Abstract
Dendritic arbor morphology is a key determinant of neuronal function. Once established, dendrite branching patterns must be maintained as the animal develops to ensure receptive field coverage. The translational repressors Nanos (Nos) and Pumilio (Pum) are required to maintain dendrite growth and branching of Drosophila larval class IV dendritic arborization (da) neurons, but their specific regulatory role remains unknown. We show that Nos-Pum-mediated repression of the pro-apoptotic gene head involution defective (hid) is required to maintain a balance of dendritic growth and retraction in class IV da neurons and that upregulation of hid results in decreased branching because of an increase in caspase activity. The temporal requirement for nos correlates with an ecdysone-triggered switch in sensitivity to apoptotic stimuli that occurs during the mid-L3 transition. We find that hid is required during pupariation for caspase-dependent pruning of class IV da neurons and that Nos and Pum delay pruning. Together, these results suggest that Nos and Pum provide a crucial neuroprotective regulatory layer to ensure that neurons behave appropriately in response to developmental cues.
KEY WORDS: Nos, hid, Caspase, da neurons, Drosophila, Non-apoptotic function
Summary: The translational repressors Nanos and Pumilio provide a crucial neuroprotective regulatory layer to ensure that Drosophila neurons behave appropriately in response to developmental cues.
INTRODUCTION
The size, shape and complexity of dendritic arbors determine how neurons receive and integrate synaptic or sensory input. Dendritic arbor morphogenesis is a highly regulated process involving initial dendrite outgrowth and branching, subsequent refinement and maintenance of branching and field coverage, and in some cases remodeling of the arbor. Although a number of transcription factors and signaling pathways that function in establishing the size and complexity of dendritic arbors have been identified, much less is known about how they are subsequently refined and maintained during neuronal development (Parrish et al., 2007).
Drosophila dendritic arborization (da) neurons have provided a powerful model system for identifying factors that regulate dendrite morphogenesis. These sensory neurons, which innervate the larval epidermis, are subdivided into four classes based on the complexity of their dendritic arbors (Grueber et al., 2002). Class IV da neurons, which form the most complex arbors, establish their dendritic territories by the end of the first larval stage, covering the body wall in a complete but non-overlapping manner. Field coverage is maintained as the larva grows dramatically between the first and third larval stages by expansion of the dendritic arbors in proportion to the growth of the larva, and in particular, the underlying epidermal epithelium (Parrish et al., 2009). Whereas most neurons are eliminated during metamorphosis, class IV da neurons undergo pruning of their dendritic arbors and distal branch retraction and are subsequently remodeled for the adult nervous system (Kuo et al., 2005, 2006; Rumpf et al., 2011; Williams et al., 2006; Williams and Truman, 2005).
The conserved RNA-binding proteins Nanos (Nos) and Pumilio (Pum) function together to regulate dendrite morphogenesis in Drosophila (Brechbiel and Gavis, 2008; Olesnicky et al., 2012; Ye et al., 2004). Class IV da neurons in both nos and pum mutant larvae exhibit a dramatic decrease in dendritic arbor complexity, characterized by a reduction of higher order branches, as well as a field coverage defect (Brechbiel and Gavis, 2008; Olesnicky et al., 2012; Ye et al., 2004). In the early embryo, Nos and Pum form a localized translational repressor complex essential for abdominal and germline development (Vardy and Orr-Weaver, 2007). Similarly, nos function in class IV da neurons requires dendritic localization of nos mRNA, suggesting that Nos and Pum act locally within dendrites to regulate target mRNAs important for dendrite morphogenesis (Brechbiel and Gavis, 2008). Because the dendritic arborization defects in nos and pum mutants arise after the initial elaboration of the dendritic arbors, Nos and Pum are thought to function in maintaining dendritic field complexity during late larval development (Brechbiel and Gavis, 2008; Olesnicky et al., 2012). Why Nos and Pum are required particularly during late larval development and their specific regulatory role in maintenance remains unknown.
We previously found that levels of the pro-apoptotic factor Hid are elevated in class IV da neurons mutant for nos or pum (Olesnicky et al., 2012). Nos and its orthologs play essential roles in preventing apoptosis of primordial germ cells (PGCs), which in Drosophila requires Nos-mediated repression of hid (Lai and King, 2013; Sato et al., 2007). However, unlike the case for PGCs, upregulation of hid in nos or pum mutant class IV da neurons does not result in apoptosis. Non-apoptotic roles in cellular reorganization and cell shaping have been identified for core apoptotic machinery components, including Hid (Arama et al., 2003; Huh et al., 2004; Oshima et al., 2006), and pruning of class IV da neuron dendrites during pupariation requires local, non-apoptotic caspase function (Kuo et al., 2006; Williams et al., 2006). Here, we investigate the physiological significance of Nos- and Pum-mediated regulation of hid and the apoptotic pathway in larval class IV da neurons. We show that failure to repress hid in these neurons results in excessive branch retraction resulting from Hid-induced caspase activity. In addition, we find that a balance of dendritic growth and retraction is required for class IV da neurons to maintain dendritic arbors at steady-state during late larval development, and that Nos is required to maintain this balance by repressing hid. We provide evidence that the specific temporal requirement for nos and pum during the late larval period coincides with an ecdysone-triggered switch in larval sensitivity to apoptotic stimuli (Kang and Bashirullah, 2014). Finally, we show that hid function during pupariation is required for caspase-dependent dendrite pruning. Taken together, our data suggest that Nos and Pum repress hid in class IV da neurons through the end of larval development to protect neurons until pupariation, when caspases are required for dendritic arbor remodeling.
RESULTS
Nos and Pum selectively repress hid to maintain dendritic arbor complexity
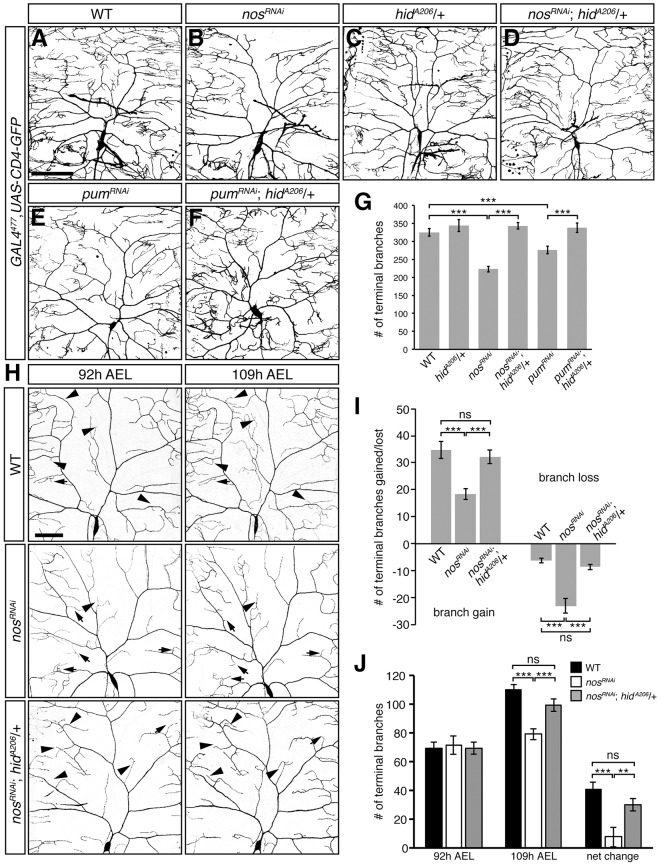
We previously found that Hid protein levels are elevated in nos and pum mutant class IV da neurons and that a chromosomal deficiency (H99) that removes hid along with two other pro-apoptotic genes, grim and reaper (rpr), rescues the loss of dendritic terminal branches in nosRNAi larvae (Olesnicky et al., 2012). To determine whether the upregulation of hid is specifically responsible for the decrease in dendrite branching observed in nos- and pum-deficient class IV da neurons, we selectively reduced hid function in these neurons by using a hypomorphic hid allele (hidA206; Abbott and Lengyel, 1991). Reduction of hid function rescued both the field coverage defect and loss of dendritic terminal branches observed in nos mutant, nosRNAi, and pumRNAi class IV da neurons (Fig. 1A-G; Fig. S1). A similar rescue was observed when hid expression was reduced by RNAi (Fig. S1). By contrast, reducing levels of Grim and Rpr in nosRNAi da neurons did not rescue either the coverage defect or loss of dendritic termini (Fig. S1). Together, these results point specifically to hid upregulation in causing the nos and pum mutant dendritic defects.
Fig. 1.
Reducing hid function rescues dendritic arborization defects of class IV da neurons lacking nos and pum. (A-F) Confocal z-series projections of representative class IV da neurons from wild-type (WT) (A), nosRNAi (B), hidA206/+ (C), nosRNAi; hidA206/+ (D), pumRNAi (E) and pumRNAi; hidA206/+ (F) larvae. (G) Quantification of the total number of terminal branches. (H) Class IV da neurons in WT, nosRNAi and nosRNAi; hidA206/+ larvae imaged at 92 h and 109 h AEL. Arrowheads indicate examples of new branch outgrowth or branch elongation and arrows designate examples of branch retraction/or loss. In A-F and H, GAL4477 was used to drive expression of UAS-RNAi transgenes and UAS-CD4-GFP (to mark the neurons). (I) Quantification of the number of new and/or elongating branches and lost or retracting branches between time points. Net change represents the difference in the average number of terminal branches observed between 92 h and 109 h AEL class IV da neurons. (J) Quantification of the number of dendritic termini in WT, nosRNAi and nosRNAi; hidA206/+ larvae at 92 h AEL and 109 h AEL. Values in G,I-J are mean±s.e.m., n≥10 for each genotype; ns, not significant; **P≤0.01, ***P≤0.001 by two-tailed Student's t-test. Scale bars: 100 μm.
The decreased dendritic branching observed in nos and pum mutants appears after the mature arborization pattern has been established, indicating a role for nos and pum in growth and maintenance rather than in the initial elaboration of dendritic arbors (Brechbiel and Gavis, 2008; Ye et al., 2004). Time-series image analysis revealed that nos and pum are required both to promote the outgrowth of new branches and to stabilize existing branches during larval growth (Olesnicky et al., 2012). Whereas wild-type neurons exhibit a net increase in the number of branches resulting from extensive branch outgrowth during the mid-to-late third larval instar (L3) period, nos and pum-deficient neurons show a net decrease as a result of reduced outgrowth and extensive branch loss (Olesnicky et al., 2012 and Fig. 1H-J). The decrease in branch growth and increase in branch retraction caused by nos RNAi is completely reversed by reducing hid function using the hidA206 allele (Fig. 1H-J) or the H99 chromosomal deficiency (data not shown). Thus, maintenance of dendritic complexity during L3 development is dependent on the ability of Nos and Pum to repress hid.
Derepression of hid results in increased dendrite retraction in class IV da neurons
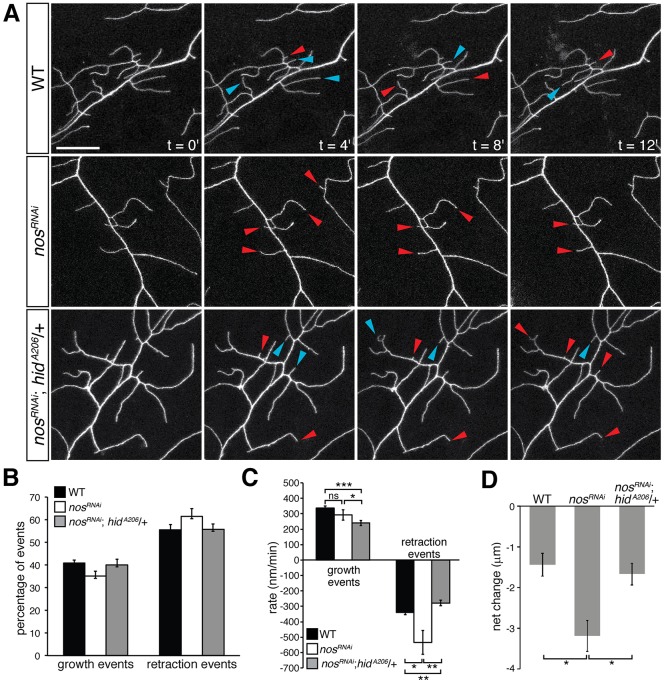
To understand better how Nos regulates the dynamics of branch growth and retraction in class IV da neurons, we performed real-time confocal imaging of third instar larvae (Fig. 2A; Movies 1-3). Wild-type and nosRNAi class IV da neurons exhibited a similar number of growth and retraction events over a span of 30 min (Fig. 2B). Although the rate of growth was similar between wild-type and nosRNAi neurons, nosRNAi neurons exhibited a 1.6-fold increase in the rate of retraction compared with wild-type (Fig. 2C). Furthermore, quantification of the net change in branch length revealed that nosRNAi neurons exhibited a 2.2-fold reduction in dendrite length (Fig. 2D).
Fig. 2.
nos-deficient class IV da neurons exhibit an increased rate of branch retraction in late L3 larvae. (A) Time-lapse confocal projections of dendrites from wild-type (WT), nosRNAi and nosRNAi; hidA206/+ larvae at 100 h AEL. GAL4477 was used to drive expression of both UAS-nosRNAi and UAS-CD4-GFP. Arrowheads indicate branches that are growing (blue) and retracting (red) with respect to the previous time point. Scale bar: 20 μm. (B) The frequency of occurrence of branch growth or retraction events as a fraction of the total number of events in WT, nosRNAi, and nosRNAi; hidA206/+ class IV da neurons. (C) Quantification of the average rate of branch growth and retraction in WT, nosRNAi, and nosRNAi; hidA206/+ class IV da neurons. (D) Mean net change in branch length in WT, nosRNAi, and nosRNAi; hidA206/+ class IV da neurons. Values are mean±s.e.m., n≥3 larvae for each genotype, 20 dendrites quantified per larva; ns, not significant; *P≤0.05, **P≤0.01, ***P≤0.0001.
To determine if the changes in branch dynamics in nosRNAi class IV da neurons could be attributed to upregulation of hid, we reduced hid function in these neurons. Reducing hid function restored the frequency of growth and retraction events, rate of retraction, and net change in dendrite length to wild-type levels (Fig. 2A-D). Interestingly, nosRNAi; hidA206/+ class IV da neurons exhibited a slower rate of branch extension compared with both wild-type and nosRNAi neurons, whereas the rate of retraction was comparable to wild-type levels (Fig. 2C). Time-series image analysis of hidA206/+ class IV da neurons also revealed a significant reduction in the number of new and/or extending branches between mid and late L3 development compared with wild type, suggesting that reducing hid function on its own during larval development slows branch growth without affecting branch retraction (Fig. S2). We do not know why reducing hid function in nosRNAi neurons decreases the instantaneous rate of dendrite growth; however, nosRNAi; hidA206/+ neurons exhibit an overall net change in branch length similar to wild-type neurons (Fig. 2D). From these experiments, we conclude that a balance of dendritic growth and retraction is required for class IV da neurons to maintain dendrite arbors at steady-state during late L3 development, and that Nos is required to maintain this balance by repressing hid.
Pum binds directly to the hid 3′UTR
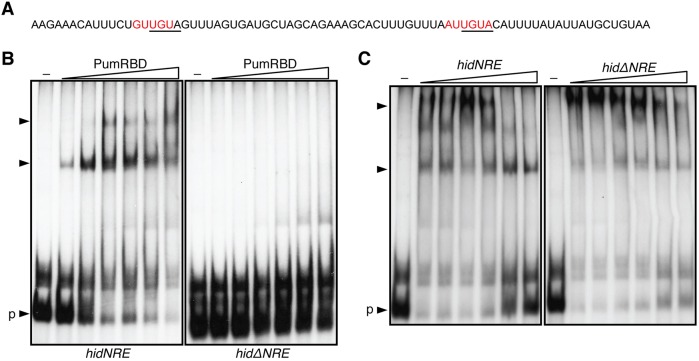
The Nos-Pum complex represses translation through the direct binding of Pum to Nos response element (NRE) motifs in the 3′UTR of target mRNAs (Sonoda and Wharton, 1999; Wharton and Struhl, 1991). An NRE within the hid 3′UTR was previously shown to be important for repression of hid in Drosophila PGCs (Fig. 3A) (Sato et al., 2007). To determine if Pum directly binds to the hid 3′UTR, we performed electrophoretic mobility shift assays (EMSAs) with purified recombinant protein encompassing the Pum RNA binding domain (PumRBD) and either a wild-type hid 3′UTR probe (hidNRE) or a hid 3′UTR probe lacking the NRE (hidΔNRE). Whereas Pum bound to the hidNRE probe, binding was abrogated by deletion of the NRE (Fig. 3B). Furthermore, binding of PumRBD to hidNRE could be competed with excess unlabeled hidNRE RNA, but not with excess unlabeled hidΔNRE RNA (Fig. 3C). These results demonstrate that Pum can directly and specifically interact with the hid 3′UTR.
Fig. 3.
The RNA-binding domain of Pum interacts with the hid 3′UTR in vitro. (A) Nucleotides 3847-3925 of the hid mRNA. The NRE sequence is in red and the nucleotides deleted in the ΔNRE probes are underlined. (B) Electrophoretic mobility shift assay to identify an interaction between Pum and the hid 3′UTR. Varying concentrations of purified PumRBD protein (0.1-500 nM) or no protein (–) was incubated with a 32P-labeled hid 3′UTR probe either containing (hidNRE) or without (hidΔNRE) the NRE. (C) Binding of PumRBD to the 32P-labeled hidNRE probe was challenged with increasing molar excess (1-, 10-, 100-, 500-, and 1000-fold) of unlabeled competitor hid RNA either containing or without the NRE (hidNRE or hidΔNRE, respectively). The binding reactions contained either no protein (–) or 300 nM purified protein. Unbound free probe indicated by ‘p’; shifts indicated by arrowheads.
Overexpression of hid in class IV da neurons recapitulates the nos mutant phenotype
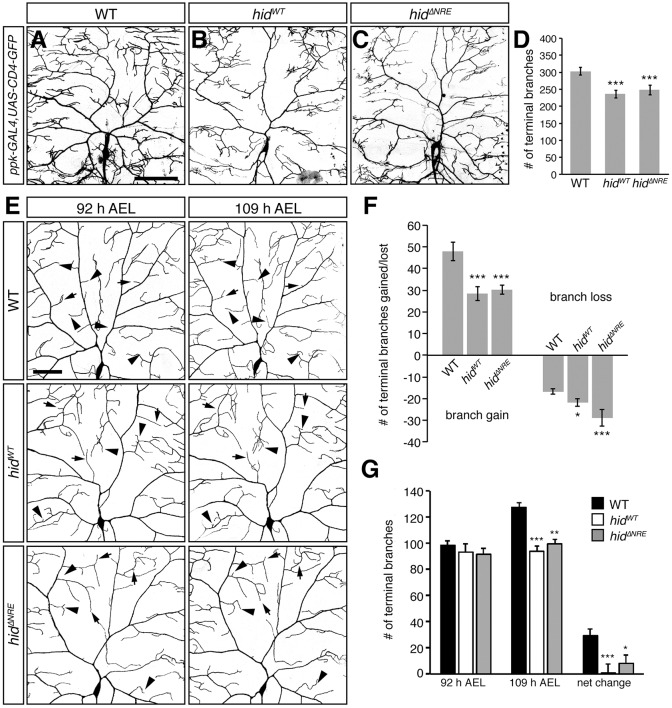
To further verify that the regulation of hid is required for dendrite morphogenesis, we tested whether overexpression of hid in larval class IV da neurons recapitulates the nos mutant phenotype. We generated a wild-type UAS-hid transgene (UAS-hidWT) and a UAS-hid transgene lacking the NRE (UAS-hidΔNRE), both of which caused 100% lethality when expressed ubiquitously using tub-GAL4 (Table S1). When expressed specifically in class IV da neurons, each caused a reduction in the number of terminal branches relative to wild-type neurons (Fig. 4A-D), which was comparable with the loss of branching observed in nosRNAi and nos mutant neurons. Expression of UAS-hid with or without the NRE did not affect the severity of the observed phenotype, suggesting that the Nos-Pum repression complex is readily saturated. The similarity between overexpression of hid and knockdown or mutation of nos in class IV da neurons extends to branch dynamics. Time-series image analysis showed that expression of either transgene resulted in an increase in branch loss and/or retraction and a decrease in branch growth as compared with wild-type neurons (Fig. 4E-G). Based on these and the experiments presented above, we conclude that repression of hid by Nos-Pum is required to maintain dendritic arbor complexity.
Fig. 4.
Overexpression of Hid recapitulates loss of nos function in larval class IV da neurons. (A-C) Confocal z-series projections of representative class IV da neurons in wild-type (WT) (A), hidWT (B), and hidΔNRE (C) larvae. (D) Quantification of the total number of dendritic terminal branches. (E) Class IV da neurons in WT, hidWT, and hidΔNRE larvae imaged at 92 h and 109 h AEL. Arrowheads indicate examples of new branch outgrowth or branch elongation and arrows designate examples of branch retraction or loss. (F) Quantification of the number of new and/or elongating branches, and lost and/or retracting branches between time points. (G) Quantification of the number of dendritic termini in WT, hidWT, and hidΔNRE larvae at 92 h AEL and 109 h AEL. Net change represents the difference in the average number of terminal branches observed between 92 h and 109 h AEL class IV da neurons. ppk-GAL4 was used to express the UAS-hid transgenes and UAS-CD4-GFP. Values in D,F,G are mean±s.e.m., n≥12 for each genotype; *P≤0.05, **P≤0.01, ***P≤0.001. Scale bars: 100 μm.
Reduction of caspase activity restores the loss of dendritic termini in nos-deficient neurons
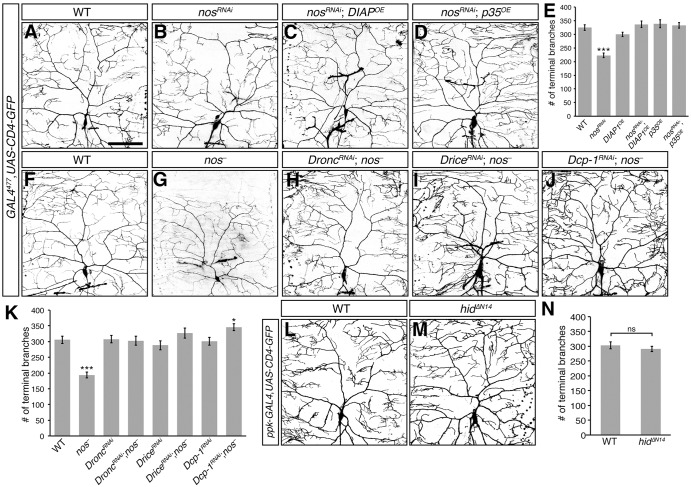
Hid is a primary regulator of apoptosis through its activation of the conserved caspase pathway (Fuchs and Steller, 2011). Hid binds to and antagonizes Drosophila inhibitor of apoptosis protein 1 (DIAP1; Diap1 – FlyBase) (Goyal et al., 2000; Hawkins et al., 1999; Wang et al., 1999), resulting in the activation of the downstream initiator caspase Dronc (Meier et al., 2000; Quinn et al., 2000) and effector caspases, Drice and Dcp-1 (Fraser et al., 1997; Song et al., 1997). Although expression of hid in class IV da neurons did not result in apoptosis (Fig. 4), non-apoptotic roles for caspases in morphogenesis led us to investigate whether the nos mutant dendritic defects could result from Hid-dependent caspase activation. We first inhibited caspase activity by overexpressing DIAP1 in nosRNAi class IV da neurons. Overexpression of DIAP1 in these neurons restored dendritic terminal branch number to wild-type (Fig. 5A-C,E). Similarly, DIAP1 overexpression rescued the dendritic defects caused by pumRNAi (data not shown). Furthermore, inhibition of effector caspase activity in nosRNAi class IV da neurons by overexpression of the baculovirus protein P35 (Hay et al., 1994) produced a similar result (Fig. 5A,B,D,E). In a complementary approach, reducing caspase activity in nos mutant class IV da neurons by RNAi-mediated knockdown of Dronc, Drice and Dcp-1 restored dendritic branching to wild-type levels (Fig. 5F-K). Taken together, these results suggest that loss of nos and pum function results in elevated caspase activity in larval class IV da neurons.
Fig. 5.
Reducing caspase activity rescues the dendritic defects of nos-deficient class IV da neurons. (A-D) Confocal z-series projections of representative class IV da neurons from wild-type (WT) (A), nosRNAi (B), nosRNAi; DIAPOE (C), and nosRNAi; p35OE (D) larvae. (E) Quantification of dendritic terminal branch number. (F-J) Class IV da neurons from WT (F), nos− (nosRC/nosRD, G), DroncRNAi; nos− (H), DriceRNAi; nos− (I), and Dcp-1RNAi; nos− (J) larvae. Knockdown of Dronc in nosRNAi da neurons resulted in a similar rescue in dendritic branching (data not shown). (K) Quantification of the total number of dendritic terminal branches. GAL4477 was used for expression of UAS-RNAi transgenes, overexpression transgenes, and UAS-CD4-GFP in A-D,F-J. (L,M) Class IV da neurons from WT (L) and hidΔN14 (M) larvae. ppk-GAL4 was used to express UAS-hidΔN14 and UAS-CD4-GFP. (N) Quantification of dendritic terminal branch number. Values in E,K,N are mean±s.e.m., n≥10 for each genotype; ns, not significant; *P≤0.05, ***P≤0.001. Scale bar: 100 μm.
The N-terminus of Hid contains a DIAP1-binding domain and deletion of this domain significantly impairs Hid function in cultured cells (Haining et al., 1999; Vucic et al., 1998). To confirm that Hid promotes caspase activation in class IV da neurons by inhibiting DIAP1, we generated a UAS-hid transgene lacking sequences encoding the N-terminal DIAP1 binding domain (UAS-hidΔN14). Stable expression of HidΔN14 was observed in transfected Drosophila S2 cells (Fig. S3). Whereas ubiquitous expression of the UAS-hidWT transgene using tub-GAL4 caused 100% lethality, ubiquitous expression of UAS-hidΔN14 resulted in partial lethality, indicating that removal of the DIAP1 binding domain significantly impairs Hid's ability to induce apoptosis (Table S1). In contrast to UAS-hidWT, expression of UAS-hidΔN14 in class IV da neurons did not affect the gross dendritic arborization pattern or the number of dendritic terminal branches as compared with wild-type neurons (Fig. 4B,D, Fig. 5L-N). Taken together, these results provide strong evidence that nos mutant dendritic defects result from elevated caspase activity as a result of upregulation of hid.
Upregulation of hid increases mitochondrial density in class IV da neurons
Permeabilization of the mitochondrial outer membrane by pro-apoptotic signals allows the release of factors crucial for caspase activation (Danial and Korsmeyer, 2004; Li et al., 1997). Hid increases mitochondrial membrane permeability when expressed in cultured Drosophila cells and early embryos (Abdelwahid et al., 2007). Because high background in surrounding tissues from reagents used to detect membrane permeability prevented us from assessing effects in da neurons, we tested if elevated Hid levels in nosRNAi da neurons generally affect mitochondria using a GFP-reporter containing a mitochondrial import sequence (mitoGFP) (Pilling et al., 2006). Whereas GFP-labeled mitochondria were observed within the soma, axon, and proximal and distal dendrites of both wild-type and nosRNAi da neurons, the density of mitochondria within the dendrites of nosRNAi neurons was significantly higher than that of wild-type neurons (Fig. S4). Reducing hid function in nosRNAi neurons restored the mitochondrial density in class IV da neuron dendrites to wild-type levels (Fig. S4), suggesting that this phenotype results from upregulation of hid.
Removal of a mitochondria localization domain within the C-terminus of Hid not only abrogates the ability of Hid to localize to mitochondria, but also impairs its function in caspase activation (Abdelwahid et al., 2007; Haining et al., 1999). To test if the localization of Hid to mitochondria is essential for its effects on dendritic branching in class IV da neurons, we expressed a mutant form of Hid lacking the mitochondria localization domain (HidΔC20). Stable expression of HidΔC20 was observed in transfected Drosophila S2 cells (Fig. S3). Ubiquitous expression of a UAS-hidΔC20 transgene using tub-GAL4 resulted in 100% lethality, indicating that in at least some tissues or at sufficient levels, HidΔC20 can promote apoptosis (Table S1). In contrast to neuronal expression of wild-type Hid, which caused a decrease in the number of terminal branches (Fig. 4), expression of HidΔC20 resulted in class IV da neurons that morphologically resembled wild-type neurons with respect to gross morphology and number of terminal branches (Fig. S4). Together, these results indicate that upregulation of Hid leads to an increase in mitochondrial density and that the ability of Hid to localize to mitochondria is required for Hid's effects on dendritic branching.
Requirement for nos coincides with the switch in sensitivity to apoptotic triggers at the mid-L3 transition
For most of their development, Drosophila larvae are resistant to apoptotic triggers. However, this resistance is lost mid-way through L3 development and larvae become highly sensitive to apoptotic activators like Hid and Rpr. This switch in sensitivity is mediated cell-autonomously by a low-level pulse of the steroid hormone ecdysone that coincides with the mid-L3 transition, when the larva initiates preparations for pupariation (Kang and Bashirullah, 2014). The striking temporal correspondence between the switch in apoptotic sensitivity and the requirement for Nos-Pum-mediated repression of hid led us to investigate the relationship between this requirement and the mid-L3 ecdysone pulse.
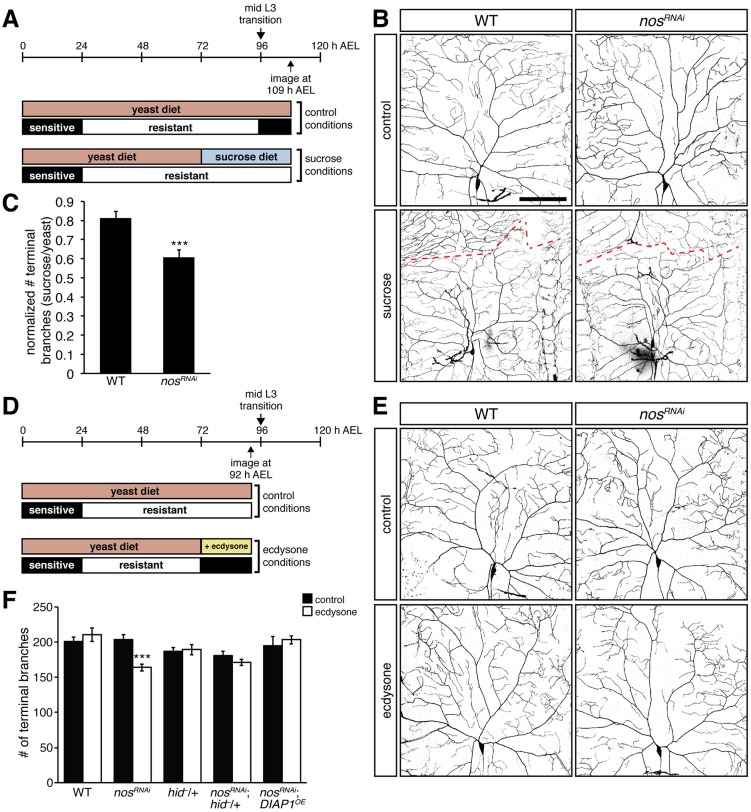
We first tested whether the temporal requirement for nos – after the mid-L3 transition – reflects the ecdysone-mediated shift in apoptotic sensitivity. Transferring early L3 larvae from a yeast diet to a sugar-only diet arrests the progression of L3 development and thus delays the mid-L3 transition (Fig. 6A) (Britton and Edgar, 1998). These early L3 larvae remain resistant to caspase activity until they are re-introduced to a complete food source (Kang and Bashirullah, 2014). As anticipated, nosRNAi larvae fed a yeast diet exhibited a significant decrease in the number of dendritic termini as compared with wild-type controls at 109 h after egg laying (AEL). By contrast, nosRNAi larvae fed a sugar-only diet showed a much less severe deficit (Fig. 6B-C), suggesting that delaying the ecdysone pulse that normally occurs during the mid-L3 transition is sufficient to inhibit the caspase-mediated effects observed in nos mutant da neurons.
Fig. 6.
The requirement for nos depends on ecdysone signaling at the mid-L3 transition. (A) Timeline shows development from egg deposition (0 h) to 120 h AEL and sensitivity or resistance to caspases. Larvae were fed a yeast diet until the beginning of the L3 stage (72 h AEL), then were either kept on yeast or transferred to a sugar-only diet. Neurons were imaged at 109 h AEL. (B) Confocal z-series projections of representative class IV da neurons in wild-type (WT) and nosRNAi larvae from the experiment shown in A. Dashed line shows the boundary of the neuron from the contralateral neuron. (C) Normalized dendritic terminal branch number in WT and nosRNAi larvae treated with a sugar-only or yeast diet. (D) Larvae were fed yeast until the beginning of the L3 stage, then either kept on yeast or transferred to yeast containing ecdysone. Neurons were imaged at 92 h AEL, prior to the mid-L3 transition. (E) Class IV da neurons in WT and nosRNAi larvae from the experiment shown in D. (F) Quantification of the dendritic terminal branch number. UAS-nosRNAi, UAS-DIAP1, and UAS-CD4-GFP were expressed using GAL4477. Values in C,F are mean±s.e.m., n≥10 for each genotype; ***P≤0.001. Scale bar: 100 μm.
We next tested whether a premature switch in apoptotic sensitivity would lead to an earlier requirement for nos function. Early L3 larvae were fed yeast containing 20-hydroxyecdysone (ecdysone) for 20 h during early L3 development and dendrite morphology was analyzed prior to the mid-L3 transition (Fig. 6D). As previously observed, class IV da neurons of untreated nosRNAi larvae were indistinguishable from wild-type larvae at 92 h AEL (Fig. 1J). By contrast, nosRNAi larvae that were administered ecdysone exhibited a significant decrease in the number of dendritic termini compared with ecdysone-treated controls (Fig. 6E-F). This loss of branching depends on caspase activation because reducing hid function or inhibiting caspase activity by overexpressing DIAP1 in nosRNAi neurons rescued branching (Fig. 6E-F). Taken together, these results explain why the nos mutant phenotype manifests late in larval development, after the ecdysone-dependent switch in caspase activation.
Overexpression of Nos and Pum delays pruning of class IV da neurons during pupariation
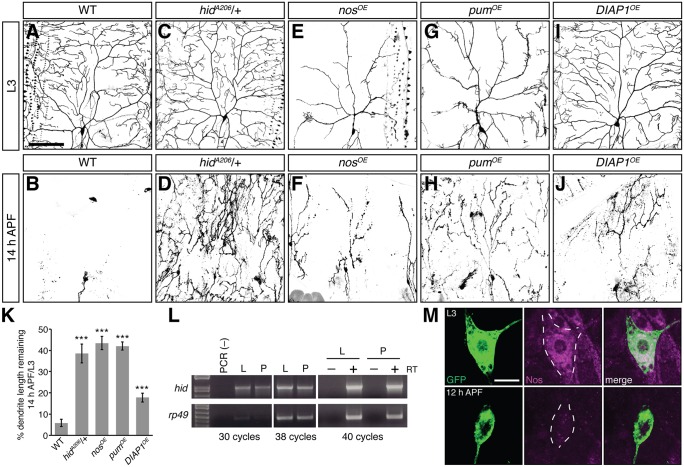
Whereas caspase activation must be suppressed for proper dendrite morphogenesis during larval stages, local non-apoptotic caspase function within the dendrites is required for pruning of class IV da neuron dendrites during metamorphosis (Kuo et al., 2006; Williams et al., 2006). Reducing caspase activity by a variety of methods including overexpression of DIAP1 delays pruning (Kuo et al., 2006; Williams et al., 2006 and Fig. 7A-B,I-J,K). However, whether Hid plays a role in regulating caspase activity during pruning has not been investigated. In wild-type class IV da neurons, the majority of dendrites are cleared by 14 h after puparium formation (APF), leaving only the soma and axon (Fig. 7A,B,K and Kuo et al., 2005, 2006; Williams et al., 2006; Williams and Truman, 2005). Reduction of hid function resulted in a delay in pruning (Fig. 7A-D,K), with a significant proportion of branch length remaining compared with wild-type animals (5.9% vs 38.5% in WT versus hidA206/+ pupae, respectively). A similar delay in pruning was observed in hidRNAi neurons (data not shown).
Fig. 7.
Reducing hid function or overexpressing Nos and Pum delays pruning during pupariation. (A-J) Confocal z-series projections of larval (A,C,E,G,I) and pupal (14 h APF; B,D,F,H,J) class IV da neurons. (A,B) WT, (C,D) hidA206/+, (E,F) nosOE, (G,H) pumOE, and (I,J) DIAP1OE. (K) The percentage of dendrite branch length remaining at 14 h APF normalized to the average branch length of neurons in late L3 larvae for each genotype. (L) RT-PCR to detect hid mRNA in wandering L3 larvae (L) and 12 h APF pupae (P). rp49 was used as a loading control. (M) Anti-GFP (green) and anti-Nos (magenta) immunostaining of class IV da neurons in wandering L3 larvae and 12 h APF pupae. Dashed line outlines the soma. All transgenes were expressed using ppk-GAL4; neurons were marked using UAS-CD4-GFP. Values in K are mean±s.e.m., n≥10 for each genotype; ***P≤0.001. Scale bar: 100 μm in A-J; 10 μm in M.
The ability of Nos-Pum to repress hid during larval development led us to test whether overexpression of Nos or Pum would also delay pruning. Previous studies have shown that overexpression of Nos and Pum in larval class IV da neurons causes severe morphological defects, including a significant decrease in terminal branch number and total branch length as compared with wild-type (Fig. 7A,E,G) (Olesnicky et al., 2012; Ye et al., 2004). We therefore normalized the dendrite length in pupal neurons overexpressing Nos (nosOE) or Pum (pumOE) to that of the mean dendrite length in larval neurons. Similarly to hidA206/+ pupae, dendrites in nosOE and pumOE pupae were still visible by 14 h APF, with ∼43.4% and 42.0% of branch length remaining, respectively (Fig. 7E-H,K). Although it has not been possible to analyze Hid protein levels reliably in class IV da neurons, results from these genetic experiments indicate that Hid is required during pupariation for caspase-mediated pruning of class IV da neurons.
Our data also suggest that repression by Nos and Pum must be overcome for the necessary production of Hid during pupariation, either through increased hid mRNA production or through downregulation of Nos and/or Pum. Semi-quantitative RT-PCR analysis of hid mRNA in larval and pupal body wall tissue containing da neurons detected little difference in hid mRNA levels between these stages (Fig. 7L). By contrast, anti-Nos immunostaining of larval and pupal class IV da neurons showed that Nos protein is dramatically reduced in pupal class IV da neurons as compared with larval neurons (Fig. 7M). These data strongly suggest that expression of Nos, which is required to prevent Hid accumulation during late larval development, must be itself downregulated to permit Hid expression and function in class IV da neurons during pupariation.
DISCUSSION
The translational repressors Nos and Pum are required to maintain dendritic complexity of class IV da neurons during late larval development. Here, we show that maintenance of the dendritic arbor requires a balance of dendrite growth and retraction and that during late larval development, this balance depends on Nos-Pum-mediated repression of hid. Together, our data indicate that upregulation of hid in larval class IV da neurons leads to excessive branch retraction resulting from an increase in caspase activity. During pupariation, caspase activity mediates the pruning of class IV da neuron dendrites. Lowering Hid levels by reducing hid gene dosage or by overexpressing Nos or Pum suppressed pruning, suggesting that the same apoptotic pathway members that promote caspase activity in pruning dendrites are also active in nos mutant larval class IV da neurons.
The dendritic arborization defects observed in nos mutant or RNAi neurons do not appear until the late L3 stage (Brechbiel and Gavis, 2008; Olesnicky et al., 2012). This timing coincides with an ecdysone pulse during the mid-L3 transition that switches the larva's sensitivity to apoptotic triggers from restrictive to permissive (Kang and Bashirullah, 2014). Moreover, we find that preventing the ecdysone pulse obviates the requirement for nos and that premature ecdysone signaling results in an earlier requirement for nos. Together, our results are consistent with a model in which Nos and Pum function after the mid-L3 transition to prevent untoward expression of Hid prior to metamorphosis.
Although Hid is a major pro-apoptotic factor in Drosophila, upregulation of Hid expression in class IV da neurons, and consequent downstream caspase activation, has non-apoptotic consequences. Why Hid expression does not result in a full apoptotic response is not yet clear. It is possible the elevated Hid levels are only sufficient for low levels of caspase activation. Alternatively, local activation of caspases in dendrites, as observed during pruning (Kuo et al., 2006; Williams et al., 2006), could alter dendrite dynamics without inducing apoptosis. Consistent with this idea, dendritic localization of nos mRNA is required for nos function in da neurons, suggesting that the Nos-Pum repressor acts locally in dendrites (Brechbiel and Gavis, 2008). We have been unable to detect local caspase activity within larval class IV da neuron dendrites using several available caspase reporters and caspase antibodies (Bardet et al., 2008; Olesnicky et al., 2012; Takemoto et al., 2003; Williams et al., 2006). In contrast to caspase activation during pruning, which is triggered by a robust ecdysone pulse prior to pupariation resulting in a potent caspase response, caspase activation in nos-deficient larval neurons does not result in dendrite fragmentation and engulfment and mainly affects higher order branches. Caspase activity in nos-deficient neurons might thus be too low to detect readily, especially in thin higher order branches.
Non-apoptotic caspase function is essential for a variety of morphogenetic events and caspases are thought to have a role in regulating actin dynamics underlying cell shape changes and cell migration (Kuranaga, 2011). During pruning, caspases have been implicated in cytoskeletal destabilization and fragmentation of dendrites as well as in clearance of severed dendrites. In nos mutants, the actin-rich higher order branches are primarily affected. Whereas the distribution of actin throughout the dendritic arbor is not grossly affected (data not shown), the observed branch retraction could result from caspase-mediated actin depolymerization. Consistent with this idea, Rac GTPases, which regulate actin dynamics, are caspase substrates and Rac1 promotes dendrite branching in class IV da neurons (Lee et al., 2003).
During metamorphosis, ecdysone-triggered transcription factors induce hid mRNA expression for apoptotic destruction of salivary glands (Jiang et al., 2000). Our data now suggest a requirement for hid in pruning of da neuron dendrites. Pruning depends on the ubiquitin-proteosome system and the E2 ubiquitin conjugating enzyme UbcD1 (Kuo et al., 2006). UbcD1 mediates DIAP1 auto-ubiquitination and degradation following apoptotic stimuli, resulting in activation of Dronc (Meier et al., 2000; Muro et al., 2002; Ryoo et al., 2002; Treier et al., 1992; Wilson et al., 2002). Consistent with a role for Hid, genetic analysis indicates that UbcD1 acts downstream of Hid to promote apoptosis (Ryoo et al., 2002). In contrast to regulation of hid in salivary glands, expression of hid mRNA does not appear to be dramatically increased in da neurons at pupariation, although we cannot rule out effects in individual neurons. This result, and the finding that overexpression of Nos and Pum suppress pruning, suggest that Nos and/or Pum must be downregulated at pupariation for hid function in pruning. The observed drop in Nos levels from larval to pupal class IV da neurons further supports this idea. How Nos protein levels are downregulated and whether this regulation is ecdysone-dependent are interesting questions for future study.
Although the overt phenotype of nos and pum mutant neurons is failure of dendrite maintenance, our results suggest that this defect is a result of untoward caspase activation. Nos and Pum might therefore function as an additional, post-transcriptional regulatory layer to prevent translation of hid mRNA present in larval da neurons and ensure that caspase activity is inhibited during late larval development following the mid-L3 ecdysone pulse. Furthermore, a need for Nos and Pum downregulation after pupariation for caspase activation during pruning creates yet another layer of control, thereby ensuring that caspases are activated only at the appropriate developmental time.
MATERIALS AND METHODS
Fly strains
The following reporters were used to visualize class IV da neurons: the GAL4477 driver (Grueber et al., 2003) was recombined with UAS-CD4-tdGFP (Han et al., 2011) to produce GAL4477, UAS-CD4-GFP; ppk-GAL4 [Bloomington Drosophila Stock Center (BDSC)] was recombined with UAS-CD4-tdGFP to produce ppk-GAL4, UAS-CD4-GFP; ppk1.9-GAL4 (Ainsley et al., 2003) was recombined with UAS-CD4-tdTom (Han et al., 2011) to produce ppk1.9-GAL4, UAS-CD4-Tom. The following transgenic lines were obtained from the BDSC: UAS-pumRNAi (P{TRiP.HMS01685}), UAS-DIAP1 (6657), UAS-p35 (Hay et al., 1994), UAS-mito-HA-GFP (Pilling et al., 2006) and tubP-GAL4 (Lee and Luo, 1999). The following RNAi lines were obtained from the Vienna Drosophila RNAi Center (VDRC; Dietzl et al., 2007): UAS-DroncRNAiVDRC (P{GD12376} and P{KK104278}), UAS-DriceRNAiVDRC (P{GD12284}), UAS-Dcp-1RNAiVDRC (P{KK106134}), and UAS-hidRNAiVDRC (P{GD1673}). The UAS-nosRNAi, UAS-nos-tub3′UTR and UAS-pum transgenic lines were previously described (Clark et al., 2002; Menon et al., 2009, 2004). The following mutant alleles were used: the strong hypormorphic allele combination nosRC/nosRD (Curtis et al., 1997), the hidA206 hypomorphic allele (Abbott and Lengyel, 1991) and the recombinant grimC15E, rpr87 double mutant, which deletes the grim and rpr loci (Tan et al., 2011).
To enhance GAL4/UAS efficiency, crosses with the different UAS-hid transgenes as well as epistasis experiments with UAS-nosRNAi and UAS-pumRNAi were performed at 29°C. All other crosses were performed at 25°C.
Plasmid construction
UAS-hid-hid3′UTR transgenes
pattB-UASt-hid-wt3′UTR and pattB-UASt-hid-ΔNRE3′UTR were generated by transferring a NotI-XbaI fragment containing the hid 5′UTR, coding region, and 3′UTR sequences from pUASp-hid5′UTR-hidCDS-FLAG-wt hid3′UTR and pUASp-hid5′UTR-hidCDS-FLAG-ΔNRE hid3′UTR (Sato et al., 2007) to pattB-UASt.
UAS-hidΔC20 transgene
A deletion removing the last 20 C-terminal amino acids of Hid was generated by joining two PCR products amplified from pUASp-hid5′UTR-hidCDS-FLAG-wt hid 3′UTR with the following pairs of primers: NotI.hid.F (5′-AAGCGGCCGCTGACAAAAAATAAAAAACGAAATC-3′) and EcoRI.hidΔ20.R (5′-AAGAATTCAGTGAAGCTCTGTGGTTTCT-3′); EcoRI.hidflagstop3′UTR.F (5′-AAGAATTCGATTATAAAGATGATGATGATAAATGA-3′) and BamHI.hid3′UTR.R (5′-CGGGATCCCTTTACACATACACATAGATGT-3′). The two PCR products were digested, respectively, with NotI and EcoRI and EcoRI and BamHI, and ligated together into pBS-SK digested with NotI and BamHI to produce pBS-HidΔC20. The hidΔC20 fragment was excised from pBS-hidΔC20 with NotI and Acc65I and inserted into pattB-UASt- to produce pattB-UAST-hidΔC20.
UAS-hidΔN14 transgene
A deletion removing the first 14 N-terminal amino acids of Hid was generated by joining two PCR products amplified from pUASp-hid5′UTR-hidCDS-FLAG-wt hid3′UTR with the following pairs of primers: NotI.hid.F (see above) and NdeI.hid.N14.R (5′-AACATATGGTGGGTGTGTGTGTG-3′); NdeI.hid.N14.F (5′-AACATATGGCGTCGAGTTCATCGG-3′) and BamHI.hid3′UTR.R (see above). Construction of pUASt-attB-hidΔN14 was performed similarly to pattB-UASt-hidΔC20.
All transgenes were inserted into the attP40 site using phiC31 integrase (Bischof et al., 2007; Markstein et al., 2008).
pBS-hidNRE and pBS-hidΔNRE
Approximately 100 bp of the 3′UTR sequences from pUASp-GFP-WT hid3′UTR or pUASp-GFP-ΔNRE hid3′UTR (Sato et al., 2007) was PCR amplified using the following primers: 5′-AAGATATCAGAAACATTTCT-3′ and 5′-AACTGCAGTAATATAAAATG-3′. After digesting with EcoRV and PstI, the PCR products were ligated into the corresponding sites of pBS-SKΔKP (Kalifa et al., 2006).
pET-SUMO-pumRBD
Sequences encompassing the Pum RBD were excised from pJC45-pumRBD (gift of K. Zinn, Caltech, Pasadena, USA) by digestion with NdeI and BamHI, end-filled, and ligated into pET-SUMO+NS (gift of P. Schedl, Princeton University, Princeton, USA) digested with XhoI and end-filled.
Immunostaining
Larvae were dissected in calcium-free saline buffer (135 mM NaCl, 5 mM KCl, 4 mM MgCl2, 2.9 mM Tris-HCl pH 7.4, 2.1 mM Tris-HCl pH 6.8, 36 mM sucrose) and fixed in 4% paraformaldehyde (PFA) for 30 min at room temperature. After fixation, larval fillets were washed three times in PBT-X (PBS, 0.3% Triton X-100) at room temperature, blocked for 1 h in PBT-X/5% normal goat serum (NGS), and then incubated in Alexa Fluor 488 rabbit anti-GFP (1:500; Life Technologies, A-21311), mouse anti-GFP (1:500; Abcam, ab1218) or rabbit anti-dsRed (1:1000; Clontech, 632496) primary antibody diluted in PBT-X/5% NGS overnight at 4°C. Fillets were washed three times in PBT-X, and either mounted in Vectashield (Vector Labs) or incubated in Alexa Fluor 488 goat anti-mouse (1:200; Life Technologies, A11001) or Alexa Fluor 568 goat anti-rabbit (1:200; Life Technologies, A11011) secondary antibody in PBT-X for 2 h at room temperature, washed three times in PBT-X and mounted in Vectashield. Pupal samples were prepared as described (Wang and Yoder, 2011) and anti-GFP immunofluorescence was performed as above. Neurons were imaged using a Leica SPE confocal microscope with a 20×/0.7 NA air objective.
For anti-Nos staining, larvae and pupae were dissected and fixed as above, followed by fixation with MeOH at −20°C. Larval and pupal samples were incubated with Image-iT (Thermo Fisher Scientific) for 30 min prior to blocking in NGS. Guinea pig anti-Nos (gift of H. Luo and H. Lipshitz, University of Toronto, Toronto, Canada) was used at 1:500, followed by Alexa Fluor 568 goat anti-guinea pig (1:200; Life Technologies, A11075). Larval and pupal neurons were imaged using a Leica SP5 confocal microscope with a 40×/1.25 NA oil objective.
Analysis of dendrite morphology
Unless otherwise noted, larval neurons were analyzed at the wandering third larval instar stage, corresponding to ∼108-120 h AEL. The dorsal-most class IV da neuron (ddaC) from abdominal segments A3-A5 was imaged, and the soma of each neuron was placed in a similar location within the field of view. The total number of dendritic terminal branches in the given field was quantified from z-series projections using ImageJ software. At least 10 neurons and 5 larvae were imaged and analyzed for each genotype.
Pupal class IV da neurons were imaged at 14 h APF (at 25°C) using the same microscopy parameters. Dendrite length was quantified using the Imaris 7.6.4 software package (Bitplane AG, Zurich, Switzerland) and the percentage of dendrite length remaining was quantified by normalizing the dendrite length remaining in 14 h APF samples to the average dendrite length in wandering L3 larvae. At least 10 neurons and five animals were imaged and analyzed for each genotype.
Live imaging of class IV da neurons
Time-series analysis was performed as previously described (Olesnicky et al., 2012). At least 10 ddaC neurons per genotype were imaged on a Leica SPE confocal microscope with a 40×/1.25 NA oil objective. For real-time analysis of branch dynamics, third instar larvae (100 h AEL) were immobilized and mounted using a 1:4 chloroform:halocarbon oil (2:1 95 Halocarbon oil:200 Halocarbon oil) mixture. One ddaC neuron per larva was imaged immediately after mounting on a Leica SP5 confocal microscope using a 40×/1.25 NA oil objective. A z-series projection was captured every 2 min for up to 30 min. For each xyzt sequence, the length of 20 terminal dendrites per frame was quantified using the Imaris 7.6.4 software. At least three larvae per genotype were imaged and quantified.
Sucrose and ecdysone feeding experiments
To delay ecdysone signaling, 72 h AEL larvae were transferred to apple juice agar plates containing 2.5% sucrose and spread with yeast paste (control) or apple juice agar plates containing 20% sucrose without yeast paste. Larvae were maintained on yeast or sucrose until they were imaged live at 109 h AEL. To prematurely activate ecdysone signaling, 72 h AEL larvae were transferred to apple juice agar plates spread with yeast (control) or yeast containing 120 μg/ml 20-hydroxyecdysone (Sigma, H5142). Larvae were maintained on yeast or yeast+ecdysone for 20 h until they were imaged live at 92 h AEL. Both experiments were performed at 25°C. At least 10 ddaC neurons and five larvae were imaged per condition and genotype.
Purification of PumRBD
His-tagged PumRBD (pET-SUMO-pumRBD) was expressed in E. coli BL21(DE3) pLysE cells for 3 h at 28° C. Cells were resuspended in lysis buffer (50 mM Tris-HCl pH 8.0, 300 mM NaCl, 0.2 mM PMSF) and sonicated four times, 30 s each treatment. Triton X-100 was added to a final concentration of 0.5% and the lysate was spun for 10 min, 14,000 g at 4°C to remove debris. The cleared lysate was passed through a column containing Ni-NTA agarose (Qiagen) twice, followed by four washes in 50 mM Tris-HCl pH 8.0, 150 mM NaCl, 10 mM Imidazole, 0.2 mM PMSF. The bound protein was eluted in 50 mM Tris-HCl, pH 8.0, 150 mM NaCl, 250 mM Imidazole, 10% glycerol, 0.2 mM PMSF, concentrated using an Amicon Ultra-15 Centrifugal Filter Unit with Ultracel-30 membrane (UFC903008, Amicon) to a concentration of 1 mg/ml and dialyzed overnight in 50 mM Tris-HCl pH 8.0, 150 mM NaCl, 10% glycerol, 0.2 mM PMSF, 1 mM DTT using a Slide-A-Lyzer Dialysis Cassette (Pierce). Purified protein was aliquoted and stored at –80° C until use.
Electrophoretic mobility shift assays
32P-labelled probes were generated by in vitro transcription of linearized pBS-hidNRE and pBS-hidΔNRE plasmids as previously described (Bergsten et al., 2001). Probes were denatured at 65°C for 5 min and placed immediately on ice. The binding reactions contained 1 mM EDTA, 10 mM Hepes, 100 mM KCl, 2 mM DTT, 0.1 mg/ml BSA, 10% glycerol, 40 U RNase Inhibitor (NEB), 32P-labeled probe, and PumRBD protein (0.1-520 nM) in a final volume of 15 μl. Binding reactions were incubated on ice for 15 min and electrophoresed on a 6% native acrylamide gel at 4°C. Competition assays were performed similarly with the addition of cold competitor RNA (up to 1000-fold) to reactions containing 300 nM PumRBD.
Cell culture, transfection, and immunoblot analysis
Drosophila S2 cells were grown in Schneider's Drosophila medium (Life Technologies) supplemented with 10% fetal bovine serum (Life Technologies), 1% GlutaMAX (Life Technologies), and 1% penicillin/streptomycin (Life Technologies), and maintained at 25°C. 100 ng each of pUAS-attB-hid and met-GAL4 (gift of T. Jongens, University of Pennsylvania, Philadelphia, USA) plasmid DNA were co-transfected into S2 cells using Effectene Transfection Reagent (Qiagen). Expression was induced with CuSO4 at a final concentration of 0.5 mM for 4 h at 25°C. Cells were harvested by centrifugation at 100 g for 5 min at room temperature. Cell extract was prepared by resuspension in 400 μl RIPA buffer (150 mM NaCl, 0.5% deoxycholate, 0.1% SDS, 50 mM Tris-HCl pH 8.0, 10 mM NaF, 0.4 mM EDTA pH 8.0, 10% glycerol) and centrifugation at 13,000 g for 30 min at 4°C. Proteins were separated by SDS-PAGE, transferred to nitrocellulose membrane, and detected by immunoblotting and chemiluminescence. The following antibodies were used: 1:3500 rabbit-anti-Hid (gift of K. White, Massachusetts General Hospital, Charlestown, USA); 1:5000 rabbit-anti-Khc (AKIN01, Cytoskeleton); 1:2500 HRP-conjugated donkey-anti-rabbit IgG (NA934, GE Healthcare Life Sciences.
RNA isolation and RT-PCR analysis
RNA from five larval fillets or from body wall tissue of five pupae (12 h APF) was extracted with Thermo Fisher Scientific TRIzol Reagent:chloroform, precipitated with isopropanol, and transferred to an RNeasy Mini Kit (Qiagen) with on-column treatment with RNase-free DNase I (Qiagen) per manufacturer's instructions. cDNA was generated using SuperScript II reverse transcriptase (Invitrogen) and oligo(dT) primer per manufacturer's instructions. Negative control samples were treated similarly except that reverse transcriptase was omitted. The following primers were used to amplify hid and rp49 (loading control): hid.F, 5'-AAGCGCAGGAGACGTGTAAT-3'; hid.R, 5'-TTTCAATCGTTTTGCCATCA-3'; rp49.F, 5'-ATGCTAAGCTGTCGCACAAA-3'; rp49.R, 5'-ATGGTGCTGCTATCCCAATC-3'.
Statistical analysis
Two-tailed Student's t-tests were used to determine significance in this study except for Table S1, in which a Fisher's exact probability test was performed.
Acknowledgements
We thank E. Olesnicky Killian, S. Little, M. Misra, and C. Tenenbaum for comments on the manuscript. We are grateful to H. Lipshitz, H. Luo, T. Jongens, S. Kobayashi (Okazaki Institute for Integrative Biosciences, Okazaki, Japan), K. Menon (Caltech, Pasadena, USA), P. Schedl, H. Steller (Rockefeller University, New York, USA), K. White, K. Zinn, the Bloomington Stock Center, the TRiP at Harvard Medical School (NIH/NIGMS R01-GM084947), and the Vienna Drosophila Resource Center (VDRC) for fly stocks and reagents. We thank Gary Laevsky for assistance with confocal microscopy.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Author contributions
B.B. and E.R.G. designed the experiments. B.B. and A.P.-J. performed the experiments. B.B. and E.R.G. analyzed the data. B.B. and E.R.G. wrote the manuscript.
Funding
This work was supported by the National Institutes of Health [R01 GM061107 to E.R.G. and F32 NS081860 to B.B.]. Deposited in PMC for release after 12 months.
Supplementary information
Supplementary information available online at http://dev.biologists.org/lookup/suppl/doi:10.1242/dev.132415/-/DC1
References
- Abbott M. K. and Lengyel J. A. (1991). Embryonic head involution and rotation of male terminalia require the Drosophila locus head involution defective. Genetics 129, 783-789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abdelwahid E., Yokokura T., Krieser R. J., Balasundaram S., Fowle W. H. and White K. (2007). Mitochondrial disruption in Drosophila apoptosis. Dev. Cell 12, 793-806. 10.1016/j.devcel.2007.04.004 [DOI] [PubMed] [Google Scholar]
- Ainsley J. A., Pettus J. M., Bosenko D., Gerstein C. E., Zinkevich N., Anderson M. G., Adams C. M., Welsh M. J. and Johnson W. A. (2003). Enhanced locomotion caused by loss of the Drosophila DEG/ENaC protein Pickpocket1. Curr. Biol. 13, 1557-1563. 10.1016/S0960-9822(03)00596-7 [DOI] [PubMed] [Google Scholar]
- Arama E., Agapite J. and Steller H. (2003). Caspase activity and a specific cytochrome C are required for sperm differentiation in Drosophila. Dev. Cell 4, 687-697. 10.1016/S1534-5807(03)00120-5 [DOI] [PubMed] [Google Scholar]
- Bardet P.-L., Kolahgar G., Mynett A., Miguel-Aliaga I., Briscoe J., Meier P. and Vincent J.-P. (2008). A fluorescent reporter of caspase activity for live imaging. Proc. Natl. Acad. Sci. USA 105, 13901-13905. 10.1073/pnas.0806983105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergsten S. E., Huang T., Chatterjee S. and Gavis, E. R. (2001). Recognition and long-range interactions of a minimal nanos RNA localization signal element. Development 128, 427-435. [DOI] [PubMed] [Google Scholar]
- Bischof J., Maeda R. K., Hediger M., Karch F. and Basler K. (2007). An optimized transgenesis system for Drosophila using germ-line-specific phiC31 integrases. Proc. Natl. Acad. Sci. USA 104, 3312-3317. 10.1073/pnas.0611511104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brechbiel J. L. and Gavis E. R. (2008). Spatial regulation of nanos is required for its function in dendrite morphogenesis. Curr. Biol. 18, 745-750. 10.1016/j.cub.2008.04.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Britton J. S. and Edgar B. A. (1998). Environmental control of the cell cycle in Drosophila: nutrition activates mitotic and endoreplicative cells by distinct mechanisms. Development 125, 2149-2158. [DOI] [PubMed] [Google Scholar]
- Clark I. E., Dobi K. C., Duchow H. K., Vlasak A. N. and Gavis E. R. (2002). A common translational control mechanism functions in axial patterning and neuroendocrine signaling in Drosophila. Development 129, 3325-3334. [DOI] [PubMed] [Google Scholar]
- Curtis D., Treiber D. K., Tao F., Zamore P. D.,Williamson J. R. and Lehmann R. (1997). A CCHC metal-binding domain in Nanos is essential for translational regulation. EMBO J. 16, 834-843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danial N. N. and Korsmeyer S. J. (2004). Cell death: critical control points. Cell 116, 205-219. 10.1016/S0092-8674(04)00046-7 [DOI] [PubMed] [Google Scholar]
- Dietzl G., Chen D., Schnorrer F., Su K.-C., Barinova Y., Fellner M., Gasser B., Kinsey K., Oppel S., Scheiblauer S. et al. (2007). A genome-wide transgenic RNAi library for conditional gene inactivation in Drosophila. Nature 448, 151-156. 10.1038/nature05954 [DOI] [PubMed] [Google Scholar]
- Fraser A. G., McCarthy N. J. and Evan G. I. (1997). drICE is an essential caspase required for apoptotic activity in Drosophila cells. EMBO J. 16, 6192-6199. 10.1093/emboj/16.20.6192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuchs Y. and Steller H. (2011). Programmed cell death in animal development and disease. Cell 147, 742-758. 10.1016/j.cell.2011.10.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goyal L., McCall K., Agapite J., Hartwieg E. and Steller H. (2000). Induction of apoptosis by Drosophila reaper, hid and grim through inhibition of IAP function. EMBO J. 19, 589-597. 10.1093/emboj/19.4.589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grueber W. B., Jan L. Y. and Jan Y. N. (2002). Tiling of the Drosophila epidermis by multidendritic sensory neurons. Development 129, 2867-2878. [DOI] [PubMed] [Google Scholar]
- Grueber W. B., Jan L. Y. and Jan Y. N. (2003). Different levels of the homeodomain protein cut regulate distinct dendrite branching patterns of Drosophila multidendritic neurons. Cell 112, 805-818. 10.1016/S0092-8674(03)00160-0 [DOI] [PubMed] [Google Scholar]
- Haining W. N., Carboy-Newcomb C., Wei C. L. and Steller H. (1999). The proapoptotic function of Drosophila Hid is conserved in mammalian cells. Proc. Natl. Acad. Sci. USA 96, 4936-4941. 10.1073/pnas.96.9.4936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han C., Jan L. Y. and Jan Y.-N. (2011). Enhancer-driven membrane markers for analysis of nonautonomous mechanisms reveal neuron-glia interactions in Drosophila. Proc. Natl. Acad. Sci. USA 108, 9673-9678. 10.1073/pnas.1106386108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawkins C. J., Wang S. L. and Hay B. A. (1999). A cloning method to identify caspases and their regulators in yeast: identification of Drosophila IAP1 as an inhibitor of the Drosophila caspase DCP-1. Proc. Natl. Acad. Sci. USA 96, 2885-2890. 10.1073/pnas.96.6.2885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hay B. A., Wolff T. and Rubin G. M. (1994). Expression of baculovirus P35 prevents cell death in Drosophila. Development 120, 2121-2129. [DOI] [PubMed] [Google Scholar]
- Huh J. R., Vernooy S. Y., Yu H., Yan N., Shi Y., Guo M. and Hay B. A. (2004). Multiple apoptotic caspase cascades are required in nonapoptotic roles for Drosophila spermatid individualization. PLoS Biol. 2, e15 10.1371/journal.pbio.0020015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang C., Lamblin A.-F. J., Steller H. and Thummel C. S. (2000). A steroid-triggered transcriptional hierarchy controls salivary gland cell death during Drosophila metamorphosis. Mol. Cell 5, 445-455. 10.1016/S1097-2765(00)80439-6 [DOI] [PubMed] [Google Scholar]
- Kalifa Y., Huang T., Rosen L. N., Chatterjee S. and Gavis E. R. (2006). Glorund, a Drosophila hnRNP F/H homolog, is an ovarian repressor of nanos translation. Dev. Cell 10, 291-301. 10.1016/j.devcel.2006.01.001 [DOI] [PubMed] [Google Scholar]
- Kang Y. and Bashirullah A. (2014). A steroid-controlled global switch in sensitivity to apoptosis during Drosophila development. Dev. Biol. 386, 34-41. 10.1016/j.ydbio.2013.12.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo C. T., Jan L. Y. and Jan Y. N. (2005). Dendrite-specific remodeling of Drosophila sensory neurons requires matrix metalloproteases, ubiquitin-proteasome, and ecdysone signaling. Proc. Natl. Acad. Sci. USA 102, 15230-15235. 10.1073/pnas.0507393102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo C. T., Zhu S., Younger S., Jan L. Y. and Jan Y. N. (2006). Identification of E2/E3 ubiquitinating enzymes and caspase activity regulating Drosophila sensory neuron dendrite pruning. Neuron 51, 283-290. 10.1016/j.neuron.2006.07.014 [DOI] [PubMed] [Google Scholar]
- Kuranaga E. (2011). Caspase signaling in animal development. Dev. Growth Differ. 53, 137-148. 10.1111/j.1440-169X.2010.01237.x [DOI] [PubMed] [Google Scholar]
- Lai F. and King M. L. (2013). Repressive translational control in germ cells. Mol. Reprod. Dev. 80, 665-676. 10.1002/mrd.22161 [DOI] [PubMed] [Google Scholar]
- Lee T. and Luo L. (1999). Mosaic analysis with a repressible cell marker for studies of gene function in neuronal morphogenesis. Neuron 22, 451-461. 10.1016/S0896-6273(00)80701-1 [DOI] [PubMed] [Google Scholar]
- Lee A., Li W., Xu K., Bogert B. A., Su K. and Gao F.-B. (2003). Control of dendritic development by the Drosophila fragile X-related gene involves the small GTPase Rac1. Development 130, 5543-5552. 10.1242/dev.00792 [DOI] [PubMed] [Google Scholar]
- Li P., Nijhawan D., Budihardjo I., Srinivasula S. M., Ahmad M., Alnemri E. S. and Wang X. (1997). Cytochrome c and dATP-dependent formation of Apaf-1/caspase-9 complex initiates an apoptotic protease cascade. Cell 91, 479-489. 10.1016/S0092-8674(00)80434-1 [DOI] [PubMed] [Google Scholar]
- Markstein M., Pitsouli C., Villalta C., Celniker S. E. and Perrimon N. (2008). Exploiting position effects and the gypsy retrovirus insulator to engineer precisely expressed transgenes. Nat. Genet. 40, 476-483. 10.1038/ng.101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meier P., Silke J., Leevers S. J. and Evan G. I. (2000). The Drosophila caspase DRONC is regulated by DIAP1. EMBO J. 19, 598-611. 10.1093/emboj/19.4.598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menon K. P., Sanyal S., Habara Y., Sanchez R., Wharton R. P., Ramaswami M. and Zinn K. (2004). The translational repressor Pumilio regulates presynaptic morphology and controls postsynaptic accumulation of translation factor eIF-4E. Neuron 44, 663-676. 10.1016/j.neuron.2004.10.028 [DOI] [PubMed] [Google Scholar]
- Menon K. P., Andrews S., Murthy M., Gavis E. R. and Zinn K. (2009). The translational repressors Nanos and Pumilio have divergent effects on presynaptic terminal growth and postsynaptic glutamate receptor subunit composition. J. Neurosci. 29, 5558-5572. 10.1523/JNEUROSCI.0520-09.2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muro I., Hay B. A. and Clem R. J. (2002). The Drosophila DIAP1 protein is required to prevent accumulation of a continuously generated, processed form of the apical caspase DRONC. J. Biol. Chem. 277, 49644-49650. 10.1074/jbc.M203464200 [DOI] [PubMed] [Google Scholar]
- Olesnicky E. C., Bhogal B. and Gavis E. R. (2012). Combinatorial use of translational co-factors for cell type-specific regulation during neuronal morphogenesis in Drosophila. Dev. Biol. 365, 208-218. 10.1016/j.ydbio.2012.02.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oshima K., Takeda M., Kuranaga E., Ueda R., Aigaki T., Miura M. and Hayashi S. (2006). IKK epsilon regulates F actin assembly and interacts with Drosophila IAP1 in cellular morphogenesis. Curr. Biol. 16, 1531-1537. 10.1016/j.cub.2006.06.032 [DOI] [PubMed] [Google Scholar]
- Parrish J. Z., Emoto K., Kim M. D. and Jan Y. N. (2007). Mechanisms that regulate establishment, maintenance, and remodeling of dendritic fields. Annu. Rev. Neurosci. 30, 399-423. 10.1146/annurev.neuro.29.051605.112907 [DOI] [PubMed] [Google Scholar]
- Parrish J. Z., Xu P., Kim C. C., Jan L. Y. and Jan Y. N. (2009). The microRNA bantam functions in epithelial cells to regulate scaling growth of dendrite arbors in Drosophila sensory neurons. Neuron 63, 788-802. 10.1016/j.neuron.2009.08.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilling A. D., Horiuchi D., Lively C. M. and Saxton W. M. (2006). Kinesin-1 and Dynein are the primary motors for fast transport of mitochondria in Drosophila motor axons. Mol. Biol. Cell 17, 2057-2068. 10.1091/mbc.E05-06-0526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinn L. M., Dorstyn L., Mills K., Colussi P. A., Chen P., Coombe M., Abrams J., Kumar S. and Richardson H. (2000). An essential role for the caspase dronc in developmentally programmed cell death in Drosophila. J. Biol. Chem. 275, 40416-40424. 10.1074/jbc.M002935200 [DOI] [PubMed] [Google Scholar]
- Rumpf S., Lee S. B., Jan L. Y. and Jan Y. N. (2011). Neuronal remodeling and apoptosis require VCP-dependent degradation of the apoptosis inhibitor DIAP1. Development 138, 1153-1160. 10.1242/dev.062703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryoo H. D., Bergmann A., Gonen H., Ciechanover A. and Steller H. (2002). Regulation of Drosophila IAP1 degradation and apoptosis by reaper and ubcD1. Nat. Cell Biol. 4, 432-438. 10.1038/ncb795 [DOI] [PubMed] [Google Scholar]
- Sato K., Hayashi Y., Ninomiya Y., Shigenobu S., Arita K., Mukai M. and Kobayashi S. (2007). Maternal Nanos represses hid/skl-dependent apoptosis to maintain the germ line in Drosophila embryos. Proc. Natl. Acad. Sci. USA 104, 7455-7460. 10.1073/pnas.0610052104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song Z., McCall K. and Steller H. (1997). DCP-1, a Drosophila cell death protease essential for development. Science 275, 536-540. 10.1126/science.275.5299.536 [DOI] [PubMed] [Google Scholar]
- Sonoda J. and Wharton R. P. (1999). Recruitment of Nanos to hunchback mRNA by Pumilio. Genes Dev. 13, 2704-2712. 10.1101/gad.13.20.2704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takemoto K., Nagai T., Miyawaki A. and Miura M. (2003). Spatio-temporal activation of caspase revealed by indicator that is insensitive to environmental effects. J. Cell Biol. 160, 235-243. 10.1083/jcb.200207111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan Y., Yamada-Mabuchi M., Arya R., St Pierre S., Tang W., Tosa M., Brachmann C. and White K. (2011). Coordinated expression of cell death genes regulates neuroblast apoptosis. Development 138, 2197-2206. 10.1242/dev.058826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treier M., Seufert W. and Jentsch S. (1992). Drosophila UbcD1 encodes a highly conserved ubiquitin-conjugating enzyme involved in selective protein degradation. EMBO J. 11, 367-372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vardy L. and Orr-Weaver T. L. (2007). Regulating translation of maternal messages: multiple repression mechanisms. Trends Cell Biol. 17, 547-554. 10.1016/j.tcb.2007.09.002 [DOI] [PubMed] [Google Scholar]
- Vucic D., Kaiser W. J. and Miller L. K. (1998). Inhibitor of apoptosis proteins physically interact with and block apoptosis induced by Drosophila proteins HID and GRIM. Mol. Cell. Biol. 18, 3300-3309. 10.1128/MCB.18.6.3300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W. and Yoder J. H. (2011). Drosophila pupal abdomen immunohistochemistry. J. Vis. Exp. 56, e3139 10.3791/3139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S. L., Hawkins C. J., Yoo S. J., Müller H.-A. and Hay B. A. (1999). The Drosophila caspase inhibitor DIAP1 is essential for cell survival and is negatively regulated by HID. Cell 98, 453-463. 10.1016/S0092-8674(00)81974-1 [DOI] [PubMed] [Google Scholar]
- Wharton R. P. and Struhl G. (1991). RNA regulatory elements mediate control of Drosophila body pattern by the posterior morphogen nanos. Cell 67, 955-967. 10.1016/0092-8674(91)90368-9 [DOI] [PubMed] [Google Scholar]
- Williams D. W. and Truman J. W. (2005). Cellular mechanisms of dendrite pruning in Drosophila: insights from in vivo time-lapse of remodeling dendritic arborizing sensory neurons. Development 132, 3631-3642. 10.1242/dev.01928 [DOI] [PubMed] [Google Scholar]
- Williams D. W., Kondo S., Krzyzanowska A., Hiromi Y. and Truman J. W. (2006). Local caspase activity directs engulfment of dendrites during pruning. Nat. Neurosci. 9, 1234-1236. 10.1038/nn1774 [DOI] [PubMed] [Google Scholar]
- Wilson R., Goyal L., Ditzel M., Zachariou A., Baker D. A., Agapite J., Steller H. and Meier P. (2002). The DIAP1 RING finger mediates ubiquitination of Dronc and is indispensable for regulating apoptosis. Nat. Cell Biol. 4, 445-450. 10.1038/ncb799 [DOI] [PubMed] [Google Scholar]
- Ye B., Petritsch C., Clark I. E., Gavis E. R., Jan L. Y. and Jan Y. N. (2004). Nanos and Pumilio are essential for dendrite morphogenesis in Drosophila peripheral neurons. Curr. Biol. 14, 314-321. 10.1016/j.cub.2004.01.052 [DOI] [PubMed] [Google Scholar]