ABSTRACT
Auxin response factors (ARFs) can function as transcriptional activators or repressors to regulate the expression of auxin response genes by specifically binding to auxin response elements (AuxREs) during plant development. Based on a genome-wide strategy using the medicinal model plant Salvia miltiorrhiza, 25 S. miltiorrhiza ARF (SmARF) gene family members in four classes (class Ia, IIa, IIb and III) were comprehensively analyzed to identify characteristics including gene structures, conserved domains, phylogenetic relationships and expression patterns. In a hybrid analysis of the phylogenetic tree, microRNA targets, and expression patterns of SmARFs in different organs, root tissues, and methyl jasmonate or indole-3-acetic acid treatment conditions, we screened for candidate SmARFs involved in various developmental processes of S. miltiorrhiza. Based on this analysis, we predicted that SmARF25, SmARF7, SmARF16 and SmARF20 are involved in flower, leaf, stem and root development, respectively. With the further insight into the targets of miR160 and miR167, specific SmARF genes in S. miltiorrhiza might encode products that participate in biological processes as described for ARF genes in Arabidopsis. Our results provide a foundation for understanding the molecular basis and regulatory mechanisms of SmARFs in S. miltiorrhiza.
KEY WORDS: Developmental processes, Auxin response factors, Auxin response elements, MicroRNA, Salvia miltiorrhiza
Summary: Genome-wide analysis identified 25 ARF gene members (seven transcriptional activators and 18 repressors) in S. miltiorrhiza. The gene structures, functional domains, miRNA targets and expression patterns were analyzed in detail.
INTRODUCTION
The phytohormone auxin, typified by indole-3-acetic acid (IAA), plays a crucial role in controlling the mechanisms by which plants grow and develop, including tropic responses, apical dominance, lateral root formation, vascular differentiation, flower and fruit development, and shoot elongation (Santner and Estelle, 2009). Auxin response factors (ARFs) are important transcription factors that can either activate or repress the transcriptional level of early/primary auxin response genes, such as Aux/IAA, Small Auxin Up RNA (SAUR) and Gretchen Hagen 3 (GH3) gene family members, by binding to auxin response elements (AuxREs, TGTCTC) or some variation of these elements (TGTCCC or TGTCAC) in their promoters (Hagen and Guilfoyle, 2002; Liu et al., 1994; Ulmasov et al., 1997, 1995, 1999b). AtARF1, which binds to the sequence TGTCTC in AuxREs, was the first cloned auxin-related transcription factor and was identified in Arabidopsis using a yeast one-hybrid system (Ulmasov et al., 1997). Recently, microarray experiments indicated that AtARF1 and AtARF5 monomers specificity prefer TGTCGG elements to the AuxRE TGTCTC (Boer et al., 2014). The complete genomic sequence of Arabidopsis provides the opportunity to identify the sequence and evolution of all members of a given gene family (Arabidopsis Genome Initiative, 2000). Genome-wide analysis identified 22 full-length ARF genes and one partial-length gene (AtARF23) containing a stop codon in its DNA-binding domain (DBD) in Arabidopsis thaliana (Okushima et al., 2005b; Remington et al., 2004). Furthermore, biochemical and genetic approaches have established crucial functions of ARF genes in the growth and development of Arabidopsis (Guilfoyle and Hagen, 2007).
Taking advantage of the genome-wide identification of A. thaliana ARFs (AtARFs), many studies have found that the ARFs AtARF1 and AtARF2 function as transcriptional repressors related to the regulation of leaf senescence, floral organ abscission and cell growth (Ellis et al., 2005; Li et al., 2004; Okushima et al., 2005a; Schruff et al., 2006); AtARF3 and AtARF4 function in developing reproductive and vegetative tissues (Pekker et al., 2005; Sessions et al., 1997; Finet et al., 2010); AtARF5 functions in Arabidopsis leaf vascular and embryo patterning (Hamann et al., 2002; Krogan et al., 2012); AtARF6 and AtARF8 function in female and male reproduction (Nagpal et al., 2005; Wu et al., 2006); AtARF7 and AtARF19 act in seedlings, roots and developing embryos (Korasick et al., 2014; Okushima et al., 2005b; Wilmoth et al., 2005); AtARF9 acts in suspensor cells to mediate hypophysis specification (Rademacher et al., 2012); and AtARF10, AtARF16 and AtARF17 function in the negative regulation of seed germination and post-germination activities (Liu et al., 2007, 2010). In Arabidopsis, certain AtARF expression patterns are controlled by miRNAs to regulate several developmental events. AtARF6 and AtARF8 are targets of miR167 (Wu et al., 2006), and AtARF10, AtARF16 and AtARF17 are targets of miR160 (Liu et al., 2007, 2010). In some cases, ARF gene expression is altered in response to exogenous auxin signals (Okushima et al., 2005b; Wang et al., 2007a).
A typical ARF contains three conserved domains: an N-terminal B3 DNA binding domain (DBD), a middle regional auxin response factor (MR), and a C-terminal PB1 protein-protein interaction domain (PB1). The DBD can recognize AuxREs or variation elements in the promoter of auxin-responsive genes (Wright and Nemhauser, 2015; Boer et al., 2014), and the PB1 domains are also found in Aux/IAAs (Guilfoyle and Hagen, 2012). Structural and biochemical studies have determined that the PB1 domains of ARFs and Aux/IAAs from AtARF7 (Korasick et al., 2014) and AtARF5 (Nanao et al., 2014) are involved in protein-protein interactions by formatting higher order oligomerization or multimerization (Wright and Nemhauser, 2015). The MR, located between the DBD and the PB1 domain, confers functions such as transcriptional activation or repression depending on its amino acid composition (Mun et al., 2012; Yu et al., 2014). Previous studies have shown that glutamine (Q)-rich MRs function as activation regions but that serine (S)-rich, serine and proline (SP)-rich, and serine and glycine (SG)-rich MRs function as repression regions in ARFs from A. thaliana (Tiwari et al., 2003; Ulmasov et al., 1999a).
Given the complete genomic sequences of many important species, there has been significant progress in the analysis and identification of the functions of ARFs. Genome-wide analysis has identified many ARFs in many other important plants, such as 25 Oryza sativa ARF (OsARF) loci (Wang et al., 2007a), 22 Solanum lycopersicum ARFs (SlARFs) (Zouine et al., 2014), 31 Brassica rapa (BrARFs) (Mun et al., 2012), 19 Vitis vinifera ARFs (VvARFs) (Wan et al., 2014), 47 Musa acuminata ARFs (MaARFs) (Hu et al., 2015), 17 Eucalyptus grandis (EgrARFs) (Yu et al., 2014), 24 Medicago truncatula ARFs (MtARFs) (Shen et al., 2015), 39 Populus trichocarpa ARFs (PtARFs) (Kalluri et al., 2007), 19 Citrus sinensis ARFs (CiARFs) (Li et al., 2015b), 11 Carica papaya ARFs (CpARFs) (Liu et al., 2015), and 35 Gossypium raimondii ARFs (GrARFs) (Sun et al., 2015). However, the ARF transcription factor family members have not been determined in Salvia miltiorrhiza, one of the most commonly used herbs in traditional Chinese medicine (TCM). S. miltiorrhiza, also referred to as danshen, belongs to the Salvia genus of the Lamiaceae family, and its dried root and rhizome are highly valued (Cheng, 2006). Danshen is well known for its use alone or in combination with other herbs in the treatment of cardiovascular diseases, as well as for its anti-inflammatory, immunomodulatory and anti-oxidative activities; the primary bioactive compounds in danshen are lipophilic diterpenoids and hydrophilic phenolic acids (Wang et al., 2007b; Dong et al., 2011). S. miltiorrhiza is also considered a good medicinal model plant in TCM research for studying the biosynthesis and regulation of active compounds (Ma et al., 2012; Xu et al., 2015). Due to the establishment of the S. miltiorrhiza genome sequence (Xu et al., 2016a) it has become feasible use in silico analysis to isolate its functional gene families such as diterpene; phenolic acid biosynthetic genes; and bHLH, AP2/ERF, WRKY, MYB and SPL transcription factors (Ji et al., 2015; Li et al., 2015a; Li and Lu, 2014; Ma et al., 2012; Wang et al., 2015; Zhang et al., 2014, 2015; Xu et al., 2016b). As ARF gene members are key factors in plant growth and development, identifying these genes in S. miltiorrhiza aid in the understanding of developmental processes and cellular responses to auxin in danshen.
Here, we isolated 25 S. miltiorrhiza ARF (SmARF) genes using a genome-wide approach. Following complete genome sequencing the sequence homology of these SmARFs and their gene expression patterns in different organs, root tissues, and methyl jasmonate (MeJA) or IAA treatment conditions, gene structures, and the phylogenetic relationships between SmARFs and AtARFs were analyzed in detail. This study provided molecular information regarding the SmARF gene family and the results will aid in selecting candidate genes related to cell growth and tissue development in S. miltiorrhiza, paving the way for further functional characterization of these SmARF genes.
RESULTS
Identification and phylogenetic analysis of danshen ARFs
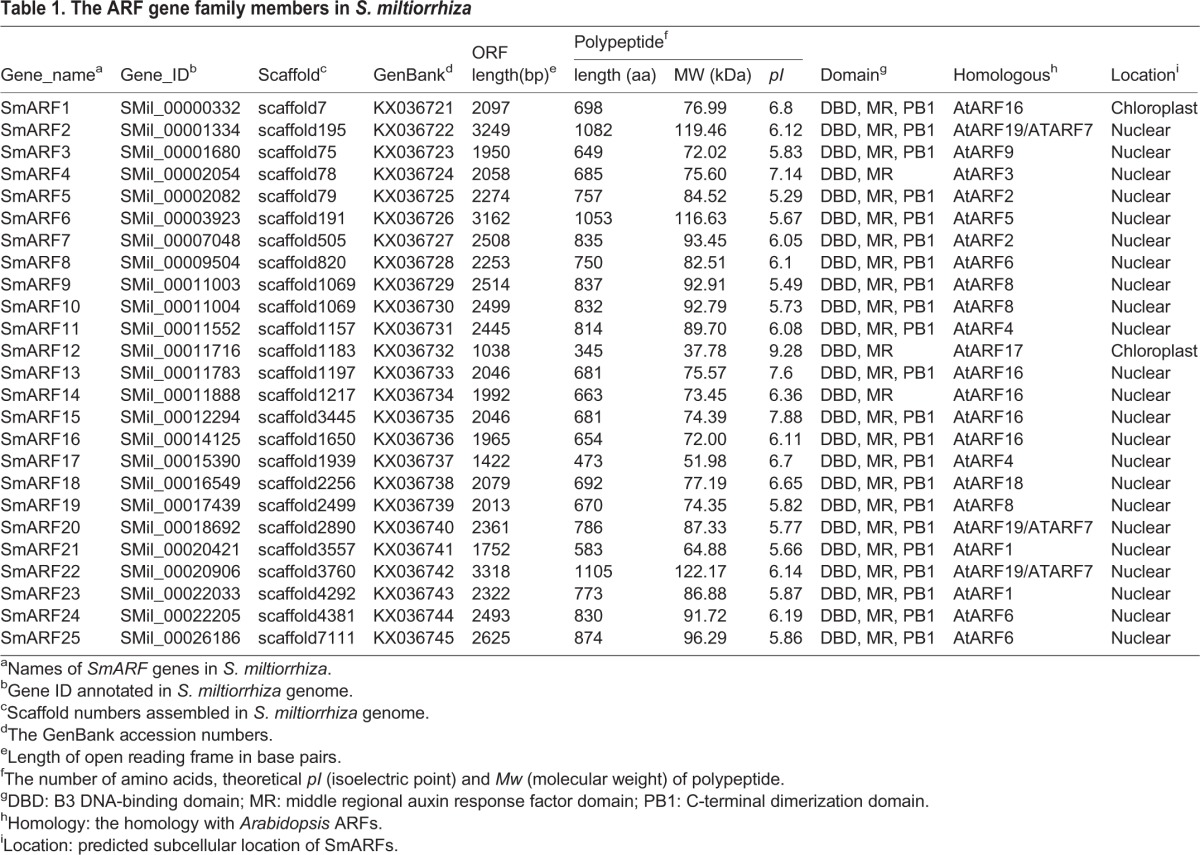
After a BLASTP search and protein domain analysis, 25 non-redundant ARF genes were identified from the genome sequences of S. miltiorrhiza. These SmARFs, located in the different scaffolds, were named SmARF1-SmARF25 according to the order of their annotated gene IDs, listed in Table 1. The number of ARF genes in S. miltiorrhiza is similar to the number in A. thaliana (23), O. sativa (25) and M. acuminata (24). The predicted proteins encoded by SmARF genes varied from 345 amino acids (SmARF12) to 1105 amino acids (SmARF22), with corresponding molecular weights from 37.78 kDa to 122.17 kDa, and the theoretical isoelectric points ranged from 5.29 (SmARF5) to 9.28 (SmARF12). Pair-wise analysis of SmARF protein homology indicated that the overall homology broadly ranged from 22% (between SmARF6 and SmARF16) to 89% (between SmARF5 and SmARF7). The SmARF9 and SmARF10 genes are located in the same scaffold1069, and the other SmARFs are distributed in different scaffolds. Most of the SmARFs were predicted to localize to the nucleus, however SmARF1 and SmARF12 were predicted to localize to chloroplasts.
Table 1.
The ARF gene family members in S. miltiorrhiza
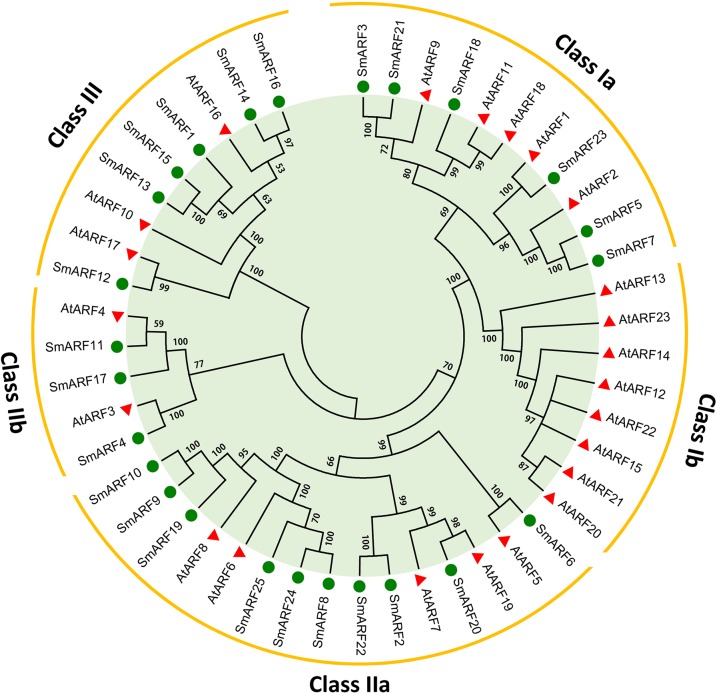
To characterize the evolutionary relationship between danshen ARF proteins and Arabidopsis ARFs, a neighbor-joining tree was constructed using the full-length amino acid sequences (Fig. 1). The results indicated that 25 SmARFs were classed together with 23 AtARFs into four clusters (classes Ia, IIa, IIb and III) according to well-supported bootstrap data. In S. miltiorrhiza, SmARF3, 5, 7, 18, 21 and 23 belong to class Ia; SmARF2, 6, 8, 9, 10, 19, 20, 22, 24 and 25 belong to the largest class IIa; SmARF4, 11 and 17 belong to class IIb; and SmARF1, 12, 13, 14, 15 and 16 belong to class III. In A. thaliana, there is another class Ib that includes AtARF12-15 and 20-23. Notably, no S. miltiorrhiza ARF proteins were clustered into class Ib from the phylogenetic tree, and this observation implies a diverging trend in the evolution of ARF genes across different plants.
Fig. 1.
Analysis of the phylogenetic relationships of ARF gene members in S. miltiorrhiza and Arabidopsis. A total of 25 SmARF proteins from S. miltiorrhiza and 23 ARF proteins from Arabidopsis were used to construct a neighbor-joining tree. Bootstrap values are presented for all branches. The 25 SmARFs and 23 AtARFs were clustered into five classes (Ia, Ib, IIa, IIb and III). Green circles denote the ARF proteins from S. miltiorrhiza, and red triangles denote the ARF proteins from Arabidopsis.
To investigate the biological processes of SmARFs, gene ontology (GO) mapping and annotation were performed using Blast2GO. The functional categorization of SmARFs as annotated by GO analysis, including their biological processes, molecular functions, and cellular components, is presented in Table S1. Regarding biological processes, eight categories met the criterion of NodeScore >2.0: cellular process (25 genes), metabolic process (25 genes), response to stimulus (25 genes), single-organism process (14 genes), biological regulation (13 genes), signaling (11 genes), developmental process (five genes), and multicellular organismal process (five genes). For developmental process, SmARF6 was related to embryo development; five SmARFs were classified into post-embryonic developmental process (SmARF1, 4, 6, 11, 25); and SmARF1, 4, 6 and 25 were predicted to participate in flower development. Notably, the biological process related to secondary metabolism was not identified. Based on the molecular function analysis, all the SmARFs were classified into DNA binding; 21 SmARFs were grouped into protein binding; and three SmARFs were categorized into sequence-specific DNA binding transcription factor activity (SmARF1, 11 and 25). According to the cellular component analysis, all SmARFs except for SmARF1 and SmARF12 were localized to the nucleus, in accordance with the subcellular localization predictions.
Gene structures and conserved domains of danshen ARFs
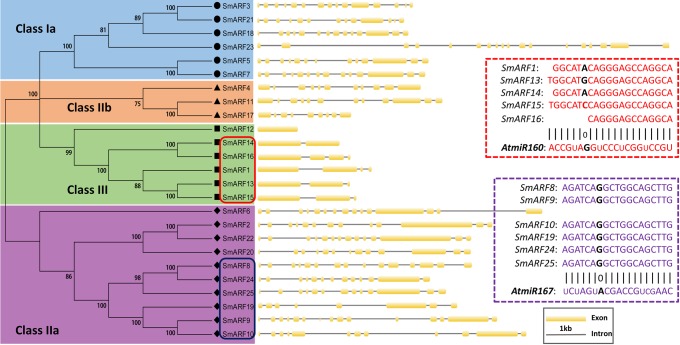
To better understand the gene structure of SmARFs, the exon-intron features among SmARFs were aligned via phylogenetic analysis (Fig. 2). The phylogenetic analysis revealed four clusters in accordance with the group data presented in Fig. 1. Gene structure analysis of all of the SmARF genes revealed that the number of exons ranges from 1 to 18, however, SmARF12 is intronless. The genes in the four groups have an average exon number ranging from three (class III) to 15 (class Ia). The results showed that the exon number of class I-II was significantly greater than that of class III; these findings were identical to the structure of AtARF genes.
Fig. 2.
Gene structure analysis of SmARF genes and prediction of their miRNA target sites according to their phylogenetic relationships. The yellow boxes represent exons; the gray lines represent introns. The red box denotes the targets of At-miR160 in SmARF genes; the purple box denotes the targets of At-miR167 in SmARF genes.
Examination of the protein homology of SmARFs to Arabidopsis ARFs showed that 10 AtARFs have no corresponding S. miltiorrhiza orthologs (AtARF10-15 and AtARF20-23). Sequence analysis and Pfam protein domain analysis showed that 92% of the identified SmARFs (23 of the 25 predicted proteins) possess the typical ARF structure, containing a highly conserved DBD, MR and PB1. In contrast to the typical ARFs, SmARF4, 12 and 14 do not contain a PB1 domain. ARFs function as transcriptional activators or repressors depending on the amino acid composition of the MR. The Q-rich MRs of seven SmARFs (SmARF2, 10, 19, 20, 22, 24 and 25) (Table S2, Fig. S1) indicate that they likely act as transcriptional activators. The SmARFs containing Q-rich MR domains belong to class IIa according to phylogenetic analysis. The other 18 SmARFs may function as transcriptional repressors based on their S-rich, SP-rich, or SG-rich MRs.
A total of 15 conserved motifs in SmARFs were characterized (motifs 1-15) using MEME software to explore their structure and functional diversity (Figs S2 and S3). SmARF2, 3, 7, 8, 9, 18, 22 and 24 in classes Ia and IIa contain the greatest number of distinct motifs (12), and SmARF12 in class III contains the fewest distinct motifs (six types). Additionally, the average motif number per SmARF varies across classes, ranging from nine (class IIb and III) to 11 (class Ia and IIa). This evidence indicated that the ARF proteins are highly conserved. Motifs 1, 3 and 12 were annotated as B3-DBDs; motifs 4, 6, 10 and 11 were annotated as auxin response superfamily MRs; and motifs 7 and 8 were annotated as the OPCA-like motif and conserved lysine motif of PB1 domain, which function in Aux/IAA-ARF multimerization. In accordance with the results of conserved domain analysis, all SmARF protein structures harbor DBD motifs (1, 3 and 12) and MR motifs (4, 6, 10 and 11); however SmARF4 and SmARF12 do not contain a PB1 (neither motif 7 nor motif 8).
Prediction of miRNA targets among SmARFs and analysis of the AuxREs in SmARF gene promoters
Using the BLASTN algorithm to identify targets of miRNA160 and miRNA167 within SmARF gene sequences, target sites of At-miRNA160 (UGCCUGGCUCCCUGGAUGCCA) were predicted within the 1300-1319 bp region of SmARF1, the 1359-1379 bp region of SmARF13, the 1348-1367 bp region of SmARF14, the 1332-1352 bp region of SmARF15 and the 1363-1376 bp region of SmARF16. Additionally, target sites of miRNA167 (UCAAGCUGCCUGCAUGAUCUA) were predicted within the 1975-1993 bp region of SmARF8, the 2302-2320 bp region of SmARF9, the 2287-2305 bp region of SmARF10, the 1777-1795 bp region of SmARF19, the 2419-2437 bp region of SmARF24 and the 2350-2368 bp region of SmARF25 (Fig. 2). The results of this analysis suggested that miR160-/167-mediated post-transcriptional regulation of ARFs is conserved between S. miltiorrhiza and Arabidopsis.
We surveyed 20 AUX/IAA and 10 GH3 primary/early auxin response gene members in S. miltiorrhiza based on a genome-wide strategy. The promoters (−1000 to −1 bp) of these two auxin response gene families were selected to screen for AuxREs. As expected, 19 of 20 AUX/IAA and 9 of 10 GH3 gene promoters contain one or more AuxREs (Table S3). These results indicated that these auxin response genes could be regulated by SmARFs in S. miltiorrhiza.
Expression patterns of SmARF genes in different plant organs or tissues
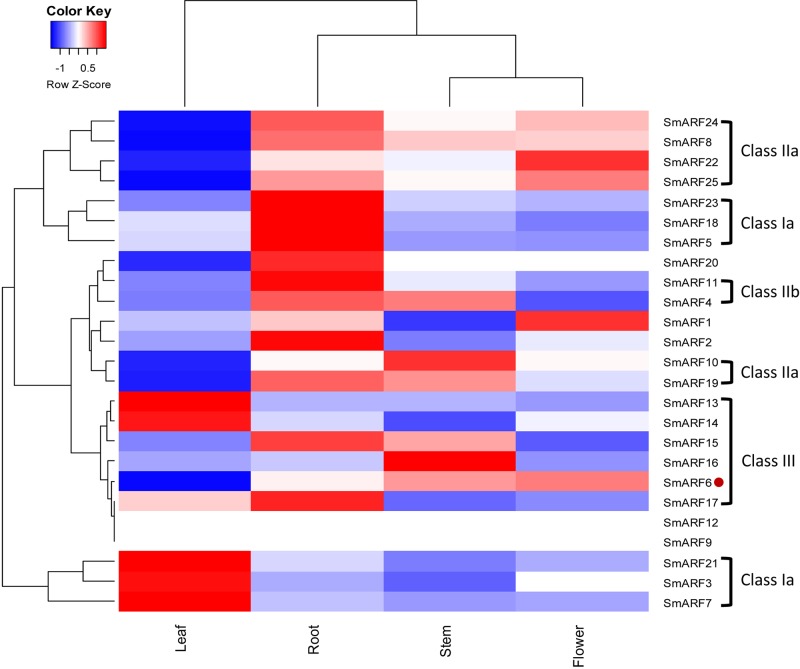
To better probe the physiological function of SmARFs, the tissue-specific expression of 25 SmARF genes in different danshen organs (leaf, root, stem and flower) was determined by analyzing the RNA-seq data (Fig. 3; Table S4). Most SmARF genes, but not SmARF9, 12 or 17 presented ubiquitous expression and high variability in all studied organs, and this result implies that these SmARFs might function in danshen growth and development. There were significant differences in SmARF expression between organs. SmARF3, 7 and 21, all of which belong to class Ia based on phylogenetic analysis, showed higher expression in danshen leaf than in other organs. SmARF4, 5, 11, 18, 20 and 23 were expressed more strongly in danshen root than in other organs, however, only SmARF16 showed stem-specific expression in S. miltiorrhiza. When comparing phylogenetic tree analysis with the expression cluster analysis, SmARF8, 10, 19, 22, 24 and 25, which belong to class IIa, showed significantly lower expression in danshen leaf than in other organs. Most of the SmARF genes from class III (SmARF13-17) also clustered in one expression branch. These results indicated that ARF genes from the same class might perform a similar physiological function in plants.
Fig. 3.
A heat map showing SmARF gene expression patterns in different organs. The red color represents upregulation of expression, the white color represents an unchanged expression level, and the blue color represents downregulation of expression. Red dot (SmARF6) is not belong to the Class III.
Previous evidence revealed that the periderm of danshen root is the primary site of biosynthesis and accumulation of tanshinones. The expression pattern of SmARF genes in different root tissues (periderm, phloem and xylem) was also examined using RNA-seq data (Fig. S4, Table S4). SmARF9, 12 and 17 displayed no expression in danshen root tissues. SmARF13 showed the greatest expression in periderm, more than four and 14 times greater than that in phloem and xylem, respectively. Furthermore, SmARF20 exhibited stronger expression in phloem and xylem than in periderm.
Expression patterns of SmARF genes upon auxin or MeJA treatment
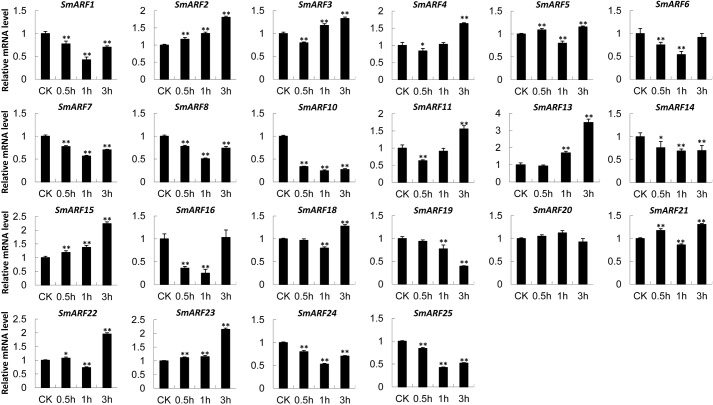
Auxin is a central regulator of plant growth and development. To investigate the response of SmARF genes to exogenous IAA stimulation, we analyzed the variation in SmARF gene expression at 0, 0.5, 1 and 3 h after 20 μM IAA treatment using qRT-PCR (Fig. 4). As expected, most SmARF genes were significantly auxin-sensitive. The overall expression patterns of SmARFs varied, with 11 SmARF mRNAs (SmARF2, 3, 4, 5, 11, 13, 15, 18, 21, 22 and 23) showing up-regulation and eight SmARF mRNAs (SmARF1, 7, 8, 10, 14, 19, 24 and 25) showing down-regulation at 3 h of IAA treatment (P<0.01 for all). One SmARF gene (SmARF20) did not display significant changes in expression (P>0.05) regardless of the treatment duration. The unmentioned SmARF6 and SmARF16 displayed significantly down-regulated expression at 0.5 and 1 h, and at 3 h, the expression of these genes returned to the same level as that for mock IAA treatment. The most strongly up-regulated SmARF genes, SmARF13, 15 and 23, were markedly induced after IAA treatment [greater than twofold increase, log (expression level) >1]. Similarly, the expression of five SmARF genes (SmARF1, 10, 16, 19, 25) showed marked down-regulation [greater than twofold decrease, log (expression level) >1]. For IAA treatment, 13 SmARF genes (SmARF1, 3, 5, 6, 7, 8, 11, 16, 18, 21, 22, 24 and 25) displayed significant up- or downregulation over the three examined time points. For example, the expression level of SmARF1 was decreased by greater than twofold at 1 h but was significantly increased at 3 h compared with the control levels.
Fig. 4.
The expression of SmARF genes in response to treatment with 20 μM IAA solution for 0.5, 1, or 3 h. CK, the untreated leaves of S. miltiorrhiza. Error bars represented variability of qRT-PCR results from three replicates. No expression of SmARF9, 12 and 17 was detected. *P<0.05; **P<0.01.
In Nicotiana benthamiana, transient silence of NbARF1 and MeJA treatment resulted in significant enrichment of leaf nicotine (Todd et al., 2010). In addition, MeJA treatment significantly alters the biosynthesis of active compounds (tanshinones or phenolic acids) in S. miltiorrhiza, hence the expression variation of SmARF genes after MeJA treatment was studied using RNA-seq data (Table S4). The results showed that SmARF24 and 25 displayed significant up-regulation and that SmARF1 exhibited downregulation after MeJA treatment. The expression of other SmARF genes showed no evident changes following MeJA treatment. This finding suggests that SmARFs might perform a small role in post-developmental processes.
DISCUSSION
The cultivation of medicinal plants has faced intense pressure due to social and environmental concerns. Studying the molecular mechanisms of medicinal plant growth processes would help resolve potential questions related to cultivation of these plants. Genome-wide characterization and analysis of SmARFs could improve the understanding of their regulatory roles in danshen growth and development. In this study, 25 ARF gene members in S. miltiorrhiza were identified, and this number was similar to that for other model plants, such as A. thaliana (23) and O. sativa (25). Protein domain analysis provided useful information for predicting the biological functions of SmARFs, which primarily depend on their characteristic DBD, MR and PB1. ARFs rely on the DBD to specifically bind to AuxREs in the promoters of auxin-responsive genes. Their PB1 is involved in homomeric and heteromeric interactions with ARFs and Aux/IAA proteins. The percentage of PB1-truncated SmARFs (8%) was much lower than that of the ARF members identified in other plants, such as Arabidopsis (17%), rice (24%) and M. truncatula (54%). ARFs can function as transcriptional activators or repressors according to the amino acid composition of the MR. The activator/repressor ratio of SmARFs was 0.39 (7/18) and this value was also much lower than the ratios for Arabidopsis (0.59) and rice (0.56). Phylogenetic analysis and divergence time estimation based on 1824 single-copy true orthologous genes indicated that S. miltiorrhiza was distantly related to Arabidopsis, with an estimated divergence time of approximately 139 million years ago (Xu et al., 2016a).We also constructed a phylogenetic tree to analyze the relationship of ARF family members between S. miltiorrhiza and Arabidopsis (Fig. 1). Phylogenetic tree analysis revealed five sister gene pairs with high bootstrap values (≥98%) between S. miltiorrhiza and Arabidopsis; this evidence supports the high homology of ARFs between species. Although there are similar numbers of ARFs between the S. miltiorrhiza and A. thaliana genomes, the absence of class Ib ARFs from S. miltiorrhiza reflects genomic expansion and rearrangements resulting from extensive duplication and deletion over a long period of evolutionary history according to the phylogenetic tree analysis. The detection of close relationships based on comparative analysis may help in the selection of candidate ARFs with specific biological functions in S. miltiorrhiza. According to the motif analysis, the motifs from different classes in SmARFs present high conservation (Fig. S2). Motif 8 and motif 7, located in the PB1 domain of the C-terminal in most SmARFs, include conserved residues (lysine motif and OPCA-like motif) of the positive and negative face found only in the ARF family, thus indicating the evolutionary conservation of ARF function (Korasick et al., 2014; Nanao et al., 2014).
Much evidence demonstrates that miRNAs play dominant roles in post-transcriptional gene regulation by binding to their complementary mRNA targets, especially to target transcription factors, in plants (Jones-Rhoades et al., 2006; Li and Zhang, 2016). In Arabidopsis, miR167 controls the expression patterns of AtARF6 and AtARF8 to regulate female and male reproduction or to promote jasmonic acid production and flower maturation (Nagpal et al., 2005; Wu et al., 2006). Phylogenetic tree analysis showed that SmARF8, 9, 10, 19, 24 and 25 were closely related to AtARF6 and AtARF8, both of which are in class IIa; all of these six SmARFs contain a target site of miR167 (Fig. 2). GO analysis also categorized SmARF25 into flower development; thus in S. miltiorrhiza we predicted that the expression of SmARF8, 10, 19, 24 and 25 might be inhibited by miR167 to regulate certain developmental processes as described for AtARF6 and AtARF8. Among them, SmARF25 was identified as the best candidate regulator of flower development. In addition, SmARF10, 19, 24 and 25 might function as transcriptional activators due to their Q-rich MRs. Additionally, miR160 was found to bind to AtARFs (AtARF10, AtARF16 and AtARF17) to negatively regulate seed germination and post-germination activities (Liu et al., 2007, 2010). These AtARFs were closely related to SmARF1, 12, 13, 14, 15 and 16 in class III, and these ARFs might function as transcriptional repressors due to the amino acid compositions of their MRs. Aside from SmARF12, other class III SmARF genes were identified to contain miR160 target sites; this finding implies that these SmARFs perform functions that are similar to the functions of AtARF10, 16 and 17.
Comprehensive analysis of SmARF gene expression patterns and the evolution of their sequences helped us screen for candidate SmARF genes with potentially distinct functions. Most SmARF genes displayed ubiquitous but highly variable expression in all studied organs, and this expression pattern suggests their functional divergence. In Arabidopsis, AtARF2 regulates leaf senescence and floral organ abscission independently of the ethylene and cytokinin response pathways (Ellis et al., 2005). In S. miltiorrhiza the expression levels of SmARF3, 7 and 21 were significantly higher in leaves than in other studied organs, and SmARF5 and SmARF7 were closely related to AtARF2 in class Ia; these findings indicate that SmARF7 might play a crucial role in leaf development. In Arabidopsis AtARF7 and AtARF19 promote leaf expansion and auxin-induced lateral root formation (Wilmoth et al., 2005). In S. miltiorrhiza, SmARF20 was grouped with AtARF7 and 19 in class IIa, and the expression of SmARF20 was much higher in danshen root than in other organs. These observations suggest that SmARF20 likely regulates auxin-induced lateral root formation. The differential expression of SmARF20 between periderm, phloem, and xylem further support its role in root development. Notably, SmARF8, 10, 19, 22, 24 and 25 also belong to class IIa with AtARF7 and 19. The expression patterns of these SmARFs were much lower in danshen leaf, reflecting that they might be involved in leaf expansion. SmARF16, a stem-specifically expressed transcription factor, likely participates in stem development.
Recently, synthetic biology, particularly the biosynthesis of natural products, has advanced by leaps and bounds. The tanshinone and phenolic acid biosynthetic pathways, which have gradually been elucidated, have attracted increasing attention (Cui et al., 2015; Guo et al., 2013; Ma et al., 2012; Xu et al., 2015); however the molecular mechanism of danshen development has been an unpopular subject despite the importance of this medicinal plant. In this study the basic functional characteristics of SmARFs, such as the presence of the conserved DBD, MR and PB1 in 88% (22/25) of the SmARFs; the significant variation in the expression of 95% (21/22) of the examined SmARFs after 0.5 h, 1 h and 3 h of IAA treatment; and the presence one or more AuxREs in 85% (17/20) of the AUX/IAA gene promoters and 90% (9/10) of the GH3 gene promoters in SmARFs indicated their regulatory roles in danshen growth and development. Further biochemical and genetic studies of candidate ARFs in S. miltiorrhiza will lead to the production of a working model for the cultivation and selective breeding of fine varieties of medicinal plants.
In summary, 25 ARF gene members (seven transcriptional activators and 18 repressors) in S. miltiorrhiza were identified, and a comprehensive account of this gene family has been performed. SmARFs were grouped into four classes with AtARFs in Arabidopsis, and the gene structures, functional domains, and miRNA targets of SmARFs were analyzed in detail. Expression patterns were used to predict candidate SmARFs involved in the regulation of various developmental processes. The results of this study will provide a basic foundation for the verification of the functions and evolution of SmARF gene family members in this model medicinal plant.
MATERIALS AND METHODS
Genome-wide survey of ARF genes in S. miltiorrhiza
The Arabidopsis ARF protein sequences (AtARF1 to AtARF23) were downloaded from the NCBI database (http://www.ncbi.nlm.nih.gov/protein/). BLASTP searches were used to identify the corresponding ARF gene members in S. miltiorrhiza using a cut-off e-value of 1.0E−10. The hidden Markov model (HMM) profiles of ARF gene family members including B3-DBD (Pfam02362), AUX_RESP (MR, Pfam06507), and AUX/IAA family (PB1, Pfam02309) members were applied to identify ARF genes based on the S. miltiorrhiza genome. The domains of all obtained ARFs were analyzed using BLAST from the Conserved Domain Database (http://www.ncbi.nlm.nih.gov/cdd). The auxin response genes, AUX/IAA gene family (Pfam02309) members and GH3 gene family (Pfam03321) members were also selected using the same approach. The Compute pI/Mw tool on the ExPASy server (http://web.expasy.org/compute_pi/) was employed to predict the theoretical isoelectric point (pI) and the molecular weight (Mw) of each SmARF protein.
Gene structure, conserved motif and subcellular localization analyses
The Gene Structure Display Server (GSDS 2.0; http://gsds.cbi.pku.edu.cn/index.php) was used to analyze the gene structure of SmARFs with the input of coding sequences (CDSs) and corresponding genomic sequences. Conserved motifs in SmARF transcription factors were identified using MEME (Suite version 4.9.1; http://meme-suite.org/tools/meme) according to the following criteria: maximum number of 15 motifs and an optimum width of 8-50 amino acids. subCELlular LOcalization predictor (CELLO v.2.5; http://cello.life.nctu.edu.tw/) was used to predict the subcellular localization of SmARF proteins.
Phylogenetic tree construction and miRNA target site analysis
All SmARF and AtARF protein sequences were pooled into MEGA6 (http://www.megasoftware.net/) to perform multiple sequence alignments. Then neighbor-joining trees were constructed using the bootstrap method with 1000 replications and pairwise deletion of gaps/missing data. The miRNA target sites of AtmiR160 and AtmiR167 in the SmARFs were searched using the PMRD database (http://bioinformatics.cau.edu.cn/PMRD/).
Plant resources
S. miltiorrhiza (line 99-3) was cultivated at the Institute of Medicinal Plant Development (IMPLAD), Chinese Academy of Medical Sciences (CAMS), in an open experimental field. Three-year-old roots, stems, and flowers were collected. The roots were peeled into three parts (periderm, phloem and xylem) (Xu et al., 2015). Leaves with or without MeJA treatment (12 h, 200 μM; Sigma-Aldrich, MO, USA) were collected from tissue culture plantlets of S. miltiorrhiza at 25°C under a long day of 16-h light/8-h dark (Zhang et al., 2015). For auxin treatment, seedlings from tissue culture plantlets were incubated for 0.5 h, 1 h, or 3 h in 20 μM IAA solution. All of the collected tissues originated from an asexual line of S. miltiorrhiza 99-3.
Sequencing data and bioinformatic analysis
The draft genome of S. miltiorrhiza was assembled and annotated in our lab [Xu et al., 2016a; Sequence Read Archive (SRA) accession number SRP051524, http://www.ncbi.nlm.nih.gov/sra]. The RNA-seq reads from different organs (root, stem and flower) were generated using Illumina HiSeq 2000 platforms (Illumina, USA; SRA accession number SRP028388). The RNA-seq reads from different root tissues (periderm, phloem and xylem) using Illumina HiSeq 2500 platforms (Illumina, USA) have been reported in our recent study (Xu et al., 2015; SRA accession number SRR1640458). The Illumina reads from leaves with or without 12 h MeJA treatment were obtained from a previous study (Luo et al., 2014; SRA accession number SRP051564). Differential SmARF gene expression in various root tissues, organs and treatment conditions was analyzed using Tophat 2.0.12 and Cufflinks 2.2.1 (Trapnell et al., 2012) by mapping Illumina-derived short reads to the S. miltiorrhiza genomic sequence. A heat map was constructed using R statistical software (Le Meur and Gentleman, 2012). GO mapping and annotation were performed using Blast2Go with a cut-off e-value of 1.0E−10.
Gene expression analysis by qRT-PCR
Four RNA samples of seedlings from tissue culture plantlets that were treated with IAA (mock, 0.5 h, 1 h, or 3 h) were isolated. Total RNA was isolated from three biological replicates for each sample using the RNeasy Plus Mini kit (Qiagen, Germany). Reverse transcription was performed using PrimeScript™ Reverse Transcriptase (TaKaRa, Japan). The qRT-PCR primers were designed using Primer Premier 6 (Table S5), and their specificity was verified by PCR. qRT-PCR analysis was conducted in triplicate using SYBR® Premix Ex Taq™ II (TaKaRa, Japan), with SmActin as a reference gene, with a LightCycler 480 real-time PCR system (Roche, Switzerland). Ct values were calculated to analyze the relative expression levels using the 2−ΔΔCt method (Livak and Schmittgen, 2001). To detect differences in the expression of candidate genes between IAA treatment durations, one-way ANOVA was performed using IBM SPSS 20 software (IBM Corporation, USA). P<0.05 (*) and P<0.01 (**) were considered to indicate significant differences in expression.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Author contributions
Z.X., J.S. and S.C. designed and coordinated the study. Z.X. and A.J. performed experiments. Z.X. analyzed the data. Z.X. and J.S. wrote the manuscript.
Funding
This work was supported by the National Natural Science Foundation of China [Grant no. 81573398] and the National Science-Technology Support Plan of China [Grant no. 2012BAI29B01].
Supplementary information
Supplementary information available online at http://bio.biologists.org/lookup/doi/10.1242/bio.017178.supplemental
References
- Arabidopsis Genome Initiative (2000). Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408, 796-815. 10.1038/35048692 [DOI] [PubMed] [Google Scholar]
- Boer D. R., Freire-Rios A., van den Berg W. A. M., Saaki T., Manfield I. W., Kepinski S., López-Vidrieo I., Franco-Zorrilla J. M., de Vries S. C., Solano R. et al. (2014). Structural basis for DNA binding specificity by the Auxin-dependent ARF transcription factors. Cell 156, 577-589. 10.1016/j.cell.2013.12.027 [DOI] [PubMed] [Google Scholar]
- Cheng T. O. (2006). Danshen: a popular Chinese cardiac herbal drug. J. Am. Coll. Cardiol. 47, 1498; author reply 1499-1500 10.1016/j.jacc.2006.01.001 [DOI] [PubMed] [Google Scholar]
- Cui G., Duan L., Jin B., Qian J., Xue Z., Shen G., Snyder J. H., Song J., Chen S., Huang L. et al. (2015). Functional divergence of diterpene syntheses in the medicinal plant Salvia miltiorrhiza Bunge. Plant Physiol. 169, 1607-1618. 10.1104/pp.15.00695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong Y., Morris-Natschke S. L. and Lee K.-H. (2011). Biosynthesis, total syntheses, and antitumor activity of tanshinones and their analogs as potential therapeutic agents. Nat. Prod. Rep. 28, 529-542. 10.1039/c0np00035c [DOI] [PubMed] [Google Scholar]
- Ellis C. M., Nagpal P., Young J. C., Hagen G., Guilfoyle T. J. and Reed J. W. (2005). AUXIN RESPONSE FACTOR1 and AUXIN RESPONSE FACTOR2 regulate senescence and floral organ abscission in Arabidopsis thaliana. Development 132, 4563-4574. 10.1242/dev.02012 [DOI] [PubMed] [Google Scholar]
- Finet C., Fourquin C., Vinauger M., Berne-Dedieu A., Chambrier P., Paindavoine S. and Scutt C. P. (2010). Parallel structural evolution of auxin response factors in the angiosperms. Plant J. 63, 952-959. 10.1111/j.1365-313X.2010.04292.x [DOI] [PubMed] [Google Scholar]
- Guilfoyle T. J. and Hagen G. (2007). Auxin response factors. Curr. Opin. Plant Biol. 10, 453-460. 10.1016/j.pbi.2007.08.014 [DOI] [PubMed] [Google Scholar]
- Guilfoyle T. J. and Hagen G. (2012). Getting a grasp on domain III/IV responsible for Auxin Response Factor–IAA protein interactions. Plant Sci. 190, 82-88. 10.1016/j.plantsci.2012.04.003 [DOI] [PubMed] [Google Scholar]
- Guo J., Zhou Y. J., Hillwig M. L., Shen Y., Yang L., Wang Y., Zhang X., Liu W., Peters R. J., Chen X. et al. (2013). CYP76AH1 catalyzes turnover of miltiradiene in tanshinones biosynthesis and enables heterologous production of ferruginol in yeasts. Proc. Natl. Acad. Sci. USA 110, 12108-12113. 10.1073/pnas.1218061110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagen G. and Guilfoyle T. (2002). Auxin-responsive gene expression: genes, promoters and regulatory factors. Plant Mol. Biol. 49, 373-385. 10.1023/A:1015207114117 [DOI] [PubMed] [Google Scholar]
- Hamann T., Benkova E., Baurle I., Kientz M. and Jürgens G. (2002). The Arabidopsis BODENLOS gene encodes an auxin response protein inhibiting MONOPTEROS-mediated embryo patterning. Genes Dev. 16, 1610-1615. 10.1101/gad.229402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu W., Zuo J., Hou X., Yan Y., Wei Y., Liu J., Li M., Xu B. and Jin Z. (2015). The auxin response factor gene family in banana: genome-wide identification and expression analyses during development, ripening, and abiotic stress. Front. Plant Sci. 6, 742 10.3389/fpls.2015.00742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji A. J., Luo H. M., Xu Z. C., Zhang X., Zhu Y. J., Liao B. S., Yao H., Song J. Y. and Chen S. L. (2015). Genome-wide identification of the AP2/ERF gene family involved in active constituent biosynthesis in Salvia miltiorrhiza. Plant Genome. 9, 1-11. 10.3835/plantgenome2015.08.0077 [DOI] [PubMed] [Google Scholar]
- Jones-Rhoades M. W., Bartel D. P. and Bartel B. (2006). MicroRNAS and their regulatory roles in plants. Annu. Rev. Plant Biol. 57, 19-53. 10.1146/annurev.arplant.57.032905.105218 [DOI] [PubMed] [Google Scholar]
- Kalluri U. C., DiFazio S. P., Brunner A. M. and Tuskan G. A. (2007). Genome-wide analysis of Aux/IAA and ARF gene families in Populus trichocarpa. BMC Plant Biol. 7, 59 10.1186/1471-2229-7-59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korasick D. A., Westfall C. S., Lee S. G., Nanao M. H., Dumas R., Hagen G., Guilfoyle T. J., Jez J. M. and Strader L. C. (2014). Molecular basis for AUXIN RESPONSE FACTOR protein interaction and the control of auxin response repression. Proc. Natl. Acad. Sci. USA 111, 5427-5432. 10.1073/pnas.1400074111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krogan N. T., Ckurshumova W., Marcos D., Caragea A. E. and Berleth T. (2012). Deletion of MP/ARF5 domains III and IV reveals a requirement for Aux/IAA regulation in Arabidopsis leaf vascular patterning. New Phytol. 194, 391-401. 10.1111/j.1469-8137.2012.04064.x [DOI] [PubMed] [Google Scholar]
- Le Meur N. and Gentleman R. (2012). Analyzing biological data using R: methods for graphs and networks. Methods Mol. Biol. 804, 343-373. 10.1007/978-1-61779-361-5_19 [DOI] [PubMed] [Google Scholar]
- Li C. and Lu S. (2014). Genome-wide characterization and comparative analysis of R2R3-MYB transcription factors shows the complexity of MYB-associated regulatory networks in Salvia miltiorrhiza. BMC Genomics 15, 277 10.1186/1471-2164-15-277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C. and Zhang B. (2016). MicroRNAs in control of plant development. J. Cell Physiol. 231, 303-313. 10.1002/jcp.25125 [DOI] [PubMed] [Google Scholar]
- Li H., Johnson P., Stepanova A., Alonso J. M. and Ecker J. R. (2004). Convergence of signaling pathways in the control of differential cell growth in Arabidopsis. Dev. Cell 7, 193-204. 10.1016/j.devcel.2004.07.002 [DOI] [PubMed] [Google Scholar]
- Li C., Li D., Shao F. and Lu S. (2015a). Molecular cloning and expression analysis of WRKY transcription factor genes in Salvia miltiorrhiza. BMC Genomics 16, 200 10.1186/s12864-015-1411-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S. B., OuYang W. Z., Hou X. J., Xie L. L., Hu C. G. and Zhang J. Z. (2015b). Genome-wide identification, isolation and expression analysis of auxin response factor (ARF) gene family in sweet orange (Citrus sinensis). Front. Plant Sci. 6, 119 10.3389/fpls.2015.00119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z. B., Ulmasov T., Shi X., Hagen G. and Guilfoyle T. J. (1994). Soybean GH3 promoter contains multiple auxin-inducible elements. Plant Cell 6, 645-657. 10.1105/tpc.6.5.645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu P.-P., Montgomery T. A., Fahlgren N., Kasschau K. D., Nonogaki H. and Carrington J. C. (2007). Repression of AUXIN RESPONSE FACTOR10 by microRNA160 is critical for seed germination and post-germination stages. Plant J. 52, 133-146. 10.1111/j.1365-313X.2007.03218.x [DOI] [PubMed] [Google Scholar]
- Liu X., Huang J., Wang Y., Khanna K., Xie Z., Owen H. A. and Zhao D. (2010). The role of floral organs in carpels, an Arabidopsis loss-of-function mutation in MicroRNA160a, in organogenesis and the mechanism regulating its expression. Plant J. 62, 416-428. 10.1111/j.1365-313X.2010.04164.x [DOI] [PubMed] [Google Scholar]
- Liu K., Yuan C., Li H., Lin W., Yang Y., Shen C. and Zheng X. (2015). Genome-wide identification and characterization of auxin response factor (ARF) family genes related to flower and fruit development in papaya (Carica papaya L.). BMC Genomics 16, 901 10.1186/s12864-015-2182-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak K. J. and Schmittgen T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT Method. Methods 25, 402-408. 10.1006/meth.2001.1262 [DOI] [PubMed] [Google Scholar]
- Luo H., Zhu Y., Song J., Xu L., Sun C., Zhang X., Xu Y., He L., Sun W., Xu H. et al. (2014). Transcriptional data mining of Salvia miltiorrhiza in response to methyl jasmonate to examine the mechanism of bioactive compound biosynthesis and regulation. Physiol. Plant 152, 241-255. 10.1111/ppl.12193 [DOI] [PubMed] [Google Scholar]
- Ma Y., Yuan L., Wu B., Li X., Chen S. and Lu S. (2012). Genome-wide identification and characterization of novel genes involved in terpenoid biosynthesis in Salvia miltiorrhiza. J. Exp. Bot. 63, 2809-2823. 10.1093/jxb/err466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mun J.-H., Yu H.-J., Shin J. Y., Oh M., Hwang H. J. and Chung H. (2012). Auxin response factor gene family in Brassica rapa: genomic organization, divergence, expression, and evolution. Mol. Genet. Genomics 287, 765-784. 10.1007/s00438-012-0718-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagpal P., Ellis C. M., Weber H., Ploense S. E., Barkawi L. S., Guilfoyle T. J., Hagen G., Alonso J. M., Cohen J. D., Farmer E. E.. et al. (2005). Auxin response factors ARF6 and ARF8 promote jasmonic acid production and flower maturation. Development 132, 4107-4118. 10.1242/dev.01955 [DOI] [PubMed] [Google Scholar]
- Nanao M. H., Vinos-Poyo T., Brunoud G., Thévenon E., Mazzoleni M., Mast D., Lainé S., Wang S., Hagen G., Li H. et al. (2014). Structural basis for oligomerization of auxin transcriptional regulators. Nat. Commun. 5, 3617 10.1038/ncomms4617 [DOI] [PubMed] [Google Scholar]
- Okushima Y., Mitina I., Quach H. L. and Theologis A. (2005a). AUXIN RESPONSE FACTOR 2 (ARF2): a pleiotropic developmental regulator. Plant J. 43, 29-46. 10.1111/j.1365-313X.2005.02426.x [DOI] [PubMed] [Google Scholar]
- Okushima Y., Overvoorde P. J., Arima K., Alonso J. M., Chan A., Chang C., Ecker J. R., Hughes B., Lui A., Nguyen D. et al. (2005b). Functional genomic analysis of the AUXIN RESPONSE FACTOR gene family members in Arabidopsis thaliana: unique and overlapping functions of ARF7 and ARF19. Plant Cell 17, 444-463. 10.1105/tpc.104.028316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pekker I., Alvarez J. P. and Eshed Y. (2005). Auxin response factors mediate Arabidopsis organ asymmetry via modulation of KANADI activity. Plant Cell 17, 2899-2910. 10.1105/tpc.105.034876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rademacher E. H., Lokerse A. S., Schlereth A., Llavata-Peris C. I., Bayer M., Kientz M., Freire Rios A., Borst J. W., Lukowitz W., Jürgens G. et al. (2012). Different auxin response machineries control distinct cell fates in the early plant embryo. Dev. Cell 22, 211-222. 10.1016/j.devcel.2011.10.026 [DOI] [PubMed] [Google Scholar]
- Remington D. L., Vision T. J., Guilfoyle T. J. and Reed J. W. (2004). Contrasting modes of diversification in the Aux/IAA and ARF gene families. Plant Physiol. 135, 1738-1752. 10.1104/pp.104.039669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santner A. and Estelle M. (2009). Recent advances and emerging trends in plant hormone signalling. Nature 459, 1071-1078. 10.1038/nature08122 [DOI] [PubMed] [Google Scholar]
- Schruff M. C., Spielman M., Tiwari S., Adams S., Fenby N. and Scott R. J. (2006). The AUXIN RESPONSE FACTOR 2 gene of Arabidopsis links auxin signalling, cell division, and the size of seeds and other organs. Development 133, 251-261. 10.1242/dev.02194 [DOI] [PubMed] [Google Scholar]
- Sessions A., Nemhauser J. L., McColl A., Roe J. L., Feldmann K. A. and Zambryski P. C. (1997). ETTIN patterns the Arabidopsis floral meristem and reproductive organs. Development 124, 4481-4491. [DOI] [PubMed] [Google Scholar]
- Shen C., Yue R., Sun T., Zhang L., Xu L., Tie S., Wang H. and Yang Y. (2015). Genome-wide identification and expression analysis of auxin response factor gene family in Medicago truncatula. Front. Plant Sci. 6, 73 10.3389/fpls.2015.00073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun R., Wang K., Guo T., Jones D. C., Cobb J., Zhang B. and Wang Q. (2015). Genome-wide identification of auxin response factor (ARF) genes and its tissue-specific prominent expression in Gossypium raimondii. Funct. Integr. Genomics 15, 481-493. 10.1007/s10142-015-0437-0 [DOI] [PubMed] [Google Scholar]
- Tiwari S. B., Hagen G. and Guilfoyle T. (2003). The roles of auxin response factor domains in auxin-responsive transcription. Plant Cell 15, 533-543. 10.1105/tpc.008417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Todd A. T., Liu E., Polvi S. L., Pammett R. T. and Page J. E. (2010). A functional genomics screen identifies diverse transcription factors that regulate alkaloid biosynthesis in Nicotiana benthamiana. Plant J. 62, 589-600. 10.1111/j.1365-313X.2010.04186.x [DOI] [PubMed] [Google Scholar]
- Trapnell C., Roberts A., Goff L., Pertea G., Kim D., Kelley D. R., Pimentel H., Salzberg S. L., Rinn J. L. and Pachter L. (2012). Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protoc. 7, 562-578. 10.1038/nprot.2012.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulmasov T., Liu Z. B., Hagen G. and Guilfoyle T. J. (1995). Composite structure of auxin response elements. Plant Cell 7, 1611-1623. 10.1105/tpc.7.10.1611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulmasov T., Hagen G. and Guilfoyle T. J. (1997). ARF1, a transcription factor that binds to auxin response elements. Science 276, 1865-1868. 10.1126/science.276.5320.1865 [DOI] [PubMed] [Google Scholar]
- Ulmasov T., Hagen G. and Guilfoyle T. J. (1999a). Activation and repression of transcription by auxin-response factors. Proc. Natl. Acad. Sci. USA 96, 5844-5849. 10.1073/pnas.96.10.5844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulmasov T., Hagen G. and Guilfoyle T. J. (1999b). Dimerization and DNA binding of auxin response factors. Plant J. 19, 309-319. 10.1046/j.1365-313X.1999.00538.x [DOI] [PubMed] [Google Scholar]
- Wan S., Li W., Zhu Y., Liu Z., Huang W. and Zhan J. (2014). Genome-wide identification, characterization and expression analysis of the auxin response factor gene family in Vitis vinifera. Plant Cell Rep. 33, 1365-1375. 10.1007/s00299-014-1622-7 [DOI] [PubMed] [Google Scholar]
- Wang D., Pei K., Fu Y., Sun Z., Li S., Liu H., Tang K., Han B. and Tao Y. (2007a). Genome-wide analysis of the auxin response factors (ARF) gene family in rice (Oryza sativa). Gene 394, 13-24. 10.1016/j.gene.2007.01.006 [DOI] [PubMed] [Google Scholar]
- Wang X., Morris-Natschke S. L. and Lee K. H. (2007b). New developments in the chemistry and biology of the bioactive constituents of Tanshen. Med. Res. Rev. 27, 133-148. 10.1002/med.20077 [DOI] [PubMed] [Google Scholar]
- Wang B., Sun W., Li Q., Li Y., Luo H., Song J., Sun C., Qian J., Zhu Y., Hayward A. et al. (2015). Genome-wide identification of phenolic acid biosynthetic genes in Salvia miltiorrhiza. Planta 241, 711-725. 10.1007/s00425-014-2212-1 [DOI] [PubMed] [Google Scholar]
- Wilmoth J. C., Wang S., Tiwari S. B., Joshi A. D., Hagen G., Guilfoyle T. J., Alonso J. M., Ecker J. R. and Reed J. W. (2005). NPH4/ARF7 and ARF19 promote leaf expansion and auxin-induced lateral root formation. Plant J. 43, 118-130. 10.1111/j.1365-313X.2005.02432.x [DOI] [PubMed] [Google Scholar]
- Wright R. C. and Nemhauser J. L. (2015). New tangles in the auxin signaling web. F1000Prime Rep. 7, 19 10.12703/P7-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu M.-F., Tian Q. and Reed J. W. (2006). Arabidopsis microRNA167 controls patterns of ARF6 and ARF8 expression, and regulates both female and male reproduction. Development 133, 4211-4218. 10.1242/dev.02602 [DOI] [PubMed] [Google Scholar]
- Xu Z., Peters R. J., Weirather J., Luo H., Liao B., Zhang X., Zhu Y., Ji A., Zhang B., Hu S. et al. (2015). Full-length transcriptome sequences and splice variants obtained by a combination of sequencing platforms applied to different root tissues of Salvia miltiorrhiza and tanshinone biosynthesis. Plant J. 82, 951-961. 10.1111/tpj.12865 [DOI] [PubMed] [Google Scholar]
- Xu H., Song J., Luo H., Zhang Y., Li Q., Zhu Y., Xu J., Li Y., Song C., Wang B. et al. (2016a). Analysis of the genome sequence of the medicinal plant Salvia miltiorrhiza. Mol. Plant. Epub ahead of print, doi:10.1016/j.molp.2016.03.010 10.1016/j.molp.2016.03.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Z.-c., Ji A.-j., Zhang X., Song J.-y. and Chen S.-l. (2016b). Biosynthesis and regulation of active compounds in medicinal model plant Salvia miltiorrhiza. Chin. Herb. Med. 8, 3-11. 10.1016/S1674-6384(16)60002-3 [DOI] [Google Scholar]
- Yu H., Soler M., Mila I., San Clemente H., Savelli B., Dunand C., Paiva J. A. P., Myburg A. A., Bouzayen M., Grima-Pettenati J. et al. (2014). Genome-wide characterization and expression profiling of the AUXIN RESPONSE FACTOR (ARF) gene family in Eucalyptus grandis. PLoS ONE 9, e108906 10.1371/journal.pone.0108906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L., Wu B., Zhao D., Li C., Shao F. and Lu S. (2014). Genome-wide analysis and molecular dissection of the SPL gene family in Salvia miltiorrhiza. J. Integr. Plant Biol. 56, 38-50. 10.1111/jipb.12111 [DOI] [PubMed] [Google Scholar]
- Zhang X., Luo H., Xu Z., Zhu Y., Ji A., Song J. and Chen S. (2015). Genome-wide characterisation and analysis of bHLH transcription factors related to tanshinone biosynthesis in Salvia miltiorrhiza. Sci. Rep. 5, 11244 10.1038/srep11244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zouine M., Fu Y., Chateigner-Boutin A.-L., Mila I., Frasse P., Wang H., Audran C., Roustan J.-P. and Bouzayen M. (2014). Characterization of the tomato ARF gene family uncovers a multi-levels post-transcriptional regulation including alternative splicing. PLoS ONE 9, e84203 10.1371/journal.pone.0084203 [DOI] [PMC free article] [PubMed] [Google Scholar]