Abstract
Recent radiations are important to evolutionary biologists, because they provide an opportunity to study the mechanisms that link micro- and macroevolution. The role of ecological speciation during adaptive radiation has been intensively studied, but radiations can arise from a diversity of evolutionary processes; in particular, on large continental landmasses where allopatric speciation might frequently precede ecological differentiation. It is therefore important to establish a phylogenetic and ecological framework for recent continental-scale radiations that are species-rich and ecologically diverse. Here, we use a genomic (approx. 1 200 loci, exon capture) approach to fit branch lengths on a summary-coalescent species tree and generate a time-calibrated phylogeny for a recent and ecologically diverse radiation of Australian scincid lizards; the genus Cryptoblepharus. We then combine the phylogeny with a comprehensive phenotypic dataset for over 800 individuals across the 26 species, and use comparative methods to test whether habitat specialization can explain current patterns of phenotypic variation in ecologically relevant traits. We find significant differences in morphology between species that occur in distinct environments and convergence in ecomorphology with repeated habitat shifts across the continent. These results suggest that isolated analogous habitats have provided parallel ecological opportunity and have repeatedly promoted adaptive diversification. By contrast, speciation processes within the same habitat have resulted in distinct lineages with relatively limited morphological variation. Overall, our study illustrates how alternative diversification processes might have jointly stimulated species proliferation across the continent and generated a remarkably diverse group of Australian lizards.
Keywords: ecomorphology, adaptive radiation, continental radiation, convergence, Cryptoblepharus, speciation
1. Introduction
Understanding the processes that promote biological diversity is a major challenge in evolutionary biology. In this context, much has been gleaned from the study of adaptive radiations; the rise of diverse ecological roles and phenotypic disparity due to role-specific adaptations within a lineage [1,2]. Adaptive radiations have drawn the attention of evolutionary biologists, because they exemplify the mechanisms that link micro- and macroevolution. During an adaptive radiation, ecological opportunity facilitates speciation and ecological diversification [3,4]. Such ecological opportunity can arise when an isolated area with a depauperate biota is colonized (i.e. islands or lakes) or when the evolution of a key trait opens a new adaptive zone (i.e. key innovation) [5,6]. Young adaptive radiations in isolated geographical entities such as islands or lakes are particularly well studied [7] and have highlighted the role of ecological speciation [8] and sexual selection [9].
However, evolutionary radiations can be triggered by a wide range of biotic and abiotic factors [10], and not all evolutionary radiations can be characterized as adaptive radiations [1,2,11]. Allopatric speciation for instance, though unlikely to occur within a small island or lake, can be an important driver of evolutionary radiation on large continental landmasses. Lineages within a new region might diversify via allopatric speciation when an ancestral species crosses a geographical barrier or when habitats become fragmented following a climatic shift [10]. Speciation in such geographical isolates can be driven via means other than ecologically mediated divergent selection and precede significant ecological differentiation. Ecological differentiation could subsequently still arise in isolation, or via character displacement between reproductively isolated lineages that come into secondary contact. Thus, processes other than ecological speciation can promote evolutionary radiation at a continental scale and even generate patterns that resemble adaptive radiation [11]. Indeed, the study of continental-scale radiations has provided wonderful examples of species proliferation and adaptive phenotypic change across major taxonomic groups [12–16]. Yet it can be challenging to identify the key evolutionary processes that have promoted speciation in such older radiations. It is therefore important to establish a phylogenetic and ecological framework for recently emerged continental clades that are widespread, species-rich, and ecologically diverse. The study of such recent radiations can ultimately provide further insight into the factors that have shaped macroevolutionary patterns across a continent [10,11,17–20].
The study of species diversification and phenotypic radiation has greatly benefited from the development of phylogeny-aware comparative methods and molecular approaches to generate large sequence datasets [4,21]. The phylogenetic structure underlying rapid radiations has been notoriously difficult to resolve due to a lack of phylogenetic signal or incongruence in phylogenetic history between loci [20–22]. This observed discordance emphasizes the need to use multi-locus datasets for species tree inference and to incorporate coalescent-based methods that account for heterogeneity in coalescent histories [23]. In addition, analysing large numbers of genetic markers will also optimize branch length estimation and this increase in accuracy is particularly relevant for comparative analyses [24]. With a complete phylogeny in place and information on contemporary phenotypic variation, patterns of adaptive diversification can be identified by comparing the fit of distinct models of phenotypic change [13]. In this study, we use a phylogenomic approach to generate a time-calibrated ultrametric tree for a recent radiation of Australian lizards, and subsequently employ comparative methods to test how habitat specialization may have influenced morphological diversification in the course of this radiation.
Skinks of the genus Cryptoblepharus have radiated across the entire Australian continent (figure 1a) while simultaneously colonizing different scansorial habitats (rocks and trees; figure 1b,d) which are largely unoccupied by other species of the rich diurnal lizard fauna. Furthermore, although terrestrial habitats are dominated by other genera of ground-dwelling skinks, there are three littoral species of Cryptoblepharus that are found in close association with rocks on beaches (figure 1c)—another unique habitat. A recent taxonomic revision using 45 (allozyme) loci and 33 morphological markers increased the number of recognized Australian species from seven to 25 [26,27]. Although this analysis was unable to resolve the phylogeny of the genus, a recent phylogenomic analysis revealed that rock and arboreal specialists are dispersed across the phylogeny [28]. Interestingly, the geographical distributions of rock and arboreal specialists are often overlapping and many instances of sympatry have been reported [27]. This genus therefore provides an excellent opportunity to study the role of habitat specialization in promoting adaptive diversification at a continental scale.
Figure 1.
Distribution of Australian Cryptoblepharus and the three habitat specialists. (a) Topographic map of Australia with the mean point of each species' distribution plotted and coloured according to habitat type (for complete distribution maps, see [25]). In situ photographs of (b) arboreal, (c) littoral, and (d) rock specialists (green, blue, and red dots on the topographic map, respectively).
The overarching goal of this study is to examine the ecological context of diversification of Australian Cryptoblepharus with a combined assessment of morphological, ecological, and phylogenetic patterns. Specifically, we focus on whether habitat specialization can explain current patterns of phenotypic variation in ecologically relevant traits. A statistical correlation between phenotype and environment is a first indication of adaptive diversification, but here we build further upon a rich literature of ecomorphological research in lizards [25,29–31]. By explicitly focusing on traits that are known to improve performance within specific habitats [3], we examine the adaptive consequences of habitat specialization and quantify convergent change across the Australian continent.
2. Material and methods
(a). Taxon sampling for phylogenetic inference
A previous allozyme and morphological analysis identified 28 lineages of Australian Cryptoblepharus [26], of which 25 were recognized as separate species. Three genetically divergent lineages were morphologically, ecologically, and geographically indistinguishable—two in C. ruber and one in C. tytthos—and were therefore not elevated to species status. We selected a single representative for each of the 28 lineages using mostly the same individuals as examined allozymically by Horner & Adams [26], except where tissues were depleted. For those species, we used recently collected field samples where species identification was verified based on morphological characteristics and a mitochondrial marker (ND2). Further details can be found in Blom et al. [28].
(b). Exon capture
We used a custom-designed exon capture approach [32], to generate a large multi-locus dataset of orthologous genetic markers suitable for phylogenetic inference. The designs of the exon capture kit, sequencing strategy, and sequencing success, are outlined in detail in Blom et al. [28] and references therein. Briefly, our capture design included exon-targets based on orthologues from seven transcriptomes of three genera closely related to Cryptoblepharus (Carlia rubrigularis, Lampropholis coggeri, and Saproscincus basiliscus; [33]). We used an in-solution hybridization capture (Roche NimbleGen) and sequenced (100 bp paired-end) the enriched libraries on a single Illumina HiSeq 2000 lane. We filtered and assembled the sequence data using a workflow that was described previously by Singhal et al. [33] and is available at https://github.com/MVZSEQ.
We have developed and applied a flexible bioinformatic workflow for alignment and alignment filtering of exonic sequences, EAPhy (v. 1.1, [34]). EAPhy automatically aligns sequences using MUSCLE (v. 3.8.31, [35]), performs checks to ensure coding of amino acids and removes missing data from the end of the alignments. EAPhy assesses each alignment individually and automatically generates either locus-specific or concatenated alignments. We only concatenated loci where each lineage with morphological data was present and the alignment of each individual locus was longer than 150 bp.
(c). Phylogenetic inference
Estimating a species tree is particularly challenging for rapid radiations [22,36]. We have previously employed summary-coalescent methods and a thorough gene tree estimation sensitivity analysis to infer the Cryptoblepharus species tree topology [28]. In brief, we first screened loci based on gene tree resolution and subsequently quantified the impact of stochastic gene tree estimation error on summary-coalescent species tree inference. Here, we use the inferred species tree topology that was well-supported across analyses. However, we excluded two lineages from the dataset for which no morphological measurements were available (sub-species: C. pulcher clarus and a divergent lineage of C. tytthos—‘carnA4’ in Horner & Adams [26]).
We generated an ultrametric tree from the concatenated alignment with BEAST v. 2.1.3 [37], while constraining the topology to that of the species tree (sensu [28]) and therefore only fitted branch lengths. We used a GTR + Γ substitution model with four Γ rate categories, a strict clock, and estimated each substitution rate from the data. In the absence of suitable fossil calibrations or a previous estimate of crown age for the genus, we scaled branch lengths from a number of expected substitutions per site to years, using an empirically obtained molecular clock estimation (0.001 substitutions/site/Myr) for another genus of lizards within the same family (Scincidae, [38]). We ran the BEAST analyses in duplicate with separate starting seeds. Each analysis was run for 10 million generations and we sampled chains every 10 000 generations. We discarded the first 10% of trees as burnin, used Tracer v. 1.5 to check for convergence, and LogCombiner v. 2.1.3 to combine the posterior sample of trees across runs. The ultrametric species tree was summarized using TreeAnnotator v. 2.1.2. Lastly, we tested whether the rate of lineage accumulation changed over time using the ltt function in the R-package ‘Phytools’ [39].
(d). Morphospace construction
To examine phenotypic changes in Australian Cryptoblepharus, we combined our species tree estimate based on genomic data with the morphological characters recorded during the last major taxonomic revision [27]. Horner [27] recorded complete metric and meristic measurements for 863 Australian Cryptoblepharus specimens. Morphometric measurements were taken under an illuminated magnifying lens, with electronic digital calipers to the nearest 0.01 mm. Across the 26 taxa for which morphological data were recorded, the 863 individuals represented an average of 33 individuals per species and all species were represented by four measurements or more (electronic supplementary material, table S1). From the suite of characters recorded for the taxonomic revision, we selected metric characters known or suspected to be relevant to the habitats used by Cryptoblepharus [30]. These include snout–vent length (SVL), forelimb length (FL), rear-limb length (RL), snout length (SE), eye to ear length (CHEEK), ear to limb length (NECK), head height (HH), and head width (HW). We used SVL as a measure for overall body size and divided the head length estimate into three separate metrics (SE, CHEEK, NECK) to account for differences that might not be appropriately captured by head length alone. To identify size-independent axes of trait variation, we calculated residual values from phylogenetic regressions of each log-transformed trait against log-SVL. For each trait, we first calculated the mean species values and then used the function phyl.resid (Phytools; [40]), to infer the size-independent trait values. We used the λ correction to avoid bias due to non-Brownian evolution, during the estimation of the phylogenetically corrected regression.
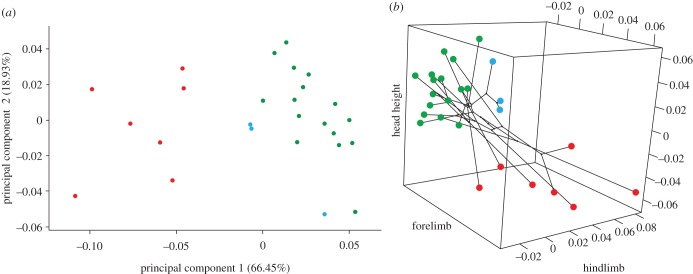
To identify the major axes of variation and reduce the multidimensionality of the data, we used a phylogenetic principal component analysis (pPCA) on the size-corrected species trait data (all traits excluding SVL), while simultaneously optimizing λ (phyl.pca, Phytools). We used a scree plot to visualize and examine the amount of variation explained by each individual component. The first two components jointly explained over 85% of the variation in the data and were retained for further in-depth analysis.
We examined the degree of morphospace occupied by each habitat type, using a three-dimensional phylomorphospace plot (phylmorphospace3d, Phytools) where each axis represents a trait that loaded strongly on PC1. By plotting the size-corrected residual scores for each species, superimposing phylogenetic relationships, and highlighting species by habitat type, the phylomorphospace plot illustrates morphological variation between and within habitat types for the traits that jointly explained most phenotypic variation. In addition to visualizing habitat specific differences in phylomorphospace, we also estimated the degree of phenotypic disparity between all species combined and within each habitat type, by calculating the average squared Euclidean distance among all pairs of PC1 scores using the disparity function from the R-package ‘Geiger’ [41].
(e). Associations between morphology and habitat
We examined whether trait values along the two major axes of trait variation differed between species occurring in different habitat types (rock, arboreal, and littoral). We used a multivariate analysis of variance (MANOVA) with habitat as a predictor variable and the species' PC scores for the first two components as the dependent variables. To conduct a MANOVA in a phylogenetic context, we used the aov.phylo function (1 000 simulations, Wilks' λ) in Geiger. We subsequently examined differences for each individual PC separately. Based on 1 000 simulations, we calculated the probability of the observed differences in PC scores between each group (phylANOVA, Phytools).
The ANOVA for each PC provides an overall view of differences along the two major axes of trait variation between species occurring in different habitats. However, to examine changes in individual morphological traits, we repeated the phylo-ANOVA's using the size-independent residual scores from phylogenetic regression for each individual trait. Lastly, we also tested differences in overall size (SVL) by comparing the log-transformed species means.
(f). Morphological evolution
To assess whether habitat shifts can explain current patterns of phenotypic diversification, we used two different approaches to estimate the evolutionary trajectory of phenotypic change. Firstly, we quantified whether morphology varies following a Brownian Motion (BM) process, where phenotypic differences accumulate at random with time, or whether morphological diversification is constrained around one or more optima (OU, Ornstein–Ühlenbeck process). Recent expansions in the class of OU models allow variable rates and strengths of selection around the trait optima [42]. However, parameter estimation in such complex models requires large numbers of taxa [42]. Because the number of Cryptoblepharus species and the frequency of habitat shifts are relatively limited, we only evaluated the presence or the absence of multiple phenotypic optima rather than estimating selection strength as well. We first estimated ancestral states for each internal node (rerootingMethod, Phytools) and then used the R-package OUwie [42] to fit three distinct models of character evolution on PC1 scores. We fitted a single rate BM model (BM1) and OU models with either a single optimum for all species (OU1) or with multiple optima, but single rates of selection (α) and stochastic motion around all optima (σ2).
Secondly, we evaluated whether independent lineages converged on similar phenotypic optima by using a comparative approach implemented in R. SURFACE [43] uses a stepwise corrected AICc (Akaike’s information criterion corrected for sample size) approach to fit Hansen models and evaluates the most optimal set of evolutionary regimes and regime shifts. SURFACE analyses consist of two distinct phases; a ‘forward’ phase during which regime shifts are added to the tree and a ‘backward’ phase during which shifts towards the same peaks are identified and collapsed. The addition and collapsing of shifts is reiterated until AICc scores cease to improve. SURFACE can identify cases of convergence across a clade without the subjective a priori designation of candidate convergent taxa, and only takes the phylogeny and multidimensional trait data as input. We ran SURFACE on the size-corrected residuals for each log-transformed trait and log-transformed species means for SVL. Finally, to visualize how species that belong to distinct regimes differ in phenotype, we plotted the size-corrected residual scores for each species in two-dimensional trait space across all traits that were inferred as significantly different between regimes.
3. Results
(a). Phylogenetic analyses
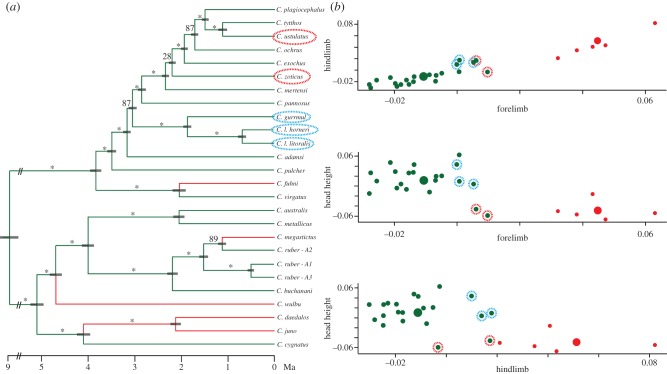
We used 1 195 loci (618 860 bp) and an empirically estimated molecular clock to generate a time-calibrated ultrametric tree (figure 3a). We fitted branch lengths on a summary-coalescent species tree that is well supported, except for the node that involves C. zoticus and C. mertensi. This analysis infers that Australian Cryptoblepharus have diversified recently, within the last 10 million years overall, across the Australian continent. Two distinct clades, with 11 and 15 taxa, respectively, have proliferated since the Pliocene and rock and arboreal specialists have emerged within each radiation. The species accumulation rate was elevated during the beginning of the continental radiation and then decreased over time (γ = −2.070, p = 0.04; electronic supplementary material, figure S1).
Figure 3.
Time-calibrated phylogeny of Australian Cryptoblepharus and phenotypic convergent regimes. (a) The branches of the phylogeny are coloured according to the convergent regime inferred with SURFACE. There are two adaptive peaks and at least four independent regime shifts. Bootstrap support is given for each bipartition [32], with asterisks indicating bootstrap support of more than 90, and grey boxes around each node representing the 95% confidence interval for the node age estimate. Non-arboreal species that belong to the arboreal regime are highlighted with a coloured dotted circle. (b) Size-corrected residual scores from phylogenetic regression are plotted for each of the traits that were identified as distinct and coloured by their inferred convergent regime. Non-arboreal species that belong to the arboreal regime are highlighted with a coloured dotted circle. Two rock specialists have not converged on the same adaptive peak as the other rock dwelling species. These species are convergent in terms of HH, but have relatively short limbs. The three littoral species only differ from the arboreal specialists in hindlimb length and are likely therefore not identified as a separate adaptive peak.
(b). Morphospace and association with ecology
The main variation in size-corrected morphology was between species from different habitats and in particular between rock and other specialists. This is shown by the separation of the rock species from all others along the first axis of the PCA (figure 2a). PC1 (66.45%) and PC2 (18.93%) jointly explain over 85% of all morphological variation, but only PC1 is significantly correlated with habitat type (phylogenetic MANOVA on both PC1 and PC2, d.f. = 2, Wilks' λ = 0.13, p < 0.01; phylogenetic ANOVAs on PC1, d.f. = 2, F = 59.19, p < 0.01, and on PC2, d.f. = 2, F = 1.43, p = 0.40). PC1 loads strongly on fore- and hindlimb length and HH (electronic supplementary material, table S2), suggesting that species that occur on rock substrates tend to have longer limbs and a more compressed head shape than species that occur on trees or in a littoral habitat. PC2 loads most strongly on features pertaining to head length but does not correlate with habitat type (electronic supplementary material, table S2).
Figure 2.
pPCA and three-dimensional phylomorphospace plot. (a) Morphological variation among species along pPC1 and pPC2, where each dot represents a species and is coloured by that species' habitat type (figure 1). PC1 separates the rock specialists from the other habitat specialists. (b) Size-corrected residual scores from phylogenetic regression are plotted for each of the traits that were identified as different between habitats. Colours correspond to the habitat type for each respective species (figure 1) and the phylogeny is mapped onto morphospace. Regardless of phylogenetic association, species are more closely clustered in morphospace by habitat.
In addition to assessing the correlation between morphology and habitat type along the two major axes of trait variation (i.e. PC1 and PC2), we also employed phylogenetic ANOVAs and post hoc t-tests to examine differences in individual traits. The overall pattern observed is similar for HH, with species that occur on rock substrates having dorsally compressed skulls (d.f. = 2, F = 28.12, p < 0.01) and no difference between arboreal and littoral specialists (d.f. = 2, T = −0.06, p = 0.97). Rock and arboreal species consistently differ in limb length (forelimb, d.f. = 2, T = −8.78, p < 0.01 and hindlimb, d.f = 2, T = −7.35, p < 0.01). Whereas (ground-dwelling) littoral specialists have similar scores for PC1 as arboreal species, phylogenetic ANOVA of individual traits show that the former have significantly longer hindlimbs than their arboreal counterparts (d.f. = 2, T = −3.24, p = 0.04), but do not differ from rock specialists (d.f. = 2, T = −1.88, p = 0.23). Arboreal and littoral species did not differ in FL (d.f. = 2, T = −2.20, p = 0.10). There was no significant correlation between habitat and any other trait (SVL, HW, SE, CHEEK, or NECK).
Significant differences between habitat types in the three divergent traits (fore-, hindlimb, and HH) are apparent when mean species scores are visualized in morphospace (figure 2b). Most notably, arboreal species tend to cluster closely and rock species are clearly distinct in terms of HH, regardless of phylogenetic association between species. These results were confirmed when comparing morphological disparity metrics, which were more than twofold lower for the arboreal species (electronic supplementary material, table S3) than for the other habitat categories.
(c). Morphological evolution
An OU model with multiple phenotypic optima (OUM: AICc score −121.54) was substantially better supported than a BM (AICc score −73.38) or OU1 model (AICc score −78.43), suggesting that there is more than one phenotypic optimum for traits that strongly load on PC1. The estimated optima were found within the values realized for the extant species (electronic supplementary material, table S4), indicating that the model is a realistic description of current morphological patterns and is not negatively biased by factors of uncertainty such as potentially spurious ancestral state reconstruction. In addition, the standard errors around the optima reflect the observed variation in morphospace and disparity metrics for each habitat category, further confirming that the inferred optima of the model represent realistic differences between habitat specialists (electronic supplementary material, table S4).
Having inferred multiple phenotypic optima, we tested whether independent lineages converged on the same optima and quantified the frequency of such shifts. The Hansen model returned by the SURFACE analysis highlights the presence of two regimes and four convergent regime shifts. The AICc score improved by 29.34 units during the forward phase and by another 25.6 units during the backward phase. The AICc score for the final model suggests that the phenotypic data match an OU model with two phenotypic optima much better than a similar model under BM or an OU model with a single optimum (electronic supplementary material, figure S2), and is therefore consistent with the result from the OUwie analysis.
The two optima identified coincide with phenotypes that match species found on either an arboreal or a rock substrate, with the four regime shifts corresponding to transitions from trees to rocks (figure 3a). However, and interestingly, two of the seven rock species did not converge in overall phenotype with the other saxicolous species, nor did littoral specialists occupy a distinct phenotypic optimum (figure 3a). Instead, the two rock (C. zoticus and C. ustulatus) species in question and all three littoral specialists (C. l. litoralis, C. l. horneri, and C. gurrmul) belong to the same regime as the arboreal specialists. When focusing on the three functional traits that are distinct between rock and other specialists, it is apparent why these species are matched to the arboreal regime by the SURFACE analysis. Whereas the two rock species are convergent in HH with other rock species, the length of their limbs is not (figure 3b). Furthermore, as previously identified with a phylogenetic ANOVA, littoral species differ in the length of their hindlimbs but not in any other trait (figure 3b).
4. Discussion
(a). Habitat specialization
Cryptoblepharus skinks are the most prominent scansorial specialists within the most species-rich family of lizards in Australia and globally (Scincidae). Species that occur on distinct substrates differ significantly in functionally relevant phenotypic traits and our analyses suggest that the evolutionary trajectories of such traits have changed in a predictable direction based on biomechanical performance tests in other lizards [30,31]. Rock species of Cryptoblepharus occupy steep sandstone escarpments, where they move rapidly on perpendicular cliffs and hide in shallow crevices between the rocks. The reduction in HH enables species to shelter in narrow cracks, sometimes just a few millimetres wide. Arboreal species however, tend to move up into trees and hide between the foliage, and likely do not experience a similar degree of selection for reduced HH. The increase in limb length of rock species aids locomotion on flat vertical surfaces by maintaining the centre of mass (and balance) close to the substrate. By contrast, long legs might present challenges for arboreal lizards by increasing the distance between the centre of mass and perch [44]. The littoral species, that tend to climb less than arboreal and rock specialists, mostly resemble arboreal species except for the length of their hindlimbs, in which they are more similar to rock species (figures 2b and 3b). Whereas scansorial species tend to rely equally on fore- and hindlimbs for locomotion, ground-dwelling lizards are expected to have relatively long hindlimbs since they mostly use their back legs to thrust forward for movement in a horizontal direction [45]. Overall, the morphology of Cryptoblepharus skinks tends to match habitat closely, such that these variants can be considered as ecomorphs.
(b). Convergence
Morphological differences are not only correlated with environment, but they also have evolved independently and resulted in at least four convergent shifts towards the same phenotypic optimum (figure 3a). Given the strong correlation between habitat and functionally relevant traits, the trait value optima of the SURFACE regimes can be interpreted as adaptive peaks for an arboreal and a rock ecomorph. Interestingly, on visual inspection of the three-dimensional phylomorphospace plot (figure 2b) and the phenotypic disparity metrics, the phenotypic variation surrounding the adaptive peaks is more limited for the arboreal type than the rock type. Even though there are less than half as many rock as arboreal lineages (7 versus 16), and both ecomorphs span the phylogeny, phenotypic disparity between the rock species is twofold greater than between the arboreal lineages (electronic supplementary material, table S3). Although we are unable to accurately estimate the strength of selection surrounding phenotypic optima with a limited number of species [42], this prominent difference in phenotypic disparity within ecomorph groups might indicate that the strength of selection is more variable towards the rock optimum and more stringent for arboreal species. This is exemplified by C. zoticus and C. ustulatus; the two rock species that were identified as belonging to the arboreal regime by the SURFACE analysis. These species are clearly convergent with other rock species for HH (figures 2b and 3b), but their fore- and hindlimbs are relatively short compared to other rock species (figures 2b and 3b) and in particular C. fuhni, the rock specialist with the most pronounced degree of limb elongation [27]. Further ecological studies and performance tests should investigate whether this difference has any consequence in terms of locomotion performance or whether this is mitigated by alternative use of a similar habitat [46]. Similarly, the three littoral species are joined with the arboreal regime because the difference in hindlimb length alone is not significant enough to identify a separate adaptive peak. Previous simulation analyses have confirmed that SURFACE performs well with datasets that include multiple convergent traits, but instances of convergence were not always identified with single (convergent) traits [43]. Thus, even though these two rock and three littoral species significantly differ along specific trait axes from arboreal types, they have not been diagnosed as distinct.
Examples of lizard species that have repeatedly adapted to rock environments and converged in limb length and head depth, have been reported previously (i.e. [47]). However, the frequency of convergence within this recent radiation of Cryptoblepharus is striking and likely correlated with the rapid spread across the Australian continent. Independent adaptive peak shifts between arboreal and rock habitat have occurred in isolated sandstone ranges that are surrounded by vast stretches of savannah woodland or desert. It is unlikely that low-dispersal rock specialists can easily migrate across such distinct habitats, such that these sandstone ranges resemble islands in a sea of unsuitable habitat where parallel evolutionary change has resulted in convergent ecomorphological lineages. These repeated convergent outcomes in functional traits strongly suggest that adaptation to different habitats has promoted an increase in ecological specialization and associated phenotypic disparity in ecologically relevant traits between sister taxa [48]. Furthermore, adaptation to distinct substrates has often facilitated the sympatric coexistence of closely related (sister) species [27]. In these respects, the recent radiation of Australian Cryptoblepharus bears strong similarity to the patterns that characterize adaptive radiations [2,3,49].
(c). Continental radiation
Our findings suggest that each of the two clades of Cryptoblepharus skinks have proliferated rapidly during the last approximately 5 Myr (figure 3a) and have repeatedly developed phenotypic traits that are known to be of functional importance within specific environments [30]. This strong correlation between habitat and morphology underlines the importance of ecologically mediated selection within this system. Repeated habitat shifts have resulted in divergent selection that has increased morphological disparity, while uniform selection across geographical isolates on similar substrates has likely limited morphological differentiation, especially within the arboreal taxa. Indeed, the taxonomy of Cryptoblepharus has traditionally been viewed as exceptionally challenging due to the limited morphological differences between species that occur in similar habitat. Hence, many of these cryptic lineages were only identified with the aid of genetic screening [26]. The presence of strong selection, either uniform or divergent, can accelerate the speciation process and is more likely to have promoted species proliferation in this genus than neutral processes [50] alone, especially given the recency of the radiation [51].
Interestingly, examination of the temporal pattern of diversification suggests that the rate of species accumulation was elevated during the beginning of the continental radiation (electronic supplementary material, figure S1). Although this is often interpreted as evidence of adaptive radiation (i.e. ‘early-burst’ signal), processes other than initial niche filling could also result in a slowdown of diversification over time [52]. Whereas a novel lineage on an isolated island or lake might rapidly have access to all available niche space, it might take a significant amount of time before a continental clade has spread across all available habitats. Indeed, our results indicate that habitat shifts have not predominantly occurred in the beginning of the radiation, but both in the deep and more recent past (figure 3a). The rapid accumulation of lineages during the beginning of the radiation could therefore simply reflect geographical isolation following an early range expansion, rather than initial diversification of niche use.
Our analyses highlight two seemingly contrasting patterns of diversification: speciation within versus between distinct habitat substrates. This suggests that the evolutionary radiation of Australian Cryptoblepharus is not solely driven by ecologically mediated divergent selection, as observed in some sympatric systems that reside on isolated islands or lakes [8,9], but rather parallels another enigmatic radiation, the Anoles of the West Indies [31]. Anolis lizards have radiated spectacularly and many distinct ecotypes occur in sympatry on islands across the Caribbean basin. However, there is no direct evidence that ecological specialists have emerged in sympatry and within-island cladogenesis appears to be limited to larger islands in the Greater Antilles, even though some of the smaller islands exhibit the same degree of environmental heterogeneity [31]. Furthermore, deep intraspecific divergence within widespread species such as A. cybotes [53] indicates that significant genetic differentiation has accrued without extensive ecomorphological divergence and highlights the potentially important role of macrohabitat in the speciation history of Anolis lizards [54]. As such, with contrasting patterns of differentiation between and within habitat types, the continental radiation of Australian Cryptoblepharus resembles the radiation of Caribbean Anoles and perhaps many other continental (or large island, e.g. [55]) systems of different ages (i.e. [17,49]).
Whereas the radiation of Caribbean Anoles might span 40–60 Myr [31], the relatively young age of the Australian Cryptoblepharus radiation invites further investigation into the mechanisms that have promoted species diversification. Of particular interest, is to understand whether the contrasting patterns of phenotypic diversification within and between habitats, also represent alternative speciation dynamics or whether reproductive isolation has developed in a similar manner and ecological differentiation has only occurred via character displacement in secondary contact [11]. By modelling demographic and divergence history, future studies can quantify the evolution of reproductive isolation between lineages in ecologically similar refugia, such as has been inferred for rainforest skinks from related genera [56]. Or alternatively, such studies can explicitly examine the geographical context of diversification and gene flow [57], between ecomorphologically distinct young sister species with a parapatric distribution (i.e. C. ruber and C. megastictus; figure 3a). Hence, the outcomes of our study will function as a phylogenetic and ecological framework, and invite further investigation into the proximate mechanisms that have driven speciation within and between habitats.
The study of adaptive radiations within isolated insular systems has provided important insights on the role of ecology in driving species proliferation. However, it is important to ask to what extent similar mechanisms promote the radiation of continental biota. Because it is challenging to address this question by focusing on older radiations, the evaluation of recent continental radiations can shed further light on how commonly speciation precedes significant ecological differentiation. Our analysis of the Cryptoblepharus radiation for instance, highlights the importance of ecological selection both within and between habitat types, but simultaneously suggests that species proliferation is not driven by divergent selection alone. The importance of examining other recent widespread radiations (e.g. the Sigmodontinae of South America [58,59]) is therefore evident and will inform our general understanding on the process of continental diversification. As such, the study of recent radiations can provide a window into the origin of biodiversity and how microevolutionary processes ultimately induce macroevolutionary change at a continental scale.
Supplementary Material
Acknowledgements
We thank Mark Adams, Jason Bragg, and Sally Potter for their ongoing support to the Cryptoblepharus project. We thank the museum curators for access to tissues and specimens, Damien Esquerré, Martha Muñoz, and Marta Vidal-García for providing helpful advice during the analyses. We thank Stewart Macdonald and Pascal Title for sharing images used in figure 1. Finally, we thank the associate editor and two anonymous reviewers for their helpful suggestions to improve the manuscript.
Ethics
The study was performed under ethics approval from the Australian National University—A2012/14.
Data accessibility
Raw data are deposited in Dryad under: http://dx.doi.org/10.5061/dryad.j4r56.
Authors' contributions
M.P.K.B. and C.M. conceived and planned the project and wrote the manuscript. M.P.K.B. and P.H. collected the data; M.P.K.B. analysed the data and prepared the figures. All authors gave final approval for the publication.
Competing interests
We declare we have no competing interests.
Funding
This work was supported by a grant from the Australian Research Council to C.M. (FL110100104).
References
- 1.Losos JB, Mahler DL. 2010. Adaptive radiation: the interaction of ecological opportunity, adaptation and speciation. In Evolution Since Darwin: The first 150 years (eds Bell MA, Futuyma DJ, Eanes WF, Levinton JS), pp. 381–420. Sunderland, MA: Sinauer Associates. [Google Scholar]
- 2.Givnish TJ. 2015. Adaptive radiation versus ‘radiation’ and ‘explosive diversification’: why conceptual distinctions are fundamental to understanding evolution. New Phytol. 207, 297–303. ( 10.1111/nph.13482) [DOI] [PubMed] [Google Scholar]
- 3.Schluter D. 2000. The ecology of adaptive radiation. Oxford, UK: Oxford University Press. [Google Scholar]
- 4.Losos JB. 2010. Adaptive radiation, ecological opportunity, and evolutionary determinism. Am. Nat. 175, 623–639. ( 10.1086/652433) [DOI] [PubMed] [Google Scholar]
- 5.Simpson GG. 1953. The major features of evolution. New York, NY: Columbia University Press. [Google Scholar]
- 6.Wright S. 1982. Character change, speciation, and the higher taxa. Evolution 36, 427–443. ( 10.2307/2408092) [DOI] [PubMed] [Google Scholar]
- 7.Losos JB, Ricklefs RE. 2009. Adaptation and diversification on islands. Nature 457, 830–836. ( 10.1038/nature07893) [DOI] [PubMed] [Google Scholar]
- 8.Rundle HD, Nagel L, Boughman JW, Schluter D. 2000. Natural selection and parallel speciation in sympatric sticklebacks. Science 287, 306–308. ( 10.1126/science.287.5451.306) [DOI] [PubMed] [Google Scholar]
- 9.Wagner CE, Harmon LJ, Seehausen O. 2012. Ecological opportunity and sexual selection together predict adaptive radiation. Nature 487, 366–369. ( 10.1038/nature11144) [DOI] [PubMed] [Google Scholar]
- 10.Simões M, Breitkreuz L, Alvarado M, Baca S, Cooper JC, Heins L, Herzog K, Lieberman BS. 2016. The evolving theory of evolutionary radiations. Trends Ecol. Evol. 31, 27–34. ( 10.1016/j.tree.2015.10.007) [DOI] [PubMed] [Google Scholar]
- 11.Rundell RJ, Price TD. 2009. Adaptive radiation, nonadaptive radiation, ecological speciation and nonecological speciation. Trends Ecol. Evol. 24, 394–399. ( 10.1016/j.tree.2009.02.007) [DOI] [PubMed] [Google Scholar]
- 12.Gould SJ. 1989. Wonderful life: the Burgess shale and the nature of history. New York, NY: WW Norton & Company. [Google Scholar]
- 13.Lapiedra O, Sol D, Carranza S, Beaulieu JM. 2013. Behavioural changes and the adaptive diversification of pigeons and doves. Proc. R. Soc. B 280, 20122893 ( 10.1098/rspb.2012.2893) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Givnish TJ, et al. 2014. Adaptive radiation, correlated and contingent evolution, and net species diversification in Bromeliaceae. Mol. Phylogenet. Evol. 71, 55–78. ( 10.1016/j.ympev.2013.10.010) [DOI] [PubMed] [Google Scholar]
- 15.Moen DS, Morlon H, Wiens JJ. 2016. Testing convergence versus history: convergence dominates phenotypic evolution for over 150 million years in frogs. Syst. Biol. 65, 146–160. ( 10.1093/sysbio/syv073) [DOI] [PubMed] [Google Scholar]
- 16.Esquerré D, Keogh JS. 2016. Parallel selective pressures drive convergent diversification of phenotypes in pythons and boas. Ecol. Lett. 19, 800–809. ( 10.1111/ele.12620) [DOI] [PubMed] [Google Scholar]
- 17.Hughes C, Eastwood R. 2006. Island radiation on a continental scale: exceptional rates of plant diversification after uplift of the Andes. Proc. Natl Acad. Sci. USA 103, 10 334–10 339. ( 10.1073/pnas.0601928103) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rowe KC, Aplin KP, Baverstock PR, Moritz C. 2011. Recent and rapid speciation with limited morphological disparity in the genus Rattus. Syst. Biol. 60, 188–203. ( 10.1093/sysbio/syq092) [DOI] [PubMed] [Google Scholar]
- 19.Rabosky DL, Donnellan SC, Grundler M, Lovette IJ. 2014. Analysis and visualization of complex macroevolutionary dynamics: an example from Australian scincid lizards. Syst. Biol. 63, 610–627. ( 10.1093/sysbio/syu025) [DOI] [PubMed] [Google Scholar]
- 20.Ebel ER, DaCosta JM, Sorenson MD, Hill RI, Briscoe AD, Willmott KR, Mullen SP. 2015. Rapid diversification associated with ecological specialization in neotropical Adelpha butterflies. Mol. Ecol. 24, 2392–2405. ( 10.1111/mec.13168) [DOI] [PubMed] [Google Scholar]
- 21.Glor RE. 2010. Phylogenetic insights on adaptive radiation. Annu. Rev. Ecol. Evol. Syst. 41, 251–270. ( 10.1146/annurev.ecolsys.39.110707.173447) [DOI] [Google Scholar]
- 22.Giarla TC, Esselstyn JA. 2015. The challenges of resolving a rapid, recent radiation: empirical and simulated phylogenomics of Philippine shrews. Syst. Biol. 64, 727–740. ( 10.1093/sysbio/syv029) [DOI] [PubMed] [Google Scholar]
- 23.Edwards SV. 2009. Is a new and general theory of molecular systematics emerging? Evolution 63, 1–19. ( 10.1111/j.1558-5646.2008.00549.x) [DOI] [PubMed] [Google Scholar]
- 24.Garamszegi LZ. 2014. Modern phylogenetic comparative methods and their application in evolutionary biology. Berlin, Germany: Springer. [Google Scholar]
- 25.Kaliontzopoulou A, Carretero MA, Llorente GA. 2010. Intraspecific ecomorphological variation: linear and geometric morphometrics reveal habitat-related patterns within Podarcis bocagei wall lizards. J. Evol. Biol. 23, 1234–1244. ( 10.1111/j.1420-9101.2010.01984.x) [DOI] [PubMed] [Google Scholar]
- 26.Horner P, Adams MA. 2007. Molecular-systematic assessment of species boundaries in Australian Cryptoblepharus (Reptilia: Squamata: Scincidae): A case study for the combined use of allozymes and morphology to explore cryptic biodiversity. Beagle Suppl. 3, 1–21. [Google Scholar]
- 27.Horner P, Adams MA. 2007. A molecular systematic assessment of species boundaries in Australian Cryptoblepharus (Reptilia: Squamata: Scincidae) – a case study for the combined use of allozymes and morphology to explore cryptic biodiversity. Beagle Suppl. 3, 1–19. [Google Scholar]
- 28.Blom MPK, Bragg JG, Potter S, Moritz C. 2016. Accounting for uncertainty in gene tree estimation: summary-coalescent species tree inference in a challenging radiation of Australian lizards. In review. (bioRxiv 10.1101/056085) [DOI] [PubMed] [Google Scholar]
- 29.Goodman B, Isaac J. 2008. Convergent body flattening in a clade of tropical rock-using lizards (Scincidae: Lygosominae). Biol. J. Linn. Soc. 94, 399–411. ( 10.1111/j.1095-8312.2008.00988.x) [DOI] [Google Scholar]
- 30.Goodman BA, Miles DB, Schwarzkopf L. 2008. Life on the rocks: habitat use drives morphological and performance evolution in lizards. Ecology 89, 3462–3471. ( 10.1890/07-2093.1) [DOI] [PubMed] [Google Scholar]
- 31.Losos JL. 2011. Lizards in an evolutionary tree: ecology and adaptive radiation of Anoles. Berkeley, CA: University of California Press. [Google Scholar]
- 32.Bragg JG, Potter S, Bi K, Moritz C. 2015. Exon capture phylogenomics: efficacy across scales of divergence. Mol. Ecol. Resour. ( 10.1111/1755-0998.12449) [DOI] [PubMed] [Google Scholar]
- 33.Singhal S. 2013. De novo transcriptomic analyses for non-model organisms: an evaluation of methods across a multi-species data set. Mol. Ecol. Resour. 13, 403–416. ( 10.1111/1755-0998.12077) [DOI] [PubMed] [Google Scholar]
- 34.Blom MPK. 2015. EAPhy: A flexible tool for high-throughput quality filtering of exon-alignments and data processing for phylogenetic methods. PLoS Curr. Tree of Life 1 ( 10.1371/currents.tol.75134257bd389c04bc1d26d42aa9089f) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32, 1792–1797. ( 10.1093/nar/gkh340) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Maddison WP. 1997. Gene trees in species trees. Syst. Biol. 46, 523–536. ( 10.1093/sysbio/46.3.523) [DOI] [Google Scholar]
- 37.Bouckaert R, Heled J, Kühnert D, Vaughan T, Wu C-H, Xie D, Suchard MA, Rambaut A, Drummond AJ. 2014. BEAST 2: A software platform for bayesian evolutionary analysis. PLoS Comput. Biol. 10, e1003537 ( 10.1371/journal.pcbi.1003537) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Brandley MC, Wang Y, Guo X, Nieto Montes de Oca A, Fería-Ortíz M, Hikida T, Ota H. 2011. Accommodating heterogenous rates of evolution in molecular divergence dating methods: an example using intercontinental dispersal of Plestiodon (Eumeces) lizards. Syst. Biol. 60, 3–15. ( 10.1093/sysbio/syq045) [DOI] [PubMed] [Google Scholar]
- 39.Revell LJ. 2011. Phytools: an R package for phylogenetic comparative biology (and other things). Methods Ecol. Evol. 3, 217–223. ( 10.1111/j.2041-210X.2011.00169.x) [DOI] [Google Scholar]
- 40.Revell LJ. 2009. Size-correction and principal components for interspecific comparative studies. Evolution 63, 3258–3268. ( 10.1111/j.1558-5646.2009.00804.x) [DOI] [PubMed] [Google Scholar]
- 41.Pennell MW, Eastman JM, Slater GJ, Brown JW, Uyeda JC, FitzJohn RG, Alfaro ME, Harmon LJ. 2014. Geiger v2.0: an expanded suite of methods for fitting macroevolutionary models to phylogenetic trees. Bioinformatics 30, 2216–2218. ( 10.1093/bioinformatics/btu181) [DOI] [PubMed] [Google Scholar]
- 42.Beaulieu JM, Jhwueng DC, Boettiger C, O'Meara BC. 2012. Modeling stabilizing selection: expanding the Ornstein-Uhlenbeck model of adaptive evolution. Evolution 66, 2369–2383. ( 10.1111/j.1558-5646.2012.01619.x) [DOI] [PubMed] [Google Scholar]
- 43.Ingram T, Mahler DL. 2013. SURFACE: detecting convergent evolution from comparative data by fitting Ornstein-Uhlenbeck models with stepwise Akaike Information Criterion. Methods Ecol. Evol. 4, 416–425. ( 10.1111/2041-210X.12034) [DOI] [Google Scholar]
- 44.Sinervo B, Losos JB. 1991. Walking the tight rope: arboreal sprint performance among Sceloporus occidentalis lizard populations. Ecology 72, 1225–1233. ( 10.2307/1941096) [DOI] [Google Scholar]
- 45.Snyder RC. 1954. The anatomy and function of the pelvic girdle and hindlimb in lizard locomotion. Am. J. Anat. 95, 1–45. ( 10.1002/aja.1000950102) [DOI] [PubMed] [Google Scholar]
- 46.Schulte JA II, Losos JB, Cruz FB, Núnez H. 2004. The relationship between morphology, escape behaviour and microhabitat occupation in the lizard clade Liolaemus (Iguanidae: Tropidurinae: Liolaemini). J. Evol. Biol. 17, 408–420. ( 10.1046/j.1420-9101.2003.00659.x) [DOI] [PubMed] [Google Scholar]
- 47.Revell LJ, Johnson MA, Schulte JA II, Kolbe JJ, Losos JB. 2007. A phylogenetic test for adaptive convergence in rock-dwelling lizards. Evolution 61, 2898–2912. ( 10.1111/j.1558-5646.2007.00225.x) [DOI] [PubMed] [Google Scholar]
- 48.Harmon LJ, Kolbe JJ, Cheverud JM, Losos JB. 2005. Convergence and the multidimensional niche. Evolution 59, 409–421. ( 10.1111/j.0014-3820.2005.tb00999.x) [DOI] [PubMed] [Google Scholar]
- 49.Pincheira-Donoso D, Harvey LP, Ruta M. 2015. What defines an adaptive radiation? Macroevolutionary diversification dynamics of an exceptionally species-rich continental lizard radiation. BMC Evol. Biol. 15, 153 ( 10.1186/s12862-015-0435-9) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mayr E. 1970. Populations, species and evolution. Cambridge, MA: Harvard University Press. [Google Scholar]
- 51.Coyne JA, Orr HA. 2004. Speciation. Sunderland, MA: Sinauer Associates. [Google Scholar]
- 52.Moen D, Morlon H. 2014. Why does diversification slow down? Trends Ecol. Evol. 29, 190–197. ( 10.1016/j.tree.2014.01.010) [DOI] [PubMed] [Google Scholar]
- 53.Glor RE, Kolbe JJ, Powell R, Larson A, Losos JB. 2003. Phylogenetic analysis of ecological and morphological diversification in Hispaniolan trunk-ground anoles (Anolis cybotes group). Evolution 57, 2383–2397. ( 10.1111/j.0014-3820.2003.tb00250.x) [DOI] [PubMed] [Google Scholar]
- 54.Wollenberg KC, Wang IJ, Glor RE, Losos JB. 2013. Determinism in the diversification of Hispaniolan trunk-ground anoles (Anolis cybotes species complex). Evolution 67, 3175–3190. ( 10.1111/evo.12184) [DOI] [PubMed] [Google Scholar]
- 55.Paun O, Turner B, Trucchi E, Munzinger J, Chase MW, Samuel R. 2016. Processes driving the adaptive radiation of a tropical tree (Diospyros, Ebenaceae) in New Caledonia, a biodiversity hotspot. Syst. Biol. 65, 212–227. ( 10.1093/sysbio/syv076) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Singhal S, Moritz C. 2013. Reproductive isolation between phylogeographic lineages scales with divergence. Proc. R. Soc. B 280, 20132246 ( 10.1098/rspb.2013.2246) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pinho C, Hey J. 2010. Divergence with gene flow: models and data. Annu. Rev. Ecol. Evol. Syst. 41, 215–230. ( 10.1146/annurev-ecolsys-102209-144644) [DOI] [Google Scholar]
- 58.Leite RN, Kolokotronis SO, Almeida FC, Werneck FP, Rogers DS, Weksler M. 2014. In the wake of invasion: tracing the historical biogeography of the South American Cricetid Radiation (Rodentia, Sigmodontinae). PLoS ONE 9, e100687 ( 10.1371/journal.pone.0110081) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Parada A, D'Elia G, Palma RE. 2015. The influence of ecological and geographical context in the radiation of Neotropical sigmodontine rodents. BMC Evol. Biol. 15, 172 ( 10.1186/s12862-015-0440-z) [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Raw data are deposited in Dryad under: http://dx.doi.org/10.5061/dryad.j4r56.