Abstract
Ocular lens morphogenesis is a model for investigating mechanisms of cellular differentiation, spatial and temporal gene expression control, and chromatin regulation. Brg1 (Smarca4) and Snf2h (Smarca5) are catalytic subunits of distinct ATP-dependent chromatin remodeling complexes implicated in transcriptional regulation. Previous studies have shown that Brg1 regulates both lens fiber cell differentiation and organized degradation of their nuclei (denucleation). Here, we employed a conditional Snf2hflox mouse model to probe the cellular and molecular mechanisms of lens formation. Depletion of Snf2h induces premature and expanded differentiation of lens precursor cells forming the lens vesicle, implicating Snf2h as a key regulator of lens vesicle polarity through spatial control of Prox1, Jag1, p27Kip1 (Cdkn1b) and p57Kip2 (Cdkn1c) gene expression. The abnormal Snf2h−/− fiber cells also retain their nuclei. RNA profiling of Snf2h−/− and Brg1−/− eyes revealed differences in multiple transcripts, including prominent downregulation of those encoding Hsf4 and DNase IIβ, which are implicated in the denucleation process. In summary, our data suggest that Snf2h is essential for the establishment of lens vesicle polarity, partitioning of prospective lens epithelial and fiber cell compartments, lens fiber cell differentiation, and lens fiber cell nuclear degradation.
KEY WORDS: Lens, Terminal differentiation, Smarca4, Brg1, Smarca5, Snf2h, Denucleation, Cataract
Summary: Depletion of Snf2h induces premature and expanded differentiation of lens precursor cells and reveals the role of Snf2h in lens morphogenesis and denucleation of lens fibers.
INTRODUCTION
ATP-dependent chromatin remodeling is required for transcription, DNA replication, DNA repair and genetic recombination (de la Serna et al., 2006). At least four families of multiprotein chromatin remodeling complexes have been identified in mammalian cells, including SWI/SNF, ISWI, CHD and INO80 (Ho and Crabtree, 2010; Sharma et al., 2010). Twenty-seven genes encode unique DEAD/H-box helicases [e.g. Brg1 (Smarca4), Brm (Smarca2), Snf2h (Smarca5) and Snf2l (Smarca1)] of these complexes. For example, ISWI/Snf2h plays roles in nucleosome sliding and assembly, while Brg1-containing SWI/SNF complexes regulate nucleosome sliding and disruption (Cairns, 2007).
Genetic studies in mice have demonstrated crucial roles for Brg1 (Bultman et al., 2000) and Snf2h (Stopka and Skoultchi, 2003) in blastocyst formation and peri-implantation development, consistent with their functions in embryonic stem cells (Ho et al., 2011; Kidder et al., 2009). Tissue-specific inactivation of Brg1 demonstrated a range of functions in multiple tissues and organs, including blood, brain, eye, lens, muscle and skin. Brg1 controls the proliferation of T-cells (Gebuhr et al., 2003), terminal differentiation in erythrocytes (Griffin et al., 2008), keratinocytes (Indra et al., 2005), lens fibers (He et al., 2010), cardiomyocytes (Hang et al., 2010), Schwann cells (Weider et al., 2012) and adult neural progenitors (Matsumoto et al., 2006; Ninkovic et al., 2013). Brg1 also controls apoptosis in T-cells (Gebuhr et al., 2003) and erythrocytes (Griffin et al., 2008). Two specific Brg1 mutant alleles were identified in model organisms. In mouse, a hypomorphic mutation in the ATPase domain was used to probe β-globin chromatin structure and expression (Bultman et al., 2005). In zebrafish, a nonsense mutation in one of two duplicated brg1 genes abrogates retinal development (Gregg et al., 2003). Compared with Brg1, less is known about the role(s) of Snf2h and of Snf2h-containing complexes (ACF, CHRAC, ISWI and WICH) during organogenesis. Snf2h regulates erythropoiesis (Stopka and Skoultchi, 2003) and neuronal progenitor cell formation and their subsequent differentiation (Alvarez-Saavedra et al., 2014).
Mammalian lens development is an advantageous system with which to study the molecular mechanisms of cellular differentiation, including the regulation of cell cycle exit, chromatin dynamics and elimination of subcellular organelles (Bassnett, 2009; Cvekl and Ashery-Padan, 2014). The lens is composed of a layer of epithelial cells that overlie a bulk of differentiated fiber cells. The mature fiber cells express and accumulate crystallin proteins, acquire a highly elongated cellular morphology, and degrade endoplasmic reticulum (ER), Golgi apparatus, mitochondria and nuclei. Lens compartmentalization into the epithelium and fibers originates from the early transitional structure termed the lens vesicle (∼E11.5 in mouse embryos). The lens vesicle is polarized. Its posterior cells exit the cell cycle in response to the BMP and FGF growth factors produced by the retina and ciliary body, and differentiate into the primary lens fiber cells (Griep and Zhang, 2004; Gunhaga, 2011). The anterior cells differentiate into a sheet of single-layered lens epithelial cells (Martinez and de Iongh, 2010). Lens epithelial cells close to the lens equator divide continually. Following cell cycle exit, these cells subsequently differentiate into secondary lens fiber cells. Between E16.5 and E18, lens fiber cell nuclei are degraded to produce an organelle-free zone (OFZ) at the center of the lens (Bassnett, 2009). DNase II-like acid nuclease DNase IIβ (Dnase2b) plays an essential role in this process. Expression of Dnase2b is downstream of transcription factors including AP-2α (Tfap2a), FoxE3, Hsf4 and Pax6 (Blixt et al., 2007; Fujimoto et al., 2004; Medina-Martinez et al., 2005; West-Mays et al., 2002; Wolf et al., 2009). Our previous studies showed that Brg1 is required for lens fiber cell differentiation, expression of DNase IIβ, and the degradation of lens nuclei (He et al., 2010).
Genetic studies have implicated retinoblastoma protein (Rb1), E2Fs and the cell cycle inhibitors p27Kip1 (Cdkn1b) and p57Kip2 (Cdkn1c) in the regulation of cell cycle exit in lens (Chen et al., 2000; McCaffrey et al., 1999; Morgenbesser et al., 1994; Wenzel et al., 2011; Zhang et al., 1998). BMP, FGF and Notch signaling pathways regulate lens fiber cell differentiation in conjunction with DNA-binding transcription factors, including FoxE3 (Blixt et al., 2007; Brownell et al., 2000; Medina-Martinez et al., 2005), Gata3 (Maeda et al., 2009), Pax6 (Shaham et al., 2009), Pitx3 (Ho et al., 2009; Medina-Martinez et al., 2009), Prox1 (Duncan et al., 2002; Wigle et al., 1999), Hey1 (Herp2) and Rbpj (Jia et al., 2007; Rowan et al., 2008). Although little is known about links between BMP and FGF signaling and these factors, disruption of Prox1 blocks expression of p27Kip1 and p57Kip2 in the posterior part of the lens vesicle, followed by arrested lens fiber cell elongation (Wigle et al., 1999). Loss of FoxE3 abrogates Prox1 expression and consequently dysregulates expression of p57Kip2 (Medina-Martinez et al., 2009). Hey1 and Rbpj DNA-binding proteins directly control p27Kip1 and p57Kip2 expression (Jia et al., 2007). Taken together, perturbation of cell cycle exit in the lens is linked to abnormal fiber cell differentiation. To expand our knowledge of chromatin remodeling during mammalian embryogenesis, we have investigated whether Snf2h regulates lens development in mouse.
RESULTS
Conditional inactivation of Snf2h disrupts lens differentiation
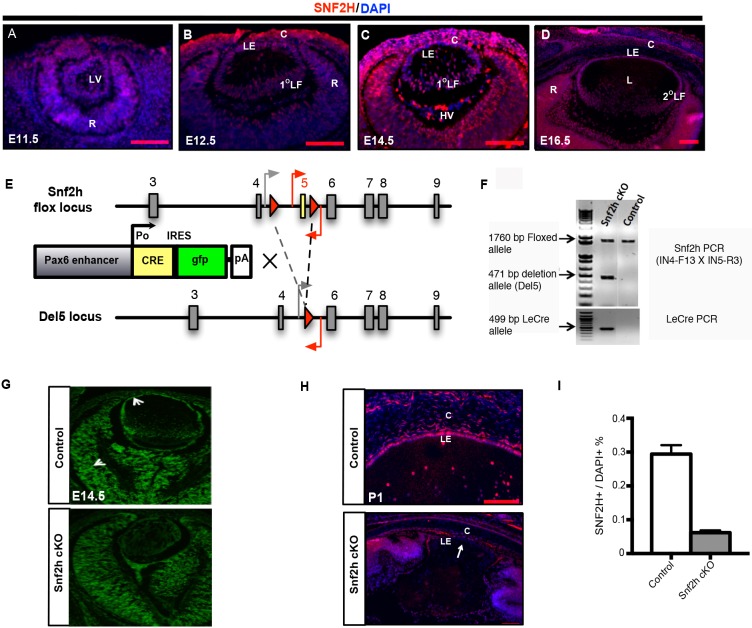
To understand the function of Snf2h in lens development, we first determined the Snf2h expression pattern during mouse eye development by immunofluorescence. We found expression of Snf2h throughout embryonic lens development (E11.5 to E16.5) (Fig. 1A-D). The data show similar levels of Snf2h expression in the anterior and posterior parts of the lens vesicle (Fig. 1B), the lens epithelium, and the primary and secondary lens fibers (Fig. 1C,D). The cornea and both inner and outer nuclear layers of the retina also express Snf2h (Fig. 1B-D). At postnatal stages, Snf2h expression continues in the lens epithelium and the differentiating secondary lens fiber cells (Fig. 1H; data not shown).
Fig. 1.
Expression of Snf2h and initial analysis of the Snf2h lens conditional knockout (Snf2h cKO) mouse. (A-D) Localization of Snf2h proteins (red) during embryonic mouse eye development as assessed by immunofluorescence. Nuclei were counterstained with DAPI (blue). HV, hyaloid vasculature; L, lens; LE, lens epithelium; 1°LF and 2°LF, primary and secondary lens fibers; C, cornea; R, retina. (E) Schematic representation of the Snf2h flox allele (showing exons 3 to 9 as gray boxes), the Le-Cre driver construct and the resulting deletion of exon 5 (Del5 locus). Two loxP sites (red triangles) were inserted to flank exon 5 (yellow box). Primers used for PCR genotyping are indicated by gray and red arrows. (F) Le-Cre-mediated recombination of the Snf2h floxed locus. The 1760, 471 and 499 bp bands correspond to the Snf2hfl, Snf2h null (Del5) and Le-Cre transgene alleles analyzed with lens/anterior segment genomic DNA, respectively. (G) Immunofluorescence analysis of Snf2h expression (green) in E14.5 control and Snf2h cKO embryos. Arrows indicate positive staining of Snf2h in the ocular tissues. (H) Immunofluorescence of Snf2h expression (red) in newborn (P1) control and Snf2h cKO eyes. Arrow indicates reduction of Snf2h expression in mutant lens but not in other regions of the eye. (I) Quantification of Snf2h-positive cells in control and Snf2h cKO P1 lenses. Error bars indicate s.d. of three different animals. There is a marked reduction of Snf2h expression in mutant lens but not in other regions of the eye (G-I). Scale bars: 100 μm.
To investigate the roles of Snf2h in mouse lens development, we inactivated Snf2h in the surface ectoderm-derived tissues using the Le-Cre transgene. The Le-Cre mouse is a transgenic line in which a 6.5 kb genomic fragment from the mouse Pax6 gene (Fig. 1E) drives the expression of Cre recombinase and green fluorescent protein (GFP) from between E8.5 and E9 (Ashery-Padan et al., 2000). Genotyping of genomic DNA samples showed bands corresponding to the Snf2h wild-type (wt), flox (fl) and null (deletion of exons 5-9) alleles (Fig. 1F). The newborn Snf2h heterozygous mice (Snf2hfl/−) appeared normal. Depletion of Snf2h proteins was confirmed in the E14.5 and newborn lens of Snf2hfl/fl; Le-Cre (Fig. 1G,H). Snf2hfl/−; Le-Crewt/+ (referred to as Snf2h cKO) exhibited a wide spectrum of ocular defects (see Fig. 2). However, their littermates were normal and served as controls in the comparative experiments.
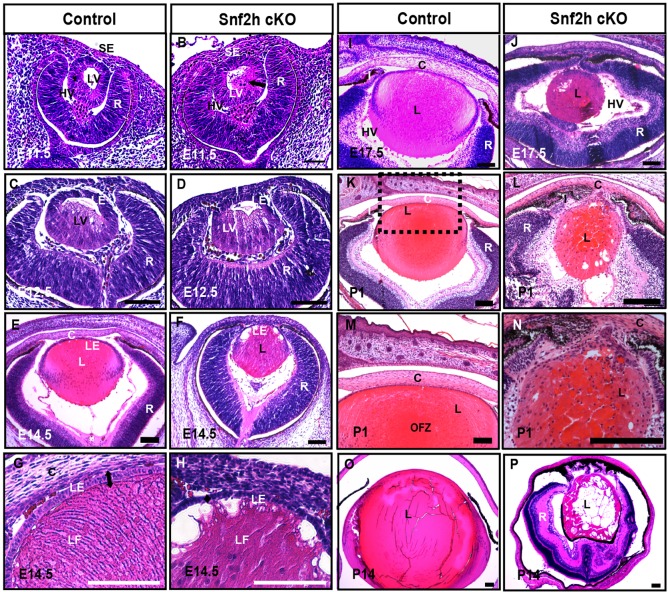
Fig. 2.
Snf2h is necessary for mouse lens development. (A,B) Lens vesicle is separated from the surface ectoderm in the Snf2h cKO at E11.5. Note that a number of cells at the posterior of the lens vesicle (LV) in the Snf2h cKO already appeared to elongate (arrow in B). (C,D) Primary lens fiber cells are elongated within the lens vesicle at E12.5. The prominent hyaloid vasculature (HV) occupies the space between the lens (L), cornea (C) and retina (R) in the Snf2h cKO eyes. (E-H) At E14.5, lens fiber cells in the Snf2h cKO were unable to form the bow region/transitional zone and the fiber-like morphology of these cells deteriorated at the anterior of the lens. (I,J) The E17.5 Snf2h cKO shows a series of defects in lens, cornea, iris (I) and retina. (K-N) The newborn Snf2h mutant lens does not form the presumptive organelle free zone (OFZ). The boxed region in K is magnified in M. (O,P) Profound ocular defects in Snf2h cKO are found at P14, including the absence of lens anterior chamber, lens vacuoles, and disorganization of the lens fiber cells. LE, lens epithelium; LF, lens fibers; SE, surface ectoderm. Scale bars: 100 μm.
The specific eye defects of the Snf2h cKO were first characterized by histology (Fig. 2). At E11.5, although the mutant lens vesicle separated normally from the surface ectoderm, a number of posterior cells started to differentiate prematurely, as evidenced by their elongation (compare Fig. 2A,B). At E12.5, both the Snf2h cKO and control lenses underwent primary lens fiber cell differentiation. However, the Snf2h cKO lens was surrounded by a hypertrophic hyaloid vasculature, leaving a narrower vitreous space between the lens and retina (compare Fig. 2C,D). Compared with the control, the Snf2h cKO lens was reduced in size at E14.5, when primary lens fiber cells normally reach the lens epithelium (compare Fig. 2E,F). The elongation of primary lens fiber cells was disturbed in Snf2h cKO lenses, as indicated by abnormal formation of transitional zones (marked by the nuclei of cells that exited the cell cycle) at the lens equator (Fig. 2E,F). In addition, the lens epithelium of the Snf2h cKO was thinner and its cuboidal morphology was compromised (Fig. 2G,H). At E17.5, the growth deficiency of the Snf2h cKO lens was very pronounced (Fig. 2I,J). At postnatal stages (P1 and P14) we detected progressive deterioration and cataractogenesis in the mutant lenses (compare Fig. 2K,M,O with L,N,P). The cornea in the Snf2h cKO failed to differentiate into its stratified layers (compare Fig. 2I,J with M,N), probably owing to loss of Snf2h from the presumptive corneal epithelial cells, which also express the Le-Cre transgene (Ashery-Padan et al., 2000). At E17.5 and P1, the abnormal lens fiber cell mass protruded towards the cornea and eventually formed iridocorneal adhesion masses at the anterior segment (Fig. 2J-L), raising questions concerning the presence of lens epithelial cells and/or their ability to establish proper contacts with the elongating lens fiber cell mass to control lens shape (see below). Notably, lens fiber cell nuclei were retained in the presumptive OFZ in the Snf2h cKO lenses (Fig. 2K,L). Taken together, deletion of Snf2h in the mouse embryo does not appear to affect early stages of lens formation; however, it leads to arrested lens growth, aberrant lens compartmentalization, perturbed fiber cell differentiation, and marked defects in lens fiber cell denucleation.
To aid data interpretation, expression of the related protein Snf2l was assessed in the mouse eye. Expression of Snf2l protein in the eye is mostly restricted to the retina, as described previously at the RNA level (Magdaleno et al., 2006). By immunofluorescence, additional expression of Snf2l was detected in the lens transitional zone (Fig. S1A-C). At the RNA level, the expression of Snf2h is much higher than that of Snf2l in both the E15.5 and newborn lens (Fig. S1D). Interestingly, depletion of Snf2h in mouse cerebral extracts was followed by increased levels of Snf2l protein (Alvarez-Saavedra et al., 2014). By contrast, western immunoblotting data showed no upregulation of Snf2l in Snf2h mutant lens/eyes (Fig. S1E).
Cellular and molecular characterization of lens differentiation defects in the Snf2h cKO model
To explain the disrupted lens growth and differentiation of Snf2h cKO embryos, we focused on lens size reduction and aberrant morphogenesis following the completion of primary lens fiber cell elongation (Fig. 2E,F). Microphthalmia suggested reduced cell growth in the anterior part of the lens vesicle/prospective lens epithelium due to the deletion of Snf2h. It has been shown previously that ISWI chromatin remodeling complexes control proliferation via the rate of S-phase progression (Arancio et al., 2010; Collins et al., 2002). To test this possibility, we evaluated cell proliferation by analysis of BrdU (5-bromo-2′-deoxyuridine) incorporation and expression of the Ki67 protein, a marker of dividing cells, in E14.5 lenses. We found that the number of proliferating presumptive lens epithelial cells was reduced in the Snf2h cKO (Fig. S2). The bilateral lens germinative zones, where active proliferating lens epithelial cells reside, normally increase in cell number towards the lens equator, where cells exit from the cell cycle and differentiate (Fig. S2A-C). However, in the Snf2h cKO lenses, reduced numbers of dividing cells were found around the lens equator at E14.5. In addition, the lens transitional zones moved anteriorly, and hence the size of the presumptive lens epithelial region was reduced (Fig. S2D-F). Quantitative analysis of BrdU-positive and Ki67-positive cells (Fig. S2G,H) confirmed these staining patterns. We conclude that the Snf2h-deficient lens cells exhibit a reduced number and disturbed pattern of dividing cells in the presumptive lens transitional zone.
The histological analysis of P1 lenses (compare Fig. 2M,N) raised a major question regarding the status of lens epithelium in Snf2h cKO lenses. Lens epithelium is marked by expression of FoxE3 and E-cadherin, and higher levels of Pax6 expression are found in lens epithelium than in lens fibers. In E12.5-E14.5 lens, expression of FoxE3 is confined to the nascent lens epithelium (Blixt et al., 2000; Medina-Martinez et al., 2005), and its inactivation accounts for the dysgenetic lens (dyl) mutant phenotype (Blixt et al., 2000; Medina-Martinez et al., 2005), which is characterized by abnormal lens fibers, defects in denucleation, vacuolization, and the structural collapse of the lens. The dyl defects are directly comparable to the present abnormalities in Snf2h mutant lens (Fig. 2). In wild-type (WT) E12.5 embryos, FoxE3 expression was found in the anterior portion of the forming lens (Fig. 3A). By contrast, expression of FoxE3 was significantly reduced in the Snf2h cKO as early as E12.5 (Fig. 3B). In E14.5 WT lens, expression of FoxE3 continued in the lens epithelial cell layer (Fig. 3C), whereas FoxE3 expression was strongly reduced in E14.5 Snf2h cKO lenses (Fig. 3D). Lens-specific expression of FoxE3 never reappeared at subsequent stages examined: E16.5 and P1 (data not shown). The DNA-binding transcription factor Pax6 is a key regulator of multiple stages of lens development (Shaham et al., 2012). At E14.5, discontinuous and moderately reduced Pax6 expression was found at the anterior of the Snf2h cKO lenses (Fig. S3C). At P1, expression of Pax6 was further reduced in mutant lenses (compare Fig. S3B,D). Thus, expression of Pax6 and its downstream target FoxE3 (Blixt et al., 2007) are reduced in lens following Snf2h depletion.
Fig. 3.
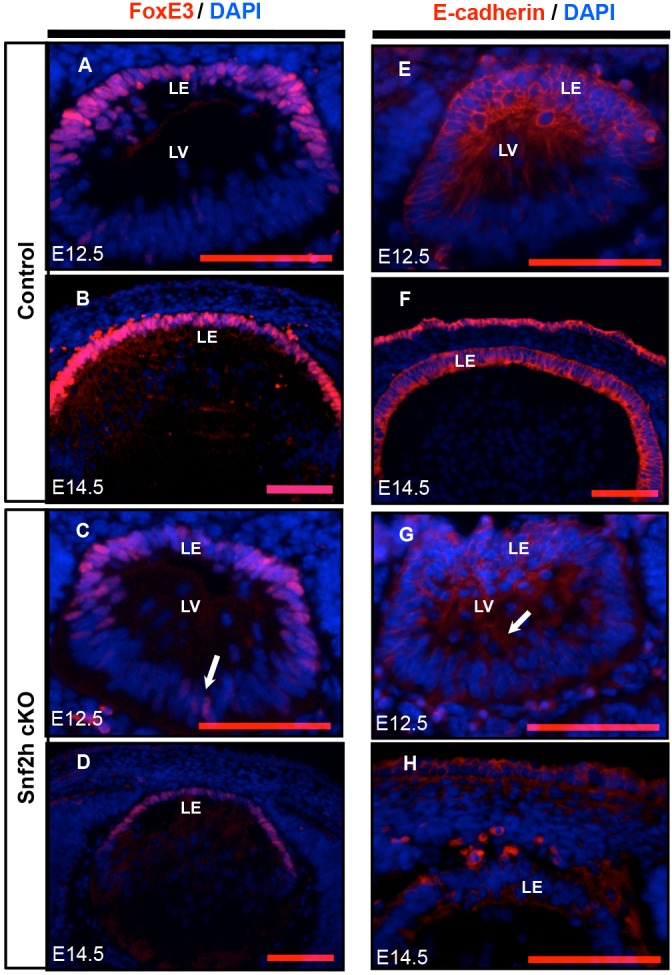
Depletion of Snf2h results in a disrupted presumptive lens epithelial compartment. (A-D) Downregulation of FoxE3 (red) in Snf2h mutant lenses. Arrow (C) indicates sparse FoxE3 protein at the posterior cells undergoing lens fiber cell elongation. (E-H) Reduced and disorganized expression of E-cadherin (red) in wild-type and Snf2h cKO lenses. Arrow (G) indicates dislocation of E-cadherin in the E12.5 Snf2h cKO lens vesicle. The nuclei were counterstained with DAPI (blue). LV, lens vesicle; LE, lens epithelium. Scale bars: 100 μm.
The defects in Snf2h mutant lens can also be assessed through expression of E-cadherin [cadherin 1 (Cdh1)], a cell-cell adhesion glycoprotein specific to epithelial cells and required for lens morphogenesis (Pontoriero et al., 2009). At E12.5 in the control, strong expression of E-cadherin was restricted to the prospective lens epithelium at the anterior of the developing lens vesicle (Fig. 3E). By contrast, in the Snf2h cKO, E-cadherin expression was reduced and its distribution was expanded towards the primary lens fiber cell compartment (Fig. 3F). Importantly, at E14.5 the Snf2h cKO lenses lost E-cadherin expression and did not establish any morphologically discernible lens epithelium (compare Fig. 3G,H). Taken together, these results show that depletion of Snf2h disrupts lens differentiation through downregulation of Pax6, FoxE3 and E-cadherin expression, and by reducing the number of cells from which the lens epithelium is normally formed.
Dysregulation of cell cycle exit control genes in Snf2h mutant lens
To probe the disrupted lens growth and differentiation we analyzed the expression of genes involved in cell cycle exit control. Prox1, regulated by FGF signaling (Zhao et al., 2008), controls expression of the cyclin kinase inhibitors p27Kip1 and p57Kip2 in differentiating lens cells (Wigle et al., 1999). In parallel, Notch signaling, as probed through conditional inactivation of the Notch2 receptor (Saravanamuthu et al., 2012), the jagged 1 (Jag1) ligand (Le et al., 2012) and the downstream Rbpj DNA-binding transcription factor (Jia et al., 2007; Rowan et al., 2008) were shown to act upstream of p27Kip1 and/or p57Kip2. We examined the expression of Prox1, Jag1, p27Kip1 and p57Kip2.
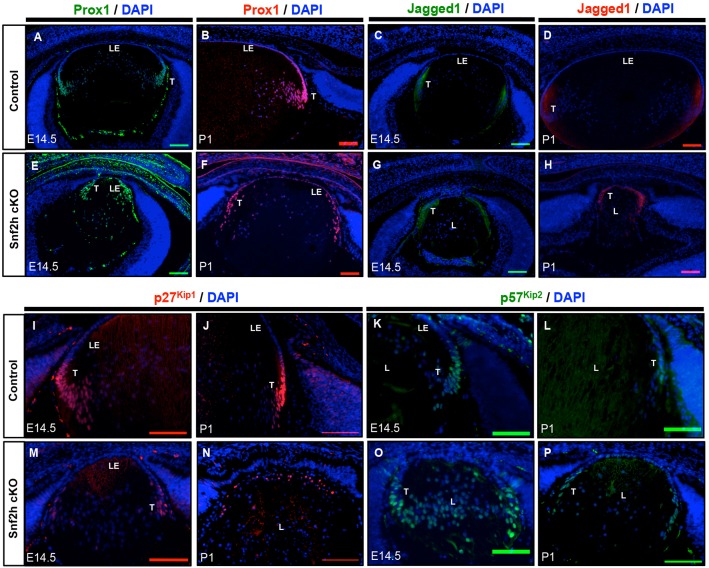
In WT E14.5 and P1 newborn lens, abundant expression of Prox1 was found in the transitional zones (Fig. 4A,C). By contrast, in Snf2h cKO lenses the expression of Prox1 was expanded into regions that included the presumptive lens epithelium (Fig. 4B,D). The Jag1 expression pattern shifted from the equatorial zone towards the lens anterior in the Snf2h cKO (Fig. 4E-H). In WT lenses, p27Kip1 and p57Kip2 (Zhang et al., 1998) were only expressed in cells localized in the lens equator transitional zone that are undergoing cell cycle exit (Fig. 4I,K,M,O). In Snf2h cKO E14.5 lenses, cells expressing p27Kip1 (Fig. 4J,L) and p57Kip2 (Fig. 4N,P) did not form the ‘compact’ transitional zone but were instead found in many abnormal positions. In P1 Snf2h cKO lenses, transitional zones were not established and the expression of p27Kip1 was found in the lens anterior compartment (Fig. 4L). These data suggest that Snf2h first regulates spatial aspects of Prox1 and Jag1 expression, and directly and/or indirectly affects the expression of p27Kip1 and p57Kip2. Hence, inactivation of Snf2h promotes cell cycle exit and thus the ‘borderline’ dividing proliferating from differentiating cells shifts towards the anterior of the lens, and cells in this region differentiate prematurely. We propose that these changes deplete the lens epithelium of progenitor cells and arrest lens growth.
Fig. 4.
Depletion of Snf2h disrupts expression of Prox1, Jag1, p27Kip1 and p57Kip2 in the lens. (A,B,E,F) In the control, Prox1 is expressed in cells undergoing cell cycle exit and at the onset of lens secondary fiber cell differentiation at the transitional zone (T) (A,B). In Snf2h cKO, Prox1 staining is expanded towards the lens anterior, including the middle of the presumptive lens epithelial layer (LE) (E,F). (C,D,G,H) Jag1 is present in cells that are starting to elongate and differentiate. Cells expressing p27Kip1 in E14.5 or P1 animals are shown in red (I,J,M,N). Cells expressing p57Kip2 in E14.5 or P1 animals are shown in green (K,L,O,P). In the control, the Jag1 staining pattern initiated at the lens equator and expanded towards the posterior of the lens (C,D). In the mutant lens, localization of Jag1 was shifted from posterior to the anterior of the lens (G,H). (I-P) Distribution of the cell cycle inhibitors p27Kip1 and p57Kip2 in control and Snf2h mutant lenses. Note disorganized localization of both p27Kip1 and p57Kip2 in the mutated lenses. Nuclei were counterstained with DAPI (blue). L, lens. Scale bars: 100 μm.
Retention of lens fiber cell nuclei, normal mitochondrial degradation, and disrupted expression of autophagy regulatory proteins in the Snf2h cKO lens
Lens fiber cell denucleation and the degradation of other subcellular organelles, including mitochondria and ER, mark the terminal differentiation of lens fibers (Bassnett et al., 2011). The persistence of nuclei in Snf2h mutant lens fibers (Fig. 2) was further examined through the detection of free 3′-OH DNA double-strand ends, which are generated by the lens-specific enzyme DNase IIβ. In the Snf2h cKO lenses, both a higher density of lens fiber nuclei and a reduced number of TUNEL-positive nuclei were observed, particularly in the lens cortical area (Fig. S4A-I). This suggested that retention of nuclei in the Snf2h cKO lens fiber cells could be caused by reduced expression and/or activity of DNase IIβ (see below).
Lens organelle degradation has recently been linked to autophagy-related processes (Basu et al., 2014) and mitophagy (Costello et al., 2013). We first examined the degradation of mitochondria (visualized by Tomm20 antibodies) and of ER (visualized by PDI antibodies) in control and Snf2h-depleted lenses. We found that both processes occurred normally in the mutant lenses (Fig. 5A,B; data not shown).
Fig. 5.
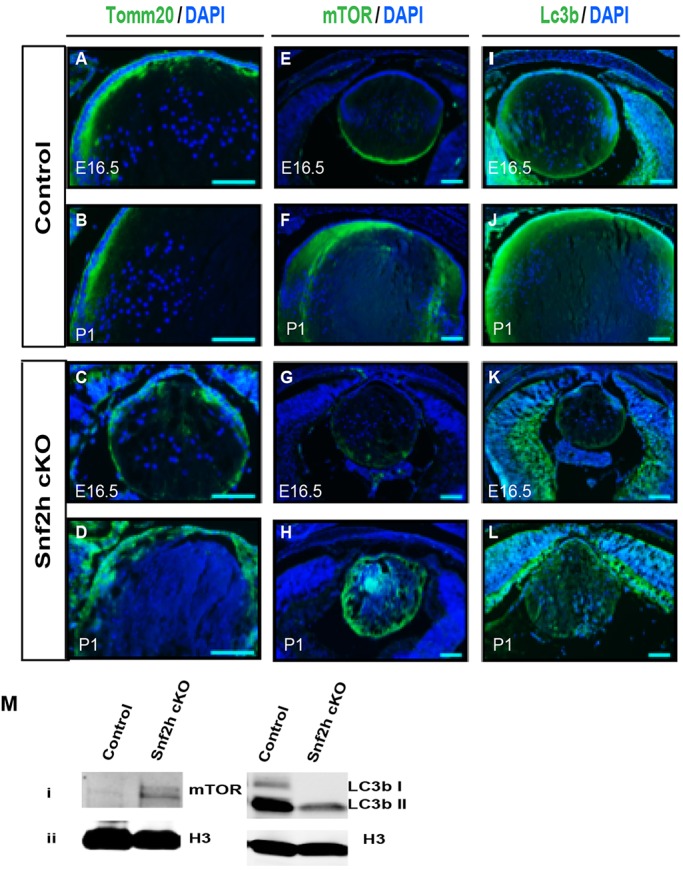
Degradation of mitochondria and expression of autophagy proteins mTOR and LC3b in E16.5 and P1 control and Snf2h cKO lenses. (A-L) Immunofluorescence analysis of Tomm20, mTOR and LC3b in control and Snf2h cKO eyes. Nuclei were counterstained with DAPI (blue). Scale bars: 100 μm. (M) Western blot analysis of mTOR and LC3b in extracts prepared from P1 tissues. LC3b I and LC3b II bands represent cytoplasmic and autophagosome forms of the protein, respectively. Histone H3 was used as a loading control.
Next, we evaluated the expression of the serine/threonine kinase mechanistic target of rapamycin (Mtor) and microtubule-associated protein 1 light chain 3 beta (LC3b; also known as Map1lc3b) autophagy proteins in control and Snf2h cKO lenses at E16.5, i.e. ∼48 h prior to the formation of the OFZ in WT mouse lens (Bassnett, 2009), as well as in newborn lens. In E16.5 control lenses, mTOR was predominantly localized near the basal ends of lens fibers (Fig. 5E). In the corresponding Snf2h-depleted lenses, expression of this kinase was both reduced and spatially perturbed (Fig. 5G). In control P1 lenses, mTOR protein was found throughout the lens fibers excluding the OFZ (Fig. 5F). By contrast, in Snf2h cKO lenses (Fig. 5H) mTOR protein was unevenly distributed throughout the entire lens fiber cell compartment, including the presumptive nuclear-free zone (NFZ). In E16.5 control lenses, LC3b was found throughout the entire lens (Fig. 5I), whereas in the Snf2h cKO there was a notable reduction of LC3b proteins in the lens (Fig. 5K). In control P1 lenses, LC3b proteins were predominantly expressed outside of the NFZ (Fig. 5J). In the absence of NFZ in Snf2h mutant lenses, LC3b proteins displayed a disorganized spatial distribution throughout the lens fiber cell mass (Fig. 5L). Finally, western immunoblotting was used to evaluate expression levels of these proteins in lens-containing cellular extracts. Less LC3b protein was present in extracts prepared from mutant tissues than from controls (Fig. 5M). Notably, expression of LC3b form II, which is associated with the autophagosome (Kabeya et al., 2000), was not found in extracts prepared from mutant newborn lens and surrounding tissues. By contrast, expression of mTOR1 appeared to be increased in the Snf2h-depleted tissues (Fig. 5M).
Taken together, these data show normal degradation of mitochondria in Snf2h-depleted lenses in the presumptive OFZ. However, degradation of nuclei in the presumptive NFZ is not executed and the autophagic flux in the Snf2h-depleted lens fiber cell compartment is disrupted.
Molecular analysis of the lens fiber cell denucleation pathway in Brg1−/− and Snf2h−/−
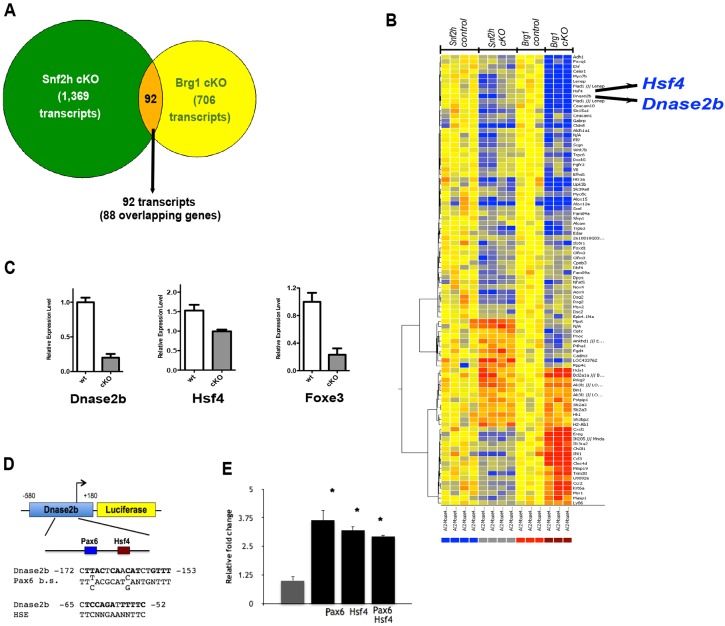
The SWI/SNF and ISWI complexes regulate gene expression through distinct molecular mechanisms (Kadam and Emerson, 2003; Narlikar et al., 2002; Tang et al., 2010). Nevertheless, similar defects in nuclear degradation were observed in both Brg1 (He et al., 2010) and Snf2h (Figs 2 and 5) cKO lenses. To clarify this, we examined differential gene expression in Snf2h cKO eyes using high-density oligonucleotide microarray hybridizations and compared the results with the earlier Brg1 null lens studies (He et al., 2010). We found 1461 differentially expressed transcripts in the Snf2h cKO eyes, including 902 upregulated and 559 downregulated genes (Fig. 6A). These genes were mostly classified into expected categories such as DNA replication, DNA damage repair, cell cycle control, transcription, and growth signaling response (Table S1). Next, we compared the Snf2h cKO differentially expressed genes with genes differentially expressed in Brg1 mutant eyes. Using our earlier data (He et al., 2010), re-analyzed using identical statistical criteria (P<0.05 and at least 1.5-fold change), we found a total of 798 differentially expressed transcripts in the Brg1 mutant eyes. The majority (96%) of the differentially expressed genes were unique to each system. Nevertheless, 92 transcripts were identified as regulated in both systems, including 88 individual genes. In this group, we found downregulation of Hsf4 and Dnase2b mRNAs (Fig. 6B,C), both of which are implicated in lens fiber cell denucleation (Cui et al., 2013; Fujimoto et al., 2004; Nishimoto et al., 2003). In addition, retention of nuclei in Foxe3 mutant lens (Medina-Martinez et al., 2005) is consistent with the reduced expression of FoxE3 observed at both the protein (Fig. 3) and mRNA (Fig. 6C) levels in Snf2h mutant lenses. Among the 1461 differentially expressed genes, dysregulated expression of p27Kip1, p57Kip2, Cdh1, Jag1, Prox1 and Rbpj in Snf2h cKO eye tissues was independently confirmed by qRT-PCR (Fig. S5).
Fig. 6.
Comparative analysis of Snf2h-dependent and Brg1-dependent transcriptomes in the eye and transcriptional regulation of Dnase2b by Hsf4 and Pax6. (A) Venn diagram comparing RNA profiling of Brg1 (1461 dysregulated transcripts) versus Snf2h (798 dysregulated transcripts) mutant mouse lens. (B) The list of 92 (∼4%) common transcripts regulated by both Snf2h and Brg1 contains 88 unique genes, including Dnase2b and Hsf4. Note that only 62 genes (52 downregulated and 10 upregulated) out of the 92 found were regulated similarly in Brg1 and Snf2h mutant eyes. (C) Reduced expression of Foxe3, Hsf4 and Dnase2b mRNAs in Snf2h mutant tissues analyzed by qRT-PCR. (D) The Dnase2b promoter (−580 to +180) showing Hsf4 and Pax6 binding sites (b.s.). HSE, heat shock element-Hsf4 binding site. (E) Hsf4 and Pax6 activate the Dnase2b promoter in cultured lens cells. Data are shown as mean±s.d. (n=3) normalized to Renilla-TK luciferase internal control, and relative fold change in luciferase activity was calculated using the empty cDNA control value set at 1 (gray bar). *P≤0.05 (paired Student's t-test), cDNA control vector versus Pax6 and/or Hsf4 cDNA.
Transcription of Dnase2b could be directly regulated by the transcription factors Hsf4 (Cui et al., 2013) and Pax6 (He et al., 2010). Hsf4 and Pax6 binding sites are located around positions −160 and −60, respectively, of the Dnase2b promoter (Fig. 6D). To test whether they directly regulate the Dnase2b promoter in cultured lens cells, we cotransfected the mouse Dnase2b promoter fragment (−580 to +180) with cDNAs encoding Hsf4 and Pax6. We found over 3-fold transcriptional activation by both factors in lens cells; however, no transcriptional synergism between these factors was detected (Fig. 6E).
Taken together, the current data show that depletion of either Brg1 or Snf2h results in the retention of nuclei in lens fiber cells, fewer TUNEL-positive nuclei, and reduced expression of Hsf4 and of Dnase2b, which is a direct target of Hsf4 and Pax6.
DISCUSSION
The goal of the present study was to dissect the in vivo roles of Snf2h during lens development using conditional gene targeting, and to compare functions of Snf2h and Brg1 in this system. We show two major roles of Snf2h in lens formation: (1) Snf2h regulates lens differentiation through maintaining the balance between epithelial and fiber cell differentiation; and (2) Snf2h and Brg1 are both independently required for lens fiber cell denucleation through regulation of at least two common genes essential for this process, namely Hsf4 and Dnase2b (Fig. 7). Comparison of the Snf2h and Brg1 loss-of-function studies in the lens demonstrate that these two chromatin remodeling enzymes play mostly distinct roles (He et al., 2010). This is consistent with the distinct biochemical modes of action of Brg1 and Snf2h ATPases (Erdel and Rippe, 2011; Kadam and Emerson, 2003; Khavari et al., 1993; Narlikar et al., 2002; Tang et al., 2010; Toiber et al., 2013). The role of Snf2h in lens differentiation is related to its recently established function in the control of Purkinje and granule cell progenitor proliferation during cerebellar development (Alvarez-Saavedra et al., 2014). In contrast to the neuronal progenitor cells, loss of Snf2h in lens was not compensated by the induction of Snf2l expression. Thus, our data demonstrate tissue-specific molecular responses following Snf2h depletion, and establish its role as a gatekeeper to assure the timely differentiation of lens fibers.
Fig. 7.
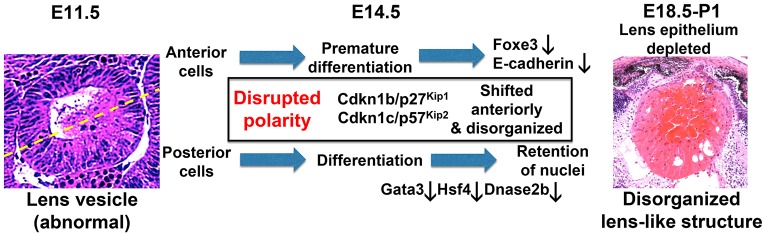
A multitude of Snf2h functions in lens development. The model proposes that, in the absence of Snf2h, disrupted polarity of the lens vesicle triggers a cascade of events resulting in abnormal lens fiber cell differentiation and premature differentiation of anterior lens epithelial cells. Yellow dashed line indicates division between the anterior and posterior compartment of the lens vesicle.
The earliest morphological abnormality that we found was disrupted polarity of the lens vesicle in Snf2h cKO embryos, as inferred from subsequent differentiation defects (Fig. 7). At E14.5 there are notable differences between control and Snf2h cKO lenses, including reduced size, vacuolization, and disruption of primary lens fiber cell differentiation in mutant lenses. In newborn lens and in the absence of the anterior epithelium, the primary lens fiber cell mass penetrates anteriorly through the bulk lens mass and reaches the cornea. This mass is distinct from corneal-lenticular bridges that originate from incomplete separation of the lens vesicle from the surface ectoderm, as caused by mutations in genes including Pax6, Foxe3 and AP-2α (Cvekl and Ashery-Padan, 2014). It is noteworthy that reduction of Snf2h expression in Xenopus by morpholinos caused similar lens growth and differentiation defects (Dirscherl et al., 2005). Analysis of lens morphology coupled with expression analysis of epithelial markers shows that the presumptive lens epithelial cell layer is markedly reduced at E14.5, and later eliminated due to the premature terminal differentiation of lens precursor cells in Snf2h mutant lenses. The ‘earlier’ cells detected at the anterior pole of the lens vesicle at E14.5 do not display the cuboidal morphology characteristic of the WT lens epithelium. Although these cells initially express E-cadherin, the expression of this crucial structural protein of lens epithelium (Pontoriero et al., 2009) is reduced at E14.5 and abolished by E16.5. In Snf2h cKO lenses, expression of FoxE3 was reduced at E12.5 and E14.5, with no detectable expression of this protein at E16.5. These findings support the idea that the lens precursor cells at the anterior portion of the Snf2h cKO lens vesicle do not differentiate properly into mature lens epithelium. Instead, these anterior cells are converted into abnormal lens fibers.
In WT lenses, the regulatory proteins Prox1 and Jag1, and their targets cyclin kinase inhibitors p27Kip1 and p57Kip2, are upregulated in the cells undergoing cell cycle exit and in the early stages of secondary lens fiber cell formation. Their unique temporal/spatial expression patterns are completely disrupted in Snf2h cKO lenses. Expression of p27Kip1 and p57Kip2 proteins is detected in scattered cells around the anterior pole of the mutant lenses. Interestingly, many features of the Snf2h cKO lenses are comparable to defects found in Rbpj lens cKO mutants (Jia et al., 2007; Rowan et al., 2008). These similarities include a disrupted lens polarization/differentiation zone boundary, loss of cell type identity of the presumptive lens epithelium, and perturbed spatial expression of p27Kip1 and p57Kip2. Since Snf2h inactivation produced more significant spatial changes in the expression of these genes than Rbpj mutants, and expression of Rbpj is strongly reduced in the Snf2h mutants, it is possible that Snf2h is genetically upstream of one or more genes encoding components of Notch signaling. It is noteworthy that retention of nuclei and downregulation of Dnase2b were also reported in Notch2 lens mutants (Saravanamuthu et al., 2012).
Our data suggest that Snf2h is required for the denucleation process. Degradation of nuclei is a process unique to lens fibers, erythrocytes and skin keratinocytes. Erythrocytes extrude their nuclei from the individual cells, which are then engulfed and degraded by macrophages (Yoshida et al., 2005). Skin keratinocytes lose their nuclei by a caspase-independent apoptosis-like process (Lippens et al., 2009). The first possibility to consider is that the abnormal differentiation in Snf2h-deficient lens fibers disrupts various ‘late’ differentiation events, including nuclear degradation. We indeed observed downregulation of Dnase2b mRNA in both Brg1 (He et al., 2010) and Snf2h (present study) mutant lenses and in Pax6+/− lenses (Wolf et al., 2009) and activation of the Dnase2b promoter by Hsf4 and Pax6 in cotransfections. Additional experiments are needed to probe the transcriptional control of Hsf4 and Dnase2b, as well as other genes (e.g. p27Kip1 and p57Kip2), by Brg1 and Snf2h via ChIP-seq.
The observation that ER and mitochondria are degraded ‘normally’ in Snf2h mutant lenses suggests that degradation of mitochondria, initiated prior degradation of nuclei (Bassnett and Beebe, 1992), is not a prerequisite for denucleation. Degradation of mitochondria in Snf2h cKO lenses indicates that mitophagy (Costello et al., 2013) is active in Snf2h mutant lens fibers. However, in Snf2h-deficient lenses the presumptive NFZ is not established, pointing to disrupted autophagy and/or autophagy-related processes (Basu et al., 2014). The reduction in phosphorylated LC3b proteins supports this possibility. In addition, lamin B phosphorylation mediated by Cdk1, a process that occurs during normal mitosis, is required for denucleation (Caceres et al., 2010; Chaffee et al., 2014). Further experiments are required to probe signaling upstream of JNK/mTOR, the phosphorylation of lamin B by Cdk1, and yet to be identified steps in the cascade of cellular and molecular events leading to nuclear degradation in lens fibers (Morishita and Mizushima, 2016). For example, a recent study has revealed a novel role of p27Kip1, upstream of Cdk1, in lens fiber cell denucleation (Lyu et al., 2016).
It is important to note that both Brg1 and Snf2h might have additional roles in the denucleation processes. ATP-dependent chromatin remodeling participates in DNA repair (Erdel and Rippe, 2011; Lans et al., 2012; Zhang et al., 2009). SWI/SNF complexes are known to be recruited to phosphorylated H2AX via the interaction between the Brg1 bromodomain and acetylated lysines in histone tails (Lee et al., 2010). Studies of the canonical DNA repair protein Nbs1 (nibrin) in lens (Park et al., 2006), DNA repair-associated proteins Ddb1 (Cang et al., 2006) and Ncoa6 (Wang et al., 2010), the identification of DNA repair foci in lens fiber cell chromatin through phosphorylated H2AX outside of the OFZ (Wang et al., 2010), and the retention of nuclei in p53 (Trp53) null lenses (Wiley et al., 2011), raise the intriguing possibility that specific components of the DNA repair machinery participate in some aspects of this process using their ‘non-canonical’ activities adopted for the lens environment. Finally, it is possible that Brg1- and Snf2h-containing complexes assist in chromatin degradation in parallel with DNase IIβ.
MATERIALS AND METHODS
Antibodies
Primary antibodies used for immunofluorescence were anti-αA-crystallin (Santa Cruz Biotechnology, sc-22743, 1:1000), anti-BrdU (BD Biosciences, 347580, 1:500), anti-E-cadherin (BD Biosciences, 610181, 1:200), anti-FoxE3 (a gift from Dr Peter Carlsson, Goteborg University, Goteborg, Sweden; 1:200), anti-histone H3 (Abcam, ab1791, 1:200), anti-jagged 1 (Santa Cruz Biotechnology, sc-8303, 1:200), anti-Ki67 (Abcam, ab15580, 1:200), anti-LC3b (Sigma-Aldrich, L7543-100UL, 1:500), anti-mTOR (Cell Signaling Technology, 7C10, 1:400), anti-p27Kip1 (Santa Cruz Biotechnology, sc-528, 1:200), anti-p57Kip2 (Santa Cruz Biotechnology, sc-8298, 1:200), anti-PDI (protein disulfide isomerase; Sigma-Aldrich, P7122-200UL, 1:100), anti-Snf2h (Bethyl Laboratories, A301-017A, 1:500), anti-Snf2l (Bethyl Laboratories, A301-086A, 1:500), anti-Pax6 (Covance, PRB-278P-100, 1:500), anti-Prox1 (Abcam, ab37128, 1:500) and anti-Tomm20 (Santa Cruz Biotechnology, sc-11415, 1:100). Secondary antibodies were Alexa Fluor 488 goat anti-rabbit IgG, Alexa Fluor 568 goat anti-rabbit IgG, Alexa Fluor 568 rabbit anti-mouse IgG (A11008, A11011, A11061, respectively, Invitrogen, 1:250) and biotin-conjugated secondary anti-rabbit IgG (Dako, E0466, 1:500).
Conditional inactivation of Snf2h in the presumptive lens ectoderm
The Snf2h flox allele (in C57BL/6 background) was created through homologous recombination as described elsewhere (Alvarez-Saavedra et al., 2014). The Snf2h null allele was obtained by deletion of exons 5 to 9. Snf2h cKOs (Snf2hfl/−; Le-Cre/+) and their control littermates were generated by crossing the Snf2hfl/fl with the Snf2h+/−; Le-Cre/+ mice. Primers used for genotyping were (5′-3′): IN4-F13, GTGCAAAGCCCAGAGACGATGGTATG; IN4-F14, ACTGAGGACTCTGATGCAAACAGTCAAG; IN5-R3, TACACAACTAAGGCAGTGGGTTATAGTGC; IN9A-R35, TCACTATATTTAGAGTCAGATGTATCAACTGGTCC. The PCR cycle was as follows: initial denaturation step at 95°C for 3 min; then 35 cycles at 95°C for 40 s, primer annealing at 60°C for 40 s, polymerization at 72°C for 50 s; and a final extension at 72°C for 10 min. The Le-Cre transgenic mouse and genotyping are described elsewhere (Ashery-Padan et al., 2000; Wolf et al., 2013). Animal husbandry and experiments were conducted in accordance with the approved protocol of the Institutional Animal Care and Use Committee and the ARVO Statement for the Use of Animals in Ophthalmic and Vision Research. Noon of the day the vaginal plug was detected and was considered E0.5.
Histological analysis, immunofluorescence, immunohistochemistry and immunoblotting
Animals were euthanized by CO2 and mouse embryos were dissected from pregnant females. In some cases, whole eyeballs were removed from the postnatal animals. Tissues were then fixed in 10% neutral buffered paraformaldehyde overnight at 4°C, processed and embedded in paraffin. Serial sections were cut at 5 μm thickness through the mid section of the lens. Slides were stained with Hematoxylin and Eosin, or used for subsequent experiments. Immunohistochemistry was performed as described elsewhere (He et al., 2010).
Immunofluorescence was performed following standard procedures. Tissues were incubated with primary antibodies overnight at 4°C in a humidified chamber and with the secondary antibody for 1 h at room temperature. Sections were mounted with VECTASHIELD Antifade Mounting Medium (Vector Laboratories). The nuclei were counterstained with 4′,6-diamidino-2-phenylindole dihydrochloride (DAPI). Whole-cell extracts were prepared from lens and surrounding remnants of the anterior segment (it was impossible to isolate the mutant lens) in homogenization buffer [10 mM Tris pH 7.9, 1 mM EDTA, 0.1% SDS and protease inhibitors (Roche)] followed by sonication. The supernatants were analyzed by SDS-PAGE and 4-15% gradient gel (Bio-Rad). Proteins were then transferred to a nitrocellulose membrane. Membranes were blocked using Odyssey blocking buffer in PBS (Li-Cor) for 1 h and incubated with primary antibody overnight at 4°C. The membrane was then washed with TBS (Tris-buffered saline) containing 0.1% Tween 20. Secondary antibody was anti-rabbit IRDye 800 CW (Li-Cor). Bound antibody was imaged using a Li-Cor Odyssey imager. For each experiment, at least two extracts from control and Snf2h mutant tissues were analyzed.
RNA expression profiling
Collection of lens and surrounding tissues from P1 eye and total RNA isolation are described elsewhere (He et al., 2010). Four biological replicates of RNAs from different Snf2h cKO embryos and their littermates were used. cDNA synthesis and amplifications were performed with Ovation RNA Amplification System V2 (Nugen) using 50 ng total RNA per sample. Amplified cDNAs were cleaned and purified with the DNA Clean & Concentrator-25 Kit (Zymo Research). Fragmentation and labeling were performed using the FL Ovation cDNA Biotin Module V2 (Nugen). The four sets of samples were subsequently hybridized on Mouse Genome 430A 2.0 Arrays (Affymetrix).
Bioinformatic tools and statistical filtering of RNA microarray results
Differentially regulated genes/mRNAs between Snf2h knockout and control littermates were identified using biological quadruplicate sets of robust multichip average (RMA)-normalized Affymetrix CEL files (Irizarry et al., 2003) by a combination of Student's t-test (P<0.05) and significance analysis of microarrays (SAM; false discovery rate set to <1%), using the TIGR Multiexperiment Viewer of the TM4 microarray software suite (Saeed et al., 2003). A similar strategy was used to identify differentially regulated genes/mRNAs in Brg1 null lenses in newborn eyes (biological triplicates, RMA normalization, by applying Student's t-test P<0.05) (He et al., 2010). Primary data were deposited in the NCBI Gene Expression Omnibus database under accession numbers GSE41608 (the Snf2h cKO part) and GSE25168 (the Brg1 cKO part). The GO and KEGG pathway functional annotations were performed using the NIH web-based Database for Annotation, Visualization and Integrated Discovery (DAVID) (Huang et al., 2009).
qRT-PCR
cDNA was diluted 10-fold and qRT-PCR was performed with the Applied Biosystems (ABI) 7900HT fast real-time PCR system with Power SYBR Green PCR Master Mix (ABI) as described elsewhere (He et al., 2010). Primers, and genes used for normalization (B2m, Sdha and Hprt), are listed in Table S2.
Transient transfections and reporter gene analysis
A conserved region (−580 to +180) of the mouse Dnase2b promoter was synthesized by GenScript and cloned into the pGL3-Basic vector (Promega). Pax6 and Hsf4 cDNAs were obtained in the CMV vectors pKW10 and pCMV6-XL6, respectively (Yang et al. 2006; OriGene). Transient co-transfections were conducted in mouse lens epithelial αTN4 cells (Yang and Cvekl, 2005). The cells were transfected using Lipofectamine 2000 (Life Technologies) with 0.5 µg firefly luciferase reporter plasmid, 200 ng empty control vector (pKW10/pCMV6-XL6) or expression plasmids. As an internal control, 0.25 ng Renilla-TK luciferase plasmid was included. The promoter activity was measured using the Dual-Luciferase Reporter Assay System (Promega) 30 h following transfection. The experiments were performed in triplicate with two independent repeats.
Acknowledgements
We thank Drs Peter Carlsson for FoxE3 antibodies; Dr Melinda Duncan for helpful advice; and Mrs Jie Zhao for help with the mouse work. Core facilities were provided by AECOM Genomics and NYU Genome Technology Center.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Author contributions
S.H., S.L., R.S.M., Q.X., W.L., J.Z., M.K., A.I.S., T.S. and A.C. designed the experiments. S.H., S.L., R.S.M., Q.X., L.A.B., W.L.K. and J.Z. performed experiments and analyzed data. J.K., R.M., H.H., W.E., R.A.-P., A.I.S. and T.S. developed the mouse models. S.H., R.A.-P., A.I.S., T.S. and A.C. conceived the project and wrote the manuscript. S.H., S.L., R.S.M., Q.X. and J.Z. prepared figures.
Funding
Grant support was from the National Institutes of Health [R01 EY012200, EY014237 and EY014237-7S1 to A.C.; EY013022 to M.K.; CA079057 to A.I.S.; EY022645 to W.L.]; The Czech Science Foundation (Grantová agentura České republiky, GACR) [P305/12/1033 to T.S. and J.K.]; an unrestricted grant from Research to Prevent Blindness to the Department of Ophthalmology and Visual Sciences, Albert Einstein College of Medicine; T.S. is supported by BIOCEV funded by the Czech Ministry of Education (Ministerstvo školství, mládeže a tělovýchovy) [LH15170, UNCE: 204021, NPU II: LQ1604 and CZ.1.05/1.1.00/02.0109 (ERDF, MEYS)]; R.A.-P. is supported by the Israel Science Foundation [228/14], the Ministry of Science and Technology, Israel [36494], the Ziegler Foundation, and the United States-Israel Binational Science Foundation [2013016]. Deposited in PMC for release after 12 months.
Data availability
Primary microarray data have been deposited in NCBI Gene Expression Omnibus under accession numbers GSE41608 (Snf2h cKO) and GSE25168 (Brg1 cKO).
Supplementary information
Supplementary information available online at http://dev.biologists.org/lookup/suppl/doi:10.1242/dev.135285/-/DC1
References
- Alvarez-Saavedra M., De Repentigny Y., Lagali P. S., Raghu Ram E. V. S., Yan K., Hashem E., Ivanochko D., Huh M. S., Yang D., Mears A. J. et al. (2014). Snf2h-mediated chromatin organization and histone H1 dynamics govern cerebellar morphogenesis and neural maturation. Nat. Commun. 5, 4181 10.1038/ncomms5181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arancio W., Onorati M. C., Burgio G., Collesano M., Ingrassia A. M. R., Genovese S. I., Fanto M. and Corona D. F. (2010). The nucleosome remodeling factor ISWI functionally interacts with an evolutionarily conserved network of cellular factors. Genetics 185, 129-140. 10.1534/genetics.110.114256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashery-Padan R., Marquardt T., Zhou X. and Gruss P. (2000). Pax6 activity in the lens primordium is required for lens formation and for correct placement of a single retina in the eye. Genes Dev. 14, 2701-2711. 10.1101/gad.184000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassnett S. (2009). On the mechanism of organelle degradation in the vertebrate lens. Exp. Eye Res. 88, 133-139. 10.1016/j.exer.2008.08.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassnett S. and Beebe D. C. (1992). Coincident loss of mitochondria and nuclei during lens fiber cell differentiation. Dev. Dyn. 194, 85-93. 10.1002/aja.1001940202 [DOI] [PubMed] [Google Scholar]
- Bassnett S., Shi Y. and Vrensen G. F. J. M. (2011). Biological glass: structural determinants of eye lens transparency. Philos. Trans. R. Soc. B Biol. Sci. 366, 1250-1264. 10.1098/rstb.2010.0302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basu S., Rajakaruna S., Reyes B., Van Bockstaele E. and Menko A. S. (2014). Suppression of MAPK/JNK-MTORC1 signaling leads to premature loss of organelles and nuclei by autophagy during terminal differentiation of lens fiber cells. Autophagy 10, 1193-1211. 10.4161/auto.28768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blixt A., Mahlapuu M., Aitola M., Pelto-Huikko M., Enerback S. and Carlsson P. (2000). A forkhead gene, FoxE3, is essential for lens epithelial proliferation and closure of the lens vesicle. Genes Dev. 14, 245-254. [PMC free article] [PubMed] [Google Scholar]
- Blixt Å., Landgren H., Johansson B. R. and Carlsson P. (2007). Foxe3 is required for morphogenesis and differentiation of the anterior segment of the eye and is sensitive to Pax6 gene dosage. Dev. Biol. 302, 218-229. 10.1016/j.ydbio.2006.09.021 [DOI] [PubMed] [Google Scholar]
- Brownell I., Dirksen M. and Jamrich M. (2000). Forkhead Foxe3 maps to the dysgenetic lens locus and is critical in lens development and differentiation. Genesis 27, 81-93. [DOI] [PubMed] [Google Scholar]
- Bultman S., Gebuhr T., Yee D., La Mantia C., Nicholson J., Gilliam A., Randazzo F., Metzger D., Chambon P., Crabtree G. et al. (2000). A Brg1 null mutation in the mouse reveals functional differences among mammalian SWI/SNF complexes. Mol. Cell 6, 1287-1295. 10.1016/S1097-2765(00)00127-1 [DOI] [PubMed] [Google Scholar]
- Bultman S. J., Gebuhr T. C. and Magnuson T. (2005). A Brg1 mutation that uncouples ATPase activity from chromatin remodeling reveals an essential role for SWI/SNF-related complexes in β-globin expression and erythroid development. Genes Dev. 19, 2849-2861. 10.1101/gad.1364105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caceres A., Shang F., Wawrousek E., Liu Q., Avidan O., Cvekl A., Yang Y., Haririnia A., Storaska A., Fushman D. et al. (2010). Perturbing the ubiquitin pathway reveals how mitosis is hijacked to denucleate and regulate cell proliferation and differentiation in vivo. PLoS ONE 5, e13331 10.1371/journal.pone.0013331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cairns B. R. (2007). Chromatin remodeling: insights and intrigue from single-molecule studies. Nat. Struct. Mol. Biol. 14, 989-996. 10.1038/nsmb1333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cang Y., Zhang J., Nicholas S. A., Bastien J., Li B., Zhou P. and Goff S. P. (2006). Deletion of DDB1 in mouse brain and lens leads to p53-dependent elimination of proliferating cells. Cell 127, 929-940. 10.1016/j.cell.2006.09.045 [DOI] [PubMed] [Google Scholar]
- Chaffee B. R., Shang F., Chang M.-L., Clement T. M., Eddy E. M., Wagner B. D., Nakahara M., Nagata S., Robinson M. L. and Taylor A. (2014). Nuclear removal during terminal lens fiber cell differentiation requires CDK1 activity: Appropriating mitosis-related nuclear disassembly. Development 141, 3388-3398. 10.1242/dev.106005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Q., Hung F. C., Fromm L. and Overbeek P. A. (2000). Induction of cell cycle entry and cell death in postmitotic lens fiber cells by overexpression of E2F1 or E2F2. Invest. Ophthalmol. Vis. Sci. 41, 4223-4231. [PubMed] [Google Scholar]
- Collins N., Poot R. A., Kukimoto I., García-Jiménez C., Dellaire G. and Varga-Weisz P. D. (2002). An ACF1–ISWI chromatin-remodeling complex is required for DNA replication through heterochromatin. Nat. Genet. 32, 627-632. 10.1038/ng1046 [DOI] [PubMed] [Google Scholar]
- Costello M. J., Brennan L. A., Basu S., Chauss D., Mohamed A., Gilliland K. O., Johnsen S., Menko A. S. and Kantorow M. (2013). Autophagy and mitophagy participate in ocular lens organelle degradation. Exp. Eye Res. 116, 141-150. 10.1016/j.exer.2013.08.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui X., Wang L., Zhang J., Du R., Liao S., Li D., Li C., Ke T., Li D. W.-C., Huang H. et al. (2013). HSF4 regulates DLAD expression and promotes lens de-nucleation. Biochim. Biophys. Acta 1832, 1167-1172. 10.1016/j.bbadis.2013.03.007 [DOI] [PubMed] [Google Scholar]
- Cvekl A. and Ashery-Padan R. (2014). The cellular and molecular mechanisms of vertebrate lens development. Development 141, 4432-4447. 10.1242/dev.107953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Serna I. L., Ohkawa Y. and Imbalzano A. N. (2006). Chromatin remodelling in mammalian differentiation: lessons from ATP-dependent remodellers. Nat. Rev. Genet. 7, 461-473. 10.1038/nrg1882 [DOI] [PubMed] [Google Scholar]
- Dirscherl S. S., Henry J. J. and Krebs J. E. (2005). Neural and eye-specific defects associated with loss of the imitation switch (ISWI) chromatin remodeler in Xenopus laevis. Mech. Dev. 122, 1157-1170. 10.1016/j.mod.2005.08.002 [DOI] [PubMed] [Google Scholar]
- Duncan M. K., Cui W., Oh D.-J. and Tomarev S. I. (2002). Prox1 is differentially localized during lens development. Mech. Dev. 112, 195-198. 10.1016/S0925-4773(01)00645-1 [DOI] [PubMed] [Google Scholar]
- Erdel F. and Rippe K. (2011). Binding kinetics of human ISWI chromatin-remodelers to DNA repair sites elucidate their target location mechanism. Nucleus 2, 105-112. 10.4161/nucl.2.2.15209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujimoto M., Izu H., Seki K., Fukuda K., Nishida T., Yamada S.-i., Kato K., Yonemura S., Inouye S. and Nakai A. (2004). HSF4 is required for normal cell growth and differentiation during mouse lens development. EMBO J. 23, 4297-4306. 10.1038/sj.emboj.7600435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gebuhr T. C., Kovalev G. I., Bultman S., Godfrey V., Su L. and Magnuson T. (2003). The role of Brg1, a catalytic subunit of mammalian chromatin-remodeling complexes, in T cell development. J. Exp. Med. 198, 1937-1949. 10.1084/jem.20030714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregg R. G., Willer G. B., Fadool J. M., Dowling J. E. and Link B. A. (2003). Positional cloning of the young mutation identifies an essential role for the Brahma chromatin remodeling complex in mediating retinal cell differentiation. Proc. Natl. Acad. Sci. USA 100, 6535-6540. 10.1073/pnas.0631813100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griep A. and Zhang P. (2004). Lens cell proliferation: the cell cycle. In Development of the Ocular Lens (ed. Lovicu F. J. and Robinson M. L.), pp. 191-213. New York: Cambridge University Press. [Google Scholar]
- Griffin C. T., Brennan J. and Magnuson T. (2008). The chromatin-remodeling enzyme BRG1 plays an essential role in primitive erythropoiesis and vascular development. Development 135, 493-500. 10.1242/dev.010090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunhaga L. (2011). The lens: a classical model of embryonic induction providing new insights into cell determination in early development. Philos. Trans. R. Soc. B Biol. Sci. 366, 1193-1203. 10.1098/rstb.2010.0175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hang C. T., Yang J., Han P., Cheng H.-L., Shang C., Ashley E., Zhou B. and Chang C. P. (2010). Chromatin regulation by Brg1 underlies heart muscle development and disease. Nature 466, 62-67. 10.1038/nature09130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He S., Pirity M. K., Wang W.-L., Wolf L., Chauhan B. K., Cveklova K., Tamm E. R., Ashery-Padan R., Metzger D., Nakai A. et al. (2010). Chromatin remodeling enzyme Brg1 is required for mouse lens fiber cell terminal differentiation and its denucleation. Epigenetics Chromatin 3, 21 10.1186/1756-8935-3-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho L. and Crabtree G. R. (2010). Chromatin remodelling during development. Nature 463, 474-484. 10.1038/nature08911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho H.-Y., Chang K.-H., Nichols J. and Li M. (2009). Homeodomain protein Pitx3 maintains the mitotic activity of lens epithelial cells. Mech. Dev. 126, 18-29. 10.1016/j.mod.2008.10.007 [DOI] [PubMed] [Google Scholar]
- Ho L., Miller E. L., Ronan J. L., Ho W. Q., Jothi R. and Crabtree G. R. (2011). esBAF facilitates pluripotency by conditioning the genome for LIF/STAT3 signalling and by regulating polycomb function. Nat. Cell Biol. 13, 903-913. 10.1038/ncb2285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang D. W., Sherman B. T. and Lempicki R. A. (2009). Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4, 44-57. 10.1038/nprot.2008.211 [DOI] [PubMed] [Google Scholar]
- Indra A. K., Dupe V., Bornert J.-M., Messaddeq N., Yaniv M., Mark M., Chambon P. and Metzger D. (2005). Temporally controlled targeted somatic mutagenesis in embryonic surface ectoderm and fetal epidermal keratinocytes unveils two distinct developmental functions of BRG1 in limb morphogenesis and skin barrier formation. Development 132, 4533-4544. 10.1242/dev.02019 [DOI] [PubMed] [Google Scholar]
- Irizarry R. A., Bolstad B. M., Collin F., Cope L. M., Hobbs B. and Speed T. P. (2003). Summaries of Affymetrix GeneChip probe level data. Nucleic Acids Res. 31, e15 10.1093/nar/gng015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia J., Lin M., Zhang L., York J. P. and Zhang P. (2007). The Notch signaling pathway controls the size of the ocular lens by directly suppressing p57Kip2 expression. Mol. Cell. Biol. 27, 7236-7247. 10.1128/MCB.00780-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabeya Y., Mizushima N., Ueno T., Yamamoto A., Kirisako T., Noda T., Kominami E., Ohsumi Y. and Yoshimori T. (2000). LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. EMBO J. 19, 5720-5728. 10.1093/emboj/19.21.5720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadam S. and Emerson B. M. (2003). Transcriptional specificity of human SWI/SNF BRG1 and BRM chromatin remodeling complexes. Mol. Cell 11, 377-389. 10.1016/S1097-2765(03)00034-0 [DOI] [PubMed] [Google Scholar]
- Khavari P. A., Peterson C. L., Tamkun J. W., Mendel D. B. and Crabtree G. R. (1993). BRG1 contains a conserved domain of the SWI2/SNF2 family necessary for normal mitotic growth and transcription. Nature 366, 170-174. 10.1038/366170a0 [DOI] [PubMed] [Google Scholar]
- Kidder B. L., Palmer S. and Knott J. G. (2009). SWI/SNF-Brg1 regulates self-renewal and occupies core pluripotency-related genes in embryonic stem cells. Stem Cells 27, 317-328. 10.1634/stemcells.2008-0710 [DOI] [PubMed] [Google Scholar]
- Lans H., Marteijn J. A. and Vermeulen W. (2012). ATP-dependent chromatin remodeling in the DNA-damage response. Epigenetics Chromatin 5, 4 10.1186/1756-8935-5-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le T. T., Conley K. W., Mead T. J., Rowan S., Yutzey K. E. and Brown N. L. (2012). Requirements for Jag1-Rbpj mediated Notch signaling during early mouse lens development. Dev. Dyn. 241, 493-504. 10.1002/dvdy.23739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H.-S., Park J.-H., Kim S.-J., Kwon S.-J. and Kwon J. (2010). A cooperative activation loop among SWI/SNF, γ-H2AX and H3 acetylation for DNA double-strand break repair. EMBO J. 29, 1434-1445. 10.1038/emboj.2010.27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lippens S., Hoste E., Vandenabeele P., Agostinis P. and Declercq W. (2009). Cell death in the skin. Apoptosis 14, 549-569. 10.1007/s10495-009-0324-z [DOI] [PubMed] [Google Scholar]
- Lyu L., Whitcomb E. A., Jiang S., Chang M.-L., Gu Y., Duncan M. K., Cvekl A., Wang W. L., Limi S., Reneker L. W. et al. (2016). Unfolded-protein response-associated stabilization of p27(Cdkn1b) interferes with lens fiber cell denucleation, leading to cataract. FASEB J. 30, 1087-1095. 10.1096/fj.15-278036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeda A., Moriguchi T., Hamada M., Kusakabe M., Fujioka Y., Nakano T., Yoh K., Lim K.-C., Engel J. D. and Takahashi S. (2009). Transcription factor GATA-3 is essential for lens development. Dev. Dyn. 238, 2280-2291. 10.1002/dvdy.22035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magdaleno S., Jensen P., Brumwell C. L., Seal A., Lehman K., Asbury A., Cheung T., Cornelius T., Batten D. M., Eden C. et al. (2006). BGEM: an in situ hybridization database of gene expression in the embryonic and adult mouse nervous system. PLoS Biol. 4, e86 10.1371/journal.pbio.0040086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez G. and de Iongh R. U. (2010). The lens epithelium in ocular health and disease. Int. J. Biochem. Cell Biol. 42, 1945-1963. 10.1016/j.biocel.2010.09.012 [DOI] [PubMed] [Google Scholar]
- Matsumoto S., Banine F., Struve J., Xing R. B., Adams C., Liu Y., Metzger D., Chambon P., Rao M. S. and Sherman L. S. (2006). Brg1 is required for murine neural stem cell maintenance and gliogenesis. Dev. Biol. 289, 372-383. 10.1016/j.ydbio.2005.10.044 [DOI] [PubMed] [Google Scholar]
- McCaffrey J., Yamasaki L., Dyson N. J., Harlow E. and Griep A. E. (1999). Disruption of retinoblastoma protein family function by human papillomavirus type 16 E7 oncoprotein inhibits lens development in part through E2F-1. Mol. Cell. Biol. 19, 6458-6468. 10.1128/MCB.19.9.6458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medina-Martinez O., Brownell I., Amaya-Manzanares F., Hu Q., Behringer R. R. and Jamrich M. (2005). Severe defects in proliferation and differentiation of lens cells in Foxe3 null mice. Mol. Cell. Biol. 25, 8854-8863. 10.1128/MCB.25.20.8854-8863.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medina-Martinez O., Shah R. and Jamrich M. (2009). Pitx3 controls multiple aspects of lens development. Dev. Dyn. 238, 2193-2201. 10.1002/dvdy.21924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgenbesser S. D., Williams B. O., Jacks T. and DePinho R. A. (1994). p53-dependent apoptosis produced by Rb-deficiency in the developing mouse lens. Nature 371, 72-74. 10.1038/371072a0 [DOI] [PubMed] [Google Scholar]
- Morishita H. and Mizushima N. (2016). Autophagy in the lens. Exp. Eye Res. 144, 22-28. 10.1016/j.exer.2015.08.019 [DOI] [PubMed] [Google Scholar]
- Narlikar G. J., Fan H.-Y. and Kingston R. E. (2002). Cooperation between complexes that regulate chromatin structure and transcription. Cell 108, 475-487. 10.1016/S0092-8674(02)00654-2 [DOI] [PubMed] [Google Scholar]
- Ninkovic J., Steiner-Mezzadri A., Jawerka M., Akinci U., Masserdotti G., Petricca S., Fischer J., von Holst A., Beckers J., Lie C. D. et al. (2013). The BAF complex interacts with Pax6 in adult neural progenitors to establish a neurogenic cross-regulatory transcriptional network. Cell Stem Cell 13, 403-418. 10.1016/j.stem.2013.07.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimoto S., Kawane K., Watanabe-Fukunaga R., Fukuyama H., Ohsawa Y., Uchiyama Y., Hashida N., Ohguro N., Tano Y., Morimoto T. et al. (2003). Nuclear cataract caused by a lack of DNA degradation in the mouse eye lens. Nature 424, 1071-1074. 10.1038/nature01895 [DOI] [PubMed] [Google Scholar]
- Park J.-H., Park E.-J., Lee H.-S., Kim S. J., Hur S.-K., Imbalzano A. N. and Kwon J. (2006). Mammalian SWI/SNF complexes facilitate DNA double-strand break repair by promoting γ-H2AX induction. EMBO J. 25, 3986-3997. 10.1038/sj.emboj.7601291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pontoriero G. F., Smith A. N., Miller L.-A. D., Radice G. L., West-Mays J. A. and Lang R. A. (2009). Co-operative roles for E-cadherin and N-cadherin during lens vesicle separation and lens epithelial cell survival. Dev. Biol. 326, 403-417. 10.1016/j.ydbio.2008.10.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowan S., Conley K. W., Le T. T., Donner A. L., Maas R. L. and Brown N. L. (2008). Notch signaling regulates growth and differentiation in the mammalian lens. Dev. Biol. 321, 111-122. 10.1016/j.ydbio.2008.06.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saeed A. I., Sharov V., White J., Li J., Liang W., Bhagabati N., Braisted J., Klapa M., Currier T., Thiagarajan M. et al. (2003). TM4: a free, open-source system for microarray data management and analysis. Biotechniques 34, 374-378. [DOI] [PubMed] [Google Scholar]
- Saravanamuthu S. S., Le T. T., Gao C. Y., Cojocaru R. I., Pandiyan P., Liu C., Zhang J., Zelenka P. S. and Brown N. L. (2012). Conditional ablation of the Notch2 receptor in the ocular lens. Dev. Biol. 362, 219-229. 10.1016/j.ydbio.2011.11.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaham O., Smith A. N., Robinson M. L., Taketo M. M., Lang R. A. and Ashery-Padan R. (2009). Pax6 is essential for lens fiber cell differentiation. Development 136, 2567-2578. 10.1242/dev.032888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaham O., Menuchin Y., Farhy C. and Ashery-Padan R. (2012). Pax6: a multi-level regulator of ocular development. Prog. Retin. Eye Res. 31, 351-376. 10.1016/j.preteyeres.2012.04.002 [DOI] [PubMed] [Google Scholar]
- Sharma S., Kelly T. K. and Jones P. A. (2010). Epigenetics in cancer. Carcinogenesis 31, 27-36. 10.1093/carcin/bgp220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stopka T. and Skoultchi A. I. (2003). The ISWI ATPase Snf2h is required for early mouse development. Proc. Natl. Acad. Sci. USA 100, 14097-14102. 10.1073/pnas.2336105100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang L., Nogales E. and Ciferri C. (2010). Structure and function of SWI/SNF chromatin remodeling complexes and mechanistic implications for transcription. Prog. Biophys. Mol. Biol. 102, 122-128. 10.1016/j.pbiomolbio.2010.05.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toiber D., Erdel F., Bouazoune K., Silberman D. M., Zhong L., Mulligan P., Sebastian C., Cosentino C., Martinez-Pastor B., Giacosa S. et al. (2013). SIRT6 recruits SNF2H to DNA break sites, preventing genomic instability through chromatin remodeling. Mol. Cell 51, 454-468. 10.1016/j.molcel.2013.06.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W.-L., Li Q., Xu J. and Cvekl A. (2010). Lens fiber cell differentiation and denucleation are disrupted through expression of the N-terminal nuclear receptor box of NCOA6 and result in p53-dependent and p53-independent apoptosis. Mol. Biol. Cell 21, 2453-2468. 10.1091/mbc.E09-12-1031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weider M., Küspert M., Bischof M., Vogl M. R., Hornig J., Loy K., Kosian T., Müller J., Hillgärtner S., Tamm E. R. et al. (2012). Chromatin-remodeling factor Brg1 is required for schwann cell differentiation and myelination. Dev. Cell 23, 193-201. 10.1016/j.devcel.2012.05.017 [DOI] [PubMed] [Google Scholar]
- Wenzel P. L., Chong J.-L., Sáenz-Robles M. T., Ferrey A., Hagan J. P., Gomez Y. M., Rajmohan R., Sharma N., Chen H. Z., Pipas J. M. et al. (2011). Cell proliferation in the absence of E2F1-3. Dev. Biol. 351, 35-45. 10.1016/j.ydbio.2010.12.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- West-Mays J. A., Coyle B. M., Piatigorsky J., Papagiotas S. and Libby D. (2002). Ectopic expression of AP-2α transcription factor in the lens disrupts fiber cell differentiation. Dev. Biol. 245, 13-27. 10.1006/dbio.2002.0624 [DOI] [PubMed] [Google Scholar]
- Wigle J. T., Chowdhury K., Gruss P. and Oliver G. (1999). Prox1 function is crucial for mouse lens-fibre elongation. Nat. Genet. 21, 318-322. 10.1038/6844 [DOI] [PubMed] [Google Scholar]
- Wiley L. A., Rajagopal R., Dattilo L. K. and Beebe D. C. (2011). The tumor suppressor gene Trp53 protects the mouse lens against posterior subcapsular cataracts and the BMP receptor Acvr1 acts as a tumor suppressor in the lens. Dis. Model. Mech. 4, 484-495. 10.1242/dmm.006593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf L. V., Yang Y., Wang J. H., Xie Q., Braunger B., Tamm E. R., Zavadil J. and Cvekl A. (2009). Identification of Pax6-dependent gene regulatory networks in the mouse lens. PLoS ONE 4, e4159 10.1371/journal.pone.0004159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf L., Harrison W., Huang J., Xie Q., Xiao N., Sun J., Kong L., Lachke S. A., Kuracha M. R., Govindarajan V. et al. (2013). Histone posttranslational modifications and cell fate determination: lens induction requires the lysine acetyltransferases CBP and p300. Nucleic Acids Res. 41, 10199-10214. 10.1093/nar/gkt824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y. and Cvekl A. (2005). Tissue-specific regulation of the mouse αA-crystallin gene in lens via recruitment of Pax6 and c-Maf to its promoter. J. Mol. Biol. 351, 453-469. 10.1016/j.jmb.2005.05.072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y., Stopka T., Golestaneh N., Wang Y., Wu K., Li A., Chauhan B. K., Gao C. Y., Cveklová K., Duncan M. K. et al. (2006). Regulation of αA-crystallin via Pax6, c-Maf, CREB and a broad domain of lens-specific chromatin. EMBO J. 25, 2107-2118. 10.1038/sj.emboj.7601114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida H., Kawane K., Koike M., Mori Y., Uchiyama Y. and Nagata S. (2005). Phosphatidylserine-dependent engulfment by macrophages of nuclei from erythroid precursor cells. Nature 437, 754-758. 10.1038/nature03964 [DOI] [PubMed] [Google Scholar]
- Zhang P. M., Wong C., DePinho R. A., Harper J. W. and Elledge S. J. (1998). Cooperation between the Cdk inhibitors p27(KIP1) and p57(KIP2) in the control of tissue growth and development. Genes Dev. 12, 3162-3167. 10.1101/gad.12.20.3162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L., Zhang Q., Jones K., Patel M. and Gong F. (2009). The chromatin remodeling factor BRG1 stimulates nucleotide excision repair by facilitating recruitment of XPC to sites of DNA damage. Cell Cycle 8, 3953-3959. 10.4161/cc.8.23.10115 [DOI] [PubMed] [Google Scholar]
- Zhao H., Yang T., Madakashira B. P., Thiels C. A., Bechtle C. A., Garcia C. M., Zhang H., Yu K., Ornitz D. M., Beebe D. C. et al. (2008). Fibroblast growth factor receptor signaling is essential for lens fiber cell differentiation. Dev. Biol. 318, 276-288. 10.1016/j.ydbio.2008.03.028 [DOI] [PMC free article] [PubMed] [Google Scholar]