C. (2015). Spermatogonial stem cells and spermatogenesis. In Knobil and Neill's Physiology of Reproduction, 4th Edn (ed. Plant T. M. and Zeleznik A. J.), pp. 595-635. Amsterdam: Academic Press. [Google Scholar]
- Yang F., Yao Y., Jiang Y., Lu L., Ma Y. and Dai W. (2012). Sumoylation is important for stability, subcellular localization, and transcriptional activity of SALL4, an essential stem cell transcription factor. J. Biol. Chem. 287, 38600-38608. 10.1074/jbc.M112.391441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Q.-E., Gwost I., Oatley M. J. and Oatley J. M. (2013a). Retinoblastoma protein (RB1) controls fate determination in stem cells and progenitors of the mouse male germline. Biol. Reprod. 89, 113 10.1095/biolreprod.113.113159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Q.-E., Racicot K. E., Kaucher A. V., Oatley M. J. and Oatley J. M. (2013b). MicroRNAs 221 and 222 regulate the undifferentiated state in mammalian male germ cells. Development 140, 280-290. 10.1242/dev.087403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon H.-G., Chan D. W., Reynolds A. B., Qin J. and Wong J. (2003). N-CoR mediates DNA methylation-dependent repression through a methyl CpG binding protein Kaiso. Mol. Cell 12, 723-734. 10.1016/j.molcel.2003.08.008 [DOI] [PubMed] [Google Scholar]
- Yoshida S., Takakura A., Ohbo K., Abe K., Wakabayashi J., Yamamoto M., Suda T. and Nabeshima Y.-I. (2004). Neurogenin3 delineates the earliest stages of spermatogenesis in the mouse testis. Dev. Biol. 269, 447-458. 10.1016/j.ydbio.2004.01.036 [DOI] [PubMed] [Google Scholar]
- Yoshida S., Sukeno M. and Nabeshima Y.-I. (2007). A vasculature-associated niche for undifferentiated spermatogonia in the mouse testis. Science 317, 1722-1726. 10.1126/science.1144885 [DOI] [PubMed] [Google Scholar]
- Yoshinaga K., Nishikawa S., Ogawa M., Hayashi S., Kunisada T., Fujimoto T. and Nishikawa S. (1991). Role of c-kit in mouse spermatogenesis: identification of spermatogonia as a specific site of c-kit expression and function. Development 113, 689-699. [DOI] [PubMed] [Google Scholar]
- Zhang Y., Liu T., Meyer C. A., Eeckhoute J., Johnson D. S., Bernstein B. E., Nusbaum C., Myers R. M., Brown M., Li W. et al. (2008). Model-based analysis of ChIP-Seq (MACS). Genome Biol. 9, R137 10.1186/gb-2008-9-9-r137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L., Tang J., Haines C. J., Feng H., Lai L., Teng X. and Han Y. (2011). c-kit and its related genes in spermatogonial differentiation. Spermatogenesis 1, 186-194. 10.4161/spmg.1.3.17760 [DOI] [PMC free article] [PubMed] [Google Scholar]