ABSTRACT
Danon disease is a rare, severe X-linked form of cardiomyopathy caused by deficiency of lysosome-associated membrane protein 2 (LAMP-2). Other clinical manifestations include skeletal myopathy, cognitive defects and visual problems. Although individuals with Danon disease have been clinically described since the early 1980s, the underlying molecular mechanisms involved in pathological progression remain poorly understood. LAMP-2 is known to be involved in autophagy, and a characteristic accumulation of autophagic vacuoles in the affected tissues further supports the idea that autophagy is disrupted in this disease. The LAMP2 gene is alternatively spliced to form three splice isoforms, which are thought to play different autophagy-related cellular roles. This Commentary explores findings from genetic, histological, functional and tissue expression studies that suggest that the specific loss of the LAMP-2B isoform, which is likely to be involved in macroautophagy, plays a crucial role in causing the Danon phenotype. We also compare findings from mouse and cellular models, which have allowed for further molecular characterization but have also shown phenotypic differences that warrant attention. Overall, there is a need to better functionally characterize the LAMP-2B isoform in order to rationally explore more effective therapeutic options for individuals with Danon disease.
KEY WORDS: Danon disease, Autophagy, Lysosome-associated membrane protein 2, LAMP-2, Cardiomyopathy, Medical genetics, Macroautophagy
Summary: Danon disease is a severe form of cardiomyopathy, and the molecular mechanisms involved in progression of the disease are poorly understood; however, perturbation of autophagy is likely to play a crucial role.
Introduction
Danon disease is a rare but severe X-linked form of cardiomyopathy that usually leads to profound hypertrophic cardiomyopathy (HCM; a thickening of the heart muscle) and causes death or a need for cardiac transplantation in males by the third decade (Fig. 1A) (D'souza et al., 2014; Taylor et al., 2007). When Danon disease was first described in 1981, ultrastructural analysis of muscle biopsies from two boys with the disease showed that their cells contained abundant glycogen particles, primarily within lysosomal vacuoles (Danon et al., 1981). Because of these observations, the disease was originally postulated to be due to a glycogen storage defect. However, although glycogen-containing autophagic vacuoles have continued to be reported in affected individuals (Cheng and Fang, 2012), genetic, histological and ultrastructural analyses have revealed that disruption of autophagy – a process by which cells disassemble dysfunctional or unneeded molecular and cellular components for reuse – is the probable underlying mechanism of Danon disease.
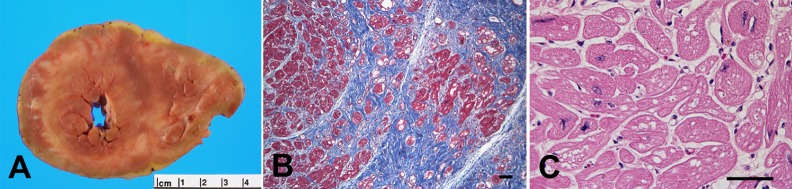
Fig. 1.
Explanted cardiac muscle from a 14-year-old male with Danon disease. Images show (A) a cross-section of explanted heart with biventricular hypertrophy and fibrosis, (B) a photomicrograph (with trichrome stain) of cardiomyocytes (red) and extensive fibrosis (blue) and (C) a photomicrograph (with hematoxylin and eosin stain) revealing extensive vacuolization. Scale bars: 100 μm. Reprinted from Taylor et al. (2007) with permission. Copyright 2007, Nature Publishing Group.
Danon disease is caused by mutations in the lysosome-associated membrane protein 2 (LAMP2) gene, usually leading to deficiency of the LAMP-2 protein (Eskelinen, 2005; Nishino et al., 2000). LAMP-2 is an abundant constituent of the membrane of the lysosome, which is a eukaryotic organelle containing digestive enzymes that break down its contents (Konecki et al., 1994). Unsurprisingly, based on its location, LAMP-2 is involved in autophagy, with its three different splice isoforms (LAMP-2A, LAMP-2B, and LAMP-2C) each playing different autophagy-related roles. There are three main types of autophagy: macroautophagy, chaperone-mediated autophagy (CMA) and microautophagy (Mrschtik and Ryan, 2015). The LAMP-2A isoform has been clearly shown to be involved in CMA (Eskelinen et al., 2005; Schneider and Cuervo, 2014), whereas the role of LAMP-2B has been characterized to a much lesser extent, but it is likely to be involved in macroautophagy (Bandyopadhyay et al., 2008; Tanaka et al., 2000; Nishino et al., 2000). Macroautophagy is the most prevalent form of autophagy and is thought to be the type of autophagy that is most affected in Danon disease (Endo et al., 2015; Tanaka et al., 2000).
In this Commentary, following a brief summary of clinical manifestations, the molecular and cellular aspects of Danon disease will be presented; these findings suggest that loss of the LAMP-2B isoform has a crucial role in causing Danon disease through mediating the disruption of macroautophagy and potentially other vesicle-trafficking processes. We will then discuss the characterizations conducted using mouse models separately owing to the phenotypic differences observed when these models are compared to the human disease. Based on these findings, we argue that a better understanding of the functions of the LAMP-2B isoform is needed before more effective therapeutics for Danon disease individuals can be developed.
Clinical manifestations of Danon disease
Males with Danon disease first show symptoms at 12 years of age, on average, and the majority (88%) develop HCM. By the time they are approximately 18 years old, the disease has led to cardiac transplantation or death (Boucek et al., 2011). Severe HCM, arrhythmias and a need for cardiac transplantation are the most morbid features. Although it is an X-linked disease, heterozygous females (i.e. carriers) are also affected but develop a more dilated cardiac phenotype and a progression of disease that occurs approximately 15 years later than in male counterparts (Prall et al., 2006; Boucek et al., 2011). Postmortem and explanted hearts display substantial fibrosis (D'souza et al., 2014) (Fig. 1B), and many affected individuals (males and females) develop arrhythmias that lead to sudden cardiac death. Specifically, ventricular (Hedberg Oldfors et al., 2015; Miani et al., 2012; Maron et al., 2009; Lobrinus et al., 2005) and atrial (Charron et al., 2004) arrhythmias have both been reported and are often severe, even in women (Miani et al., 2012). Additionally, individuals with Danon disease can also develop (usually relatively mild) skeletal myopathy (80–90% of males), primarily mild cognitive defects and learning disabilities (70–100% of males), and visual problems (69% of males) (Thiadens et al., 2012; Schorderet et al., 2007; Prall et al., 2006; D'souza et al., 2014). For a complete and recent review of clinical descriptions of affected individuals, see D'souza et al. (2014). For reviews discussing other diseases that share clinical similarities with Danon disease, including Pompe disease, Niemann-Pick type C and other lysosomal storage diseases, see Ruivo et al. (2009), D'souza et al. (2014) and Pastores and Hughes (2015). Although Danon disease shares clinical manifestations with other diseases, the primary distinguishing feature of Danon disease is a probable disruption of autophagy, as evidenced by the molecular phenotype, which will be explored next.
Molecular phenotype of Danon disease
Since its original description in 1981, subsequent findings have revealed that disrupted autophagy is the probable underlying mechanism of Danon disease, caused by mutations in the LAMP2 gene that usually lead to a loss of expression of LAMP-2. Given the roles of LAMP-2 in autophagy – which will be discussed in detail later – disruption of autophagy is not surprising. Accordingly, in cardiac tissue of individuals with Danon disease, dramatic increases in cytoplasmic vacuolation are apparent (Endo et al., 2015; He et al., 2014) (Fig. 1C). In these cells, expression of microtubule-associated protein 1 light chain 3 proteins (encoded by MAP1LC3A, MAP1LC3B, MAP1LC3C; collectively referred to here as LC3), markers of autophagy that are discussed more later, as well as ubiquitin, which comprises the ubiquitin–proteasome system, are increased in myocardium autophagic vacuoles, suggesting that the maturation and clearance of autophagosomes is stunted (Endo et al., 2015). The number of lipid droplets in these cells is also increased. In addition, the diseased cardiomyocytes are hypertrophic, and the myocardium undergoes degeneration, as evidenced by lipofuscin accumulation, which can be indicative of damage to cellular organelles, including lysosomes and disarray of myofibrils (He et al., 2014; Endo et al., 2015).
Recently, fibroblast biopsies taken from two males with Danon disease were reprogrammed into induced pluripotent stem cells (iPSCs) and differentiated into cardiomyocytes (iPSC-CMs); this has allowed for further phenotypic and functional studies of the disease on a controlled molecular level (Hashem et al., 2015). These derived cardiomyocytes displayed molecular and cellular phenotypes that were similar to those observed in Danon disease tissues and included accumulation of intracytoplasmic vacuoles, hypertrophy and impaired autophagic flux with a nearly complete deficiency of mature autophagic vacuoles, as assessed using an LC3 reporter (discussed further in the Autophagy section) (Hashem et al., 2015). Interestingly, both skin fibroblasts and iPSC-CMs from the Danon disease individuals had significant accumulations of early autophagic vesicles (compared to wild-type cells), and starvation (which induces autophagy) increased the number of such vesicles in the iPSC-CMs, but not in the fibroblasts; this might be related to the metabolically active nature of cardiac cells. Longer Ca2+-decay times were also observed in the Danon iPSC-CMs, which is consistent with a decline in systolic and diastolic function (seen in heart failure), and in other iPSC-CMs used to model heart failure phenotypes. Furthermore, the iPSC-CMs displayed mitochondrial defects, including increased levels of mitochondrial oxidative stress, and increased levels of apoptosis (Hashem et al., 2015). These effects could potentially be related because mitochondria normally produce reactive oxygen species (ROS) as oxidative phosphorylation by-products; therefore, dysfunctional mitochondria can produce excessive amounts of ROS and might be more prone to activate apoptosis (Shires and Gustafsson, 2015).
Histological examinations of skeletal muscle biopsies from Danon individuals, which have been more frequently reported owing to the relative ease of obtaining these tissues, have resulted in similar findings and are also indicative of autophagy disruption. For instance, in skeletal muscle biopsies, an accumulation of vacuolar structures are consistently found; these contain acetylcholine esterase, proteins of the sarcolemma (the muscle fiber cell membrane) and extracellular matrix proteins (typically those of the basal lamina, such as dystrophin, laminin and spectrin) (Dougu et al., 2009; Endo et al., 2015; Yang and Vatta, 2007), and are known as autophagic vacuoles with unique sarcolemmal features (AVSFs). AVSFs should be located near the peripheral sarcolemma if they are sarcolemma-derived but, because they are instead found scattered throughout the cytoplasm, it is thought that these AVSFs are generated through a poorly understood de novo biogenesis process (Endo et al., 2015). Disease-associated skeletal muscle vacuoles have also been reported to accumulate glycogen (Bertini et al., 2005; He et al., 2014; Yang and Vatta, 2007), degenerating mitochondria (Yang and Vatta, 2007), lipids (Bertini et al., 2005) and basophilic granules (implicating buildup of lysosomal organelles in the myofibers) (He et al., 2014; Sugie et al., 2005; Endo et al., 2015; Dougu et al., 2009), and to express increased levels of LC3 and ubiquitin (Endo et al., 2015).
Pathological studies of brain tissue from individuals with Danon disease have been limited, although an autopsy study on one individual with intellectual disability has been reported (Furuta et al., 2013). Here, histological and microscopic analysis of the brain tissue revealed similar findings to those reported for skeletal and cardiac muscle biopsies, namely the striking presence of lipofuscin-like granules containing glycogen, as well as lysosomal storage abnormalities as revealed by electron microscopy analyses and, moreover, a lack of normal autophagic vacuoles (Furuta et al., 2013; Endo et al., 2015). However, unlike the observations made in skeletal and cardiac muscle biopsies, the studied neurons did not appear to express LC3 and exhibited only low levels of ubiquitin. Clearly, further investigation is needed to better understand the pathological phenotype in the brain.
Some clinical studies have described the ophthalmic manifestations of individuals with Danon disease (Thiadens et al., 2012; Prall et al., 2006; Lobrinus et al., 2005). Both male individuals with Danon disease and female carriers have been found to have retinopathy, including peripheral pigmentary retinopathy (Prall et al., 2006) and peripheral ‘salt-and-pepper’ retinopathy (Schorderet et al., 2007; Thiadens et al., 2012), although sometimes the condition is milder in females. In one study, both enrolled males had a near-complete loss of pigmentation of the retinal pigment epithelium (RPE) in the macula (Prall et al., 2006), whereas a boy from another study had RPE hypopigmentation (Schorderet et al., 2007). Another study of a family found that two out of four male members with Danon disease developed cone-rod dystrophy, with thinning of the RPE and overlying photoreceptor layers (Thiadens et al., 2012). Photoreceptor cells (comprised of rods and cones) absorb photons of light through components called photoreceptor outer segments (POSs). Each day, at least 2000–4000 POSs are shed by photoreceptors and broken down by the underlying RPE layer through phagocytosis (Thiadens et al., 2012). Unlike autophagy, phagocytosis requires import of its cargo, but similar to autophagy, its cargo must be degraded through localization to, and fusion with, the lysosome (Nandrot, 2014). Some key phagocytic factors have been identified, including myosin VIIA – a mechanochemical protein thought to transport nascent POS-based vesicles (i.e. phagosomes) into the cell (Gibbs et al., 2003) – as well as proteins and proteases (e.g. βA3-crystallin, A1-crystallin and cathepsin D) that are involved in lysosomal degradation following fusion of the phagosome with the lysosome (Nandrot, 2014; Valapala et al., 2016). However, it is likely that other factors with an active role in retinal phagocytosis are yet to be identified, particularly because these molecular mechanisms have only been characterized using nocturnal rodent models, and phagocytotic cycles are different in diurnal animals, such as humans (Nandrot, 2014). Interestingly, disruption of retinal phagocytosis leads to lipofuscin accumulation (Nandrot, 2014), which is also seen in Danon individuals, as described above. LAMP-2 is highly expressed in RPE lysosomes (Schorderet et al., 2007), indicative of a role in the retinal phagocytotic process. Indeed, mutation of the LAMP2 gene could ultimately lead to the loss of RPE and photoreceptors, and thus to a loss of visual acuity (Thiadens et al., 2012).
LAMP-2 structure and isoforms
Understanding the structure of LAMP-2 and its splice isoforms is important for determining their varying roles in autophagy, and how these might be disrupted in Danon disease. The human LAMP-2 protein contains nine exons, with exons 1 to 8 and part of exon 9 making up the lysosome lumenal domain. In this large lumenal domain, four loops are predicted to form through disulfide bridges and flank a proline-rich ‘hinge’ region (Eskelinen et al., 2005). The hinge region acts as a linker for two similar glycosylated domains, each of which contains two of the disulfide bridges (Wilke et al., 2012). The lumenal domain of LAMP-2 is heavily glycosylated to avoid degradation due to the low pH of the lysosome (Mrschtik and Ryan, 2015). For additional details on the recently solved crystal structure of the LAMP-2 protein – with a focus on the lumenal domain – see Wilke et al. (2012). The remaining part of exon 9 comprises both a single transmembrane domain (24 amino acids long) and a short cytoplasmic tail (11 amino acids long) at the C-terminal end (Eskelinen et al., 2005; Konecki et al., 1994, 1995). The cytoplasmic tail is likely to function as a receptor for the uptake of proteins and, occasionally, other molecules (such as RNA and DNA) into the lysosome for their degradation (D'souza et al., 2014; Nishino et al., 2000). It is worth noting that although LAMP-1, which belongs to the same family as LAMP-2, shares 37% amino acid sequence homology with LAMP-2, these proteins have been found to be distinct and to have largely separate functions (Eskelinen, 2006). Overall, the LAMP1-knockout mouse phenotype is relatively mild (Eskelinen, 2006), and although there might be shared functions between LAMP-1 and LAMP-2, LAMP-1 levels have not been found to increase to compensate for blockage of CMA, which is the best characterized function of LAMP-2 (Massey et al., 2006), nor does transient expression of LAMP-1 alleviate cholesterol accumulation (Schneede et al., 2011). For a detailed comparison of LAMP-1 and LAMP-2 functions, see Eskelinen (2006).
As mentioned previously, the LAMP2 gene can be alternatively spliced to give rise to three splice isoforms, LAMP-2A, LAMP-2B and LAMP-2C. For a thorough review of the isoform nomenclature and standardization across different species, see Eskelinen et al. (2005). Exon 9 of LAMP-2 pre-messenger RNA (mRNA) is alternatively spliced to create the three splice isoforms, which are identical in their lysosome lumenal domains but differ in the transmembrane domain and cytoplasmic tail (Fig. 2C). LAMP-2A plays a role in CMA, whereas the other isoforms do not appear to be involved in CMA (Eskelinen et al., 2005). Studies suggest that LAMP-2B could instead be involved in macroautophagy, primarily based on the impaired fusion seen in individuals with mutations in LAMP-2B only (Bandyopadhyay et al., 2008; Tanaka et al., 2000; Nishino et al., 2000). Both CMA and macroautophagy will be discussed in detail later. LAMP-2C is less characterized but is thought to be primarily, and potentially solely, involved in the degradation of RNA and DNA (RNautophagy and DNautophagy, respectively) based on pull-down assays identifying interacting proteins as being RNA- or DNA-binding proteins (Fujiwara et al., 2013, 2014). Owing to these differences in function and tissue expression, which will be explored more in the following sections, LAMP-2A and LAMP-2B are thought to be more relevant to Danon disease and, consequently, these isoforms will receive more attention than LAMP-2C in this Commentary.
Fig. 2.
Amino acid sequence alignments of the LAMP-2A and LAMP-2B isoforms in human and mouse. Alignments show (A) human and mouse LAMP-2 (exons 1–8), (B) human and mouse LAMP-2A and LAMP-2B isoforms (exon 9 only), which are the LAMP-2 isoforms most relevant to Danon disease, and (C) human LAMP-2A and LAMP-2B isoforms (exon 9 only). Blue and yellow backgrounds indicate identical and similar amino acids, respectively. Gray backgrounds indicate cysteine residues thought to be involved in disulfide bridges. Black, red and green font colors indicate lumenal, transmembrane and cytoplasmic residues, respectively (Eskelinen et al., 2005). Amino acid sequences were obtained from Ensembl, and alignments were generated using the Bioinformatics Organization Multiple Align Show tool.
In their cytoplasmic tails, LAMP-2A and LAMP-2B share 27% (3 out of 11) identical amino acids, and an additional 27% (3 out of 11) of their amino acid residues are similar, with 45% (5 out of 11) of residues not showing any similarity. Such differences in the cytoplasmic tail might enable the isoforms to perform separate receptor-mediated functions (i.e. CMA and macroautophagy) and make this region, possibly, of most relevance to Danon disease (D'souza et al., 2014; Nishino et al., 2000).
Role of LAMP-2B in Danon disease
Although most LAMP2 mutations leading to Danon disease are predicted to result in deficiency of all three of the LAMP-2 isoforms, isoform-specific Danon disease mutations have only so far been found in the LAMP-2B isoform. This suggests that LAMP-2B deficiency is potentially sufficient (owing to the reporting of isoform-specific mutations) and necessary (all mutations affect LAMP-2B) to cause Danon disease (D'souza et al., 2014; Boucek et al., 2011; Nishino et al., 2000) (Table 1). For extensive lists of reported LAMP-2 mutations in individuals with Danon disease, see Cheng and Fang (2012), Dougu et al. (2009) and D'souza et al., (2014). Accordingly, deficiency of LAMP-2B alone has been found to cause ‘the full Danon syndrome’, as stated by the authors, including HCM, skeletal myopathy and the cognitive defects that have been detected in one individual (Nishino et al., 2000). In one of the individuals that we have studied, LAMP-2B deficiency was also found to result in a typical case of Danon disease, requiring cardiac transplantation (Boucek et al., 2011). However, in another individual, LAMP-2B deficiency alone was found to only cause a milder presentation Danon syndrome, with mild left ventricular diastolic dysfunction, limb-girdle muscle weakness and sub-clinical neuropathy (Hong et al., 2012). It is also worth noting that a LAMP-2B missense mutation that results in apparently normal levels (but irregular distribution) of LAMP-2B expression in skeletal muscle biopsies from multiple members of one family has been associated with different Danon-disease-like symptoms, including a potentially much milder cardiac phenotype (although there was sudden cardiac death at the age of 28 of one female family member), potentially more severe and progressive muscle weakness (which is uncommon in Danon disease individuals), decreased visual acuity and normal intellectual abilities (van der Kooi et al., 2008). Interestingly, upon ultrastructural examination, accumulation of autophagic vacuoles containing glycogen, lipid droplets and lipofuscin were also observed in this family (van der Kooi et al., 2008). It is possible that compared to loss of expression of all three isoforms, LAMP-2B deficiency or dysfunction alone might typically result in milder Danon-disease-like cardiac and cognitive phenotypes but still give rise to similar or more severe skeletal myopathy. This could be explained by the tissue-specific expression levels of the normal LAMP-2 isoforms, their roles in these tissues, and/or cross-talk and compensatory mechanisms occurring between the different forms of autophagy (Kaushik et al., 2008).
Table 1.
Evidence for the involvement of the LAMP-2A and LAMP-2B isoforms in the development of Danon disease
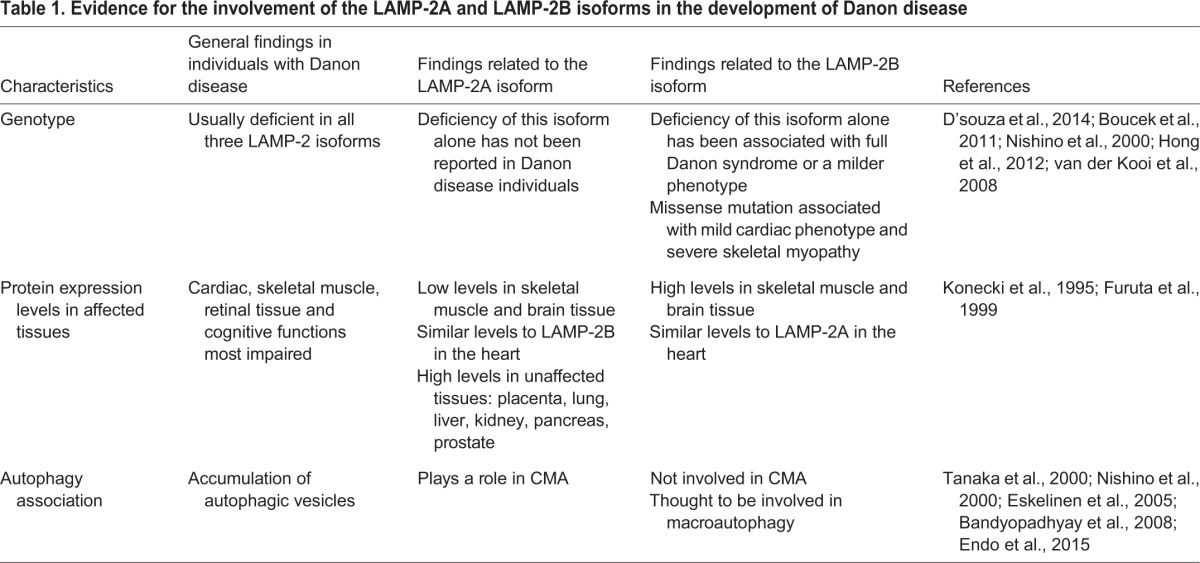
Supporting the idea of a key role of LAMP-2B in the causality of Danon disease is the tissue-specific expression of the different LAMP-2 isoforms; LAMP-2B appears to be more highly expressed in the most affected tissues compared to LAMP-2A and LAMP-2C under normal conditions (Table 1). Human LAMP-2B has specifically been found to be more abundantly expressed at the mRNA level in skeletal muscle and brain tissue than the LAMP-2A isoform, which is present at only low levels in these tissues (Konecki et al., 1995; Furuta et al., 1999; Rothaug et al., 2015). LAMP-2B is most highly expressed in skeletal muscle (Konecki et al., 1995), which could be the reason why the most striking disease symptom reported in members of a family with a LAMP-2B missense mutation is severe and progressive skeletal muscle weakness (van der Kooi et al., 2008). Human LAMP-2A, by contrast, is more ubiquitously expressed; it is expressed in placenta, lung, liver, kidney, pancreas and prostate tissues (Konecki et al., 1995; Furuta et al., 1999). Both LAMP-2A and LAMP-2B are thought to be expressed at similar levels in the human heart, although this is based on limited studies (Konecki et al., 1995; Yang and Vatta, 2007). Investigations in human tissues have not been reported; however, in 2013, a tissue expression study of LAMP-2C mRNA in mice found this isoform to be most highly expressed in brain tissue and present at lower levels in other tissues, including detectable levels in skeletal muscle and the heart (Fujiwara et al., 2013), although a later study detected no LAMP-2C mRNA in the mouse brain (Rothaug et al., 2015). In agreement with some of these findings, we have conducted RNA sequencing (RNA-Seq) analysis of normal human iPSC-CMs and normal explanted human hearts, and found that expression levels of the LAMP-2A and LAMP-2B isoforms are similar in these cells and tissues, with both isoforms significantly more highly expressed than LAMP-2C (unpublished data; T.J.R., M.E.S., L.M., M.R.G.T., Eric Adler, Sherin Hashem and Carmen Sucharov). It is possible that the relative expression levels of the isoforms are altered depending on certain metabolic conditions, such as starvation (which induces autophagy); this could affect the relevance of the different isoforms to the Danon disease phenotype. Additional expression studies have been conducted in mice, and these will be explored separately later owing to phenotypic differences observed between individuals with Danon disease and mouse models. The molecular phenotype of individuals with Danon disease described above, wherein macroautophagy specifically appears to be impacted, is also supportive of a key role for LAMP-2B (Table 1).
Autophagy
Macroautophagy is thought to be the type of autophagy most affected in Danon disease, owing to the accumulation of autophagic vacuoles in muscle biopsies and the idea that the disease is largely caused by loss of expression of the LAMP-2B isoform, which is thought to be involved in macroautophagy (Table 1) (Endo et al., 2015; Tanaka et al., 2000). In macroautophagy, cargo is engulfed by double-membraned vesicles called autophagosomes, which fuse with the lysosome to degrade their contents; this typically results in free fatty acids and amino acids for the cell to reuse (Nishida et al., 2015; Mrschtik and Ryan, 2015; Massey et al., 2008) (Fig. 3). Macroautophagy can be selective or non-selective. CMA, by contrast, is receptor mediated and generally a more selective process where vesicles are not needed for shuttling cargo to the lysosome (Schneider and Cuervo, 2014). The cytoplasmic tail of LAMP-2A is thought to function as a receptor in CMA (Endo et al., 2015). In the recently discovered degradation processes of RNautophagy and DNautophagy, which are mediated by LAMP-2C, this isoform acts as a receptor by interacting with RNA- or DNA-binding proteins, respectively, to allow RNA or DNA to be directly taken up into lysosomes (Fujiwara et al., 2013, 2014). Because nucleic acids have not been reported in the autophagic vacuoles that are characteristic of Danon disease, these degradation processes will not be discussed; please see Fujiwara et al. (2013, 2014) for details. Macroautophagy and CMA are explored in more detail below with a focus on macroautophagy because it appears to be the most affected in Danon disease (Endo et al., 2015; Tanaka et al., 2000).
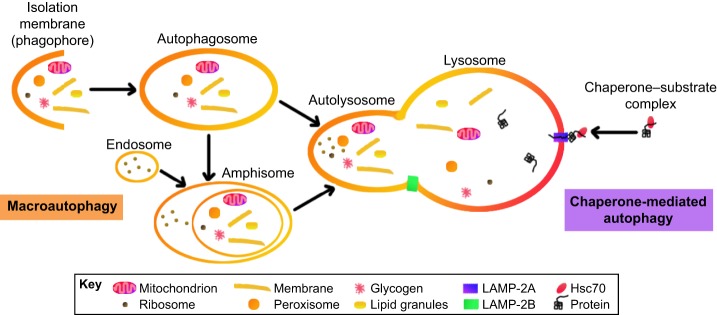
Fig. 3.
Mammalian pathways of macroautophagy and chaperone-mediated autophagy (CMA). In macroautophagy, a variety of cargo can be engulfed by an autophagosome, which fuses with a lysosome to form an autolysosome to then degrade its contents. Alternatively, an autophagosome can fuse with an endosome to form an amphisome, which then fuses with the lysosome. The processes of autophagosome and amphisome fusion with the lysosome are thought to require LAMP-2B, although any direct binding partners of LAMP-2B, which are important for these processes, are unknown. In CMA, target proteins are recognized and bound by heat shock cognate protein 70 (Hsc70; also known as HSPA8), which forms a chaperone–substrate complex that is transported to the lysosome, where interaction with LAMP-2A transport the target protein across the lysosome membrane.
In macroautophagy, initially a small flat membrane sack of cisterna, called an isolation membrane (or phagophore), forms in the cytoplasm (Hamasaki et al., 2013; Nishida et al., 2015). Several autophagy-related (Atg) proteins are involved in forming, expanding and elongating the isolation membrane. (For in-depth discussions of these Atg proteins, see Nemchenko et al., 2011; Mizushima et al., 2011; Wesselborg and Stork, 2015; Nishida et al., 2015.) The membrane becomes enclosed around its cargo to create a double-membraned autophagosome (Nishida et al., 2015; Nemchenko et al., 2011; Eskelinen et al., 2005). Material degraded through macroautophagy includes defective mitochondria, ribosomes, peroxisomes, endoplasmic reticulum and additional membrane structures, glycogen, lipid granules and other cytosolic aggregates (Kovsan et al., 2010; Kundu et al., 2008; Kuma and Mizushima, 2010; Schneider and Cuervo, 2014; Shires and Gustafsson, 2015; Nishida et al., 2015). The targeted material might be identified by cargo-recognizing proteins, which bind to the cargo and a membrane component of the developing autophagosome such as LC3 (Schneider and Cuervo, 2014; Wild et al., 2014). For example, PTEN-induced kinase 1 (PINK1), Parkin and p62 (also known as SQSTM1) are involved in identifying mitochondria for degradation, whereas p62 is additionally involved in recognizing lipid droplets (Jin and Youle, 2012; Schneider and Cuervo, 2014). Mutations in PINK1 and Parkin are associated with Parkinson's disease, in which cells exhibit defective mitochondrial turnover (Jin and Youle, 2012; Schneider and Cuervo, 2014). LC3, a molecule frequently used as a marker of autophagy (and the mammalian homolog of Atg8), has been thought to be required for expansion of the isolation membrane and autophagosome formation, although exceptions have been found (Kishi-Itakura et al., 2014; Nishida et al., 2009). During autophagy, LC3 is typically converted from the soluble form LC3-I to the lipidated form LC3-II (Klionsky et al., 2007). However, certain Atg-knockout cells (ATG5 and ATG3 knockouts) can accumulate expanded isolation membranes without converting LC3-I to LC3-II (Kishi-Itakura et al., 2014), and it has also been found that in some alternative macroautophagy processes the LC3-I to LC3-II conversion does not necessarily occur (Nishida et al., 2009). Additionally, findings arising from measurement of the conversion of LC3-I to LC3-II to determine autophagy activity and rates have been controversial (Klionsky et al., 2007), altogether making LC3 conversion a questionable marker of autophagy.
Following this, the outer membrane of an autophagosome fuses with a lysosome membrane in a LAMP-2-dependent manner to form an autolysosome (Endo et al., 2015); therein, the cargo of the inner membrane is degraded by the digestive enzymes of the lysosome, generating amino acids and lipids that are transported to the cytosol to be reused (Nishida et al., 2015). Alternatively, an autophagosome might fuse with an endosome (a vesicle formed from the plasma membrane of the cell) in a LAMP-2-dependent manner to form an amphisome, which then fuses with a lysosome. Because the LAMP-2B isoform is thought to be involved in macroautophagy, these lysosome fusion processes – which ensure proper autophagosome function and maturation – are likely to be perturbed upon loss of function of LAMP-2B. It is worth noting, however, that the presence of excessive autophagic vacuoles in individuals with Danon disease does not necessarily indicate that LAMP-2B deficiency is truly causal. Although LAMP-2 is primarily localized to the lysosome membrane, it is additionally found in the membranes of early endosomes, late endosomes and amphisomes, as well as in plasma membranes (Endo et al., 2015). The presence of LAMP-2 on the inner surface of lysosome membranes might also help to maintain an acidic environment in the lysosome and prevent lysosomal digestive enzymes from auto-digesting cellular cytoplasmic components (Endo et al., 2015; Saftig et al., 2001). Interestingly, through its cytoplasmic tail, LAMP-2B (but not LAMP-2A) has been found to interact with the transporter associated with antigen processing like (TAPL; also known as ABCB9) protein, which transports peptides in the cytosol to the lysosomal lumen (Demirel et al., 2012). Although this interaction does not appear to affect the subcellular localization or transportation of the peptides, LAMP-2B might instead act to increase the half-life of TAPL (Demirel et al., 2012). Other major players in the fusion of autophagosomes to lysosomes are the soluble NSF attachment protein receptor (SNARE) proteins; these are not known to interact with LAMP-2. In particular, the autophagosomal SNARE protein syntaxin 17 (STX17) has a key role in this fusion process and localizes to the outer membrane of autophagosomes (but is not found in isolation membranes); for details on these and other fusion factors whose discussion is beyond the scope of this Commentary, see Nishida et al. (2015), Itakura et al. (2012), Schneider and Cuervo (2014), Shen and Mizushima (2014). It is possible that the ubiquitin–proteasome system and microautophagy are upregulated to compensate for macroautophagy and/or CMA blockage, which could be a potential therapeutic option to explore for Danon disease individuals (Massey et al., 2008).
Like macroautophagy, CMA can be activated by cellular stress, such as oxidative stress, starvation and UV exposure, but CMA is much more selective than macroautophagy because it only targets specific proteins (Massey et al., 2008; Mrschtik and Ryan, 2015; Schneider and Cuervo, 2014). CMA uses the cytosolic chaperone heat shock cognate protein 70 (Hsc70; also known as HSPA8), which recognizes proteins containing a specific amino acid targeting motif (KFERQ-like). Hsc70 binds to these proteins, and the chaperone–substrate complex is transported to the lysosome, where it interacts with LAMP-2A on the lysosomal membrane to transport the target protein across the membrane and into the lysosomal lumen for degradation. For more details on CMA that are beyond the scope of this Commentary, see Kaushik and Cuervo (2012), Schneider and Cuervo (2014).
Mouse models of Danon disease – similarities and differences
A LAMP2 knockout mouse model of Danon disease has been used in multiple studies with the aim to better characterize the disease; however, although these mice display many features similar to those seen in humans with Danon disease, clear differences have also been reported. Based on amino acid sequence homology, the short cytoplasmic tail, which might be involved in receptor-mediated functions (i.e. CMA or macroautophagy), is highly conserved between humans and mice for the LAMP-2A and LAMP-2B isoforms [with 82% (9 out of 11) residues being identical for the LAMP-2A isoforms, and 91% (10 out of 11) residues being identical for the LAMP-2B isoforms] (Fig. 2B). However, the lumenal portion of LAMP-2 (exons 1–8) is less conserved between species [with 61% (226 out of 368) identical and 75% (276 out of 368) similar or identical residues, and 2% (10 out of 368) gaps] (Fig. 2A). Like humans, mice also have a LAMP-2C isoform (Eskelinen et al., 2005).
Phenotypically, both humans with Danon disease and Lamp2-knockout mice have increased mortality rates, with 50% of mice dying between the age of 20 and 40 days (Saftig et al., 2001; Tanaka et al., 2000); all mice show an accumulation of autophagic vacuoles in cardiac and skeletal muscle cells (Tanaka et al., 2000; Stypmann et al., 2006; Saftig et al., 2001; Yang and Vatta, 2007; Eskelinen, 2006). Also, similar to the human pathology, Lamp2-knockout mice develop severely reduced cardiac contractility, with a reduced ejection fraction and cardiac output, as well as increased ratios of heart weight to total body weight, important features for recapitulating the hypertrophic cardiac phenotype characteristic of Danon disease (Tanaka et al., 2000; Stypmann et al., 2006; Eskelinen, 2006). However, the cardiac phenotype in humans seems more severe overall; the vacuolation in human cardiomyocytes appears to be more pronounced than it is in Lamp2-knockout mice. This is relevant because cardiomyopathy is usually the major defining characteristic of the human disease (Saftig et al., 2001). By contrast, skeletal muscle physiologically appears to be affected to similar extents in the mouse model and human disease; myofibers in both systems are degenerative, with proximal muscles being more severely impacted than distal ones (Saftig et al., 2001). Similar to individuals with Danon disease who develop vision problems and mild cognitive defects, some neuropathological studies of Lamp2-knockout mice have also suggested that the central nervous system is impacted, specifically manifesting as impaired learning (likely to be caused by hippocampal dysfunction due to disrupted lysosomal activity) and motor deficits (Rothaug et al., 2015). However, earlier mouse studies have not reported abnormalities of the central nervous system (Saftig et al., 2001; Endo et al., 2015).
Beyond the cardiac and skeletal muscle phenotypes, there are potential differences in other organs affected. Lamp2-knockout mice have been reported to have excessive accumulations of autophagic vacuoles in organs that are not typically affected in individuals with Danon disease, specifically the liver, pancreas, spleen, thymus and kidneys (Saftig et al., 2001; Tanaka et al., 2000; Yang and Vatta, 2007). Lamp2-knockout mice also display periodontitis due to impaired neutrophils being unable to effectively eliminate bacterial pathogens, which is likely to be caused by reduced fusion of late endosomes and/or lysosomes with phagosomes (Beersten et al., 2007). Although in some Danon disease individuals, increases in the activity of liver enzymes and autophagosome accumulations in hepatocytes have been reported, the autophagic function of hepatocytes is much more severely impacted in mice (Saftig et al., 2001; Yang and Vatta, 2007; Tanaka et al., 2000). Overall, the mouse model might represent a more severe pathology than that seen in individuals with Danon disease in terms of the number of different organs affected, although the cardiac phenotype in humans appears to be more severe, which is noteworthy because this is the primary cause of mortality in people (Saftig et al., 2001; Endo et al., 2015). However, it has been proposed that defects in other tissues of individuals with Danon disease might have been overlooked as the focus was on examining the severe cardiac dysfunction (Endo et al., 2015).
In terms of molecular and cellular characterization, affected tissues of the Lamp2-knockout mice share many similarities with those of individuals with Danon disease, and examinations of cell lines derived from mice have afforded insights into the molecular pathogenic mechanisms of Danon disease. As seen in humans, glycogen accumulates within the affected mouse model cells, both inside and outside of autophagic vacuoles (Tanaka et al., 2000). Affected mouse cardiac cells have been reported to frequently contain a single mitochondrion in their autophagic vacuoles (Eskelinen, 2006), which reflect the dysfunctional mitochondrial behavior observed in human iPSC-CMs derived from individuals with Danon disease (Hashem et al., 2015). In the accumulated autophagosomes in pathogenic mouse hepatocytes, fusion between the autophagosomes and lysosomes appears to be impaired, whereas fusion between endosomes and autophagosomes does not appear to be altered (Eskelinen, 2005; Tanaka et al., 2000). Of note, Lamp2-deficient mice accumulate cholesterol in their liver (Schneede et al., 2011) and brain (Rothaug et al., 2015). This cholesterol defect has been further characterized in Lamp2-deficient mouse embryonic fibroblasts (MEFs), which accumulate unesterified cholesterol in late endosomes and/or lysosomes (Eskelinen et al., 2004; Eskelinen, 2006). This accumulation could be rescued by the lumenal domain and membrane-proximal part of LAMP-2 (in a mechanism independent of CMA), suggesting that these regions of LAMP-2 might have a crucial role in the transport of unesterified cholesterol. Specifically, unesterified cholesterol is generated by endogenous synthesis in the cytosol or through low-density lipoprotein (LDL) uptake, and becomes esterified in the endoplasmic reticulum. LAMP-2 might be important for cholesterol transportation from the cytosol to the endoplasmic reticulum (Schneede et al., 2011).
Concluding remarks
Although the progression of Danon disease in individuals has become better characterized clinically, our understanding of the underlying pathological molecular mechanisms remains limited. In particular, improving our knowledge of the role of LAMP-2 in macroautophagy, and potentially in other intracellular vesicle transport mechanisms that the LAMP-2B isoform might be involved in, might help us to better understand the disease and, consequently, to develop more targeted and effective treatment plans. Lamp2-deficient mice have been valuable tools in investigating this disease, although as discussed here, caution should be taken when drawing parallels between these models and the human pathology because there are clear differences, such as a potentially more severe cardiac phenotype in humans. The recent development of human-derived iPSC-CMs offers a promising way to investigate the molecular mechanisms under controlled conditions, although this cell-based system is clearly limited in recapitulating whole-animal physiological conditions. Moving forward, researchers are likely to need to utilize both animal and cellular models in order to best understand the molecular aspects of Danon disease and rationally develop effective treatments based on the knowledge gained.
Acknowledgements
The authors would like to thank Andrew J. Bonham (Metropolitan State University of Denver, Denver, CO), for critical reading of the manuscript. The authors would like to also thank Carmen C. Sucharov (University of Colorado Anschutz Medical Campus, Aurora, CO) and Eric D. Adler and Sherin I. Hashem (University of California San Diego, San Diego, CA) for their unpublished data contributions.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Funding
Supported by National Institutes of Health [grant number 1R01HL109209-01A1]; and a Trans-Atlantic Network of Excellence grant from Fondation Leducq (Leducq Foundation) [grant number 14-CVD 03]. Deposited in PMC for release after 12 months.
References
- Bandyopadhyay U., Kaushik S., Varticovski L. and Cuervo A. M. (2008). The chaperone-mediated autophagy receptor organizes in dynamic protein complexes at the lysosomal membrane. Mol. Cell. Biol. 28, 5747-5763. 10.1128/MCB.02070-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beersten W., Willenborg M., Everts V., Zirogianni A., Podschun R., Schroder B., Eskelinen E.-L. and Saftig P. (2007). Impaired phagosomal maturation in neutrophils leads to periodontitis in lysosomal-associated membrane protein-2 knockout mice. J. Immunol. 180, 475-482. [DOI] [PubMed] [Google Scholar]
- Bertini E., Donati M. A., Broda P., Cassandrini D., Petrini S., Dionisi-Vici C., Ballerini L., D'Amico A., Pasquini E., Minetti C. et al. (2005). Phenotypic heterogeneity in two unrelated Danon patients associated with the same LAMP-2 gene mutation. Neuropediatrics 36, 309-313. 10.1055/s-2005-872844 [DOI] [PubMed] [Google Scholar]
- Boucek D., Jirikowic J. and Taylor M. (2011). Natural history of Danon disease. Gen. Med. Off. J. Am. Coll. Med. Gen. 13, 563-568. 10.1097/gim.0b013e31820ad795 [DOI] [PubMed] [Google Scholar]
- Charron P., Villard E., Sebillon P., Laforet P., Maisonobe T., Duboscq-Bidot L., Romero N., Drouin-Garraud V., Frebourg T., Richard P. et al. (2004). Danon's disease as a cause of hypertrophic cardiomyopathy: a systematic survey. Heart 90, 842-846. 10.1136/hrt.2003.029504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Z. and Fang Q. (2012). Danon disease: focusing on heart. J. Hum. Genet. 57, 407-410. 10.1038/jhg.2012.72 [DOI] [PubMed] [Google Scholar]
- Danon M. J., Oh S. J., DiMauro S., Manaligod J. R., Eastwood A., Naidu S. and Schliselfeld L. H. (1981). Lysosomal glycogen storage disease with normal acid maltase. Neurology 31, 51-51 10.1212/WNL.31.1.51 [DOI] [PubMed] [Google Scholar]
- Demirel O., Jan I., Wolters D., Blanz J., Saftig P., Tempe R. and Abele R. (2012). The lysosomal polypeptide transporter TAPL is stabilized by interaction with LAMP-1 and LAMP-2. J. Cell Sci. 125, 4230-4240. 10.1242/jcs.087346 [DOI] [PubMed] [Google Scholar]
- Dougu N., Joho S., Shan L., Shida T., Matsuki A., Uese K., Hirono K., Ichida F., Tanaka K., Nishino I. et al. (2009). Novel LAMP-2 mutation in a family with Danon disease presenting with hypertrophic cardiomyopathy. Circ. J. Off. J. Jpn. Circ. Soc. 73, 376-380. 10.1253/circj.CJ-08-0241 [DOI] [PubMed] [Google Scholar]
- D'souza R. S., Levandowski C., Slavov D., Graw S. L., Allen L. A., Adler E., Mestroni L. and Taylor M. R. G. (2014). Danon disease: clinical features, evaluation, and management. Circ. Heart Fail. 7, 843-849. 10.1161/CIRCHEARTFAILURE.114.001105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endo Y., Furuta A. and Nishino I. (2015). Danon disease: a phenotypic expression of LAMP-2 deficiency. Acta Neuropathol. 129, 391-398. 10.1007/s00401-015-1385-4 [DOI] [PubMed] [Google Scholar]
- Eskelinen E.-L. (2005). Maturation of autophagic vacuoles in mammalian cells. Autophagy 1, 1-10. 10.4161/auto.1.1.1270 [DOI] [PubMed] [Google Scholar]
- Eskelinen E.-L. (2006). Roles of LAMP-1 and LAMP-2 in lysosome biogenesis and autophagy. Mol. Asp. Med. 27, 495-502. 10.1016/j.mam.2006.08.005 [DOI] [PubMed] [Google Scholar]
- Eskelinen E.-L., Schmidt C. K., Neu S., Willenborg M., Fuertes G., Salvador N., Tanaka Y., Lullmann-Rauch R., Hartmann D., Heeren J. et al. (2004). Disturbed cholesterol traffic but normal proteolytic function in LAMP-1/LAMP-2 double-deficient fibroblasts. Mol. Biol. Cell 15, 3132-3145. 10.1091/mbc.E04-02-0103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eskelinen E.-L., Cuervo A. M., Taylor M. R. G., Nishino I., Blum J. S., Dice J. F., Sandoval I. V., Lippincott-Schwartz J., August J. T. and Saftig P. (2005). Unifying nomenclature for the isoforms of the lysosomal membrane protein LAMP-2. Traffic 6, 1058-1061. 10.1111/j.1600-0854.2005.00337.x [DOI] [PubMed] [Google Scholar]
- Fujiwara Y., Furuta A., Kikuchi H., Aizawa S., Hatanaka Y., Konya C., Uchida K., Yoshimura A., Tamai Y., Wada K. et al. (2013). Discovery of a novel type of autophagy targeting RNA. Autophagy 9, 403-409. 10.4161/auto.23002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujiwara Y., Kikuchi H., Aizawa S., Furuta A., Hatanaka Y., Konya C., Uchida K., Wada K. and Kabuta T. (2014). Direct uptake and degradation of DNA by lysosomes. Autophagy 9, 1167-1171. 10.4161/auto.24880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furuta K., Yang X.-L., Chen J.-S., Hamilton S. R. and August J. T. (1999). Differential expression of the lysosome-associated membrane proteins in normal human tissues. Arch. Biochem. Biophys. 365, 75-82. 10.1006/abbi.1999.1147 [DOI] [PubMed] [Google Scholar]
- Furuta A., Wakabayashi K., Haratake J., Kikuchi H., Kabuta T., Mori F., Tokonami F., Katsumi Y., Tanioka F., Uchiyama Y. et al. (2013). Lysosomal storage and advanced senescence in the brain of LAMP-2-deficient Danon disease. Acta Neuropathol. 125, 459-461. 10.1007/s00401-012-1075-4 [DOI] [PubMed] [Google Scholar]
- Gibbs D., Kitamoto J. and Williams D. S. (2003). Abnormal phagocytosis by retinal pigmented epithelium that lacks myosin VIIa, the Usher syndrome 1B protein. Proc. Natl. Acad. Sci. USA 100, 6481-6486. 10.1073/pnas.1130432100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamasaki M., Furuta N., Matsuda A., Nezu A., Yamamoto A., Fujita N., Oomori H., Noda T., Haraguchi T., Hiraoka Y. et al. (2013). Autophagosomes form at ER-mitochondria contact sites. Nature 495, 389-393. 10.1038/nature11910 [DOI] [PubMed] [Google Scholar]
- Hashem S. I., Perry C. N., Bauer M., Han S., Clegg S. D., Ouyang K., Deacon D. C., Spinharney M., Panopoulos A. D., Izpisua Belmonte J. C. et al. (2015). Brief report: oxidative stress mediates cardiomyocyte apoptosis in a human model of Danon disease and heart failure. Stem Cells 33, 2343-2350. 10.1002/stem.2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He J., Wang Y. and Jiang T. (2014). Danon disease. Herz 39, 877-879. 10.1007/s00059-013-3900-5 [DOI] [PubMed] [Google Scholar]
- Hedberg Oldfors C., Máthé G., Thomson K., Tulinius M., Karason K., Östman-Smith I. and Oldfors A. (2015). Early onset cardiomyopathy in females with Danon disease. Neuromusc. Disord. 25, 493-501. 10.1016/j.nmd.2015.03.005 [DOI] [PubMed] [Google Scholar]
- Hong D., Shi Z., Zhang W., Wang Z. and Yuan Y. (2012). Danon disease caused by two novel mutations of the LAMP2 gene: implications for two ends of the clinical spectrum. Clin. Neuropathol. 31, 224-231. 10.5414/NP300465 [DOI] [PubMed] [Google Scholar]
- Itakura E., Kishi-Itakura C. and Mizushima N. (2012). The hairpin-type tail-anchored SNARE syntaxin 17 targets to autophagosomes for fusion with endosomes/lysosomes. Cell 151, 1256-1269. 10.1016/j.cell.2012.11.001 [DOI] [PubMed] [Google Scholar]
- Jin S. M. and Youle R. J. (2012). PINK1- and Parkin-mediated mitophagy at a glance. J. Cell Sci. 125, 795-799. 10.1242/jcs.093849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaushik S. and Cuervo A. M. (2012). Chaperone-mediated autophagy: a unique way to enter the lysosome world. Trends Cell Biol. 22, 407-417. 10.1016/j.tcb.2012.05.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaushik S., Massey A. C., Mizushima N. and Cuervo A. M. (2008). Constitutive activation of chaperone-mediated autophagy in cells with impaired macroautophagy. Mol. Biol. Cell 19, 2179-2192. 10.1091/mbc.E07-11-1155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kishi-Itakura C., Koyama-Honda I., Itakura E. and Mizushima N. (2014). Ultrastructural analysis of autophagosome organization using mammalian autophagy-deficient cells. J. Cell. Sci. 127, 4089-4102. 10.1242/jcs.156034 [DOI] [PubMed] [Google Scholar]
- Klionsky D. J., Cuvero A. M. and Seglen P. O. (2007). Methods for monitoring autophagy from yeast to humans. Autophagy 3, 181-206. 10.4161/auto.3678 [DOI] [PubMed] [Google Scholar]
- Konecki D. S., Foetisch K., Schlotter M. and Lichter-Konecki U. (1994). Complete cDNA sequence of human lysosome-associated membrane protein-2. Biochem. Biophys. Res. Comm. 205, 1-5. 10.1006/bbrc.1994.2620 [DOI] [PubMed] [Google Scholar]
- Konecki D. S., Foetisch K., Zimmer K. P., Schlotter M. and Lichter-Konecki U. (1995). An alternatively spliced form of the human lysosome-associated membrane protein-2 gene is expressed in a tissue-specific manner. Biochem. Biophys. Res. Comm. 215, 757-767. 10.1006/bbrc.1995.2528 [DOI] [PubMed] [Google Scholar]
- Kovsan J., Bashan N., Greenberg A. S. and Rudich A. (2010). Potential role of autophagy in modulation of lipid metabolism. Am. J. Physiol. Endocrinol. Metab. 298, E1-E7. 10.1152/ajpendo.00562.2009 [DOI] [PubMed] [Google Scholar]
- Kuma A. and Mizushima N. (2010). Physiological role of autophagy as an intracellular recycling system: with an emphasis on nutrient metabolism. Sem. Cell Dev. Biol. 21, 683-690. 10.1016/j.semcdb.2010.03.002 [DOI] [PubMed] [Google Scholar]
- Kundu M., Lindsten T., Yang C.-Y., Wu J., Zhao F., Zhang J., Selak M. A., Ney P. A. and Thompson C. B. (2008). Ulk1 plays a critical role in the autophagic clearance of mitochondria and ribosomes during reticulocyte maturation. Blood 112, 1493-1502. 10.1182/blood-2008-02-137398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lobrinus J. A., Schorderet D. F., Payot M., Jeanrenaud X., Bottani A., Superti-Furga A., Schlaepfer J., Fromer M. and Jeannet P.-Y. (2005). Morphological, clinical and genetic aspects in a family with a novel LAMP-2 gene mutation (Danon disease). Neuromuscul. Disord. 15, 293-298. 10.1016/j.nmd.2004.12.007 [DOI] [PubMed] [Google Scholar]
- Maron B. J., Roberts W. C., Arad M., Haas T. S., Spirito P., Wright G. B., Almquist A. K., Baffa J. M., Saul J. P., Ho C. Y. et al. (2009). Clinical outcome and phenotypic expression in LAMP2 Cardiomyopathy. JAMA 301, 1253-1259. 10.1001/jama.2009.371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massey A. C., Kaushik S., Sovak G., Kiffin R. and Cuervo A. M. (2006). Consequences of the selective blockage of chaperone-mediated autophagy. Proc. Natl. Acad. Sci. USA 103, 5805-5810. 10.1073/pnas.0507436103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massey A. C., Follenzi A., Kiffin R., Zhang C. and Cuervo A. M. (2008). Early cellular changes after blockage of chaperone-mediated autophagy. Autophagy 4, 442-456. 10.4161/auto.5654 [DOI] [PubMed] [Google Scholar]
- Miani D., Taylor M., Mestroni L., D'Aurizio F., Finato N., Fanin M., Brigido S. and Proclemer A. (2012). Sudden death associated with danon disease in women. Am. J. Cardiol. 109, 406-411. 10.1016/j.amjcard.2011.09.024 [DOI] [PubMed] [Google Scholar]
- Mizushima N., Yoshimori T. and Ohsumi Y. (2011). The role of ATG proteins in autophagosome formation. Annu. Rev. Cell Dev. Biol. 27, 107-132. 10.1146/annurev-cellbio-092910-154005 [DOI] [PubMed] [Google Scholar]
- Mrschtik M. and Ryan K. M. (2015). Lysosomal proteins in cell death and autophagy. FEBS J. 282, 1858-1870. 10.1111/febs.13253 [DOI] [PubMed] [Google Scholar]
- Nandrot E. F. (2014). Animal models, in “The Quest to Decipher RPE Phagocytosis”. In Retinal Degenerative Diseases (ed. Ash J. D., Grimm C., Hollyfield J. G., Anderson R. E., LaVail M. M. and Rickman C. B.), pp. 77-83. New York: Springer New York. [DOI] [PubMed] [Google Scholar]
- Nemchenko A., Chiong M., Turer A., Lavandero S. and Hill J. A. (2011). Autophagy as a therapeutic target in cardiovascular disease. J. Mol. Cell. Cardiol. 51, 584-593. 10.1016/j.yjmcc.2011.06.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishida Y., Arakawa S., Fujitani K., Yamaguchi H., Mizuta T., Kanaseki T., Komatsu M., Otsu K., Tsujimoto Y. and Shimizu S. (2009). Discovery of Atg5/Atg7-independent alternative macroautophagy. Nature 461, 654-658. 10.1038/nature08455 [DOI] [PubMed] [Google Scholar]
- Nishida K., Kyoi S., Yamaguchi O., Sadoshima J. and Otsu K. (2015). The role of autophagy in the heart. Cell Death Differ. 16, 31-38. 10.1038/cdd.2008.163 [DOI] [PubMed] [Google Scholar]
- Nishino I., Fu J., Tanji K., Yamada T., Shimojo S., Koori T., Mora M., Riggs J. E., Oh S. J., Koga Y. et al. (2000). Primary LAMP-2 deficiency causes X-linked vacuolar cardiomyopathy and myopathy (Danon disease). Nature 406, 906-910. 10.1038/35022604 [DOI] [PubMed] [Google Scholar]
- Pastores G. M. and Hughes D. A. (2015). Non-neuropathic lysosomal storage disorders: Disease spectrum and treatments. Best Pract. Res. Cl. En. 29, 173-182. 10.1016/j.beem.2014.08.005 [DOI] [PubMed] [Google Scholar]
- Prall F. R., Drack A., Taylor M., Ku L., Olson J. L., Gregory D., Mestroni L. and Mandava N. (2006). Ophthalmic manifestations of Danon disease. Ophthalmology 113, 1010-1013. 10.1016/j.ophtha.2006.02.030 [DOI] [PubMed] [Google Scholar]
- Rothaug M., Stroobants S., Schweizer M., Peters J., Zunke F., Allerding M., D'Hooge R., Saftig P. and Blanz J. (2015). LAMP-2 deficiency leads to hippocampal dysfunction but normal clearance of neuronal substrates of chaperone-mediated autophagy in a mouse model for Danon disease. Acta Neuropathol. Commun. 3, 6 10.1186/s40478-014-0182-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruivo R., Anne C., Sagne C. and Gasnier B. (2009). Molecular and cellular basis of lysosomal transmembrane protein dysfunction. BBA-Mol. Cell Res. 1793, 636-649 10.1016/j.bbamcr.2008.12.008 [DOI] [PubMed] [Google Scholar]
- Saftig P., von Figura K., Tanaka Y. and Lüllmann-Rauch R. (2001). Disease model: LAMP-2 enlightens Danon disease. Trends Mol. Med. 7, 37-39. 10.1016/S1471-4914(00)01868-2 [DOI] [PubMed] [Google Scholar]
- Schneede A., Schmidt C. K., Hölttä-Vuori M., Heeren J., Willenborg M., Blanz J., Domanskyy M., Breiden B., Brodesser S., Landgrebe J. et al. (2011). Role for LAMP-2 in endosomal cholesterol transport. J. Cell. Mol. Med. 15, 280-295. 10.1111/j.1582-4934.2009.00973.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider J. L. and Cuervo A. M. (2014). Autophagy and human disease: emerging themes. Curr. Opin. Genet. Dev. 26, 16-23. 10.1016/j.gde.2014.04.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schorderet D. F., Cottet S., Lobrinus J. A., Borruat F.-X., Balmer A. and Munier F. L. (2007). Retinopathy in Danon disease. JAMA Ophthal. 125, 231-236. 10.1001/archopht.125.2.231 [DOI] [PubMed] [Google Scholar]
- Shen H.-M. and Mizushima N. (2014). At the end of the autophagic road: an emerging understanding of lysosomal functions in autophagy. Trends Biochem. Sci. 39, 61-71. 10.1016/j.tibs.2013.12.001 [DOI] [PubMed] [Google Scholar]
- Shires S. E. and Gustafsson A. B. (2015). Mitophagy and heart failure. J. Mol. Med. 93, 253-262. 10.1007/s00109-015-1254-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stypmann J., Janssen P. M. L., Prestle J., Engelen M. A., Kögler H., Lüllmann-Rauch R., Eckardt L., von Figura K., Langrebe J., Mleczko A. et al. (2006). LAMP-2 deficient mice show depressed cardiac contractile function without significant changes in calcium handling. Basic Res. Cardiol. 101, 281-291. 10.1007/s00395-006-0591-6 [DOI] [PubMed] [Google Scholar]
- Sugie K., Noguchi S., Kozuka Y., Arikawa-Hirasawa E., Tanaka M., Yan C., Saftig P., von Figura K., Hirano M., Ueno S. et al. (2005). Autophagic vacuoles with sarcolemmal features delineate Danon disease and related myopathies. J. Neuropathol. Exp. Neurol. 64, 513-522. 10.1093/jnen/64.6.513 [DOI] [PubMed] [Google Scholar]
- Tanaka Y., Guhde G., Suter A., Eskelinen E.-L., Hartmann D., Lullmann-Rauch R., Janssen P. M. L., Blanz J., von Figura K. and Saftig P. (2000). Accumulation of autophagic vacuoles and cardiomyopathy in LAMP-2-deficient mice. Nature 406, 902-906. 10.1038/35022595 [DOI] [PubMed] [Google Scholar]
- Taylor M. R. G., Ku L., Slavov D., Cavanaugh J., Boucek M., Zhu X., Graw S., Carniel E., Barnes C., Quan D. et al. (2007). Danon disease presenting with dilated cardiomyopathy and a complex phenotype. J. Hum. Gen. 52, 830-835. 10.1007/s10038-007-0184-8 [DOI] [PubMed] [Google Scholar]
- Thiadens A. A. H. J., Slingerland N. W. R., Florijn R. J., Visser G. H., Riemslag F. C. and Klaver C. C. W. (2012). Cone-rod dystrophy can be a manifestation of Danon disease. Graefes Arch. Clin. Exp. Ophthalmol. 250, 769-774. 10.1007/s00417-011-1857-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valapala M., Sergeev U., Wawrousek E., Hose E., Zigler J. S. and Sinha D. (2016). Modulation of V-ATPase by βA3/A1-Crystallin in retinal pigment epithelial cells. Adv. Exp. Med. Biol. 854, 779-784. 10.1007/978-3-319-17121-0_104 [DOI] [PubMed] [Google Scholar]
- Van der Kooi A. J., van Langen I. M., Aronica E., van Doorn P. A., Wokke J. H. J., Brusse E., Langerhorst C. T., Bergin P., Dekker L. R. C., Lekanne dit Deprez R. H. et al. (2008). Extension of the clinical spectrum of Danon disease. Neurology 70, 1358-1359. 10.1212/01.wnl.0000309219.61785.b3 [DOI] [PubMed] [Google Scholar]
- Wesselborg S. and Stork B. (2015). Autophagy signal transduction by ATG proteins: from hierarchies to networks. Cell. Mol. Life Sci. 72, 4721-4757. 10.1007/s00018-015-2034-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wild P., McEwan D. G. and Dikic I. (2014). The LC3 interactome at a glance. J. Cell Sci. 127, 3-9. 10.1242/jcs.140426 [DOI] [PubMed] [Google Scholar]
- Wilke S., Krausze J. and Büssow K. (2012). Crystal structure of the conserved domain of the DC lysosomal associated membrane protein: implications for the lysosomal glycocalyx. BMC Biol. 10, 62 10.1186/1741-7007-10-62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z. and Vatta M. (2007). Danon disease as a cause of autophagic vacuolar myopathy. Congenit. Heart Dis. 2, 404-409. 10.1111/j.1747-0803.2007.00132.x [DOI] [PubMed] [Google Scholar]