Abstract
Vertebrate fossils have been collected for hundreds of years and are stored in museum collections around the world. These remains provide a readily available resource to search for preserved proteins; however, the vast majority of palaeoproteomic studies have focused on relatively recently collected bones with a well-known handling history. Here, we characterize proteins from the nasal turbinates of the first Castoroides ohioensis skull ever discovered. Collected in 1845, this is the oldest museum-curated specimen characterized using palaeoproteomic tools. Our mass spectrometry analysis detected many collagen I peptides, a peptide from haemoglobin beta, and in vivo and diagenetic post-translational modifications. Additionally, the identified collagen I sequences provide enough resolution to place C. ohioensis within Rodentia. This study illustrates the utility of archived museum specimens for both the recovery of preserved proteins and phylogenetic analyses.
Keywords: collagen I, palaeoproteomics, museum specimens, nasal turbinates, palaeoproteomics
1. Introduction
Palaeoproteomics is a rapidly growing subfield of molecular palaeontology, because it has been shown that proteins can provide phylogenetically informative sequence data, which persists well beyond the limit of DNA preservation [1–5]. Consequently, examination of protein sequences from vertebrate remains has become an attractive way to resolve parts of the vertebrate evolutionary tree that remain ambiguous after morphology-based phylogenetic inference [5–7]. However, the vast majority of these studies have relied on protein extraction from specimens that were recently collected (e.g. less than 30 years ago), limiting the diversity of taxa that have been characterized [1,2,4,5,7–12]. This fails to use the extensive collections of vertebrate remains possessed by many museums that were collected by naturalists in the nineteenth and early to mid-twentieth centuries. These specimens provide a readily available resource for proteomic investigation, and the sheer diversity of these collections holds great potential for the breadth of questions that may be elucidated by in-depth study.
However, proteomic investigations of long-archived museum specimens present many challenges, and care must be taken in interpretation of sequences derived from them. Typically, many of these specimens have an unknown and undocumented handling history, and thus critical information such as the application of consolidants, storage conditions and excavation details are missing. The lack of these data is one of the primary reasons why many palaeoproteomic studies have focused on recently collected remains; indeed, a known collection record has been a criterion for specimen selection that we (T.P.C. and E.R.S.) have applied to our own investigations [1,10]. Additionally, recent evidence suggests that molecules begin to degrade once a skeleton is removed from the equilibrium state of its burial environment [13–15], which may lead to loss of molecular information that could otherwise be gleaned from a freshly collected specimen of the same species. Together, these challenges contribute to the perception that historically collected specimens are simply too problematic to study using palaeoproteomic methodology.
To test whether we could obtain endogenous, phylogenetically informative peptide sequences from historical museum collections, we conducted proteomic analyses on protein extracts from the first known skull of Castoroides ohioensis (New York State Museum, NYSM VP-47), which was collected in 1845. Here, we characterize proteins and protein modifications obtained from a specimen collected more than 170 years ago, and use them to conduct phylogenetic analyses that support their endogeneity and utility for evolutionary investigations.
2. Material and methods
(a). Castoroides ohioensis skull
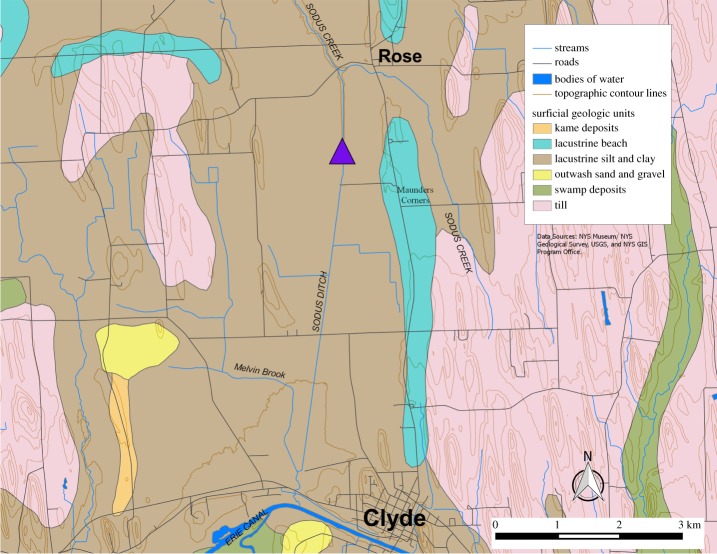
This skull (NYSM VP-47) was recovered in 1845 on the farm of General W. H. Adams of Clyde, NY, during excavation of a canal that connected Lake Ontario to the Erie Canal (figure 1) [16–18]. It was found approximately 2.5 m below the surface in a layer of fine sand (about 0.75 m thick) containing Planorbis, Valvata and Cyclas shell fragments [16]. Three layers occur above the skull, which were originally described as follows from the surface: (i) ‘vegetable soil’ (0.5–0.75 m thick) with heavy tree growth; (ii) a plant-rich (i.e. twigs, leaves, plant fragments) layer of fine sand with some clay (up to 1 m thick); and (iii) peat (over 1 m thick) including wood, bark, leaves and tree trunks [16]. Recently, the specimen has been carbon dated to 10 150 ± 50 14C yr BP (12 050–11 420 cal yr BP, 2σ; NOSAMS OS-73632; [19]).
Figure 1.
Approximate discovery location (centre of the purple triangle) of the skull based on historical accounts. (Online version in colour.)
(b). Protein extraction
Two sets of fragments from the nasal turbinates (sample 1, 19.5 mg; sample 2, 19.8 mg) of NYSM VP-47 were homogenized with a Bead Ruptor 24 (Omni International) using the non-demineralization buffer from [20] to extract proteins. The turbinates were chosen because this skull has an apparent varnish on the entire exterior (figure 2) that does not appear to have been applied within the nasal cavity. In short, the bone proteins were extracted in 600 µl of 400 mM ammonium phosphate dibasic/200 mM ammonium bicarbonate/4 M guanidine HCl pH 8.2 with the following homogenization parameters: five cycles of 45 s homogenization at 6.95 m s−1 followed by 5 min dwell time on ice. After extraction, all samples were centrifuged at 13 200 r.p.m. for 10 min and the protein concentration was measured with the Pierce Protein Plus (Coomassie) Assay. Subsequently, the samples were microdialysed [21] for 2 days at 4°C, then stored at −20°C until use. A buffer control (a sample that contains no bone material, only buffer reagents) was produced in tandem with the fossil sample, using the same procedure above.
Figure 2.

Dorsal view of the Castoroides ohioensis skull showing the apparent varnish. (Online version in colour.)
(c). Protein digestion
Dialysed protein (30 µg) and a dialysed buffer control (equal volume to the protein sample) were reduced with 10 mM dithiothreitol for 1 h at room temperature, alkylated with 30 mM iodoacetamide for 1 h in the dark, then digested with Promega Trypsin Gold (1 : 100, trypsin: bone protein) overnight at 37°C. The resultant peptides were desalted using Empore C18 (3 M) stage tips [22].
(d). Mass spectrometry
Desalted C. ohioensis peptides (10 µg/injection) and an equal volume of buffer blank were separated on a ThermoScientific BioBasic C18 column using Agilent 1200 series HPLC with the following gradient: 2% B 0–5 min, 30% B 5–35 min, 60% B 35–60 min, 95% B 60.01–64 min, 2% B 64.01–75 min. Buffer A is 0.2% formic acid and buffer B is 100% acetonitrile, 0.2% formic acid. Eluted peptides were characterized in positive mode on an LTQ-Orbitrap XL (Thermo Scientific) with the following parameters: top five peaks fragmented with CID (35 normalized collision energy) with a 2 m z−1 isolation window; 30 000 resolution for precursor; precursor scan range 300–2000 m z−1. The five most abundant peaks were fragmented using CID and analysed in the ion trap.
(e). Data analysis
All RAW files were searched using PEAKS 7.5 [23] with the following parameters: precursor mass tolerance was set to 10 ppm and fragment ion mass tolerance to 0.5 Da. Three missed cleavages were allowed, and non-specific cleavage was allowed at one end of each peptide. The following post-translational modifications (PTMs) were allowed: fixed—carbamidomethylation [C]; variable—oxidation [M], oxidation or hydroxylation [RYFPNKD], [G] @C-term, carboxymethylation [KW, X@N-term] and deamidation [NQ]. Up to 7 PTMs were allowed per peptide to accommodate for large collagen I peptides with abundant proline hydroxylations. Spectra were searched against the UniProt Mammalia and cRAP (The Global Proteome Machine) databases, and PEAKS PTM and SPIDER were enabled. FDR filtering was set to less than or equal to 1% for PSMs, and either less than or equal to 1% FDR for proteins or a protein score of −101gP ≥ 20 (whichever was more strict) plus 1 unique peptide. Neither database contains bacterial nor fungal proteins.
(f). Phylogenetic analysis
Peptides were aligned using Seaview 4.5.4 × 64 against various Euarchontoglire collagen I sequences derived from UniProt (electronic supplementary material, table S1). A basic consensus method was used to generate final C. ohioensis collagen I alpha 1 and alpha 2 sequences. For leucine (L) and isoleucine (I) determination if the consensus method resulted in ambiguity during alignment, we chose the majority sequence homology with the aligned species to determine which amino acid was chosen (sensu [6]). This was applied to the following residues (chain, residue, I/L): Co1a1547-L, Co1a11013-L, Co1a11125-L, Co1a11164-L, Co1a11167-L, Co1a2255-I, Co1a2320-L, Co1a2329-L, Co1a2460-L, Co1a2589-L, Co1a2876-L. After manual concatenation, MrBayes files were generated in Mesquite v. 3.04 [24]. Using MrBayes v. 3.2.2 × 64 [25], the collagen I matrix was searched with the following parameters: outgroup Homo sapiens, prset aamodelpr = mixed, 100 000 generations with sampling frequency 10, and burnin of 25 000 generations. Trees were output in FigTree v. 1.4.0.
3. Results and discussion
Palaeoproteomic studies have focused on relatively recently collected vertebrate remains because of their unknown sampling history. Our selection of nasal turbinates from this historically collected C. ohioensis skull overcomes this unknown history for the first time and has allowed us to detect many endogenous peptides from collagen I and a single peptide from haemoglobin beta (see below). Nasal turbinates provide a relatively protected area for sampling in many mammalian skulls, reducing the likelihood of having consolidants applied to them, and offer a position to destructively sample that has limited or no impact on the external morphology of the skull.
(a). Mass spectrometry
The mass spectrometry analysis of sample 1 (20.15 ± 1.11 mg protein g−1 bone) resulted in 3405 MS1 scans and 14 582 MS2 scans (2234 peptide spectrum matches (PSMs), 446 unique peptides). Sample 2 (19.95 ± 2.27 mg protein g−1 bone) resulted in 3381 MS1 scans and 14 458 MS2 scans (2427 PSMs, 462 unique peptides). For both samples, the vast majority of the detected peptides were from collagen I alpha 1 and alpha 2, with the exception of one haemoglobin beta peptide (electronic supplementary material, figure S1). A consensus of collagen peptides from both samples resulted in 49.7% of the mature sequence of collagen I alpha 1 (when compared with mouse mature collagen I alpha 1), and 44.0% collagen I alpha 2. The prevalence of collagen I in these samples is consistent with bone tissue, where collagen I represents 90% of the total bone protein [26]. Further, the recovery of over 40% of both alpha 1 and alpha 2 suggests that the molecular preservation in this historically collected specimen is substantial enough to support basic phylogenetic analyses and evaluate PTM preservation.
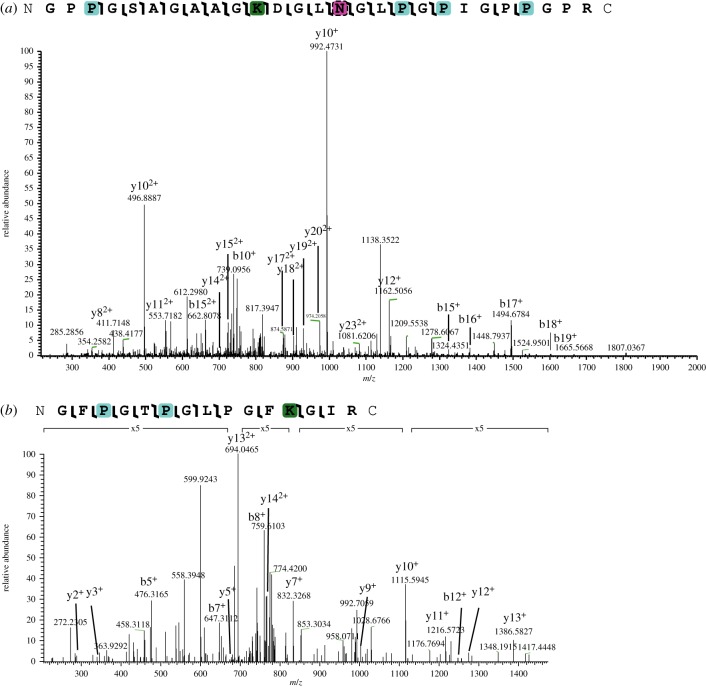
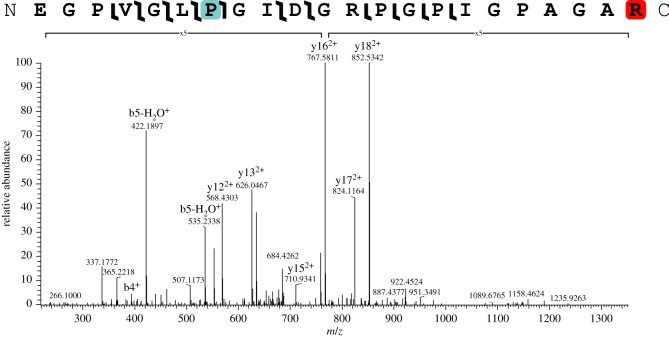
We detected a variety of both in vivo and diagenetic PTMs in these samples, but did not detect glucosylgalactosyllysine (contra [11]). The lack of glucosylgalactosyl modification may be related to non-detection of the cross-linking region within the collagen triple helix [27] or neutral loss of the sugar molecules and limited peptide fragmentation during CID fragmentation [28]. For NYSM VP-47, the majority of in vivo PTMs identified were hydroxylation of proline (figures 3 and 4). Hydroxyprolines are essential for forming the tertiary structure of collagen I into a triple helix [29]; thus, the abundance of hydroxyproline is consistent with almost exclusive detection of collagen I peptides. We also identified methylation on arginine residues, consistent with [10] and glutamic acid residues (figure 4). Arginine methylation was localized to one position on the collagen I alpha 2 chain (EGPVGLPGIDGRPGPIGPAGAR, position 492 (compared to Mus musculus)) and one position on the collagen I alpha 2 chain (AGDRGETGPSGPAGPAGPTGAR, position 1043 (compared to M. musculus)), which are probably endogenous to NYSM VP-47 because no known diagenetic modifications have been shown to change PTMs with this specificity. However, these collagen I arginine methylations have unknown roles in extant collagen I, so further investigation is warranted to understand their function in bone. Both hydroxylation and methylation have been previously identified in palaeoproteomic studies of bone collagen I [5,8,10–12], so this specimen further supports biological PTM preservation in fossils. For in vivo PTMs, we hypothesize that the hydroxylation, methylation and acetylation we observe on these collagen I peptides are derived from NYSM VP-47 because they (i) require enzymes to produce with specificity [10,30,31] and (ii) are observed on expected residues compared to extant PTM profiles [10], especially for hydroxylation of proline on the third residue in the Gly-X-Y motif [29,32]. However, other novel PTMs of collagen I are still being localized and detected, so identification of previously unknown PTMs does not preclude them being endogenous.
Figure 3.
(a) Collagen I alpha 1 peptide with hydroxyproline (teal), the AGE CML (green) and variable deamidation (pink with hatched border). (b) Collagen I alpha 2 peptide with hydroxyproline and CML. Magnified regions of the spectrum are denoted with a bracket and magnification level. (Online version in colour.)
Figure 4.
Collagen I alpha 2 peptide with hydroxyproline (teal) and methylated arginine (red) on the C-terminus. Magnified regions of the spectrum are denoted with a bracket and magnification level. (Online version in colour.)
We also observe a number of previously identified diagenetic changes to the sequences. Some of these modifications may be the result of sample preparation (e.g. deamidation); however, our non-demineralization-based, slightly basic buffer extraction method (in contrast to acidic methods) should limit the introduction of artefacts. These diagenetic PTMs (electronic supplementary material, table S2) include at least three examples of protein truncation (i.e. non-tryptic N-terminal or C-terminal ends [10]), the advanced glycation end-product (AGE) carboxymethyllysine (CML; figure 3a,b), and variable deamidation (figure 3a). The following truncations were observed, and are denoted by | or |. indicating the amino acid following a truncation occurs at the end of the peptide: (i) collagen I alpha 2 GPAGEPGTAGPPGTP|GPQG|L|LGAPGILGLPGSR.; (ii) collagen I alpha 2 GSDGSVGPVGPAGPIGSAGPPGFPGAPGPKG|. and (iii) collagen I alpha 1 G|VSVPGPMGPSGPR. These truncations and the CML-modified peptides are located at different positions than those from previously detected from non-mammalian collagen I sequences [10], supporting their diagenetic, non-enzymatic origin. These truncations, which would result in many collagen sequences of variable, unpredictable lengths, may be a source of the smearing frequently observed in palaeoproteomics SDS-PAGE gels [12]. Future analyses of protein truncations and other diagenetic changes may allow for a deeper understanding of how collagen I degrades and/or preserves in the fossil record [33]. The combination and prevalence of AGEs and protein truncations in these samples support the endogeneity of these sequences to C. ohioensis, as recent protein contamination of the skull would not show characteristics of degradation, and that the collagen is well preserved.
(b). Phylogenetic analysis
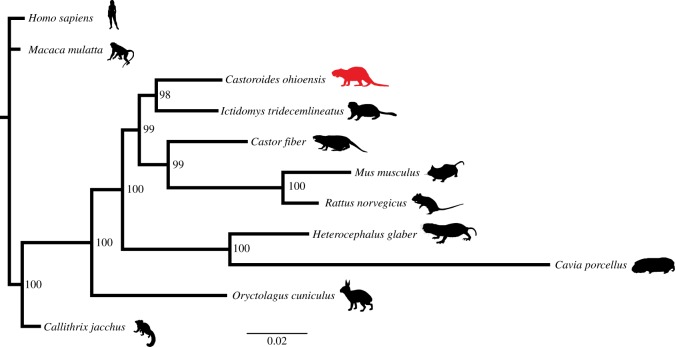
To test whether the degraded peptide sequences of collagen I recovered from this specimen were truly from C. ohioensis and not an exogenous source, we performed phylogenetic analyses using these peptides. Collagen I is a largely conserved protein; however, it has recently been used to resolve the phylogenetic position of multiple taxa with unresolved phylogenetic positions [5–7]. Castoroides ohioensis has a relatively constrained phylogenetic position within Castoroidinae (a subclade within Rodentia) [34,35]; however, no previous attempt to apply molecular phylogenetic inference has been attempted for this species. Here, we searched an aligned, concatenated collagen I alpha 1 and alpha 2 matrix (2856 amino acid sites, 910 complete sites (no gaps, no X), 195 variable sites) using MrBayes allowing for optimized model selection based on the results. The Dayhoff model [36] was the only model used for this analysis and was the only model found to have a model probability of more than 0.050. The resultant tree (figure 5) from the analysis of the consensus C. ohioensis collagen I sequences resulted in a tree topology with C. ohioensis as a sister taxon to Ictomys tridecemlineatus within Rodentia (figure 5), with Oryctolagus cuniculus (Lagomorpha) representing the sister taxon to the Rodentia clade ((Heterocephalus glaber, Cavia porcellus), ((C. ohioensis, I. tridecemlineatus), (Castor fiber, (Mus musculus, Rattus norvegicus)))). This topology generally reflects what has been observed for extant taxa using nuclear genes [37] with the exception of the relationship between C. ohioensis and I. tridecemlineatus. Synthesizing morphological trees containing C. ohioensis [34] and the molecular Rodentia tree [37] indicates that I. tridecemlineatus should be phylogenetically closer to O. cuniculus than C. ohioensis, and C. ohioensis should be the sister taxon to C. fiber, but this may be the result of the conserved nature of collagen I. Regardless of minor topological differences compared with previous analyses, the well-supported placement of C. ohioensis within Rodentia based solely on collagen I sequences supports the endogeneity of these sequences. Future addition of other collagen I sequences and other protein sequences will further refine the molecular placement of C. ohioensis within Rodentia.
Figure 5.
Bayesian phylogeny of C. ohioensis collagen I sequences compared to other Euarchontoglire collagen I sequences. (Online version in colour.)
4. Conclusion
Recent advances in palaeoproteomics have allowed for the investigation of new questions about the kinds of proteins [8,12] and in vivo PTMs [10] that preserve, as well as the types of PTMs that can arise from diagenesis [10]. However, the restriction of previous studies to recently collected specimens has limited the scope of such investigations. For the first time, we have been able to detect peptide sequences from skeletal remains collected over 170 years ago. This C. ohioensis skull was not collected for molecular analyses or stored, for much of its history, in a way that would promote preservation of biomolecules; however, it has yielded a large amount of peptide sequence information and PTM data that are both endogenous and place C. ohioensis within Rodentia. These data support that historically collected vertebrate remains can retain proteins and that they can be used extensively in future palaeoproteomic studies to better cover greater portions of the phylogenetic tree.
Supplementary Material
Acknowledgements
We would like to thank E. Cleland for generating the map in figure 1 using QGIS 2.8.1 with publicly accessible data from the New York Geological Survey, D. Zagorevski for access to the proteomics core at RPI and Omni International for access to the Bead Ruptor 24. We would also like to thank F. Welker for providing the Castor fiber collagen I sequence. We would also like to thank the following people for access to silhouettes and images for adaptation used in figure 5: Greg Goebel (adapted Ictidoys tridecemlineatus; CCBY-SA 2.0); Virginie Moerenhout (adapted Cavia porcellus; CCBY 2.0); Rebecca Groom (Rattus norvegicus; CCA-SA 3.0 unported); Steven Traver (Heterocephalus glaber, Oryctolagus cuniculus, Castor fiber; Public Domain Dedication 1.0); Madeleine Price-Ball (Mus musculus; Public Domain Mark 1.0); Phylopic (Macaca mulatta; Public Domain 1.0); NASA (Homo sapiens; Public Domain Mark 1.0); Yan Wong (Callithrix jacchus; Public Domain Dedication 1.0); and Zimices (Castoroides ohioensis; CCA-SA 3.0 unported).
Data accessibility
All RAW mass spectrometry data, PEAKS results and C. ohioensis collagen I sequence fasta are available at Dryad (http://dx.doi.org/10.5061/dryad.326fv).
Authors' contributions
All authors conceived and designed this study. T.P.C. acquired the data, R.S.F. provided access to NYSM VP-47, T.P.C. and E.R.S. performed bioinformatics, and all authors made interpretations. T.P.C., E.R.S. and R.S.F. wrote the manuscript, and all authors revised it leading to the final version.
Competing interests
We declare we have no competing interests.
Funding
This project was funded by NSF CCMI 1363526 (D.V.) and NSF INSPIRE (E.R.S.).
References
- 1.Cleland TP, et al. 2015. Mass spectrometry and antibody-based characterization of blood vessels from Brachylophosaurus canadensis. J. Proteome Res. 14, 5252–5262. ( 10.1021/acs.jproteome.5b00675) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Asara JM, Schweitzer MH, Freimark LM, Phillips M, Cantley LC. 2007. Protein sequences from mastodon and Tyrannosaurus rex revealed by mass spectrometry. Science 316, 280–285. ( 10.1126/science.1137614) [DOI] [PubMed] [Google Scholar]
- 3.Organ CL, Schweitzer MH, Zheng W, Freimark LM, Cantley LC, Asara JM. 2008. Molecular phylogenetics of mastodon and Tyrannosaurus rex. Science 320, 499 ( 10.1126/science.1154284) [DOI] [PubMed] [Google Scholar]
- 4.Schweitzer MH, et al. 2009. Biomolecular characterization and protein sequences of the Campanian Hadrosaur B. canadensis. Science 324, 626–631. ( 10.1126/science.1165069) [DOI] [PubMed] [Google Scholar]
- 5.Welker F, et al. 2015. Ancient proteins resolve the evolutionary history of Darwin's South American ungulates. Nature 522, 81–84. ( 10.1038/nature14249) [DOI] [PubMed] [Google Scholar]
- 6.Buckley M. 2013. A molecular phylogeny of Plesiorycteropus reassigns the extinct mammalian order ‘Bibymalagasia’. PLoS ONE 8, e59614 ( 10.1371/journal.pone.0059614) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Buckley M. 2015. Ancient collagen reveals evolutionary history of the endemic South American ‘ungulates’. Proc. R. Soc. B 282, 20142671 ( 10.1098/rspb.2014.2671) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cappellini E, et al. 2012. Proteomic analysis of a Pleistocene mammoth femur reveals more than one hundred ancient bone proteins. J. Proteome Res. 11, 917–926. ( 10.1021/pr200721u) [DOI] [PubMed] [Google Scholar]
- 9.Orlando L, et al. 2013. Recalibrating Equus evolution using the genome sequence of an early Middle Pleistocene horse. Nature 499, 74–78. ( 10.1038/nature12323) [DOI] [PubMed] [Google Scholar]
- 10.Cleland TP, Schroeter ER, Schweitzer MH. 2015. Biologically and diagenetically derived peptide modifications in moa collagens. Proc. R. Soc. B 282, 20150015 ( 10.1098/rspb.2015.0015) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hill RC, Wither MJ, Nemkov T, Barrett A, D'Alessandro A, Dzieciatkowska M, Hansen KC. 2015. Preserved proteins from extinct bison latifrons identified by tandem mass spectrometry; hydroxylysine glycosides are a common feature of ancient collagen. Mol. Cell Proteomics 14, 1946–1958. ( 10.1074/mcp.M114.047787) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wadsworth C, Buckley M. 2014. Proteome degradation in fossils: investigating the longevity of protein survival in ancient bone. Rapid Commun. Mass Spectrometry 28, 605–615. ( 10.1002/rcm.6821) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pruvost M, Schwarz R, Correia VB, Champlot S, Braguier S, Morel N, Fernandez-Jalvo Y, Grange T, Geigl EM. 2007. Freshly excavated fossil bones are best for amplification of ancient DNA. Proc. Natl Acad. Sci. USA 104, 739–744. ( 10.1073/pnas.0610257104) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bollongino R, Tresset A, Vigne JD. 2008. Environment and excavation: pre-lab impacts on ancient DNA analyses. C. R. Palevol. 7, 91–98. ( 10.1016/j.crpv.2008.02.002) [DOI] [Google Scholar]
- 15.Lopez-Polin L, Olle A, Caceres I, Carbonell E, de Castro JMB. 2008. Pleistocene human remains and conservation treatments: the case of a mandible from Atapuerca (Spain). J. Hum. Evol. 54, 539–545. ( 10.1016/j.jhevol.2007.07.011) [DOI] [PubMed] [Google Scholar]
- 16.Hall J, Wyman J. 1846. Notice on the geological position of the cranium of the Castoroides ohioensis. Boston Journ. Nat. Hist 5, 385–401. [Google Scholar]
- 17.Hall J. 1846. On the geological relations of the fossil Castoroides ohioensis. Proc. Boston Soc. Nat. Hist. 2, 167–168. [Google Scholar]
- 18.Hartnagel CA, Bishop SC. 1921. The mastodons, mammoths and other Pleistocene mammals of New York State: being a descriptive record of all known occurrences. Albany, NY: University of the State of New York. [Google Scholar]
- 19.Feranec RS, Kozlowski AL. 2010. Ams radiocarbon dates from Pleistocene and Holocene mammals housed in the New York State Museum, Albany, New York, USA. Radiocarbon 52, 205–208. [Google Scholar]
- 20.Cleland TP, Vashishth D. 2015. Bone protein extraction without demineralization using principles from hydroxyapatite chromatography. Anal. Biochem. 472, 62–66. ( 10.1016/j.ab.2014.12.006) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sroga GE, Karim L, Colón W, Vashishth D. 2011. Biochemical characterization of major bone-matrix proteins using nanoscale-size bone samples and proteomics methodology. Mol. Cell. Proteomics 10, M110006718. ( 10.1074/mcp.M110.006718) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rappsilber J, Mann M, Ishihama Y. 2007. Protocol for micro-purification, enrichment, pre-fractionation and storage of peptides for proteomics using StageTips. Nat. Protocols 2, 1896–1906. ( 10.1038/nprot.2007.261) [DOI] [PubMed] [Google Scholar]
- 23.Ma B, Zhang K, Hendrie C, Liang C, Li M, Doherty-Kirby A, Lajoie G. 2003. PEAKS: powerful software for peptide de novo sequencing by tandem mass spectrometry. Rapid Commun. Mass Spectrometry 17, 2337–2342. ( 10.1002/rcm.1196) [DOI] [PubMed] [Google Scholar]
- 24.Maddison WP, Maddison DR.2015. Mesquite: a modular system for evolutionary analysis. Version 3.04. See http://mesquiteproject.org .
- 25.Ronquist F, Huelsenbeck JP. 2003. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19, 1572–1574. ( 10.1093/bioinformatics/btg180) [DOI] [PubMed] [Google Scholar]
- 26.Hauschka PV, Wians FH. 1989. Osteocalcin-hydroxyapatite interaction in the extracellular organic matrix of bone. Anat. Rec. 224, 180–188. ( 10.1002/ar.1092240208) [DOI] [PubMed] [Google Scholar]
- 27.Eyre DR, Weis MA, Wu J-J. 2008. Advances in collagen cross-link analysis. Methods 45, 65–74. ( 10.1016/j.ymeth.2008.01.002) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mechref Y. 2001. Use of CID/ETD mass spectrometry to analyze glycopeptides. In Current protocols in protein science (eds JE Coligan, BM Dunn, DW Speicher, PT Wingfield), pp. 12.11.1–12.11.11. New York, NY: John Wiley & Sons, Inc. [Google Scholar]
- 29.Prockop DJ, Kivirikko KI. 1995. Collagens: molecular biology, diseases, and potentials for therapy. Annu. Rev. Biochem. 64, 403–434. ( 10.1146/annurev.bi.64.070195.002155) [DOI] [PubMed] [Google Scholar]
- 30.Bedford MT, Richard S. 2005. Arginine methylation: an emerging regulator of protein function. Mol. Cell 18, 263–272. ( 10.1016/j.molcel.2005.04.003) [DOI] [PubMed] [Google Scholar]
- 31.Kouzarides T. 2000. Acetylation: a regulatory modification to rival phosphorylation? EMBO J. 19, 1176–1179. ( 10.1093/emboj/19.6.1176) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kalluri R. 2003. Basement membranes: structure, assembly and role in tumour angiogenesis. Nat. Rev. Cancer 3, 422–433. ( 10.1038/nrc1094) [DOI] [PubMed] [Google Scholar]
- 33.San Antonio JD, Schweitzer MH, Jensen ST, Kalluri R, Buckley M, Orgel JPRO. 2011. Dinosaur peptides suggest mechanisms of protein survival. PLoS ONE 6, e20381 ( 10.1371/journal.pone.0020381) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rybczynski N. 2006. Castorid phylogenetics: implications for the evolution of swimming and tree-exploitation in beavers. J. Mamm. Evol. 14, 1–35. ( 10.1007/s10914-006-9017-3) [DOI] [Google Scholar]
- 35.Korth WW. 2001. Comments on the systematics and classification of the beavers (Rodentia, Castoridae). J. Mamm. Evol. 8, 279–296. ( 10.1023/a:1014468732231) [DOI] [Google Scholar]
- 36.Dayhoff MO, Schwartz R, Orcutt B. 1978. A model of evolutionary change in proteins. In Atlas of protein sequence and structure, vol. 5, suppl. 3 (ed. Dayhoff MO.), pp. 345–352. Washington, DC: National Biomedical Research Foundation. [Google Scholar]
- 37.Blanga-Kanfi S, Miranda H, Penn O, Pupko T, DeBry RW, Huchon D. 2009. Rodent phylogeny revised: analysis of six nuclear genes from all major rodent clades. BMC Evol. Biol. 9, 1–12. ( 10.1186/1471-2148-9-71) [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All RAW mass spectrometry data, PEAKS results and C. ohioensis collagen I sequence fasta are available at Dryad (http://dx.doi.org/10.5061/dryad.326fv).