Abstract
Modelling the spatial spread of vector-borne zoonotic pathogens maintained in enzootic transmission cycles remains a major challenge. The best available spatio-temporal data on pathogen spread often take the form of human disease surveillance data. By applying a classic ecological approach—occupancy modelling—to an epidemiological question of disease spread, we used surveillance data to examine the latent ecological invasion of tick-borne pathogens. Over the last half-century, previously undescribed tick-borne pathogens including the agents of Lyme disease and human babesiosis have rapidly spread across the northeast United States. Despite their epidemiological importance, the mechanisms of tick-borne pathogen invasion and drivers underlying the distinct invasion trajectories of the co-vectored pathogens remain unresolved. Our approach allowed us to estimate the unobserved ecological processes underlying pathogen spread while accounting for imperfect detection of human cases. Our model predicts that tick-borne diseases spread in a diffusion-like manner with occasional long-distance dispersal and that babesiosis spread exhibits strong dependence on Lyme disease.
Keywords: invasion, Lyme disease, babesiosis, tick-borne disease, occupancy model
1. Introduction
Tick-borne diseases (TBDs) are responsible for over 300 000 human cases per year in the USA alone [1] and are globally emerging owing to the expanding ranges of tick vectors, reservoir hosts and pathogens [2–4]. Understanding the ecological process of TBD invasion is fundamental for developing effective surveillance systems and designing measures to limit further geographical spread. However, quantifying the spatial spread of zoonotic TBD maintained in enzootic transmission cycles remains a major challenge [5] because longitudinal data from sampling vectors and wildlife host species at wide spatial scales are not currently available. For this reason, human surveillance data offer us the best spatially explicit time series available. However, these data represent only the observed epidemiological manifestation of a complex constellation of processes: introduction and establishment of tick-borne pathogens in enzootic transmission cycles (generating entomological risk), human exposure to infected tick vectors, human infection, progression from infection to disease, case diagnosis and case reporting.
Occupancy and metapopulation models, commonly used in population ecology, can be applied to model the spread of TBD. Occupancy models explicitly consider the observation process (in this case, diagnosing and reporting human cases) as contingent on the underlying distribution of a species (in this case, a tick-borne pathogen) and have been used to examine the prevalence of imperfectly detected human and wildlife pathogens and disease vectors [6–10]. However, their use in modelling the spatio-temporal distribution of invading pathogens is relatively new [11,12] and to the best of our knowledge, they have not been applied to explore the processes underlying spatial spread of disease. Metapopulation models consider the dynamics of spatially separated but interacting populations (such as the patchy distribution of a tick-borne pathogen) [13]. Metapopulation models have been used to model infectious disease dynamics [13–16], but have not been applied within an occupancy framework to dissect the processes underlying spatial disease spread.
Lyme disease and babesiosis are recently emerging in North America: Lyme disease was first described in Lyme, Connecticut in 1976 and babesiosis on Nantucket Island, Massachusetts in 1969 [17,18]. Both the Lyme disease bacteria, Borrelia burgdorferi, and the babesiosis parasite, Babesia microti, share the same tick vector (Ixodes scapularis) and an overlapping community of vertebrate reservoir hosts. Despite their similarities, Lyme disease has rapidly spread through most of New England and the Midwest, and over 30 000 cases of Lyme disease are reported each year in the USA, although the true burden of disease is estimated to be 10 times greater [1,4]. By contrast, babesiosis expansion appears to follow that of Lyme disease and fewer than 2000 babesiosis cases are reported yearly [19–21].
How do the two co-vectored pathogens move across the landscape and what explains their markedly different invasion trajectories? The spread of tick-borne pathogens and diseases coarsely followed the spread of I. scapularis ticks out of southern New England; however, little is known about the latent processes underlying rapid emergence [20,22]. Apparent spread of reported disease may reflect the underlying ecological invasion, epidemiological (exposure) and/or observation processes (human case diagnosis and surveillance).
Ixodes scapularis only move a few metres during each life stage [23,24]; thus, tick-borne pathogen dispersal is dependent on movement of mammalian and avian hosts, which differ greatly both in their dispersal capacity, proportion of the tick population fed and reservoir competence for both B. burgdorferi and B. microti [19,25–27]. It remains unclear whether the risk of TBD propagates locally, generating a rabies disease-like invasion front [11,16] or if TBD risk is driven by long-distance dispersal as occurs for West Nile Virus fever [28]. Although several studies have identified possible ecological correlates of TBD prevalence in humans, including density of infected ticks [29], and correlates of the distribution of infected ticks, including proximity to water bodies [30] and, for babesiosis, prevalence of Lyme disease [31], the ecological factors driving spread of reported TBD at the scale of the northeast United States remain unresolved. Spread differences between pathogens may reflect biological differences or may be in part an artefact of the better-defined symptomatology and/or heightened awareness of Lyme disease compared with babesiosis [32–34].
We use an occupancy model to test the following ecological and epidemiological hypotheses about the processes of TBD spread and the observed differences in Lyme disease and babesiosis emergence.
Processes underlying spread.
(i) Both local spread and long-distance dispersal contribute to the spread of TBD;
(ii) high tick density facilitates introductions of TBD; and
(iii) proximity to water (a major river or the Atlantic coastline) facilitates spread of TBD.
Contrasting spread trajectories.
(iv) Lyme disease presence facilitates the colonization and persistence of babesiosis; and
(v) Lyme disease is more likely to be reported than babesiosis.
Our modelling approach allows us to quantify the spatial dynamics of pathogen invasion and provides insights into the future distribution of Lyme disease and babesiosis in the northeast United States.
2. Material and methods
(a). Data
As Lyme disease and babesiosis are nationally notifiable diseases, states are required to collect and report surveillance data. We compiled human case data available at the town level from the Connecticut, Massachusetts, New Hampshire and Rhode Island state health departments from 1984 to 2014 for Lyme disease and from 1985 to 2014 for babesiosis (figure 1 and electronic supplementary material, figure S1 and table S1). Data from Vermont and Maine were not available at the town level. We included both probable and confirmed cases and excluded imported cases. Reporting effort varied across states and over time [35,36] (electronic supplementary material, table S1). Here, we are primarily interested in the invasion process and thus modelled spatio-temporal occupancy dynamics rather than the abundance of cases. The disease status of a town was defined as either 0 (no cases reported) or 1 (at least one case reported) based on the yearly surveillance data as in a typical Levins-type metapopulation model, which does not consider the internal dynamics of a subpopulation [13,16].
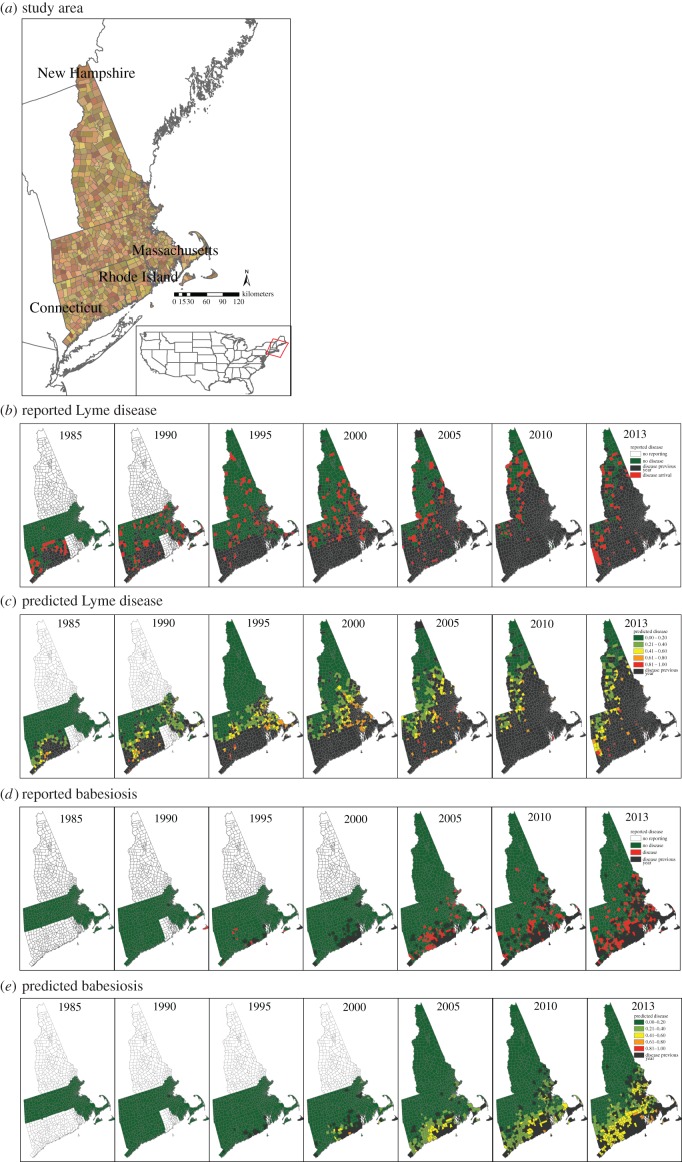
Figure 1.
Study area, (a) shown with available surveillance data and model predictions. (b) Lyme disease reports and (c) model predicted town-level disease status from 1985 to 2013. (d) Babesiosis reports and (e) model predictions. Town colours in (a) are for visualization only. In (b–d), towns in which disease was reported in the previous year are indicated in black. Predictions are one-step-ahead predictions conditional on the previous years' reports.
Density data for infected I. scapularis were derived from a previously published field- and climate-based model for density of infected nymphal I. scapularis ticks based on sampling from 2004 to 2006 [37]. Infected tick density is thus a static spatial predictor; however, habitat suitability for I. scapularis and thus infected I. scapularis density is dynamic [38] and exhibits spatio-temporal variation not captured by this covariate.
We determined the minimum Cartesian distance from the town geographical centroid to one of the three major rivers in our study area—the Hudson, Connecticut and Merrimack Rivers—or the Atlantic coastline in R v. 3.1.1 [39] as a measure of proximity to a major water body.
(b). Model
To test hypotheses regarding the patterns and drivers of TBD spread, we fitted an occupancy model including observation (disease reporting) and ecological processes. This allowed us to quantify the contribution of latent processes to disease spread and test the significance of covariates to each spread process independently. We compare this ecological process model with a null model that assumes the probability of at least one reported human case in a town depends only on its disease status and that of its adjacent neighbours in the previous time step (electronic supplementary material, text S1, see neighbourhood definitions, below).
Town-level case reports are available for each of 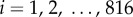 towns. Let
towns. Let 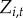 be the disease status of town i at time t (a random variable). The disease status of town i at time t (
be the disease status of town i at time t (a random variable). The disease status of town i at time t (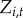 ) arises from an underlying Bernoulli process with the probability of at least one reported human case occurring in town i at time t
) arises from an underlying Bernoulli process with the probability of at least one reported human case occurring in town i at time t
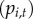 , where
, where 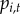 depends on the infection status of town i and its neighbours, defined below, in the previous year
depends on the infection status of town i and its neighbours, defined below, in the previous year  .
.
(i). Observation process
TBDs are substantially under-reported [1,35], and heterogeneities in reporting across space and time are difficult to quantify. Our data include reported cases; therefore, we modelled 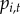 as the probability of at least one reported human case occurring in town i at time t. Data on B. burgdorferi or B. microti incidence (e.g. information from serosurveys) are not available at regional spatial scales and therefore, we cannot estimate incidence rates of symptomatic and asymptomatic disease nor reporting rates for the two diseases. Reporting occurs at the state-level and states have differing case definitions, reporting efforts and surveillance infrastructure. Therefore, we introduced a state reporting effect
as the probability of at least one reported human case occurring in town i at time t. Data on B. burgdorferi or B. microti incidence (e.g. information from serosurveys) are not available at regional spatial scales and therefore, we cannot estimate incidence rates of symptomatic and asymptomatic disease nor reporting rates for the two diseases. Reporting occurs at the state-level and states have differing case definitions, reporting efforts and surveillance infrastructure. Therefore, we introduced a state reporting effect 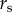 , which captures state-level variation in surveillance. More specifically, we defined state reporting effect
, which captures state-level variation in surveillance. More specifically, we defined state reporting effect 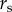 as the probability cases in any town in state s will be documented by the public health department of their state (constant probability across towns) given the modelled probability of case occurrence
as the probability cases in any town in state s will be documented by the public health department of their state (constant probability across towns) given the modelled probability of case occurrence 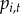 :
:
| 2.1 |
While surveillance probably varies across towns within the same state, available data do not include site-time replicates; therefore, a model including town-varying reporting effects is not identifiable [11,40]. To test the hypothesis that the Lyme disease surveillance is more intensive than babesiosis (hypothesis (v)), we compared 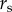 estimated for each disease.
estimated for each disease.
(ii). Ecological process
We adapted a previously described occupancy model [41] to model the latent ecological processes underlying TBD spread—initial colonization of a previously unoccupied area, local persistence or extinction, and potentially, recolonization. (Parameters are defined in the electronic supplementary material, table S2.)
(i) Initial colonization. If town i was not diseased at the previous time step
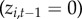 and the town has never before reported disease
and the town has never before reported disease 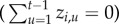 , then the town is available for initial colonization with probability
, then the town is available for initial colonization with probability 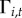 .
.(ii) Persistence. If town i was diseased at the previous time step
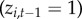 , then disease may persist locally with probability
, then disease may persist locally with probability 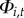 . (The probability of local extinction is
. (The probability of local extinction is 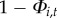 )
)(iii) Re-colonization. If town i was not diseased at the previous time step
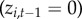 , but the town previously reported disease at some point during the surveillance period
, but the town previously reported disease at some point during the surveillance period 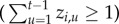 , then the town can be re-colonized with probability
, then the town can be re-colonized with probability 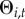 . This allowed us to distinguish between primary and subsequent introductions.
. This allowed us to distinguish between primary and subsequent introductions.
The indicator variable 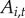 represents local disease history and the availability of town i for initial disease introduction. If a town has never reported disease, it is available to be colonized by disease,
represents local disease history and the availability of town i for initial disease introduction. If a town has never reported disease, it is available to be colonized by disease, 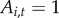 . If the town has previously reported disease in any year,
. If the town has previously reported disease in any year, 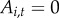 :
:
| 2.2 |
The probability of reported disease in town i at time t is the sum of the three processes:
| 2.3 |
We included town population size 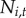 as a covariate at the highest level of the model because it may influence both the observation process (owing to increased local reporting effort in larger towns) as well as the latent process of disease spread, as larger towns have more susceptible hosts who may be infected and diseased.
as a covariate at the highest level of the model because it may influence both the observation process (owing to increased local reporting effort in larger towns) as well as the latent process of disease spread, as larger towns have more susceptible hosts who may be infected and diseased.
Because both Lyme and babesiosis cases may have occurred before reporting commenced, there is a non-zero probability of human cases occurring for town i in the year case reports were initiated, 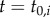 . We treated the latent probability of reported cases occurring during this first year of reporting in town i as a random variable:
. We treated the latent probability of reported cases occurring during this first year of reporting in town i as a random variable:
| 2.4 |
(iii). Spatial structure
Disease may persist or colonize each town owing to spread from neighbouring towns, probably owing to pathogen movement via dispersal of small mammals or birds [42–44]. We explored a variety of models for how a town's risk for initial colonization, persistence or re-colonization may depend on neighbouring towns' disease status (hypothesis (i)). To determine an appropriate definition for the spatial neighbourhood (the spatial extent of towns that exert disease pressure on the focal town), we fitted a series of models with different measures of neighbourhood disease intensity 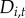 and evaluated model fit with Δ deviance information criterion (ΔDIC) compared with the non-spatial process-based model (electronic supplementary material, table S3) [45].
and evaluated model fit with Δ deviance information criterion (ΔDIC) compared with the non-spatial process-based model (electronic supplementary material, table S3) [45].
We model each ecological process (initial colonization, persistence and re-colonization) as a function of spatially dependent disease spread in addition to spread independent of spatial context. For example, following previously described notation [41], initial colonization is modelled as
| 2.5 |
Here, the logit-scaled probability of initial colonization is a function of the spatial neighbourhood; the coefficients 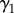 and
and 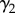 represent first- and second-order dependence of initial colonization on neighbourhood disease intensity
represent first- and second-order dependence of initial colonization on neighbourhood disease intensity 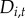 . Evidence of a second-order relationship between colonization and neighbourhood disease intensity could, for example, represent a saturating effect of increased pathogen pressure from nearby towns [41]. Alternatively, a quadratic dependence on neighbourhood disease intensity may reveal a surveillance effect analogous to an Allee effect [46]. For instance, colonization probability may have a positive relationship with
. Evidence of a second-order relationship between colonization and neighbourhood disease intensity could, for example, represent a saturating effect of increased pathogen pressure from nearby towns [41]. Alternatively, a quadratic dependence on neighbourhood disease intensity may reveal a surveillance effect analogous to an Allee effect [46]. For instance, colonization probability may have a positive relationship with 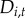 at low levels, but in towns/years with high
at low levels, but in towns/years with high 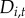 , population awareness may be high, leading to changes in human behaviours and thus exposure patterns, potentially resulting in a decreased probability of colonization of reported disease. The intercept,
, population awareness may be high, leading to changes in human behaviours and thus exposure patterns, potentially resulting in a decreased probability of colonization of reported disease. The intercept, 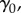 represents an additional colonization pressure, independent of spatial neighbourhood.
represents an additional colonization pressure, independent of spatial neighbourhood.
(iv). Ecological covariates
To test the hypotheses about drivers of tick-borne pathogen spread outlined in the Introduction, we included covariates at the process level (initial colonization, persistence or re-colonization) (equations (2.10)–(2.12)).
To test if high tick density facilitates disease spread (hypothesis (ii)), we included density of infected nymphal ticks, 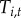 , as a covariate; the coefficients
, as a covariate; the coefficients 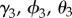 represent the effect of tick density on initial colonization, persistence and re-colonization, respectively. To test if proximity to water facilitates movement of tick-borne pathogens (hypothesis (iii)), we included distance to a major water body,
represent the effect of tick density on initial colonization, persistence and re-colonization, respectively. To test if proximity to water facilitates movement of tick-borne pathogens (hypothesis (iii)), we included distance to a major water body, 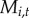 , as a covariate; the coefficients
, as a covariate; the coefficients 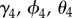 represent the effect of proximity to water on initial colonization, persistence and re-colonization, respectively. To test if Lyme disease facilitates spread of babesiosis (hypothesis (iv)), we included Lyme endemicity,
represent the effect of proximity to water on initial colonization, persistence and re-colonization, respectively. To test if Lyme disease facilitates spread of babesiosis (hypothesis (iv)), we included Lyme endemicity, 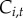 , defined as the number of years a town has reported cases of Lyme disease prior to year t, as a predictor for babesiosis status; the coefficients
, defined as the number of years a town has reported cases of Lyme disease prior to year t, as a predictor for babesiosis status; the coefficients 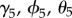 represent the effect of Lyme endemicity on initial colonization, persistence and re-colonization, respectively.
represent the effect of Lyme endemicity on initial colonization, persistence and re-colonization, respectively.
The equations describing the saturated model for the presence of disease reports in reported disease in town i at time t (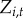 ) are as follows:
) are as follows:
| 2.6 |
probability of reported disease (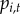 ):
):
| 2.7 |
probability of initial colonization (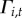 ):
):
| 2.8 |
probability of persistence (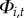 ):
):
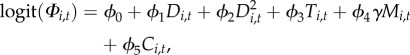 |
2.9 |
probability of re-colonization (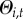 ):
):
| 2.10 |
(v). Model implementation and validation
We fitted the models using a Markov chain Monte Carlo (MCMC) algorithm with Gibbs sampling to simulate sequences of dependent samples from the posterior distribution of model parameters, implemented in JAGS v. 3.4.0 [47] (model code in electronic supplementary material, text S2). All priors were non-informative (electronic supplementary material, table S2). We ran three independent chains, and inference was based on 100 000 samples after discarding a burn-in of 10 000 iterations and thinning chains every 10 draws. MCMC convergence was assessed with the Brooks–Gelman–Rubin convergence diagnostic [48].
Our primary objectives were to test evidence in the data for our a priori hypotheses and model-based prediction. Thus, we generated the model set described above and used model selection to determine support in the data for the model/hypothesis. We used the DIC to evaluate whether increasing model complexity decreased model deviance while penalizing for overfitting (electronic supplementary material, table S4). Parameter estimates from a model that includes non-significant parameters (i.e. the full covariate model) should not be biased but may introduce excess noise to predictions [49]. Accordingly, in the Results and Discussion, we present parameter estimates from what we call the parsimonious model—the best-fit model under DIC stripped of covariates whose parameter estimate credible intervals overlap 0. Parameter estimates change little between the full covariate and parsimonious models (compare figure 2 with electronic supplementary material, figure S2 and figure 3 with electronic supplementary material, figure S3), confirming that bias is not introduced by exclusion of non-significant parameters.
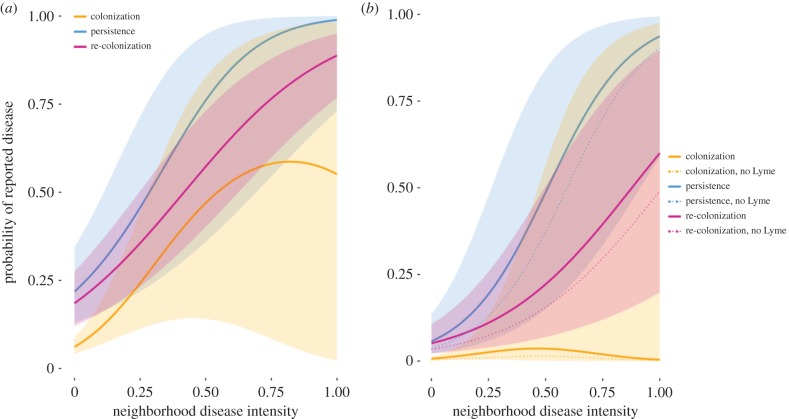
Figure 2.
Probability of disease reports plotted against the fraction of neighbouring towns reporting cases for (a) Lyme disease and (b) babesiosis estimated by the parsimonious model. Solid lines indicate effect of spatial neighbourhood on the mean probability of reported disease; shaded regions represent the 95% credible intervals. Plots depict the effect of neighbourhood disease intensity and associated uncertainty while holding other covariates at their mean value. In (b), thin lines indicate the effect of neighbourhood disease intensity on probability of babesiosis disease reports in towns in which we set Lyme disease endemicity to 0 years. (Online version in colour.)
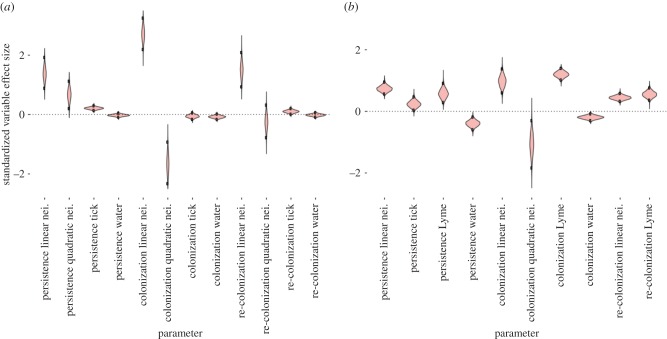
Figure 3.
Violin plot of variable effect size for parameters included in the parsimonious (a) Lyme disease and (b) babesiosis models. Variable effect size can be interpreted as the increase in probability of reported disease (logit-scaled) when the explanatory variable increases by one standard deviation. Black points on each violin indicate the 95% credible interval of the parameter estimate. Parameters with a variable effect size 95% credible interval spanning 0 (dotted line) in the full covariate model were removed in the parsimonious model. Parameter abbreviations are defined in the electronic supplementary material, table S2. (Online version in colour.)
We validate the parsimonious models in two ways. First, we evaluate one-step-ahead predictions (i.e. model predictions conditional on data observed for the previous year with area under the curve (AUC), determined using the R package ‘pROC’) [50]. Second, to test forecasting capacity, we partitioned the data into a ‘training’ period including case reports through 2011 used to fit the model parameters, and a ‘validation’ period including data from 2012 to 2014. We calculated neighbourhood disease intensity 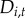 for each year as defined in equation (2.5), so that forecasts in 2013 and 2014 were a function of forecasted neighbourhood disease status in the previous year, not reported status. Out-of-fit forecasts were assessed with AUC.
for each year as defined in equation (2.5), so that forecasts in 2013 and 2014 were a function of forecasted neighbourhood disease status in the previous year, not reported status. Out-of-fit forecasts were assessed with AUC.
To convert continuous predicted probabilities of disease into binary disease forecasts, we identified a probability threshold that minimizes the difference between model sensitivity and specificity [51].
3. Results
(a). Lyme disease
The spread of Lyme disease exhibits strong spatial dependence and is associated with I. scapularis density, as evidenced by the best-fitting Lyme disease model (electronic supplementary material, table S4). One-step-ahead model predictions of disease had a mean AUC of 0.93 (AUC > 0.7 is generally considered a good fit) [52]. When reporting began in Connecticut in 1984, Lyme disease cases were reported only in eastern coastal Connecticut. The model predicts the diffusion-like spread of cases from neighbours northwards along two seemingly independent corridors: along the Atlantic coast through Rhode Island, eastern Massachusetts and into southeastern New Hampshire; and along the New York state border, with an apparent lag further inland (figure 1). By 2014, the model predicts most New England towns report Lyme disease cases, with the exception of a corridor of uninfected towns in west central Massachusetts and northwestern New Hampshire.
The occupancy model distinguishes the three processes contributing to the spread of reported disease and the contribution of spatial and environmental covariates (figures 2a and 3a, electronic supplementary material, table S3). We find a significant spatial effect for each disease process, evidence that disease risk propagates from neighbours (figure 2a and electronic supplementary material, table S3). The spatial neighbourhood which best predicts spread of disease is an inverse distance weighted sum of the disease status of all towns in the study area rather than only adjacent towns, evidence that both local spread (from adjacent towns) as well as spread at longer distances contribute to movement of Lyme disease (electronic supplementary material, table S3). As neighbourhood disease intensity increases, the probability of each invasion process increases (figure 2a). We estimated a mean dispersal velocity for Lyme disease of 11.4 km per year by fitting a connectivity function (though this model of spatial neighbourhood did not provide the best fit to the data, electronic supplementary material, table S5).
Lyme disease persistence and re-colonization but not initial colonization are positively associated with tick habitat suitability (figure 3a). Lyme disease spread is not associated with proximity to a water body. State reporting effects were high (greater than 0.95) across all states, which suggests a lack of variability in reporting between states (electronic supplementary material, figure S4).
(b). Babesiosis
The spread of babesiosis exhibits strong spatial dependence in addition to dependence on Lyme disease endemicity, tick density and proximity to water, as evidenced by the best-fitting babesiosis model (electronic supplementary material, table S4). One-step-ahead predictions had a mean AUC of 0.92. The babesiosis model predicts the slow spread of babesiosis north through eastern coastal Connecticut and into a few isolated towns on Cape Cod and Nantucket Island (south of Cape Cod) in the 1990s (figure 1). During the 2000s, the model predicts continuous, slow spread of babesiosis northeast along the Atlantic coast. Much of Connecticut, eastern Massachusetts and southern coastal New Hampshire have a moderate probability of reporting cases by 2010.
Again, the occupancy model distinguishes the three processes underlying spread of babesiosis. We find a significant spatial effect for each disease process, evidence that babesiosis risk propagates from neighbours (figure 2b and electronic supplementary material, table S3). Similar to Lyme disease, the best-fitting definition of spatial neighbourhood indicates that spread is best predicted by an inverse distance weighted average of the reporting status of all towns in the study area in the previous year, suggesting the disease pressure comes from all towns in New England but particularly nearby towns (electronic supplementary material, table S3). As neighbourhood disease intensity increases, the probability of each invasion process increases (figure 2b). We estimated a mean dispersal velocity for babesiosis of 10.1 km year−1 by fitting a connectivity function (electronic supplementary material, table S5), although again this was not the best-fitting spatial model.
Babesiosis spread exhibits strong dependence on Lyme disease; initial colonization, persistence and re-colonization of babesiosis are each positively associated with a history of Lyme disease endemicity (figure 3b), and no towns report babesiosis without a prior history of Lyme disease reporting. A hypothetical scenario of babesiosis invasion in the absence of Lyme disease was modelled by setting Lyme history to 0 for all towns. Babesiosis spread is suppressed in such a scenario (figure 2b, dotted lines). Babesiosis persistence and initial colonization are negatively associated with distance to a major water body (figure 3b). Babesiosis persistence is additionally positively associated with tick habitat suitability (figure 3a). State reporting effect varied across states (electronic supplementary material, figure S4).
(c). Comparison of Lyme and babesiosis trajectories
When holding all covariates at their mean values, Lyme disease has significantly higher rates of initial colonization and re-colonization compared with babesiosis across all neighbourhood disease intensities (figure 2). Independent of spatial neighbourhood, the probability of Lyme disease initial colonization is predicted to be 10 times that of babesiosis (figure 2a,b yellow line intercepts).
(d). Model forecasting
To validate the parsimonious models, we re-fitted the model to case reports through 2011 in order to forecast disease in each of 3 years between 2012 and 2014 [53].
Out-of-fit Lyme disease model forecasts have a mean AUC of 0.93. We identify a probability threshold that minimizes the difference between sensitivity and specificity (electronic supplementary material, figure S5); forecasted probabilities of disease greater than 88.6% for Lyme disease can be interpreted as disease forecasts. The true positive rate or sensitivity (the probability the model correctly forecasts disease) is 86.1% (1596 out of 1854); the true negative rate or specificity (the probability the model correctly forecasts absence of disease) 86.0% (289 out of 336; electronic supplementary material, figure S6). The model performs well (mean AUC of 0.94) ahead of the ‘invasion front’, i.e. in towns with no infected adjacent neighbours in the previous year. Forecasts ahead of the invasion front have a sensitivity of 52.9% (9 out of 17) and specificity of 100% (60 out of 60).
Modelling the reporting (observation) process improved Lyme disease model fit as measured by ΔDIC compared with the model without the observation process (electronic supplementary material, table S4). However, Lyme disease forecasting AUC, sensitivity and specificity did not differ between the parsimonious models with and without the reporting process.
Out-of-fit babesiosis model forecasts have a mean AUC of 0.85. Probabilities of disease greater than 14.0% for babesiosis (electronic supplementary material, figure S5) can be interpreted as disease forecasts. The true positive rate or sensitivity (the probability the model correctly forecasts disease) is 74.7% (510 out of 682); the true negative rate or specificity (the probability the model correctly forecasts absence of disease) 74.7% (1126 out of 1508; electronic supplementary material, figure S6). The model performs well (mean AUC of 0.87) ahead of the ‘invasion front’. Forecasts ahead of the invasion front have a sensitivity of 75.6% (65 out of 86) and specificity of 84.4% (696 out of 824).
Modelling the reporting (observation) process similarly improved babesiosis model fit as measured by ΔDIC compared with the model without the observation process (electronic supplementary material, table S4). Similar to Lyme disease forecasting, overall babesiosis forecasting performance as measured by AUC, sensitivity and specificity did not differ between the parsimonious models with or without the reporting process. Forecasting sensitivity ahead of the invasion front is sacrificed if the reporting process is not modelled: sensitivity drops from 75.6% to 46.5% when reporting is not considered. Forecasting specificity changes from 84.4% to 92.1% when reporting is not modelled.
4. Discussion
Modelling the spatial spread of vector-borne zoonotic pathogens maintained in enzootic transmission cycles remains a major challenge [5]. Human surveillance data often constitute the best available spatio-temporal data on pathogen spread. Over the past 40 years, TBD have rapidly spread across the northeast and midwest United States and pose a significant and growing public health burden [2,3,20,21,32,54,55]. Although the increase in human cases of TBD is striking, the crawl of ticks and tick-borne pathogens across the landscape is largely unobserved. By applying classic ecological approaches of occupancy and metapopulation modelling to an epidemiological question of disease spread, we harnessed human surveillance data to model the invasion of tick-borne pathogens. We tested hypotheses about how pathogens move (i.e. the spatial dependence of invasion) and why pathogens are spreading (i.e. environmental and ecological drivers of spread).
(a). Processes underlying spread
As hypothesized, the processes underlying TBD invasion—colonization, persistence and re-colonization—each exhibit strong spatial dependence. Defining neighbourhood disease intensity as the inverse-distance weighted sum of the disease status of all towns in the study area rather than by adjacency provides the best fit to the data, indicating that spatial spread of both diseases is a combination of both local spread from adjacent neighbours as well as spread at longer distances. In this definition of spatial neighbourhood, closer towns are given greater weight, indicating that local spread contributes greatly to the spread of TBD.
Local spread is probably attributable to mammalian hosts that often disperse less than 5 km in addition to avian hosts moving short distances [42–44,56,57]. Long-distance colonization plays a smaller, but nonetheless significant role in colonization of TBD. Long-distance introductions of disease may be attributable to dispersal on migratory passerines [56,58]. Avian dispersal of I. scapularis populations has been described in geographically isolated sites in Canada where mammalian introductions were unlikely [43,59]. Alternatively, observed long-distance introductions ahead of the ‘invasion front’ may reflect differences between enzootic infection prevalence, human exposures, human disease incidence or reporting. For babesiosis, there are several apparent long-distance ‘colonization’ events in which towns with no infected neighbours report disease and then disease appears to go extinct. This reflects the low prevalence of babesiosis: in many towns in which cases are reported, there is only a single case. The apparent colonization and extinction probably reflect the epidemiological process of human infection and disease reporting rather than reflect the ecological process of B. microti spread across the landscape (i.e. in these colonization/extinction events, B. microti does not go extinct locally, but rather no cases are reported owing to stochasticity in human infection per surveillance).
Field studies have demonstrated that tick populations need to be established prior to successful pathogen colonization and that density of infected nymphal ticks is associated with Lyme disease prevalence in humans [29,37,60–62]. In contrast to our hypotheses, we found that tick density was positively associated with Lyme disease persistence and re-colonization, but not initial colonization. This may be owing to the lack of data on spread of infected I. scapularis over the reporting period. Increased tick density is associated with persistence of babesiosis, but not initial or re-colonization, probably owing to collinearity with Lyme disease endemicity.
As predicted, initial colonization and persistence of babesiosis is associated with proximity to a major water body. River corridors and the Atlantic Coast provide suitable, humid microhabitats for ticks and are known corridors of ticks and reservoir hosts [30,58]. The lack of an association between Lyme disease spread and proximity to water may be an artefact of the reporting time series available. The Lyme disease bacteria, B. burgdorferi, had already spread up the Hudson River and much of the Atlantic Coast prior to the start of the reporting time series, making it difficult to capture any association between water corridors of dispersal and disease. By contrast, the reporting time series captures the early invasion of the babesiosis parasite, B. microti, along the Atlantic coastline and up the Hudson River Valley (figure 1).
(b). Contrasting spread trajectories
Our model captures differences in invasion trajectories for B. burgdorferi and B. microti. While B. burgdorferi appears to have spread rapidly and became endemic across most of New England by the late 2010s, B. microti appears to have spread slowly and became endemic only in southern coastal New England (figure 1). Babesiosis spread is characterized by lower rates of initial colonization and re-colonization compared with Lyme disease and, importantly, strong dependence on Lyme disease endemicity (figure 2). However, the mean model-estimated rate of spread of Lyme disease and babesiosis did not differ substantially over the reporting period (11.36 and 10.10 km year−1, respectively). This reveals that differences in current distributions of the two diseases may reflect a temporal lag in spread of babesiosis with respect to Lyme disease owing to differences in the ecological invasion processes, pathogenesis in humans and/or owing to differential surveillance measures for the two diseases, rather than a difference in the speed of invasion.
The association of babesiosis invasion processes with Lyme disease endemicity is consistent with laboratory studies which demonstrate that co-infection with B. burgdorferi enhances transmission of B. microti in reservoir hosts and thus lowers the ecological threshold for B. microti establishment in proportion to B. burgdorferi infection in the mouse population [31]. Lyme disease endemicity also probably serves as a proxy for a suite of other unmeasured environmental, ecological or health system variables associated with high incidence of TBD, including spatio-temporal variation in I. scapularis density. The strong dependence on Lyme disease suggests that spread of babesiosis is likely to continue until it approximates the range of Lyme disease. However, our results suggest that Lyme disease, once reported, is more likely to persist than babesiosis, consistent with previous studies that have identified a higher basic reproductive number, R0, for B. burgdorferi compared with B. microti [31].
Consistent with hypotheses, the probability of long-distance introduction of Lyme disease to towns independent of spatial context is 10 times more likely than long-distance movement of babesiosis. This may reflect a smaller propagule pressure of B. microti-infected ticks owing to the lower prevalence of B. microti across the study area as well as the fact that birds are more competent hosts for B. burgdorferi than for B. microti [19,58].
Mean state reporting effects are higher for Lyme disease compared with babesiosis, probably reflecting the higher prevalence, greater population and physician awareness, and more distinct symptomatology of Lyme disease compared with babesiosis [32,34]. Variation in state reporting effects is higher for babesiosis than for Lyme disease, probably reflecting heterogeneities in awareness and surveillance efforts across states [29].
Modelling variation in imperfect reporting is critical for babesiosis, because disease reports ahead of the apparent invasion front probably reflect reporting variation rather than reflect parasite invasion (e.g. the parasite is already established in the enzootic cycle ahead of the invasion front observed by human cases). Our results suggest that an occupancy modelling framework may contribute most to epidemiological models when there is substantial spatial variation in the reporting process.
(c). Study limitations
Here, we modelled the spatial spread of reported cases of TBD. Owing to widespread and differential [33] under- and over-reporting of Lyme disease and babesiosis, evolving case definitions [35], and changing population awareness, raw epidemiological data may not represent the true distribution of cases and subclinical infections. However, modelling variation in reporting across states allows us to address the issue of imperfect detection of human cases.
5. Conclusion
We harness epidemiological surveillance data to examine an unobserved stochastic process of pathogen spread in an enzootic cycle. The spatio-temporal framework can be readily modified and adapted for study of other emerging zoonotic pathogens [34].
Supplementary Material
Supplementary Material
Supplementary Material
Acknowledgements
The authors thank Marina Antillon, Linda Capewell, Molly Rosenberg, Fu-Chi Hsieh and Kimberly Tsao for their contributions to data collection. Models were run on Yale University's High Performance Computing cluster, Louise (supported by NIH grant nos. RR19895 and RR029676-01). The authors thank Dr Robert Bjornson for bioinformatics assistance.
Data accessibility
Town-level Lyme disease and babesiosis surveillance data for Connecticut, Rhode Island and Massachusetts are available at Dryad: http://dx.doi.org/10.5061/dryad.fs348.
Authors' contributions
M.D.W., P.J.K., V.E.P., K.M.P., C.T.W., H.D.G. and K.S.W. conceived the eco-epidemiological question, K.M.P. formalized the modelling approach. K.S.W. collected data, implemented model, and analysed output data. K.M.P. and V.E.P. contributed to model construction and fitting. K.S.W. wrote the first draft of the manuscript, and all authors contributed substantially to revisions.
Competing interests
We have no competing interests.
Funding
K.S.W. was supported by the NIH pre-doctoral training grant 5T32AI007404-23 and the NIH Ruth L. Kirschstein National Research Service Award F31 AI118233-01A1. K.M.P. was partly supported by the Research and Policy in Disease Dynamics (RAPIDD) programme, Fogarty International Center, NIH and Department of Homeland Security and partly by USDA-APHIS-WS. C.T.W. and V.E.P. were funded by RAPIDD. P.J.K. was supported by the Gordon and Laura Gund Foundation. M.D.W. was supported by the National Institutes of Health, Ecology and Evolution of Infectious Disease Programme (R01 GM105246).
References
- 1.Centers for Disease Control and Prevention. 2015 How many people get Lyme disease? Atlanta, GA: Centers for Disease Control and Prevention.
- 2.Telford SR, Goethert HK. 2004. Emerging tick-borne infections: rediscovered and better characterized, or truly ‘new’? Parasitology 129, S301–S327. [DOI] [PubMed] [Google Scholar]
- 3.Piesman J, Eisen L. 2008. Prevention of tick-borne diseases. Annu. Rev. Entomol. 53, 323–343. ( 10.1146/annurev.ento.53.103106.093429) [DOI] [PubMed] [Google Scholar]
- 4.Kugeler KJ, Farley GM, Forrester JD, Mead PS. 2015. Geographic distribution and expansion of human Lyme disease, United States. Emerg. Infect. Dis. 21, 1455–1457. ( 10.3201/eid2108.141878) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kilpatrick AM, Randolph SE. 2012. Drivers, dynamics, and control of emerging vector-borne zoonotic diseases. Lancet 380, 1946–1955. ( 10.1016/S0140-6736(12)61151-9) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Abad-Franch F, Ferraz G, Campos C, Palomeque FS, Grijalva MJ, Aguilar HM, Miles MA. 2010. Modeling disease vector occurrence when detection is imperfect: infestation of Amazonian palm trees by triatomine bugs at three spatial scales. PLoS Negl. Trop. Dis. 4, e620 ( 10.1371/journal.pntd.0000620) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Adams MJ, et al. 2010. Using occupancy models to understand the distribution of an amphibian pathogen, Batrachochytrium dendrobatidis. Ecol. Appl. Ecol. Soc. Am. 20, 289–302. ( 10.1890/08-2319.1) [DOI] [PubMed] [Google Scholar]
- 8.Lachish S, Gopalaswamy AM, Knowles SCL, Sheldon BC. 2012. Site-occupancy modelling as a novel framework for assessing test sensitivity and estimating wildlife disease prevalence from imperfect diagnostic tests. Methods Ecol. Evol. 3, 339–348. ( 10.1111/j.2041-210X.2011.00156.x) [DOI] [Google Scholar]
- 9.Elmore SA, Huyvaert KP, Bailey LL, Milhous J, Alisauskas RT, Gajadhar AA, Jenkins EJ. 2014. Toxoplasma gondii exposure in Arctic-nesting geese: a multi-state occupancy framework and comparison of serological assays. Int. J. Parasitol. Parasites Wildl. 3, 147–153. ( 10.1016/j.ijppaw.2014.05.005) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Colvin ME, Peterson JT, Kent ML, Schreck CB. 2015. Occupancy modeling for improved accuracy and understanding of pathogen prevalence and dynamics. PLoS ONE 10, e0116605 Public Library of Science ( 10.1371/journal.pone.0116605) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hooten MB, Wikle CK. 2010. Statistical agent-based models for discrete spatio-temporal systems. J. Am. Stat. Assoc. 105, 236–248. ( 10.1198/jasa.2009.tm09036) [DOI] [Google Scholar]
- 12.Beyer HL, Hampson K, Lembo T, Cleaveland S, Kaare M, Haydon DT. 2011. Metapopulation dynamics of rabies and the efficacy of vaccination. Proc. R. Soc. B 278, 2182–2190. ( 10.1098/rspb.2010.2312) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Levins R. 1969. Some demographic and genetic consequences of environmental heterogeneity for biological control. Bull. Entomol. Soc. Am. 15, 237–240. ( 10.1093/besa/15.3.237) [DOI] [Google Scholar]
- 14.George DB, Webb CT, Pepin KM, Savage LT, Antolin MF. 2013. Persistence of black-tailed prairie-dog populations affected by plague in northern Colorado, USA. Ecology 94, 1572–1583. ( 10.1890/12-0719.1) [DOI] [PubMed] [Google Scholar]
- 15.Riley S. 2007. Large-scale spatial-transmission models of infectious disease. Science 316, 1298–1301. ( 10.1126/science.1134695) [DOI] [PubMed] [Google Scholar]
- 16.Smith DL, Lucey B, Waller LA, Childs JE, Real LA. 2002. Predicting the spatial dynamics of rabies epidemics on heterogeneous landscapes. Proc. Natl Acad. Sci. USA 99, 3668–3672. ( 10.1073/pnas.042400799) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Steere ACC, Malawista SE, Snydman DR, Andiman WA. 1976. A cluster of arthritis in children and adults in Lyme, Connecticut. Arthritis Rheum. 19, 824. [DOI] [PubMed] [Google Scholar]
- 18.Western K, Benson G. 1970. Babesiosis in a Massachusetts resident. N. Engl. J. Med. 283, 854–856. ( 10.1056/NEJM197010152831607) [DOI] [PubMed] [Google Scholar]
- 19.Hersh MH, Tibbetts M, Strauss M, Ostfeld RS, Keesing F. 2012. Reservoir competence of wildlife host species for Babesia microti. Emerg. Infect. Dis. 18, 1951–1957. ( 10.3201/eid1812.111392) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Spielman A, Wilson M. 1985. Ecology of Ixodes Dammini-borne human babesiosis and Lyme disease. Annu. Rev. Entomol. 30, 439–460. ( 10.1146/annurev.en.30.010185.002255) [DOI] [PubMed] [Google Scholar]
- 21.Krause PJ, Telford SR, Ryan R, Hurta AB, Kwasnik I, Luger S, Niederman J, Gerber M, Spielman A. 1991. Geographical and temporal distribution of babesial infection in Connecticut. J. Clin. Microbiol. 29, 1–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Matuschka F, Spielman A. 1986. The emergence of Lyme disease in a changing environment in North America and central Europe. Exp. Appl. Acarol. 2, 337–353. ( 10.1007/BF01193900) [DOI] [PubMed] [Google Scholar]
- 23.Daniels TJ, Fish D. 1990. Spatial distribution and dispersal of unfed larval Ixodes dammini (Acari: Ixodidae) in Southern New York. Environ. Entomol. Entomol. Soc. Am. 19, 5. [Google Scholar]
- 24.Falco RC, Fish D. 1991. Horizontal movement of adult Ixodes dammini (Acari: Ixodidae) attracted to CO2-baited traps. J. Med. Entomol. 28, 726–729. ( 10.1093/jmedent/28.5.726) [DOI] [PubMed] [Google Scholar]
- 25.Spielman A, Etkind P. 1981. Reservoir hosts of human babesiosis on Nantucket Island. Am. J. Trop. Med. Hyg. 30, 560–565. [DOI] [PubMed] [Google Scholar]
- 26.Mather TN, Wilson ML, Moore SI, Ribeiro JM, Spielman A. 1989. Comparing the relative potential of rodents as reservoirs of the Lyme disease spirochete (Borrelia burgdorferi). Am. J. Epidemiol. 130, 143–150. [DOI] [PubMed] [Google Scholar]
- 27.LoGiudice K, Ostfeld RS, Schmidt KA, Keesing F. 2003. The ecology of infectious disease: effects of host diversity and community composition on Lyme disease risk. Proc. Natl Acad. Sci. USA 100, 567–571. ( 10.1073/pnas.0233733100) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pybus O, Suchard M. 2012. Unifying the spatial epidemiology and molecular evolution of emerging epidemics. Proc. R. Soc. B 109, 15 066– 15 071 ( 10.1073/pnas.1206598109) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pepin KM, Eisen RJ, Mead PS, Piesman J, Fish D, Hoen AG, Barbour AG, Hamer S, Diuk-Wasser MA. 2012. Geographic variation in the relationship between human Lyme disease incidence and density of infected host-seeking Ixodes scapularis nymphs in the eastern United States. Am. J. Trop. Med. Hyg. 86, 1062–1071. ( 10.4269/ajtmh.2012.11-0630) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wilson ML, Ducey AM, Litwin TS, Gavin TA, Spielman A. 1990. Microgeographic distribution of immature Ixodes dammini ticks correlated with that of deer. Med. Vet. Entomol. 4, 151–159. ( 10.1111/j.1365-2915.1990.tb00273.x) [DOI] [PubMed] [Google Scholar]
- 31.Dunn JM, et al. 2014. Borrelia burgdorferi promotes the establishment of Babesia microti in the northeastern United States. PLoS ONE 9, e115494 ( 10.1371/journal.pone.0115494) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vannier E, Krause PJ. 2012. Human babesiosis. N. Engl. J. Med. 366, 2397–2407. ( 10.1056/NEJMra1202018) [DOI] [PubMed] [Google Scholar]
- 33.Diuk-Wasser MA, et al. 2014. Monitoring human babesiosis emergence through vector. Emerg. Infect. Dis. 20, 225–231. ( 10.3201/eid2002.130644) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Diuk-Wasser MA, Vannier E, Krause PJ. 2015. Coinfection by Ixodes tick-borne pathogens: ecological, epidemiological, and clinical consequences. Trends Parasitol. 32, 30–42. ( 10.1016/j.pt.2015.09.008) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Young J. 1998. Underreporting of Lyme disease. N. Engl. J. Med. 338, 1622–1623. ( 10.1056/NEJM199805283382211) [DOI] [PubMed] [Google Scholar]
- 36.White DJ, et al. 1991. The geographic spread and temporal increase of the Lyme disease epidemic. J. Am. Med. Assoc. 266, 1230–1236. ( 10.1001/jama.1991.03470090064033) [DOI] [PubMed] [Google Scholar]
- 37.Diuk-Wasser MA, et al. 2010. Field and climate-based model for predicting the density of host-seeking nymphal Ixodes scapularis, an important vector of tick-borne disease agents in the eastern United States. Glob. Ecol. Biogeogr. 19, 504–5014. ( 10.1111/j.1466-8238.2010.00526.x) [DOI] [Google Scholar]
- 38.Ostfeld RS, Canham CD, Oggenfuss K, Winchcombe RJ, Keesing F. 2006. Climate, deer, rodents, and acorns as determinants of variation in Lyme disease risk. PLoS Biol. 4, e145 ( 10.1371/journal.pbio.0040145) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.R Core Team. 2014. R: a language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing. [Google Scholar]
- 40.Royle J, Dorazio RM. 2008. Hierarchical modeling and inference in ecology: the analysis of data from populations, metapopulations and communities. London, UK: Elsevier. [Google Scholar]
- 41.Bled F, Royle JA, Cam E. 2011. Hierarchical modeling of an invasive spread: the Eurasian collared-dove Streptopelia decaocto in the United States. Ecol. Appl. 21, 290–302. ( 10.1890/09-1877.1) [DOI] [PubMed] [Google Scholar]
- 42.Madhav N, Brownstein J, Tsao J, Fish D. 2004. A dispersal model for the range expansion of blacklegged tick (Acari: Ixodidae). J. Med. Entomol. 41, 842–852. ( 10.1603/0022-2585-41.5.842) [DOI] [PubMed] [Google Scholar]
- 43.Ogden NH, Mechai S, Margos G. 2013. Changing geographic ranges of ticks and tick-borne pathogens: drivers, mechanisms and consequences for pathogen diversity. Front. Cell Infect. Microbiol. 3, 46 ( 10.3389/fcimb.2013.00046) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hamer SA, Tsao JI, Walker ED, Hickling GJ. 2010. Invasion of the Lyme disease vector Ixodes scapularis: implications for Borrelia burgdorferi endemicity. Ecohealth 7, 47–63. ( 10.1007/s10393-010-0287-0) [DOI] [PubMed] [Google Scholar]
- 45.Hanski I. 1994. A practical model of metapopulation dynamics. J. Anim. Ecol. 63, 151–162. ( 10.2307/5591) [DOI] [Google Scholar]
- 46.Courchamp F, Clutton-Brock T, Grenfell B. 1999. Inverse density dependence and the Allee effect. Trends Ecol. Evol. 14, 405–410. ( 10.1016/S0169-5347(99)01683-3) [DOI] [PubMed] [Google Scholar]
- 47.Plummer M. 2012. JAGS version 3.3.0 user manual. (October): http://people.math.aau.dk/~kkb/Undervisning/Bayes14/sorenh/docs/jags_user_manual.pdf. Publication date is October 1 2012.
- 48.Gelman A, Rubin DB. 1992. Inference from iterative simulation using multiple sequences. Stat. Sci. 7, 457–472. ( 10.1214/ss/1177011136) [DOI] [Google Scholar]
- 49.Whittingham MJ, Stephens PA, Bradbury RB, Freckleton RP. 2006. Why do we still use stepwise modelling in ecology and behaviour? J. Anim. Ecol. 75, 1182–1189. ( 10.1111/j.1365-2656.2006.01141.x) [DOI] [PubMed] [Google Scholar]
- 50.Robin X, Turck N, Hainard A, Tiberti N, Lisacek F, Sanchez J-C, Müller M. 2011. pROC: an open-source package for R and S+ to analyze and compare ROC curves. BMC Bioinformatics 12, 77 ( 10.1186/1471-2105-12-77) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lobo JM, Jiménez-Valverde A, Real R. 2008. AUC: a misleading measure of the performance of predictive distribution models. Glob. Ecol. Biogeogr. 17, 145–151. ( 10.1111/j.1466-8238.2007.00358.x) [DOI] [Google Scholar]
- 52.Hosmer DW Jr, Lemeshow S, Sturdivant RX. 2013. Applied logistic regression (ed. Sons JW.), 3rd edn Hoboken, NJ: Wiley. [Google Scholar]
- 53.Reiner RC, King AA, Emch M, Yunus M, Faruque AS, Pascual M. 2012. Highly localized sensitivity to climate forcing drives endemic cholera in a megacity. Proc. Natl Acad. Sci. USA 109, 2033–2036. ( 10.1073/pnas.1108438109) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Spielman A, Clifford C. 1979. Human babesiosis on Nantucket Island, USA: description of the vector, Ixodes (Ixodes) dammini, n. sp.(Acarina: Ixodidae). J. Med. Entomol. 15, 218–234. ( 10.1093/jmedent/15.3.218) [DOI] [PubMed] [Google Scholar]
- 55.Piesman J, Mather TNN, Donahue JGG, Levine J, Campbell JDD, Karakashian SJJ, Spielman A. 1986. Comparative prevalence of Babesia microti and Borrelia burgdorferi in four populations of Ixodes dammini in eastern Massachusetts. Acta Trop. 43, 263–270. [PubMed] [Google Scholar]
- 56.Ogden NH, Barker IK, Francis CM, Heagy A, Lindsay LR, Hobson KA. 2015. How far north are migrant birds transporting the tick Ixodes scapularis in Canada? Insights from stable hydrogen isotope analyses of feathers. Ticks Tick Borne Dis. 6, 715–720. ( 10.1016/j.ttbdis.2015.06.004) [DOI] [PubMed] [Google Scholar]
- 57.Hamer S, Goldberg T, Kitron U. 2012. Wild birds and urban ecology of ticks and tick-borne pathogens, Chicago, Illinois, USA, 2005–2010. Locus 18, 2005–2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ogden NH, et al. 2008. Role of migratory birds in introduction and range expansion of Ixodes scapularis ticks and of Borrelia burgdorferi and Anaplasma phagocytophilum in Canada. Appl. Environ. Microbiol. 74, 1780–1790. ( 10.1128/AEM.01982-07) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Leighton PA, Koffi JK, Pelcat Y, Lindsay LR, Ogden NH. 2012. Predicting the speed of tick invasion: an empirical model of range expansion for the Lyme disease vector Ixodes scapularis in Canada. J. Appl. Ecol. 49, 457–464. ( 10.1111/j.1365-2664.2012.02112.x) [DOI] [Google Scholar]
- 60.Stafford KC, Cartter ML, Magnarelli LA, Ertel SH, Mshar PA. 1998. Temporal correlations between tick abundance and prevalence of ticks infected with Borrelia burgdorferi and increasing incidence of Lyme disease. J. Clin. Microbiol. 36, 1240–1244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Falco RC, McKenna DF, Daniels TJ, Nadelman RB, Nowakowski J, Fish D, Wormser GP. 1999. Temporal relation between Ixodes scapularis abundance and risk for Lyme disease associated with erythema migrans. Am. J. Epidemiol. 149, 771–776. ( 10.1093/oxfordjournals.aje.a009886) [DOI] [PubMed] [Google Scholar]
- 62.Rand PW, Lacombe EH, Dearborn R, Cahill B, Elias S, Lubelczyk CB, Beckett GA, Smith RP. 2007. Passive surveillance in Maine, an area emergent for tick-borne diseases. J. Med. Entomol. 44, 1118–1129. ( 10.1093/jmedent/44.6.1118) [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Town-level Lyme disease and babesiosis surveillance data for Connecticut, Rhode Island and Massachusetts are available at Dryad: http://dx.doi.org/10.5061/dryad.fs348.