Abstract
The concept of optimizing health care by understanding and generating knowledge from previous evidence, ie, the Learning Health-care System (LHS), has gained momentum and now has national prominence. Meanwhile, the rapid adoption of electronic health records (EHRs) enables the data collection required to form the basis for facilitating LHS. A prerequisite for using EHR data within the LHS is an infrastructure that enables access to EHR data longitudinally for health-care analytics and real time for knowledge delivery. Additionally, significant clinical information is embedded in the free text, making natural language processing (NLP) an essential component in implementing an LHS. Herein, we share our institutional implementation of a big data-empowered clinical NLP infrastructure, which not only enables health-care analytics but also has real-time NLP processing capability. The infrastructure has been utilized for multiple institutional projects including the MayoExpertAdvisor, an individualized care recommendation solution for clinical care. We compared the advantages of big data over two other environments. Big data infrastructure significantly outperformed other infrastructure in terms of computing speed, demonstrating its value in making the LHS a possibility in the near future.
Keywords: health-care analytics, big data, natural language processing, learning health-care system
Introduction
The rapid adoption of electronic health records (EHRs) as a result of the HITECH act of American Recovery and Reinvestment Act has ushered significant changes in the quality of clinical practice in the United States.1 Besides the use of EHR for clinical practice, the EHR data have the potential to advance health care by generating and implementing new knowledge through informatics and analytics. Informatics-driven analytics has recently gained national prominence and momentum, thereby enabling the evolution of a novel field known as the Learning Health-care System (LHS).2 The Institute of Medicine defines the LHS as a system that generates new knowledge and embeds it into the clinical practice. This is done by utilizing clinical data and a robust technology infrastructure to enable seamless refinement and delivery of best practices for continuous improvement in health care.3 Secondary use of EHR data has facilitated the learning cycles in discovery, implementation, and the evaluation of new knowledge toward better health care. One prerequisite to implement LHS is an infrastructure, which allows access to both longitudinal and near real-time patient EHR data in order to facilitate discovery and delivery of best practices.
EHR data consist of information in both structured data elements and unstructured formats. Much clinical information is embedded in clinical narratives, which pose significant challenges in streamlining the information to be utilized for the LHS.4 For example, detailed information about patient conditions, interventions, clinical progress, and treatment outcomes is often captured in clinical notes. Natural language processing (NLP) offers opportunities to tap into clinical narratives to extract the information needed for various clinical applications.5–7 Estimates indicate that around 80% of the clinical information resides in the unstructured narrative.8 NLP solutions, which automatically extract discrete, actionable data from clinical narratives, pave the way to data-driven health care, the key development toward outcome-based care and payment models.
In general, NLP can be computationally very intensive not only due to the sheer volume of the unstructured clinical data but also due to the complexity of NLP where traditional computing infrastructure does not have the inherent capacity for implementing scalable NLP solutions.9,10
In this study, we introduce a big data-empowered NLP infrastructure, which delivers high-performance, scalable, and real-time NLP solutions at the Mayo Clinic. In the following sections, we provide background and related work followed by a summary of various NLP initiatives at the Mayo Clinic. We then describe the compelling need and the implementation of a big data-empowered NLP infrastructure. We finally discuss an application MayoExpertAdvisor (MEA) that delivers near real-time care recommendation to clinicians at the point of care and the significant role played by big data-empowered NLP infrastructure in this process.
Background and Related Work
Figure 1 illustrates the learning cycle in an LHS: practice, data, research, and knowledge. With the rapid adoption of EHRs, clinical practice generates large amounts of clinical data.11 Researchers have been extensively utilizing EHR data for secondary purposes including clinical decision support, outcomes improvement, biomedical research, and epidemiologic monitoring of the nation’s health. Knowledge discovered through research can then be utilized to improve patient care. The most significant initiative related to the LHS is The National Patient-Centered Clinical Research Network (PCORnet) formed in 2013 by Patient-Centered Outcomes Research Institute (PCORI), which consists of 11 clinical data research networks (CDRNs) and 18 patient-powered research networks. These organizations have made significant progress toward analyzing the data within these networks focusing on common conditions, rare conditions, and genetic disorders. There are nationwide networks other than PCORnet that also play positive role in facilitating LHS. For instance the collaboration between Kaiser Permanente and Strategic Partners, Patient Outcomes Research To Advance Learning,12 Scalable Collaborative Infrastructure for a Learning Health-care System,13 PaTH (University of Pittsburgh/UPMC, Penn State, College of Medicine, Temple University Hospital, and Johns Hopkins University), leading four academic health centers,14 and PEDSnet, another consortium of eight children’s hospitals, are initiatives involving multiple institutions.15 Large initiatives such as the PCORnet provide an infrastructure for a national LHS.
Figure 1.

Learning cycle in an LHS. Analytics experts enable the cycle. Domain pragmatics provides the contextual information related to the domain, which is needed for discovering knowledge. Users are people who consume the knowledge.
NLP has been an integral component in the LHS, as evidenced by one of the review criteria in the recent CDRN phase II request for application, being the demonstration of NLP capability for phenotyping.16 Figure 2 provides an overview of clinical NLP. At a high level, NLP generally consists of the following components: tokenization, syntactic parsing, semantic parsing, and pragmatic interpretation. It may also include upstream components such as speech recognition or optical character recognition or downstream components of data mining, text analytics, visualization, and summarization of the NLP results. In health care, a critical additional component is required – terminology mapping. This component takes the content that is clinically relevant and produces codes for unified semantic representations of clinical concepts. These codes are subsequently used in various applications such as billing, compliance, quality measurement, clinical decision support, and others.
Figure 2.
Generic clinical NLP process. Clinical NLP involves processing textual data obtained from clinical notes and voice dictated text. The process includes both syntactic and semantic processing. While syntactic components identify the grammatical structure of the text, the semantic components identify clinical concepts and its context such as experiencer, certainty, and negation.
Since the 1980s, NLP has been utilized to harness the information embedded in clinical narratives. One of the oldest and most studied clinical NLP systems is the Medical Language Extraction and Encoding System (MedLEE) developed by Friedman et al at the Columbia University in the mid-1990s.17 MedLEE was initially developed on chest radiology reports18 but further extended to work on any kind of clinical notes. An NIH-funded national center, Informatics for Integrating Biology, and the Bedside (i2b2), has organized both challenges and shared tasks focusing on problems less studied in clinical NLP and sharing annotated clinical notes that removed some of the barriers to the development of clinical NLP systems.19–25
However, one of the major bottlenecks in integrating NLP into clinical workflow has been the lack of computing infrastructure to implement real-time NLP solutions. With the recent advances in big data, it becomes apparent that the streaming and distributed computing capacity in the big data technology stack makes the implementation of NLP in the LHS possible.
One example of big data-empowered NLP solution is IBM Watson, a cognitive system developed by IBM Research Center with the capability of analyzing natural language content.26 Watson incorporates multiple layers of NLP technologies including machine learning and a question answering system.27–32 Recently, building upon the technologies behind Watson, IBM has invested in health-care analytics by improving clinical NLP capability. For example, Wang et al improved the performance of medical relation extraction in Watson.33 IBM Watson is an independent analytical application that needs to be integrated into an EHR workflow for an effective use in clinical practice. In the recent past, EHRs do have some inbuilt NLP capabilities in their workflow. Cerner corporation has developed a sophisticated EHR that is not only safer and easier to use but smart enough to decipher the contextual meaning behind the descriptions in clinical text.34 Chart Search, a search platform within Cerner has the capabilities to understand the intent of a query to perform semantic search.
Besides these big corporation initiatives, there are few efforts in academic institutions where big data-empowered NLP solutions have significantly advanced the clinical care by reducing the processing times of clinical data. Agerri et al (2015)10 have demonstrated that the big data infrastructure can help scale NLP analytics to provide near real-time solutions.35 On the other hand, Divita et al (2015)9 explored an alternative approach for scaling NLP solutions through multithreading and running NLP modules concurrently. They took software engineering approach instead of assembling a robust hardware infrastructure for scaling the computing performance.
Essentially we have two models for performance scaling, as discussed above. One option is to have the right choice of robust hardware infrastructure, while the other is to engineer a robust software solution. At the Mayo Clinic, we took a middle path for scaling NLP applications, striking a fine balance between engineering a robust software solution and choosing a sophisticated big data infrastructure for deploying software. While big data-empowered NLP offers the best hardware infrastructure, MedTagger is a suite of best-of-breed NLP modules developed based on rigorous software engineering models. We believe that this combination will help us realize the LHS as a possibility in the near future at Mayo Clinic.
NLP Implementation Prior to the Big Data Era at Mayo Clinic
Mayo Clinic has a long history of using patient records as an organized resource to support research and quality improvement.36 Early efforts to create an EHR for surgical recording system associated with ICD coding of diagnosis and procedures began in the late 1980s. The deployment of the first EHR project introducing clinical notes launched in the mid-1990 and introduced semiautomated coding of master sheet diagnosis in Medical Index using NLP. These efforts evolved into creating a data warehouse, a joint development with IBM to create a comprehensive clinical data repository derived from EHR.37 The Mayo–IBM collaboration resulted in the first version of the Mayo Clinic Life Science System, which provided search capability for structured data, unstructured text, and NLP annotated text. However, a systematic utilization of the data (ie, normalization, extraction, transform, and load) was recognized as a critical need for a semantically integrated data store at the enterprise level. Hence, in 2007, Mayo initiated work on a data repository, the Enterprise Data Trust (EDT), built on industry standard and optimized for business intelligence and flexible data utilization.38 This integrated source of data across the Mayo enterprise, as shown in Table 1, has served as a foundation for many aspects of clinical research and practice using NLP.
Table 1.
Unstructured text in EDT.
| DOCUMENT TYPE | DESCRIPTION | VOLUME |
|---|---|---|
| Clinical notes | Defacto medical description for patient interactions. Documents not codified. | 78M |
| Pathology reports | Confirmed diagnosis, partially codified. | 5M |
| Radiology | Examination type | 5M |
| Surgery notes | Codified using Mayo modified ICD9 Procedure Codes. 4M | |
| LAB interpretive reports | Detailed interpretive reports on lab results (eg, Genetic tests) | <1M |
Since 2001, Mayo Clinic has invested in NLP for processing clinical notes using syntactic and semantic features of the language. Mayo has pioneered clinical NLP research in multiple aspects. As part of Mayo–IBM collaboration, Mayo has released an open-source clinical NLP system: clinical Text Analysis and Knowledge Extraction System (cTAKES)39 built on the Unstructured Information Management Architecture (UIMA). cTAKES has been recognized by many clinical informatics communities and has improved its functionality and portability through collaborations. cTAKES became an Apache project in 2012. Additional NLP pipelines have been developed by the Mayo Clinic and released in open source through the Open Health Natural Language Processing Consortium.* MedTagger for indexing medical concepts, information extraction, and named entity recognition,40 MedXN for medication extraction and normalization,41 MedTime for clinical temporal information extraction,42 and MedCoref for coreference resolution in clinical text43 are some of the tools available in open source.
Currently, we provide two main types of NLP services in clinical practice. One is concept indexing, and the other is high-level information extraction. An NLP pipeline identifies the clinical concept mentions and the contextual information such as negation, certainty, and experiencer mentioned in a document. The concepts were also subsequently indexed as a separate field in a database to facilitate better cohort retrieval. The NLP program also provides a framework for high-level information extraction that is very use-case driven. MedTaggerIE, an information extraction component in MedTagger, has been used for various use-case driven information extraction tasks. It is a pattern-based information extraction framework developed under UIMA. There are three knowledge components for an information extraction engine, ie, dictionary, normalization, and regular expression. Dictionary specifies the terms or patterns to extract the concept mentioned in the text, normalization defines the target concept to which the textual extractions needs to be mapped, and regular expressions define the overall rules based on the other two components. These knowledge components are all externalized in order to maximize customizability and maintenance.40 Given a large collection of clinical narratives, we have conducted multiple large-scale research studies yielding NLP processing knowledge that is ready to be part of NLP engines used in production.
Implementation of Big Data-Empowered Clinical NLP at Mayo Clinic
One of the major reasons for the big data initiative at the Mayo Clinic is the ability to extract information from the EHR near real time to meet the information needs at the point of care. Figure 3 shows a high-level architecture of the Mayo big data implementation.
The bottom layer of Figure 3 represents EHR data sources, which generate data during clinical care. The data can be in diverse formats including structured fields, problem lists, laboratory values, unstructured notes, images, and other information.
- In the middle layer, data from the primary sources are streamed to the big data environment via messaging queues. The big data layer itself consists of the following components:
- Data ingestion (messaging system) – accepts data from multiple sources, both streaming data as well as archived data;
- Data analytics (NLP) – enables stream and batch processing of data;
- Data storage and retrieval – consists of next-generation approaches for storing and indexing intermediate or final datasets.
The topmost layer represents various applications, which consume the semantically enriched data provided by the analytical processing layer.
Figure 3.
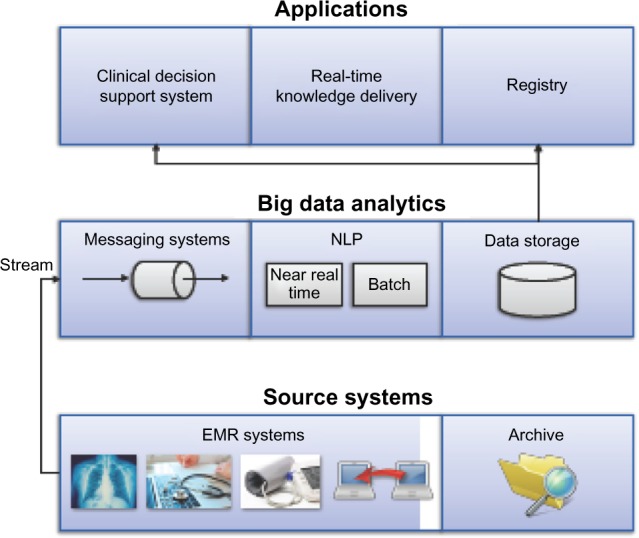
A high-level architecture of big data-empowered analytics in LHS. Big data architecture at Mayo consists of three layers: (i) data ingestion layer that reads data from real-time feeds from the EMR and archived data, (ii) big data analytics layer that does stream processing for analyzing the data, and (iii) data storage and retrieval that stores the information and knowledge that are generated through big data analytics and facilitate retrieval at the appropriate time for clinical use.
Big Data Technologies Adopted
The big data implementation at Mayo has been designed for both analytical processing and new storage methodologies to facilitate faster retrieval. We chose Apache Hadoop as the big data platform, which includes components such as Apache Storm to provide real-time distributed computation environment, HBase for fast key-based data retrieval, and Elastic-search for efficient indexing and querying of information. In the following, we briefly describe these technologies.
Apache Storm is a programming model agnostic stream-processing environment, which we used for streaming and scalable computing. Storm architecture consists of a cluster, where a master node distributes jobs to the slave nodes. The underlying structure of Storm is a graph topology, which consists of nodes that serve as the processing environment while the edges serve as the message broker communicating between the nodes. Nodes in the Storm topology essentially fall into two categories as follows: (i) Spouts to stream data from sources and (ii) Bolts to perform processing on data stream emitted by a Spout. A Bolt in turn emits a stream that can be utilized by other bolts. Apache Storm enables real-time analytics environment and data delivery through multiple processing streams of data effectively increasing the throughput.
Apache Hadoop is inherently designed for large-scale processing, predominantly in batch-processing mode across multiple, horizontally scaled server nodes built from commodity hardware.44 Apache Hadoop allows the data to be processed faster and more efficiently than it would be in conventional supercomputer architecture. It relies on a parallel file system where computation and data are connected via high-speed networking. The Hadoop framework relies on map reduce formalism to deliver fault-tolerant scaling. In our big data implementation, we use MapReduce jobs to quickly search across clinical documentation to extract particular subsets of information that need to be processed through our Storm infrastructure. Currently, we are not using MapReduce jobs to scale the processing of any process in the big data implementation at the Mayo Clinic. We continue to investigate and develop more MapReduce and Spark capabilities in our infrastructure.
Apache HBase is an open-source distributed, nonrelational database modeled after Google’s Big Table.45 HBase does not support SQL as a query language, instead HBase provides a rich Java API. It is built on HDFS and hence can be deployed on commodity hardware. HBase is meant for a large amount of data in the range of billions of rows. An instance of HBase has a collection of tables. Each table contains rows with row-keys and arbitrary number of columns. These columns contain key–value pairs, which are versioned by timestamp by default. HBase was chosen to enable very fast key-based retrieval of documents stored in the big data environment.
Elasticsearch is a distributed full-text search engine that is built on Apache Lucene. Elasticsearch can handle large-scale real-time data to perform real-time analytics, which will enable the application layers to access the semantically enriched data in big data in near real time. Elastic search provides mechanisms to horizontally scale the retrieval by adding additional nodes for processing. It is fairly resilient that it can auto detect failure nodes and perform load balancing to ensure both data safety and accessibility. It also supports the notion of multitenancy where multiple indices can be housed on an instance of Elasticsearch.
NLP Implementation in Big Data
The implementation of big data-empowered NLP infrastructure is a critical component in the Mayo Clinic Unified Data Platform (UDP) initiative. The mission of UDP is to provide a centralized, yet collaborative and community-oriented data services framework that enables and facilitates all data-driven projects across the Mayo Clinic enterprise. The big data infrastructure itself plays a significant role in data enrichment, discovery, and delivery. Prior efforts on clinical data have empowered retrospective studies and research. Before the big data initiative, the UDP had been limited in its ability to provide real-time data analytics and delivery, which had hampered the ability to implement decision support systems at the point of care.
The current big data infrastructure at the Mayo Clinic has the following configurations: one production cluster, one integration cluster, one development cluster, and a discovery cluster. The discovery cluster is utilized for research work and other analytical purposes.
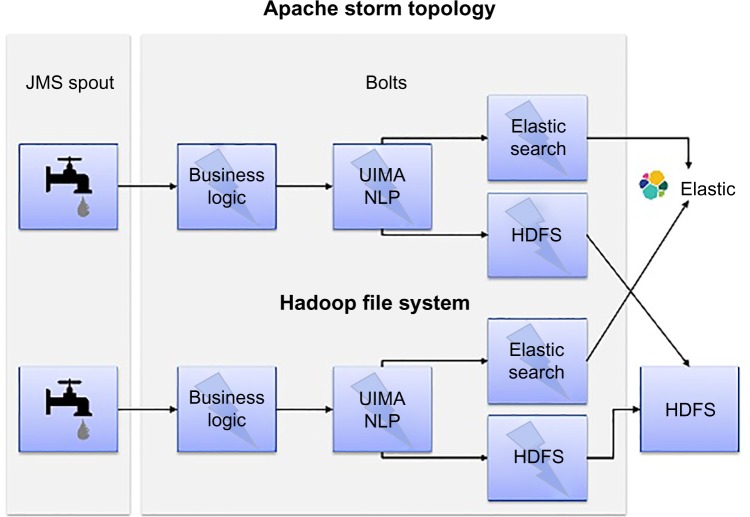
Figure 4 shows the big data-empowered NLP architecture that uses Apache Storm to realize real-time NLP processing. Specifically, we implement a bolt for NLP processing in the Storm topology, which can instantiate the required NLP pipeline configurable through a configuration file. Clinical documents in HL7 format (input data) are read from message queues using the Java Messaging System. We implemented multiple bolts related to the NLP processing in our Storm Topology. The “Parse-Bolt” parses the HL7 messages utilizing the open-source HL7-HAPI-based parser.** The “NLP-Bolt” initializes the required NLP pipeline to be used for processing based on the project-specific configuration. We have two storage-related bolts: “Elastic Search-Bolt” and “HDFS-Bolt”. While the former is used to index the HL7 messages and the NLP results, the latter is used to store them in HDFS.
Figure 4.
Data-empowered NLP architecture. Apache storm topology consists of the following components: (i) Spout: streamlines data from their respective data sources; (ii) Bolts: Processing unit often dedicated to a single type of process; (iii) HDFS – currently used for archive; (iv) Elasticsearch – retrieve data from archive.
Big Data Implementation – a Clinical Use Case
Very recently, we implemented a decision support system, namely, MEA, an application that delivers individualized care recommendations for clinical practice using the big data infrastructure. This section describes the broad goal of MEA and the specific big data implementation. We, also describe the performance of NLP modules in terms of computational time in three different server environments including the big data. We summarize the performance of each infrastructure especially on how the reduced processing times of big data infrastructure will impact the overall goal of the LHS implementation.
Mayo Expert Advisor
To deliver the optimal care to individual patients, Mayo Clinic has made a significant investment in developing knowledge assets regarding best practice pathways, guidelines, and the associated tools to manage these guidelines. Currently, Mayo-vetted best practices are being authored by Specialty Councils, Centers, or others and are available through a web-based application, AskMayoExpert (AME).46 AME currently has over 115 Care Process Models (CPMs) and 40 risk factor scoring tools. These risk factors are taken into account when choosing the right intervention as outlined in the CPMs. However, the provider has to manually enter patient data into scoring tools and then review the protocols in AME to see what care recommendation or intervention is best suited for the patient. For a given patient, there might be many CPMs that are applicable to enable individualized knowledge delivery; all applicable recommendations have to be taken into account. Thus, to fully realize the value of all these knowledge assets in AME, we need to incorporate the knowledge into the clinical workflow of the providers in the context of individual patient care.
However, there is no consistent mechanism to present relevant knowledge in the context of patient-specific data. Therefore, knowledge delivery at the point of care requires real-time information extraction to populate data elements needed for delivering patient-specific screening reminders, follow-up recommendations, shared decision-making tools, patient-specific links to AME, and other resources.
MEA was implemented through a multiunit collaborative effort, leveraging the work that was already done in another web-based solution (Generic Disease Management System).47 An NLP pipeline deployed on big data provided unstructured text analysis, which was then integrated with the information from structured resources such as coded problem list, laboratory tests, procedures, and medications to understand the patient context and deliver relevant knowledge for patient care at the point of care.
We successfully implemented the knowledge workflow outlined in three CPMs namely, atrial fibrillation, hyperlipidemia, and congestive heart failure (CHF) during the pilot phase of MEA. The domain experts identified nearly 25 data elements (refer Table2) to be relevant in order to make a care recommendation. According to the New York Heart Association Classification, some of the information such as (for CHF CPM) implanted cardiac devices, CHF diagnosis, and the heart failure stage can only be obtained reliably from clinical notes. The NLP pipeline extracts the concepts/data elements outlined in Table 2 from the clinical notes. Besides detecting the mention level of each concept in a clinical note, the pipeline also detects additional metadata of the concept such as its certainty/affirmation, negation status, and the context such as experiencer to identify whether the concept was mentioned in the context of the patient or family. As previously described in the NLP Implementation prior to big data era at Mayo Clinic section, the whole pipeline was implemented using Apache UIMA framework.
Table 2.
CPMs and data elements.
| CARE PROCESS MODEL (CPM) | SAMPLE DATA ELEMENTS |
|---|---|
| Atrial fibrillation | Abdominal aortic aneurysm (AAA), alcohol abuse, atrial fibrillation diagnosis, biventricular ICD, biventricular pacemaker, bleeding disorder, central nervous system bleeding, cirrhosis diagnosis, congestive heart failure diagnosis, diabetes status, hematuria, implantable cardioverter defibrillator (ICD), left ventricular assist device (LVAD), myocardial infarction diagnosis |
| Hyperlipidemia | Alcohol abuse, congestive heart failure diagnosis, diabetes status, hypertension diagnosis, pancreatitis, myocardial infarction diagnosis, peripheral artery disease (PAD), stroke, thromboembolism, transient ischemic attack diagnosis, Statin induced myopathy, Eruptive Xanthomata |
| Heart Failure | Heart Failure stage (New York Heart Association Functional Classification), congestive heart failure diagnosis |
One of the main challenges for MEA is the rapid processing of historical clinical notes, radiology notes, and other unstructured data resources in order to deliver real-time, personalized clinical care recommendations. For some of the patients, the required information may occur in multiple documents at different time points. The information extracted from the individual documents of a patient needs to be synthesized across documents to check if a particular patient disease condition is consistent and trust worthy. The information extracted by the NLP pipeline is reconciled with the information from structured resources to infer at patient level whether the concept/data element is relevant to the patient. By combining the information from both the structured and unstructured sources, the resulting information is fed to a decision rule system that generates the care recommendation to be delivered to the clinician at the point of care. Figure 5 gives the outline of the overall workflow architecture of MEA. In this study, we restrict our focus to only the specific role of big data infrastructure-empowered NLP system in making care recommendations as outlined in CPMs to the clinicians.
Figure 5.
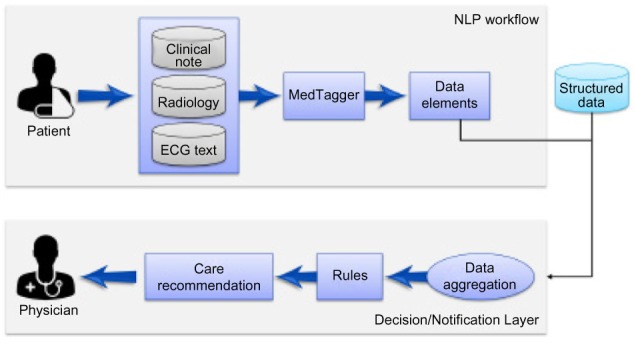
MEA workflow architecture. MEA workflow consists of three components: (i) MedTagger, a clinical NLP pipeline reads data from clinical notes, radiology notes, ECG text, and other reports and identifies data elements; (ii) Webservices aggregate the information from both the NLP pipeline and structured data sources such as laboratory values, patient provided information to synthesize concept assertion at patient level; and (iii) synthesized information is fed to a decision rule system that generates care recommendation for the clinician at the point of care.
Results and Discussion
In our pilot study, we benchmarked the performance of the same NLP pipeline in a fixed number of documents (20,000 clinical notes) in three different environments, namely, a standalone server, a data stage server, and the big data environment. Table 3 gives a broad outline on the hardware configuration of the three server environments.
Table 3.
Hardware configuration of three different server environments at Mayo Clinic.
| BIG DATA | DATA STAGE | SINGLE SERVER | |
|---|---|---|---|
| Processor | 2 of E5–26702.6 GHz | X5672 3.2 GHz | AMD Opteron (tm) Processor 2439 SE 2793 MHz |
| # of cores | 8 | 4 | 6 |
| Memory | 256 GB (Shared) 1333 MHz | 16 GB (Dedicated) | 32 GB (Shared) |
| HDD | HDFS (10 nodes) | NAS (Network Attached Storage) | RAID |
Figure 6 shows the average processing time taken to process 20,000 clinical documents in the three server environments. The standalone server had an average processing time of 23.97 minutes; data stage averaged 85.67 minutes, while the big data averaged 20.13 minutes to process 20,000 clinical notes. The data stage had significantly higher processing times when compared with the other two environments. After further investigation, there may be two reasons for this low performance of such a very powerful server: (i) the specific configuration of the data stage server was not optimal for high throughput and (ii) the data stage server was configured to run in a shared environment. It was not possible for us to schedule a job for MEA processing in a controlled and isolated data stage environment at this time. On the big data server, all the computations during this run were concentrated on a single node. There was not any significant difference in the performance of the big data (shown in Fig. 6) when compared with the standalone server, while there is a significant performance gain over the data stage.
Figure 6.
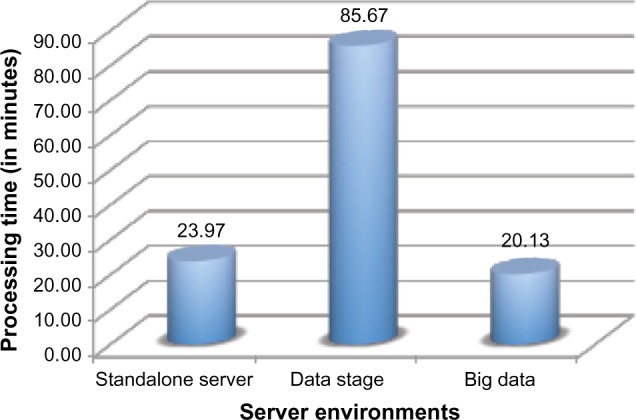
Average processing time of different server environments. Time taken for big data-empowered NLP to process 20,000 documents in three different server environments. On an average, (i) standalone server takes 23.97 minutes to complete the NLP process, (ii) data stage takes the maximum time of 85.67 minutes for the same, while (iii) big data take 20.03 minutes for the same task.
Further experiments were performed with the Storm architecture by increasing the number of parallel instances to 2, 4, 8, and 16, essentially doubling the parallel instances each time. Figure 7 clearly demonstrates the significant improvement in the performance due to increased parallelism. A substantial drop in the processing time was observed while increasing parallel instances. A configuration of 16 parallel instances took only 1.01 minutes (approximately 3 milliseconds to process a single document) to process 20,000 documents considered for the earlier experiment. The parallel system was 20 times faster than the configuration with a single instance (Fig. 6). The resilience of parallel processing power is one factor that gave big data the edge over other environments. While the drop in the processing time was very steep initially, the gain in the time tapers with increase in parallelism. This shows that there is a threshold to the number of the parallel instances beyond which there is no significant gain in the processing times. However, the optimal threshold may vary depending on the size of the data.
Figure 7.

Time taken to process 20,000 documents with varying number of parallel threads in big data. With increasing parallelism, there is a significant drop in the processing times of NLP empowered MEA algorithm.
We also studied the performance behavior of the big data with varying data size. We did this in order to ascertain the limits of gain in performance due to parallelism with increasing data. In Figure 7, we saw that for a fixed number of documents (20,000) the system achieved the best performance at 16 parallel instances. Keeping the parallel instance constant (at 16), we computed the time taken by the MEA algorithm to process 20,000 (335.08 MB of data), 40,000 (635.22 MB of data), 80,000 (1,270 MB of data), and 160,000 (2,792.99 MB of data) documents, essentially doubling the number of documents. Figure 8 shows the performance of the MEA algorithm while increasing the number of documents in big data. From Figure 8, we can infer that the processing time increases linearly with increasing number of documents. We can infer that further optimization of the number of parallel instances may be required with increasing amounts of data. At a fixed parallel instance (16 in this case), the performance of the MEA algorithm in big data may become a rate-limiting one. We believe that by adding additional nodes in big data infrastructure and increased parallelism of the Storm architecture the performance of MEA algorithm will ramp up to appreciable levels (as seen in Fig. 7).
Figure 8.
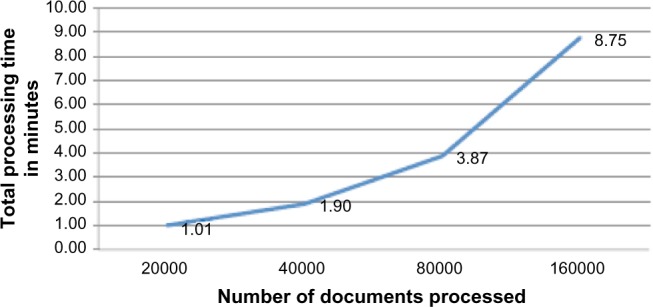
Processing time using 16 threads on varying number of documents. As the number of documents processing doubles, the processing time increases almost linearly.
For the MEA project, we piloted the implementation on 14,000 patients, which requires running NLP on 1.6 million documents. NLP in big data (empowered by 16 parallel instances) allowed the entire set to be processed within 90 minutes, which would not have been possible in the traditional Mayo Clinic infrastructure.
In summary, we have shown that the big data implementation, with the ability to run algorithms in parallel using the storm architecture, offers immense potential to realize our goal in delivering near real-time information, thereby enabling the delivery of optimal care in practice.
Limitations and Future Work
In this work, we explored the scaling performance of NLP through big data computing. Some of the technology choices that were made for this work were based on the technology stack available at the time of the project and the maturity of the software packages. Specifically, Apache Storm was chosen over Spark due to its more reliable and mature code base compared with the relatively new and not fully vetted functionality of Spark streaming. As Spark matures, it will be worthwhile to compare the technologies and see what benefits can be leve raged from each.
As far as the MedTagger NLP processing, the current NLP modules in MedTagger can be redesigned to run asynchronously using UIMA-AS or run in a nonlinear fashion. For example, Divita et al, 20159 explored multithreading to scale the performance of NLP. Additionally, we are currently exploring the use of Map Reduce/Spark to scale up some NLP module processing, which can be another way to improve the performance of NLP on a Hadoop environment. We can also remove certain overheads in preprocessing by adopting messaging and documentation standards.
More work is needed to fully leverage the potential of this infrastructure to deliver NLP solutions. There are some challenges identified in the process of our implementation that are listed as follows.
Data challenges
The uneven and complex nature of clinical documentation is a challenge in analyzing EHR data. Detailed patient treatment and outcome information is scattered in heterogeneous formats in various EHR platforms and standards in both structured and unstructured formats. Although extensive efforts has been dedicated at Mayo Clinic and other organizations to develop advanced NLP technologies, common problems in observational data such as confounding variables, bias, and missing data add to the complexity of the analytic problem.
Resource challenges
The investigation, development, testing, and deployment of NLP analysis pipelines are not trivial tasks. Based on the needs of each project, varying levels of sensitivity and specificity are required. Also, based on the clinical project requirements, there can be specific definitions or context required for NLP extracted concepts. This can make the development cycle and production implementation of NLP an expensive process, both in terms of time and money invested. Efforts are underway to build standardized processes and development tools to ensure that projects can be completed in an efficient and cost-effective manner.
Domain expertise challenges
Often, the end users of NLP analytics solutions are not experts themselves in all aspects of the health-care domain. It takes partnerships with subject-matter experts in various subspecialties of health care to build analysis pipelines that provide accurate and relevant NLP extracted information. Without the time and effort required to foster these collaborations, the level of precision required by consumers of NLP data would not be met.
Awareness challenges
Even though NLP has been around at Mayo Clinic for many years, the ability to leverage its potential in clinical settings is relatively new. The use case listed in this study shows the initial investment and success of utilizing the big data platform to provide clinically focused NLP solutions. There are many opportunities to utilize this type of big data-empowered NLP solution. Adoption of big data technology is still in its infancy at the Mayo Clinic and has not penetrated different practice divisions across the Mayo Clinic. As successful projects leverage the information provided by NLP solutions, they can serve as models for future endeavors.
Conclusions
In this study, we demonstrated the benefits of a big data-empowered NLP computing infrastructure, for the processing and delivery of NLP solutions, which enable knowledge delivery in an LHS. We have clearly shown that using big data architecture significantly reduces the processing time of clinical narratives. It enables real-time NLP processing of clinical documents to deliver care recommendations for clinical practice. This is a significant step toward implementing an LHS. Big data computing paves way for building a robust and fault-tolerant NLP infrastructure.
Acknowledgments
We acknowledge Brian N. Brownlow for making the big data and data stage server available for conducting the experiments.
Footnotes
ACADEMIC EDITOR: John P. Pestian, Editor in Chief
PEER REVIEW: Five peer reviewers contributed to the peer review report. Reviewers’ reports totaled 1,136 words, excluding any confidential comments to the academic editor.
FUNDING: This work was supported in part by the National Institute of General Medical Sciences (NIGMS) R01 GM102282 and National Library of Medicine (NLM) R01 LM011934. The authors confirm that the funder had no influence over the study design, content of the article, or selection of this journal.
COMPETING INTERESTS: Authors disclose no potential conflicts of interest.
Paper subject to independent expert blind peer review. All editorial decisions made by independent academic editor. Upon submission manuscript was subject to anti-plagiarism scanning. Prior to publication all authors have given signed confirmation of agreement to article publication and compliance with all applicable ethical and legal requirements, including the accuracy of author and contributor information, disclosure of competing interests and funding sources, compliance with ethical requirements relating to human and animal study participants, and compliance with any copyright requirements of third parties. This journal is a member of the Committee on Publication Ethics (COPE).
Author Contributions
Involved in designing the experiments and interpretation of the results for use case: HL, RKE, VCK, SM, SS, JJP. Performed the big data implementation and experiments: VCK, JJP. Participated in the discussion of big data implementation: VCK, RKE, SM, JJP, SS, YW, DL, MMR, SPM, JLR, RC, JDB, HL. Contributed toward writing the article: VCK, RKE, SM, JJP, SS, YW, DL, MMR, SPM, JLR, RC, JDB, HL. All authors reviewed and approved of the final manuscript.
REFERENCES
- 1.Health Information Technology (HITECH) Act 2009. Index for Excerpts from the American Recovery and Reinvestment Act of 2009.
- 2.Friedman C, Rubin J, Brown J, et al. Toward a science of learning systems: a research agenda for the high-functioning Learning Health System. J Am Med Inform Assoc. 2015;22:43–50. doi: 10.1136/amiajnl-2014-002977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.The Foundation for Continuous Improvement in Health and Health Care . Digital Infrastructure for the Learning Health System: Institute of Medicine. 2011. [PubMed] [Google Scholar]
- 4.Fernandes L, O’Connor M, Weaver V. Big data, bigger outcomes: healthcare is embracing the big data movement, hoping to revolutionize HIM by distilling vast collection of data for specific analysis. J AHIMA. 2012;83(10):38–43. quiz 44. [PubMed] [Google Scholar]
- 5.Demner-Fushman D, Chapman W, McDonald CJ. What can natural language processing do for clinical decision support? J Biomed Inform. 2009;42(5):760–72. doi: 10.1016/j.jbi.2009.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cimino JJ, Bright TJ, Li J. Medication reconciliation using natural language processing and controlled terminologies. Stud Health Technol Inform. 2007;129(pt 1):679–83. [PubMed] [Google Scholar]
- 7.Uzuner O, Stubbs A. Practical applications for natural language processing in clinical research: the 2014 i2b2/UTHealth shared tasks. J Biomed Inform. 2015;58:S1–5. doi: 10.1016/j.jbi.2015.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Meystre SM, Savova GK, Kipper-Schuler KC, Hurdle JF. Extracting information from textual documents in the electronic health record: a review of recent research. Yearb Med Inform. 2008:128–44. [PubMed] [Google Scholar]
- 9.Divita G, Carter M, Redd A, et al. Scaling-up NLP pipelines to process large corpora of clinical notes. Methods Inf Med. 2015;54:548–52. doi: 10.3414/ME14-02-0018. [DOI] [PubMed] [Google Scholar]
- 10.Agerri R, Bermudez J, Rigau G. IXA pipeline: efficient and ready to use multilingual NLP tools; Language Resources and Evaluation Conference (LREC2014); 2014; Reykjavik, Iceland. [Google Scholar]
- 11.Ross MK, Wei W, Ohno-Machado L. Big data and the electronic health record. Yearb Med Inform. 2014;9(1):97–104. doi: 10.15265/IY-2014-0003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McGlynn EA, Lieu TA, Durham ML, et al. Developing a data infrastructure for a learning health system: the PORTAL network. J Am Med Inform Assoc. 2014;21:596–601. doi: 10.1136/amiajnl-2014-002746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mandl K, Kohane IS, McFadden D, et al. Scalable collaborative infrastructure for a learning healthcare system (SCILHS): architecture. J Am Med Inform Assoc. 2014;21:615–20. doi: 10.1136/amiajnl-2014-002727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Amin W, Tsui F, Borromeo C, et al. The PaTH Network Team. PaTH: towards a learning health system in the Mid-Atlantic region. J Am Med Inform Assoc. 2014;21:633–6. doi: 10.1136/amiajnl-2014-002759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Forrest CB, Margolis P, Bailey LC, et al. PEDSnet: a national pediatric learning health system. J Am Med Inform Assoc. 2014;21:602–6. doi: 10.1136/amiajnl-2014-002743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.PCORI PCORI Funding Announcement: Improving Infrastructure for Conducting Patient-Centered Outcomes Research. [Accessed December 22, 2014]. Available at: http://www.pcori.org/sites/default/files/PCORI-PFA-CDRN.pdf.
- 17.Friedman C, Hripcsak G, DuMouchel W, Johnson SB, Clayton PD. Natural language processing in an operational clinical information system. J Nat Lang Eng. 1995;1(1):83–108. [Google Scholar]
- 18.Friedman C, Alderson PO, Austin JH, Cimino JJ, Johnson SB. A general natural-language text processor for clinical radiology. J Am Med Inform Assoc. 1994;1(2):161–74. doi: 10.1136/jamia.1994.95236146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chapman WW, Nadkarni PM, Hirschman L, D’Avolio LW, Savova GK, Uzuner O. Overcoming barriers to NLP for clinical text: the role of shared tasks and the need for additional creative solutions. J Am Med Inform Assoc. 2011;18(5):540–3. doi: 10.1136/amiajnl-2011-000465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Uzuner O, Bodnari A, Shen S, Forbush T, Pestian J, South BR. Evaluating the state of the art in coreference resolution for electronic medical records. J Am Med Inform Assoc. 2012;19(5):786–91. doi: 10.1136/amiajnl-2011-000784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Uzuner Ö, Solti I, Cadag E. Extracting medication information from clinical text. J Am Med Inform Assoc. 2010;17:514–8. doi: 10.1136/jamia.2010.003947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Uzuner Ö, South BR, Shen S, DuVall SL. 2010 i2b2/VA challenge on concepts, assertions, and relations in clinical text. J Am Med Inform Assoc. 2011;18(5):552–6. doi: 10.1136/amiajnl-2011-000203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Uzuner O. Recognizing obesity and comorbidities in sparse data. J Am Med Inform Assoc. 2009;16:561–70. doi: 10.1197/jamia.M3115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Uzuner O, Goldstein I, Luo Y, Kohane I. Identifying patient smoking status from medical discharge records. J Am Med Inform Assoc. 2008;15(1):14–24. doi: 10.1197/jamia.M2408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sun W, Rumshisky A, Uzuner O. Annotating temporal information in clinical narratives. J Biomed Inform. 2013;46:S5–12. doi: 10.1016/j.jbi.2013.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ferrucci D, Brown E, Chu-Carroll J, et al. Building Watson: an overview of the DeepQA project. AI Magazine. 2010;31:59–79. [Google Scholar]
- 27.Moschitti A, Chu-Carroll J, Patwardhan S, Fan J, Riccardi G. Using syntactic and semantic structural kernels for classifying definition questions in Jeopardy. Emp Met Nat Lang Proc. 2011:712–24. [Google Scholar]
- 28.McCord MC, Murdock JW, Boguraev BK. Deep parsing in Watson. IBM J Res Dev. 2012;56(3.4):3:1–3:15. [Google Scholar]
- 29.Gondek D, Lally A, Kalyanpur A, et al. A framework for merging and ranking of answers in DeepQA. IBM J Res Dev. 2012;56(3.4):14:1–14:12. [Google Scholar]
- 30.Wang C, Mahadevan S. Multiscale manifold learning; Paper presented at: AAAI; July 14–8, 2013; Bellevue, WA. [Google Scholar]
- 31.Ferrucci DA. ACM SIGARCH Computer Architecture News. New York, NY: Jun, 2011. IBM’s Watson/DeepQA. [Google Scholar]
- 32.Kalyanpur A, Murdock JW, Fan J, Welty C. Leveraging community-built knowledge for type coercion in question answering; Semantic Web–ISWC; October 23–7, 2011; Berlin, Heidelberg: [Google Scholar]
- 33.Wang C, Fan J. Medical relation extraction with manifold models. ACL. 2014:828–38. [Google Scholar]
- 34.Cerner Corporation Make your Cerner EMR smarter, safer and easier to use. 2014 [Google Scholar]
- 35.Agerri R, Artola X, Beloki Z, Rigau G, Soroa A. Big data for natural language processing: a streaming approach. Knowledge-Based Syst. 2015;79:36–42. [Google Scholar]
- 36.Kurland LT, Molgaard CA. The patient record in epidemiology. Sci Am. 1981;245(4):54. doi: 10.1038/scientificamerican1081-54. [DOI] [PubMed] [Google Scholar]
- 37.Rhodes R. Healthy approach to data: IBM and Mayo Clinic team up to create massive patient database. IBM Systems Magazine. 2002 [Google Scholar]
- 38.Chute CG, Beck SA, Fisk TB, Mohr DN. The enterprise data trust at Mayo Clinic: a semantically integrated warehouse of biomedical data. J Am Med Inform Assoc. 2010;17(2):131–5. doi: 10.1136/jamia.2009.002691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Savova GK, Masanz JJ, Ogren PV, et al. Mayo Clinical text analysis and knowledge extraction system (cTAKES): architecture, component evaluation and applications. J Am Med Inform Assoc. 2010;17(5):507–13. doi: 10.1136/jamia.2009.001560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liu H, Bielinski SJ, Sohn S, et al. AMIA Summits Transl Sci Proc. San Francisco, CA: 2013. An information extraction framework for cohort identification using electronic health records. [PMC free article] [PubMed] [Google Scholar]
- 41.Sohn S, Clark C, Halgrim S, Murphy S, Chute C, Liu H. MedXN: an open source medication extraction and normalization tool for clinical text. J Am Med Inform Assoc. 2014;21(5):858–65. doi: 10.1136/amiajnl-2013-002190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sohn S, Wagholikar KB, Li D, et al. Comprehensive temporal information detection from clinical text: medical events, time, and TLINK identification. J Am Med Inform Assoc. 2013;20(5):836–42. doi: 10.1136/amiajnl-2013-001622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jonnalagadda SR, Li D, Sohn S, et al. Coreference analysis in clinical notes: a multipass sieve with alternate anaphora resolution modules. J Am Med Inform Assoc. 2012;19(5):867–74. doi: 10.1136/amiajnl-2011-000766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dean J, Ghemawat S. MapReduce: simplified data processing on large clusters. Commun ACM. 2008;51(1):107–13. [Google Scholar]
- 45.Chang F, Dean J, Ghemawat S, et al. Bigtable: A Distributed Storage System for Structured Data. ACM Transactions on Computer Systems (TOCS) 2008 Jun;26(2):1–26. [Google Scholar]
- 46.Li DC, Liu H, Chute CG, Jonnalagadda SR. Towards assigning references using semantic, journal and citation relevance; IEEE International Conference on Bioinformatics and Biomedicine (BIBM); 2013; Shanghai. [Google Scholar]
- 47.Chaudhry Rajeev, et al. Innovations in the delivery of primary care services using a software solution: the Mayo Clinics Generic Disease Management System. International Journal of Person Centered Medicine. 2012;2(3):361–7. [Google Scholar]