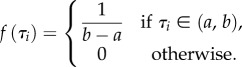Abstract
Bayesian inference of species divergence times is an unusual statistical problem, because the divergence time parameters are not identifiable unless both fossil calibrations and sequence data are available. Commonly used marginal priors on divergence times derived from fossil calibrations may conflict with node order on the phylogenetic tree causing a change in the prior on divergence times for a particular topology. Care should be taken to avoid confusing this effect with changes due to informative sequence data. This effect is illustrated with examples. A topology-consistent prior that preserves the marginal priors is defined and examples are constructed. Conflicts between fossil calibrations and relative branch lengths (based on sequence data) can cause estimates of divergence times that are grossly incorrect, yet have a narrow posterior distribution. An example of this effect is given; it is recommended that overly narrow posterior distributions of divergence times should be carefully scrutinized.
This article is part of the themed issue ‘Dating species divergences using rocks and clocks’.
Keywords: Bayesian inference, fossil calibrations, divergence time estimation
1. Introduction
Integrated Bayesian divergence time estimation combines information about the absolute ages of direct ancestors (or ancestors on side-branches), inferred from the palaeontological dating of fossils, with information about the relative ages of direct ancestors—inferred from patterns of substitution among molecular sequences of extant species [1]. The fossil and molecular data are co-dependent, because some ancestors in a phylogeny are not found in the fossil record (and therefore cannot be dated directly) and because the relative ages of the direct ancestors of extant species inferred from sequence data cannot be translated into estimates of absolute ages without information (usually from fossil calibrations) about the substitution rates per unit time on each lineage, only the product of rate and time is identifiable [2]. By combining the two sources of information, precise estimates of absolute ages can potentially be obtained for all nodes in a phylogenetic tree; this may not be possible using either fossils or sequences alone. Integrating molecular and palaeontological data in a combined Bayesian analysis is a highly challenging statistical problem.
There are multiple layers of complexity to the divergence time inference problem. To study the core problem, I will focus on the relationship between two sets of variables that in reality are never directly observed: (i) the rooted phylogenetic tree of species (with branch lengths measured in units of expected substitutions) and (ii) the set of fossil calibrations for direct ancestors of the sampled species (assumed to have a specified age distribution). In practice, these are unobserved variables that are either integrated over in a Bayesian analysis or indirectly inferred, and the level of information about the variables is highly dataset dependent. A ‘strict’ molecular clock and an infinite number of sites are needed to exactly determine relative branch lengths. In reality, the sequence data are finite, although the relative information content may sometimes approach that of ‘infinite data’ [2] and the molecular clock, needed to infer relative branch lengths, is generally imperfect—especially for more distantly related species. ‘Relaxed clock’ models are often used to infer relative branch lengths, introducing additional uncertainty (reviewed in [3]).
The placement of fossils on ancestral lineages must also be inferred based on very limited morphological information and errors in assigning fossils to ancestral lineages can have catastrophic effects on inferences [2]. Recently developed ‘tip-dating’ methods (often referred to as total-evidence dating) attempt to model morphological evolution and infer fossil placements as part of the inference process [4,5]. However, morphological evolution can be difficult to model [6] and the information content of morphological characters is often hard to judge. Finally, fossil ages are prone to uncertainties that may be difficult to capture in a parametric prior distribution (reviewed in [7]).
Perhaps surprisingly, even in the idealized situation outlined above, where the species tree, fossil lineage assignments and calibration priors are treated as known, the problem of divergence time estimation is complex. In this paper, I focus on the problem of how to specify a prior for divergence times (based on the calibration data and the species tree) that will lead to reasonable posterior inferences. I begin by discussing the effect of the node ordering in a tree topology on the divergence time prior. I consider particular cases in which conflicts of marginal calibrations and node ranks may (perhaps inadvertently) lead to a reduction of the prior variances of divergence times (a more informative prior). Next, I consider the effect of combining a species tree, with fixed relative branch lengths, with a fossil-calibration prior. Here, I step out of the ‘ideal world’ to examine the effect of a misplaced calibration on the posterior density of divergence times. Such conflicts can be difficult to identify and are not always reflected by increased variance in the posterior as would be the case in a more normal Bayesian inference scenario. This is the so-called ‘infinite sites’ case, in which sequence data are maximally informative about the products of branch lengths and rates, fixing the relative lengths of branches on the species tree. The effect of conflicts between fossil calibrations and sequence data can in this case lead to extreme outcomes in terms of both bias of point estimates and underestimation of the posterior uncertainty of parameter estimates.
2. The target of inference
It is illuminating to begin by considering the nature of differences in the information available from molecular sequences versus fossils and the targets of inference for each. Two limitations of the molecular data are: (i) it only allows us to date speciation events that gave rise to species that are currently extant and (ii) it only allows relative ages of divergence events to be inferred unless the substitution rate is known from other sources. The combination of morphology and absolute fossil ages via calibrations, on the other hand, in principle allows absolute ages to be inferred for speciation events that give rise to both extant and extinct species. However, to do so, morphological data must allow us to accurately infer both the species tree topology (including the placement of fossils that are direct ancestors) and the absolute ages of speciation events for which no fossils have been sampled. This would require both clock-like rates of morphological evolution and an adequate model of morphological change to allow the species tree to be accurately inferred. By combining disparate sources of information (fossil ages, morphology and sequences), and assuming that morphological evolution can be adequately modelled, it is possible (in principle) to estimate everything. Such ‘total-evidence’ methods are only now being developed (e.g. [5]) and it is not yet clear how well they will work. Alternative approaches model both the fossilization process and cladogenesis, using morphology only indirectly [8,9]. For example, Heath et al. [9] place a weaker constraint on fossils—that they descend from a particular node—but allow them to attach as either direct ancestors, or extinct side-branches, to any lineages descended from the node. This method appears promising based on the authors' simulations, although the effect of an incorrect assignment of a fossil to a crown group needs to be further explored, as does the amount of information actually available to infer divergence times in such a weakly constrained model.
One difference between existing methods for divergence time estimation is the treatment of species tree topology. One class of methods [2,10,11] assumes that the topology is fixed and focuses on estimating divergence times for a specific topology. A second class of methods treats the species tree topology as uncertain and focuses on jointly estimating both the topology and the divergence times [12]. The above dichotomy is similar to the general statistical problem of whether one should ‘model average’ to obtain parameter estimates because a tree topology appears more like a statistical model than a parameter [13]. Model averaging is appropriate if a parameter has the same statistical interpretation under different models. In this context, different topologies can be viewed as different statistical models with different sets of partially overlapping divergence time parameters. In some cases, the meaning of the divergence times may be very different under different topologies, suggesting model averaging may not be appropriate. Divergence time estimation is clearly sensible within the context of a particular fully specified species tree topology. Thus, this paper focuses exclusively on the problem of estimating divergence times when the species tree topology is fixed.
3. The priors
The prior on divergence times encapsulates information from dated fossils and a model of cladogenesis applied to divergence events for which no fossil-based age distributions are available. As indicated earlier, I will assume here that the topology of the species tree is fixed and only the ages of divergence events in the tree are unknown. I will also assume that the fossils are direct ancestors that can be assigned to lineages [2,10]. Newer methods [4,5,8,9] allow fossils to be on ‘side-branches’ which makes the problem more complex but potentially avoids the conflicts between tree topology and calibration priors as outlined below.
(a). Effect of ordered tree nodes
The tree topology imposes a rank order on the fossil ages. Any nodes that are direct ancestors (or descendents) in the tree have an order relative to one another. In a completely asymmetrical tree with s tips, for example (figure 1), the s – 1 divergence times are rank ordered 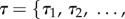
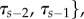 such that τj > τi if j > i. If the topology is not altered, this rank order must be preserved when prior probability distributions are applied for variables in τ. If the prior on τ is not applied jointly (with the ordering determined) then the prior will be altered once the constraints are introduced via the tree topology. In the mcmctree program [2,11], for example, the prior combining fossil calibrations and tree node ages is
such that τj > τi if j > i. If the topology is not altered, this rank order must be preserved when prior probability distributions are applied for variables in τ. If the prior on τ is not applied jointly (with the ordering determined) then the prior will be altered once the constraints are introduced via the tree topology. In the mcmctree program [2,11], for example, the prior combining fossil calibrations and tree node ages is
| 3.1 |
where  is the set of divergence times for nodes with fossil-calibration-based priors and
is the set of divergence times for nodes with fossil-calibration-based priors and 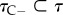 is the set of divergence times for nodes without calibrations. The density
is the set of divergence times for nodes without calibrations. The density 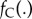 is the prior for calibration nodes (see equation (3.2)). The density
is the prior for calibration nodes (see equation (3.2)). The density 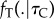 specifies the joint distribution of nodes without calibrations given a set of calibrated nodes. If
specifies the joint distribution of nodes without calibrations given a set of calibrated nodes. If 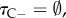 then τC = τ and the only effect of the prior is to constrain the rank order of the nodes (see below).
then τC = τ and the only effect of the prior is to constrain the rank order of the nodes (see below).
Figure 1.
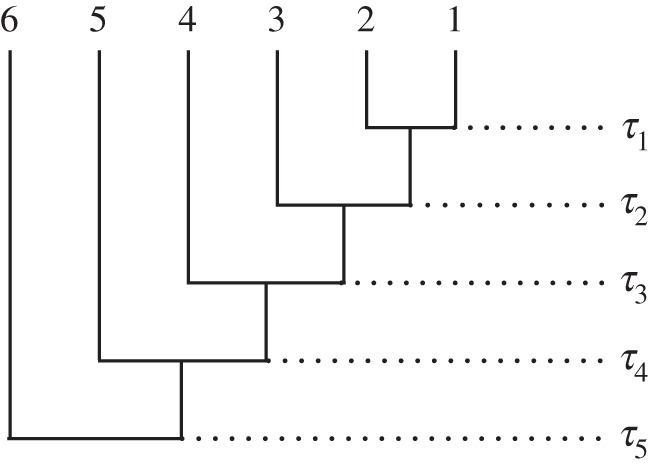
Fully asymmetrical phylogenetic tree of six species. τi denotes species divergence time i.
What is perhaps not obvious from this formulation is that 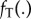 both maps the calibration variables to nodes on the tree T and imposes a rank order among them. Thus, even if all nodes have calibrations, the density
both maps the calibration variables to nodes on the tree T and imposes a rank order among them. Thus, even if all nodes have calibrations, the density 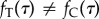 except in the particular case of a topology-consistent prior on node calibrations (see below). In some cases, there may be a negligible difference between the two densities, however, because the calibrations are in general agreement with the rank ordering of nodes in the tree. Conflicts of fossil dates with tree-based node orderings cause differences between the marginal calibration prior specified by the program user and the joint prior actually used by the program. This can cause a user to misunderstand the effect of the data versus the prior on inferences. Because the realized prior is different from the prior originally specified by the user, this observed change could be erroneously interpreted as due to the influence of the data.
except in the particular case of a topology-consistent prior on node calibrations (see below). In some cases, there may be a negligible difference between the two densities, however, because the calibrations are in general agreement with the rank ordering of nodes in the tree. Conflicts of fossil dates with tree-based node orderings cause differences between the marginal calibration prior specified by the program user and the joint prior actually used by the program. This can cause a user to misunderstand the effect of the data versus the prior on inferences. Because the realized prior is different from the prior originally specified by the user, this observed change could be erroneously interpreted as due to the influence of the data.
The formulation of equation (2.1) was applied by Yang & Rannala [2] using a birth–death process prior to obtain the density of divergence times by conditioning on the ages of nodes for which fossil calibrations are available. Namely, one can derive a conditioned birth–death prior for which particular node times are fixed variables. The remaining uncalibrated nodes are determined by a birth–death process conditioned on the ages of the fixed nodes. Drummond et al. [12] also used a birth–death prior but did not condition on the fossil calibrations; this is technically incorrect as it effectively applies two priors to nodes with calibrations [11].
In existing programs, such as mcmctree [2,11] and BEAST [14] that implement flexible calibration priors, the constraints on the ordering of the divergence times are implicitly imposed by the MCMC program. These constraints can be represented explicitly by the use of an indicator function
where IR(τ) = 1 if the set τ satisfies the order constraints of the topology and 0 otherwise. We assume here that a region of state space A exists such that IR(τ*) = 1 for any 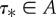 ensuring that the ratio is well defined. In most existing programs, the calibration priors are specified marginally and treated as independent. Thus, if fi(τi) is the prior density of the divergence time for calibrated node i, the joint density of the k calibrated nodes is
ensuring that the ratio is well defined. In most existing programs, the calibration priors are specified marginally and treated as independent. Thus, if fi(τi) is the prior density of the divergence time for calibrated node i, the joint density of the k calibrated nodes is
| 3.2 |
The ‘soft-tailed’ marginal densities (such as the γ distribution) considered by Yang & Rannala [2] have positive density on (0, ∞) and thus assign positive probability density to combinations of variables that are incompatible with the rank order imposed by topology. The total probability of these incompatible outcomes is
If Δ is small, then there is little difference between the calibration priors and the priors realized on the particular tree topology. If Δ is large, this indicates a conflict between fossil-calibration priors and the node ordering implied by the topology.
Example 3.1. Overlapping uniform divergence times. —
Consider the phylogeny of three species shown in figure 2. The nodes are rank ordered so that τ1 < τ2. Now, suppose that both divergence times have a uniform prior density
Figure 2.
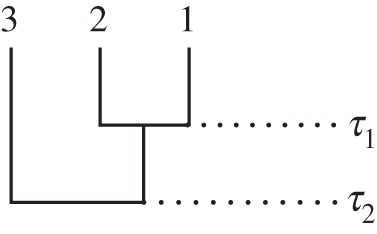
Phylogenetic tree of three species. τi denotes species divergence time i.
The expectation and variance are
The probability associated with topology-incompatible outcomes (i.e. τ1 ≥ τ2) is
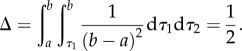 |
Thus, half the probability associated with the original priors has been eliminated by the rank order constraint.
The original state space is a square region in two dimensions and the constrained state space is a right triangle. It is interesting to consider the marginal distributions in this case. The expected values are
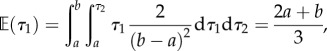 |
 |
and the variances are
Thus, the rank order constrain has caused the mean divergence time to increase for τ2 and decrease for τ1. Moreover, the variance is decreased for both variables.
(b). Topology-consistent priors
The condition  can be achieved if the fossil-calibration prior itself imposes an appropriate rank order on nodes. In that case,
can be achieved if the fossil-calibration prior itself imposes an appropriate rank order on nodes. In that case, 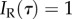 is always insured. A general definition of a topology-consistent prior is thus any prior density on divergence times for which Δ = 0.
is always insured. A general definition of a topology-consistent prior is thus any prior density on divergence times for which Δ = 0.
Example 3.2. Non-overlapping uniform divergence times. —
Consider again the three species tree of figure 2. Suppose the prior densities on divergence times τ1 and τ2 are uniform on non-overlapping intervals
and
satisfying
, where
denotes the positive real number line. The prior probability densities
and
with b2 > b1 have Δ = 0 and are topology consistent. This is possible because the uniform distribution has positive density for only a subset of
.
Theorem 3.3. —
A joint density for a set of rank-ordered nodes constructed as an n-tuple of independent random variables, each with non-zero density on
is not topology consistent.
By definition, a topology-consistent prior on the rank-ordered node ages τ must satisfy
Suppose that we construct a prior as an n-tuple of independent continuous random variables where fi(τi) is the density of variable τi with fi(τi) > 0 for all 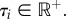 The joint density is
The joint density is
which is strictly positive on 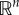 . By the definition of rank-ordered variables, a set of values
. By the definition of rank-ordered variables, a set of values 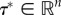 always exists that violate the inequalities such that
always exists that violate the inequalities such that 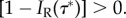 Thus, both
Thus, both  and
and 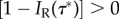 hold over some region, so that Δ > 0. The prior is therefore not topology consistent according to the definition.
hold over some region, so that Δ > 0. The prior is therefore not topology consistent according to the definition.
Example 3.4. Partially overlapping exponential divergence times. —
To illustrate theorem 3.3 with a simple example, consider again the three species tree of figure 2. Suppose the priors’ densities on divergence times τ1 and τ2 are exponential with densities
where τ2 > τ1. The region of conflict has density
Clearly, any choice of parametrizations for the exponential priors will give positive density to regions that conflict with the node rank orders and so no topology-consistent prior exists that can be constructed from duplets of independent exponential densities. The expected values of the variables under the constraint τ2 > τ1 are
and
Note that 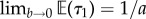 and
and 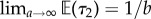 , so with a large prior mean for τ2 or a small prior mean for τ1 the marginals are little altered by the rank constraints as expected.
, so with a large prior mean for τ2 or a small prior mean for τ1 the marginals are little altered by the rank constraints as expected.
Example 3.5. Prior constructed as a convolution. —
Although topology-consistent priors cannot be constructed as n-tuples of independent random variables, it is always possible to construct a topology-consistent prior using other functional operations on independent random variables. A simple example is the joint density obtained as a convolution of independent random variables.
Consider the particular example of a topology-consistent convolution prior for the fully asymmetrical tree of figure 1. The basic idea is to model the time intervals between divergence events in the tree as random variables rather than the absolute ages of the divergences. For example, if we measure time going backwards into the past and define y1 to be the time at which the most recent speciation event occurred, y2 to be the time elapsed between the first and second speciation event, and so on, the age of the ith speciation event is 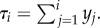 Clearly, τj < τi if j > i since waiting times between speciation events are assumed to be positive.
Clearly, τj < τi if j > i since waiting times between speciation events are assumed to be positive.
Any probability density on 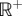 could be applied to yi so the procedure generates a family of distributions. As a simple example, one could choose the yi to be independent and identically distributed with density f(yi). More generally, one could assume independence but have a density function that varies according to the node rank. This is a rather awkward prior as it imposes a requirement that the prior variance of calibration-based divergence times is a strictly increasing function of the node rank. This may not fit the biological reality of the calibration data. A second requirement, that the ages of ranked nodes are strictly increasing, is desirable, given the node constraints, and insures that the prior is topology consistent.
could be applied to yi so the procedure generates a family of distributions. As a simple example, one could choose the yi to be independent and identically distributed with density f(yi). More generally, one could assume independence but have a density function that varies according to the node rank. This is a rather awkward prior as it imposes a requirement that the prior variance of calibration-based divergence times is a strictly increasing function of the node rank. This may not fit the biological reality of the calibration data. A second requirement, that the ages of ranked nodes are strictly increasing, is desirable, given the node constraints, and insures that the prior is topology consistent.
More effort should be directed toward deriving flexible joint prior distributions for node ages that are topology consistent, allowing phylogeneticists to propose prior distributions that do not conflict with relative age constraints imposed by the tree topology. In lieu of this, it is important to examine the form of the realized joint prior in an analysis by running the MCMC program with a fixed likelihood to estimate the prior. In general, topology constraints that are not imposed by the user-specified prior will reduce the variance of the realized prior and this can confound the influence of data versus the prior.
4. The combined information
In a ‘typical’ Bayesian analysis a parameter θ is identifiable in the sense that the likelihood of the observed data, X, is uniquely determined by the parameter,
and therefore,
is eventually dominated by the likelihood, which depends on the data rather than the prior on θ. The posterior density of θ becomes a degenerate point mass with increasing data when θ is identifiable. In the case of a non-identifiable parameter, θ, the posterior density of θ no longer converges to a point mass, even when the amount of data tends to infinity, and will instead become a density on a line, plane, etc., in hyperspace. The form of the asymptotic density is completely determined by the prior.
The divergence age estimation problem is not typical of most Bayesian inference problems because the ‘prior’ on ages (based on fossil data) is more akin to a likelihood function than a prior. The complete likelihood can be partitioned into two components: the first is the likelihood of the sequence data, D, which we denote as f(D|τ,r), given the fixed topology, divergence ages, τ, and per-site substitution rate r; the second is the likelihood of the fossil ages, C, given their lineage assignments and the ages of the divergence events of those lineages f(C|τC). The second likelihood component comes into the analysis in a posterior form
The above density includes the uncertainties of fossil age estimates as well as the intuitions of the palaeontologist about sampling effort, preservation rates, lineage assignments, etc., that are either formally, or informally, translated into the posterior on τC. This posterior is then incorporated into a prior on τ. A peculiarity is that the first likelihood component (based on D alone) does not allow τ to be inferred when the substitution rate r is unknown—the parameter is not identifiable.
The likelihood of the sequence data, D, depends on the product of divergence time and substitution rate and is, therefore, not identifiable because an uncountably infinite number of combinations of divergence times and substitution rates can be chosen that yield the same branch length (in units of expected substitutions) and therefore the same likelihood [15]. The implication is that without fossil calibrations the absolute times will be entirely determined by the prior on substitution rates and only the relative divergence times are influenced by the data. By adding fossil ages, the model becomes identifiable and the likelihoods will dominate inferences as we add both more sequence data and more precise fossil calibrations. The posterior can be formulated as
Yang & Rannala [2,11] considered the infinite sites limit
in which the branch lengths in units of expected substitutions become essentially fixed at their true values. These are referred to as the distances, d = {di}, where di is the expected number of substitutions per site on the path leading to any extant species descending from node i. Yang & Rannala [2] showed that this distribution is one-dimensional (determining the posterior density of the rate, r). One implication of this result is that an error in any fossil placement will affect all estimated divergence times. Another implication is that estimated divergence times remain uncertain even with infinite sequence data—this makes sense in the light of the non-identifiability of absolute ages of divergence events given the sequence likelihood.
(a). An example with three species
Yang & Rannala [2] derived the general form of the posterior density of τ for an infinite sequence length and a strict molecular clock. Here, I apply this result to a simple example with three species (figure 2). There are two divergence event ages in the tree τ = {τ1, τ2} and we assume that both have fossil calibration priors. Let the prior on the substitution rate be g(r) and the prior on ages be f(τ1, τ2). Using the relationship di = rτi and applying a transformation of variables,
Let the prior on r be an exponential density with mean 1/λ,
and let the prior on τi be
with the joint prior on τ being a product of the marginal priors. Now, consider the specific case where 1/λ = 10−9, μ1 = 1 × 107, μ2 = 2 × 107 and σ = 1 × 104. If the true ages are τ1 = μ1 and τ2 = μ2, and the mean of the rate prior is equal to the true substitution rate, r = 10−9, then the posterior of τ1 is as shown in figure 3. If the calibration prior for τ1 is instead grossly incorrect with true τ1 = 1.5 × 10−7 the resulting posterior is shown in figure 3. In this case, the incorrect calibration prior leads to a posterior that is apparently very precise but grossly incorrect.
Figure 3.
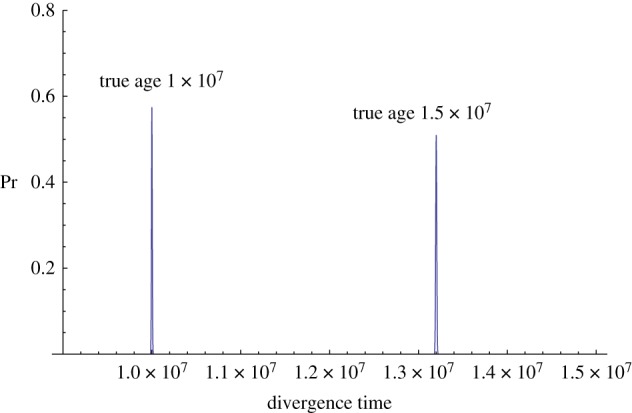
Posterior densities of τ1 with either true age of τ1 = 1 × 107 (peak at left) or τ1 = 1.5 × 107 (peak at right). (Online version in colour.)
5. Concluding remarks
The species divergence time inference problem has unusual statistical properties owing to the fact that the two distinct sources of data (sequence and fossil calibrations) will only lead to an identifiable model when used in combination. Correct assignment of fossils to ancestral lineages appears critical. Incorrect fossil assignments can result in posterior densities that are highly inaccurate, yet appear very precise. The specification of the prior on calibrations should also be done with care as the combination of marginal fossil calibrations with rank order constraints of phylogenetic trees can cause a change in the prior that may reduce the apparent uncertainty of divergence time estimates. This can be checked by running an MCMC program using a fixed likelihood. Given the complexity of this problem, even in the simplest scenarios, caution should be used when interpreting the results of empirical analyses, and overly narrow credible intervals for divergence times should be scrutinized for evidence of fossil conflicts and possible lineage mis-assignments of fossils.
Heled & Drummond [16] considered the more difficult problem of finding a joint prior for both topology and divergence times that preserves the marginal distributions of all calibration priors. They were able to formulate a solution for the case of a single calibration. In the case of a fixed topology, their requirement is equivalent to what is referred to as a topology-consistent prior in this paper. Theorem 3.3 therefore shows that for multiple calibrations, each with density on the real line, the objective of preserving marginals for calibrations is impossible to attain.
Competing interests
I declare I have no competing interests.
Funding
I received no funding for this study.
References
- 1.Rannala B, Yang Z. 2013. Molecular clock dating. In The Princeton guide to evolution (ed. JB Losos), pp. 67–74. Princeton, NJ: Princeton University Press. [Google Scholar]
- 2.Yang Z, Rannala B. 2006. Bayesian estimation of species divergence times under a molecular clock using multiple fossil calibrations with soft bounds. Mol. Biol. Evol. 23, 212–226. ( 10.1093/molbev/msj024) [DOI] [PubMed] [Google Scholar]
- 3.Heath T, Moore B. . In Bayesian phylogenetics: methods, algorithms, and applications. 2014. Bayesian inference of species divergence times (eds M-H Chen, L Kuo, PO Lewis), pp. 277–316. Boca Raton, FL: CRC Press. [Google Scholar]
- 4.Ronquist F, Klopfstein S, Vilhelmsen L, Schulmeister S, Murray DL, Rasnitsyn AP. 2012. A total-evidence approach to dating with fossils, applied to the early radiation of the Hymenoptera. Syst. Biol. 61, 973–999. ( 10.1093/sysbio/sys058) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang C, Stadler T, Klopfstein S, Heath TA, Ronquist F. 2015. Total-evidence dating under the fossilized birth–death process. Syst. Biol. 65, 228–249. ( 10.1093/sysbio/syv080) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ronquist F. 2004. Bayesian inference of character evolution. Trends Ecol. Evol. 19, 475–481. ( 10.1016/j.tree.2004.07.002) [DOI] [PubMed] [Google Scholar]
- 7.Donoghue PCJ, Benton MJ. 2007. Rocks and clocks: calibrating the Tree of Life using fossils and molecules. Trends Ecol. Evol. 22, 424–431. ( 10.1016/j.tree.2007.05.005) [DOI] [PubMed] [Google Scholar]
- 8.Wilkinson RD, Steiper ME, Soligo C, Martin RD, Yang Z, Tavaré S. 2010. Dating primate divergences through an integrated analysis of palaeontological and molecular data. Syst. Biol. 60, 16–31. ( 10.1093/sysbio/syq054) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Heath TA, Huelsenbeck JP, Stadler T. 2014. The fossilized birth–death process for coherent calibration of divergence-time estimates. Proc. Natl Acad. Sci. USA 111, E2957–E2966. ( 10.1073/pnas.1319091111) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Thorne JL, Kishino H, Painter IS. 1998. Estimating the rate of evolution of the rate of molecular evolution. Mol. Biol. Evol. 15, 1647–1657. ( 10.1093/oxfordjournals.molbev.a025892) [DOI] [PubMed] [Google Scholar]
- 11.Rannala B, Yang Z. 2007. Inferring speciation times under an episodic molecular clock. Syst. Biol., 56, 453–466. ( 10.1080/10635150701420643) [DOI] [PubMed] [Google Scholar]
- 12.Drummond AJ, Ho SYW, Phillips MJ, Rambaut A. 2006. Relaxed phylogenetics and dating with confidence. PLoS Biol. 4, 699 ( 10.1371/journal.pbio.0040088) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yang Z. 1994. Maximum likelihood phylogenetic estimation from DNA sequences with variable rates over sites: approximate methods. J. Mol. Evol. 39, 306–314. ( 10.1007/BF00160154) [DOI] [PubMed] [Google Scholar]
- 14.Bouckaert R, Heled J, Kühnert D, Vaughan T, Wu C-H, Xie D, Suchard MA, Rambaut A, Drummond AJ. 2014. BEAST 2: a software platform for Bayesian evolutionary analysis. PLoS Comput. Biol. 10, e1003537 ( 10.1371/journal.pcbi.1003537) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rannala B. 2002. Identifiability of parameters in MCMC Bayesian inference of phylogeny. Syst. Biol. 51, 754–760. ( 10.1080/10635150290102429) [DOI] [PubMed] [Google Scholar]
- 16.Heled J, Drummond AJ. 2012. Calibrated tree priors for relaxed phylogenetics and divergence time estimation. Syst. Biol. 61, 138–149. ( 10.1093/sysbio/syr087) [DOI] [PMC free article] [PubMed] [Google Scholar]