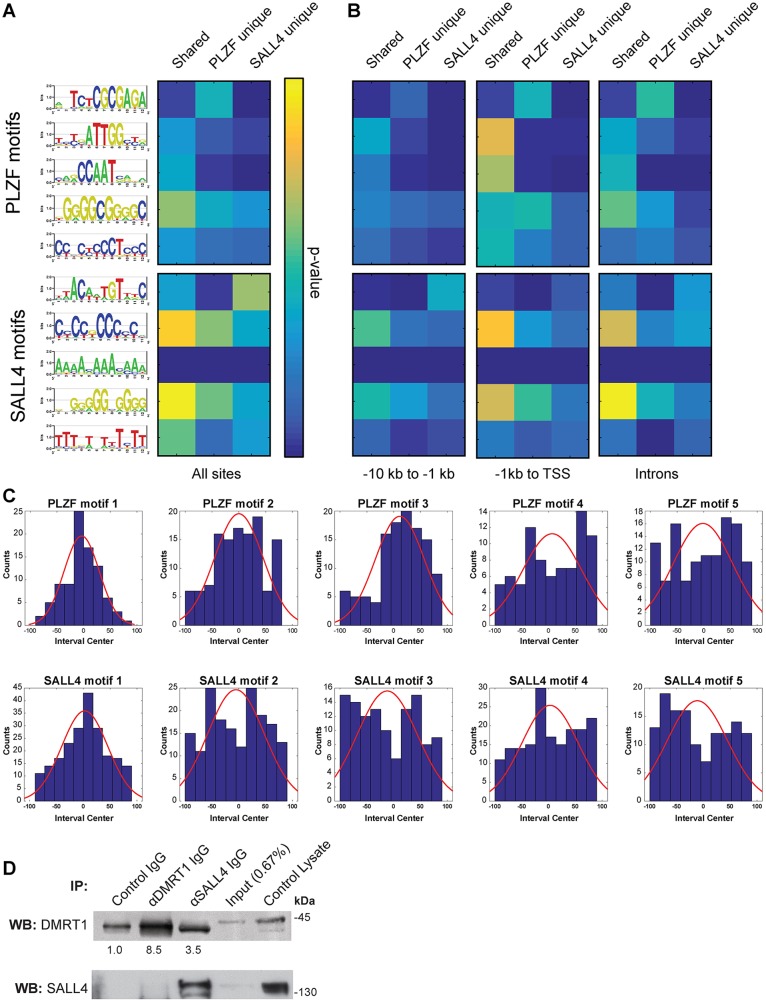
Fig. 2.
SALL4 and PLZF binding motifs in undifferentiated spermatogonia. (A) MEME-derived binding motifs for PLZF (top) and SALL4 (bottom) are shown as DNA base stack logos at each motif position, with their relative size indicative of frequency and total letter stack height reflecting information content at each position (bits). Motif scanning analysis (MAST) was performed with PLZF unique, SALL4 unique and shared binding sites and resulting hypergeometric P-values (Tables S3, S4) are presented as a heat map (yellow indicates lower P-value, see scale). (B) MAST using promoter and intron subsets of unique and shared binding sites (see Fig. 1F). (C) Histograms showing the position of the top five motifs in PLZF (top) and SALL4 (bottom) intervals normalized to the interval center (red curve shows regression). (D) Western blots for DMRT1 (top) and SALL4 (bottom) using control THY1+ spermatogonial lysate, IP input lysate, and eluates from control IgG, DMRT1 IgG or SALL4 IgG IPs. Average band intensities from three replicates are shown beneath the DMRT1 blots.