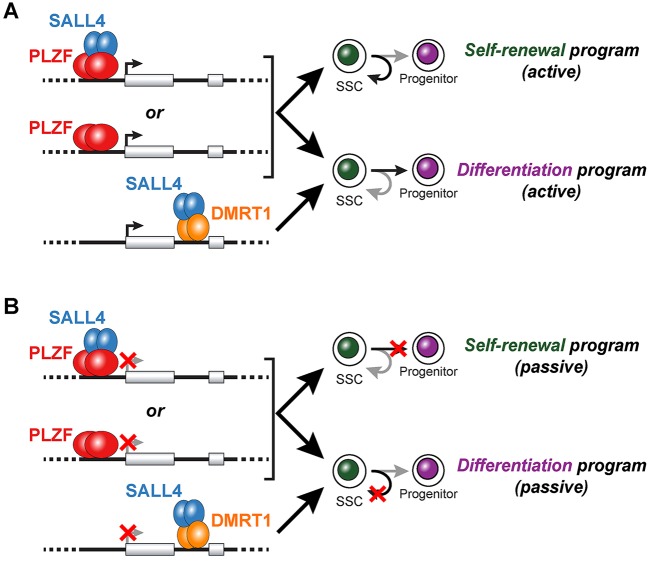
Fig. 6.
Models of PLZF-SALL4 regulation of spermatogonial fate. Binding activities and putative regulatory roles of PLZF and SALL4 in undifferentiated spermatogonia. (A) PLZF dimers (with and without SALL4 interaction) bind to promoters and drive positive gene regulation to initiate gene expression programs associated with self-renewal (curved arrow, top) and differentiation (straight arrow, bottom). Prototypical targets are those involved in SSC self-renewal (Bcl6b, Etv5, Fos) and differentiation (Foxo1). SALL4 binding to introns might also be targeted by DMRT1 and promote the expression of genes involved in spermatogonial differentiation. (B) Passively, PLZF and SALL4 might also drive self-renewal and differentiation gene expression programs by repressing genes involved in the reciprocal cell fate. That is, as above, PLZF dimers (with and without SALL4) interact with gene promoters and SALL4 is targeted to introns by DMRT1. But in this case, the transcriptional effect is repression (red X) of both differentiation (top) and self-renewal (bottom) genes, blocking these fate pathways and leading to reciprocal passive increases in the opposing fate outcome (self-renewal, top; differentiation, bottom). Repression of later spermatogenic genes to prevent differentiation and passively enhance self-renewal would fit into this category. The extent to which these active and passive regulatory mechanisms, via PLZF alone, PLZF-SALL4 or SALL4-DMRT1 complexes occur simultaneously or separately is not known. Dimers are shown throughout, but the precise stoichiometry of DNA binding remains an open question.