Abstract
Accurate sequence and assembly of genomes is a critical first step for studies of genetic variation. We generated a high-quality assembly of the gorilla genome using single-molecule, real-time sequence technology and a string graph de novo assembly algorithm. The new assembly improves contiguity by two to three orders of magnitude with respect to previously released assemblies, recovering 87% of missing reference exons and incomplete gene models. Although regions of large, high-identity segmental duplications remain largely unresolved, this comprehensive assembly provides new biological insight into genetic diversity, structural variation, gene loss, and representation of repeat structures within the gorilla genome. The approach provides a path forward for the routine assembly of mammalian genomes at a level approaching that of the current quality of the human genome.
High-quality sequence and assembly of genomes is a lynchpin to our understanding of the genetic diversity and evolution of species. The development of massively parallel sequencing technologies has drastically reduced the cost and increased the throughput of genome sequencing (1, 2). Although these advances have enabled sequencing of many more species and individuals, assemblies are left incomplete and fragmented in large part because the underlying sequence reads are too short [<200 base pairs (bp)] to traverse complex repeat structures (3). This has led to incomplete gene models, less accurate representation of repeats, and biases in our understanding of genome biology. The published western lowland gorilla genome assembly, for example, was generated by a mix of ABI capillary sequence and whole-genome shotgun Illumina short sequencing read pairs (4). Although the accuracy is high and many important inferences regarding the evolution of the species could be made, the resulting assembly contains more than 400,000 sequence gaps (Table 1) (4). Moreover, the use of the human genome to help guide the assembly of the gorilla genome created an artificially low number of structural rearrangements (5). The effect of these misassemblies and missing data becomes exacerbated when genomic comparisons are made among multiple primate genomes. For example, nearly 20% of the human genome could not be readily aligned in a four-way comparison among apes in large part because of the draft nature of nonhuman primate genomes (2). The recent development and application of long-read sequencing technologies has shown considerable promise in improving human genome assemblies as well as our understanding of genetic variation (3, 6, 7). We applied this long-read sequencing technology to generate an alternate genome assembly of the western lowland gorilla.
Table 1.
Gorilla assembly statistics.
| gorGor3* | Susie3 | |
|---|---|---|
| Individual | Kamilah | Susie |
| Total genome length (bp) | 3,035,660,144 | 3,080,414,926 |
| Number of contigs | 464,874 | 16,073 |
| Total sequence (bp) | 2,828,866,575 | 3,080,414,926 |
| Placed contig length (bp) | 2,718,960,062 | 2,790,620,487 |
| Unplaced contig length (bp) | 109,906,513 | 289,794,439 |
| Maximum contig length (bp) | 191,556 | 36,219,563 |
| Contig N50 (bp) | 11,661 | 9,558,608 |
| Number of scaffolds | 57,196 | 554 |
| Maximum scaffold length | 10,247,101* | 110,018,866 |
| Scaffold N50 (bp) | 913,458 | 23,141,960 |
Values are taken from previously published gorilla genome paper (4).
SMRT sequence and assembly of the gorilla genome
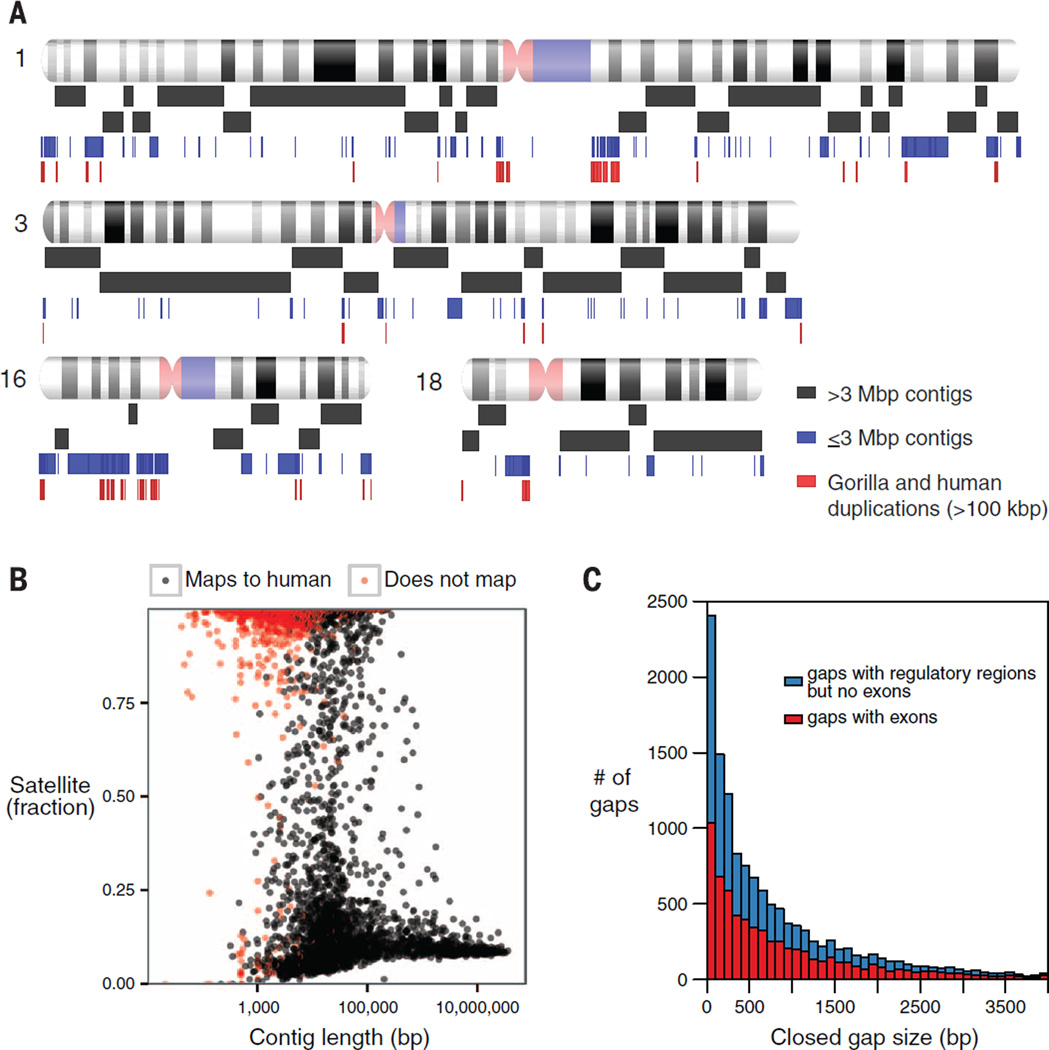
We generated 74.8-fold whole-genome shotgun sequence coverage using a single-molecule, real-time (SMRT) sequencing platform from peripheral blood DNA isolated from a western lowland gorilla (Gorilla gorilla gorilla) named Susie. All data were generated by using P6-C4 sequence chemistry from genomic libraries (>20 kbp in length), with an average subread length of 12.9 kbp. We applied a string graph assembly algorithm, Falcon (v.0.3.0), and consensus algorithm, Quiver (8), to generate a 3.1-Gbp assembly with a contig N50 of 9.6 Mbp (Table 1 and Figs. 1 and 2). Falcon leverages error-corrected long “super-reads” to generate a string graph representation of the genome that is subsequently refined by using a series of operations designed to break spurious edges and bridge across repetitive regions. The assembly produced 16,073 sequence contigs, including 889 contigs >100 kbp. Compared with a recent diploid assembly of a human genome (NA12878; N50 = 906 kbp) (7), the Falcon assembly represents a 10-fold improvement. We estimate that 98.9% of the gorilla euchromatin assembled into 1854 sequence contigs on the basis of alignment to human (GRCh38). The contigs were ordered and oriented into scaffolds (scaffoldN50 = 23.1 Mbp) with bacterial artificial chromosome (BAC)– and fosmid-end sequences. Sequence analysis reveals that most of the smaller contigs (<100 kbp) consist of either centromeric or telomeric satellite sequence or collapsed segmental duplications (Fig. 1B). Chromosomal regions with higher segmental duplication content tended to be enriched for shorter sequence contigs. In fact, we observed a strong correlation [correlation coefficient (r) = 0.76] between the remaining euchromatic gaps and the presence of gorilla or human segmental duplications (fig. S15).
Fig. 1. Gorilla genome assembly.
(A) Schematic depicting assembly contig lengths (contig N50 = 9.6 Mbp) mapped to human GRCh38 chromosomes. The first two rows of black rectangles represent contigs >3 Mbp, the blue rectangles correspond to contigs ≤3 Mbp, and red rectangles correspond to blocks of human/gorilla segmental duplications >100 kbp. (B) Mappability and satellite content of Susie3 contigs. Satellite content defined by use of RepeatMasker (28) and Tandem Repeats Finder (29). Contigs that are unable to map to GRCh38 by using BLASR (colored red) (30) contain a high fraction of satellite sequence. (C) Length distribution of gaps in the published gorilla assembly gorGor3 closed by Susie3 and containing exons or regulatory regions. Of the gaps in gorGor3, 94% were closed in Susie3, with thousands corresponding to missing exons (red) and putative noncoding regulatory DNA (blue).
Fig. 2. Gorilla genome ideogram.
Schematic depicting assembly contig lengths mapped to gorilla chromosomes. The first two rows of black rectangles represent contigs >3 Mbp, the green rectangles correspond to contigs >1 Mbp and ≤3 Mbp, and blue rectangles correspond to contigs ≤1 Mbp.
Compared with a previous gorilla genome assembly (gorGor3), this assembly represents a substantial decrease in assembly fragmentation (433,861 versus 16,073 contigs, >96% reduction in total contig number) (Fig. 3). Using the N50 contig length as a metric, we estimate a contiguity improvement of >819-fold with respect to the published gorilla genome assembly and >180-fold with respect to a more recently released upgrade of this Illumina-based assembly (Table 1 and table S4). When we aligned our sequence to the published gorilla genome reference (gorGor3), we closed 94% of the 433,861 gorGor3 gaps (fig. S13), resulting in the addition of least 164 Mbp of euchromatic sequence. This additional sequence dramatically improved gene annotation, including the discovery of thousands of exons and putative regulatory elements (Fig. 1C).
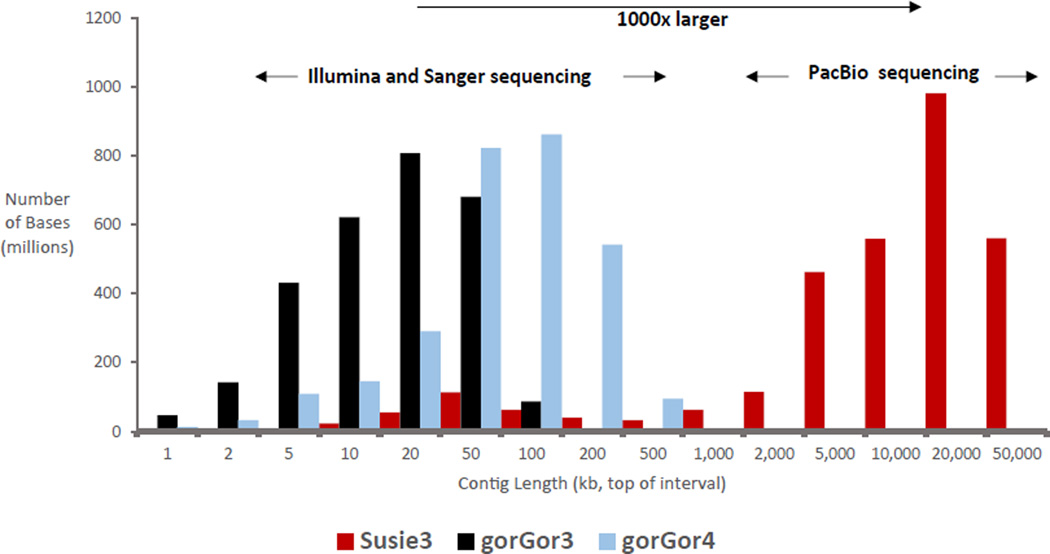
Fig. 3. Comparison of gorilla genome assemblies.
The contig length distribution for the resulting long-read assembly (Susie3) is 2 to 3 orders of magnitude larger when compared with previous gorilla genome assemblies (gorGor3 and gorGor4) that were generated by using Illumina and Sanger sequencing technology.
An analysis of the gaps in gorGor3 showed that they were enriched 3.8-fold for Alu short interspersed nuclear element (SINE) repeats, revealing that this G+C–rich primate repeat was particularly problematic in the initial gorilla assembly. Overall, there was a positive correlation between gap size and repeat content, especially for segmental duplications (three- to fivefold enrichment). For example, 10,959 gorGor3 gaps were in excess of 2 kbp; of these, 21% (2298) mapped to segmental duplications (table S12). Although heterochromatic regions could still not be resolved in the new gorilla assembly, our analysis of the underlying sequence data indicates that 10% of the gorilla genome consists of satellite repeats (table S15). A 32-bp satellite (pCht7) (9) associated with subterminal caps of gorilla chromosomes was more abundant than α-satellite repeats (4.0 versus 2.3%) within the genome sequence data (table S15). In contrast to other apes, these data suggest that telomeric-associated heterochromatic repeats are more abundant than centromeric repeats in gorilla.
Quality assessment
We assessed the quality of the assembly using two independent sources of data. First, we used paired-end sequence data generated during the sequencing and assembly of the first western lowland gorilla (Kamilah) to assess the assembly integrity. Our analysis showed that 98.6% of the new gorilla assembly was supported by concordant paired-end sequence data generated from either large-insert BAC or fosmid gorilla clones (table S2). An analysis of aligned high-quality Sanger sequence data (54.5 Mbp) revealed high sequence identity (99.71%) within the potential range of allelic diversity expected among western lowland gorillas (10) (table S10). In order to more precisely assess sequencing accuracy, we generated and aligned Illumina whole-genome sequence data from the same gorilla (Susie) and used sequence differences to estimate an initial error of one per 1000 base pairs [quality value (QV) = 30]. The most frequent errors observed were a threefold excess of single base pair insertions. We used Illumina whole-genome sequence data from Susie as well as six additional western lowland gorilla genomes (10) to correct these indel errors. Using these additional genomes, we created a “pan” gorilla reference genome (Susie3), correcting indel errors and identifying the most common single-nucleotide polymorphism variants. After error correction, we estimate that Susie3 has less than one error per 5000 bp (QV > 35) (table S7).
Comparative analyses between human and gorilla genomes
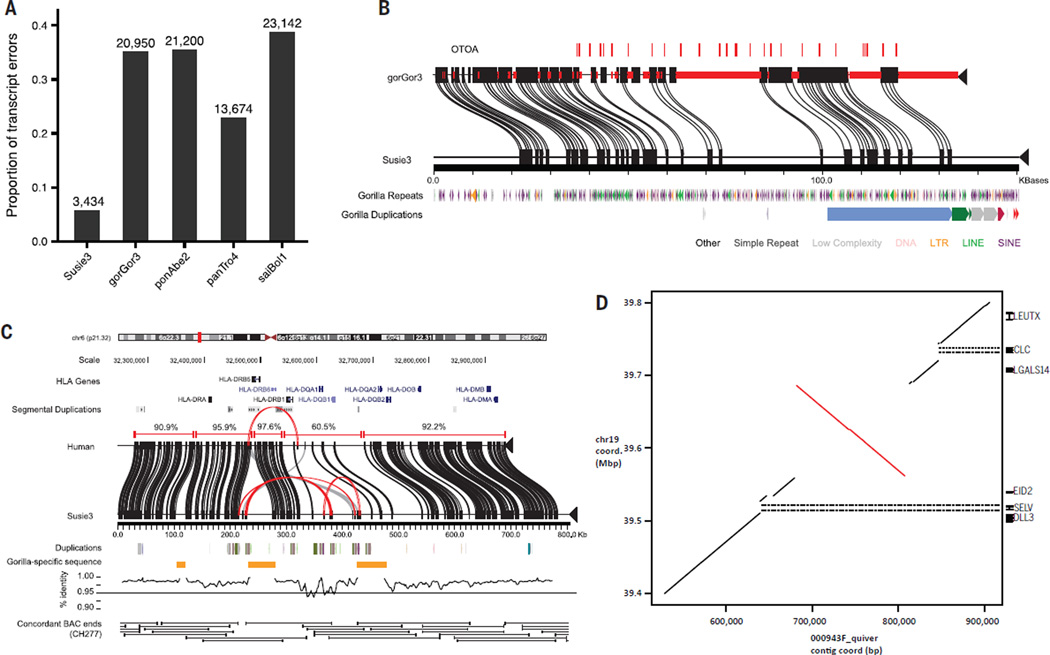
To determine the potential biological utility of this new reference gorilla genome, we measured the increased yield in gene content from gap closures in the gorilla reference by identifying human RefSeq (11) exons that map within gap regions of gorGor3 that are closed in Susie3. Using this projection, we estimate that 87%(11,105 of 12,754) of the missing RefSeq exons are recovered and that 94% of the incomplete gorilla genes are resolved for at least one isoform in Susie3. More generally, we found that an additional 1 to 3% of Illumina RNA-sequencing (RNA-seq) data and an additional 6 to 7% of gorilla ESTs are mapped by standard mapping methods to this more complete genome assembly (table S13). The assembly also improves the contiguous representation of genes predicted by human GENCODE (12). Compared with other nonhuman primate genomes, Susie3 has far fewer assembly errors, making it one of the most complete primate genomes after human (Fig. 4). We found that 88% of protein-coding transcripts (GENCODE) have >99% identity against Susie3 compared with ~55% in gorGor3 (fig. S18). We leveraged the new assembly, available RNA-seq data, and Augustus TransMap (13–15) alignments to define a new set of 45,087 consensus gene models for protein-coding transcripts (isoforms from 19,633 genes) in the gorilla genome.
Fig. 4. Gene annotation and structural variation.
(A) Proportion of GENCODE transcripts with assembly errors when aligned with gorilla assemblies Susie3 and gorGor3, and three reference assemblies, including orangutan (ponAbe2), chimpanzee (panTro4), and squirrel monkey (saiBol1). Examples of assembly errors include transcript mappings extending off the end of contigs/scaffolds, containing unknown bases, or incomplete transcript mapping. (B) An example of a gene, otoancorin (OTOA), with complete exon representation (red ticks) resolved in the new assembly. Red bars on gorGor3 sequence indicate gaps in the assembly. Alignments between gorilla assemblies are based on Miropeats (31). (C) Alignment of MHC Class II locus in Susie3 against GRCh37 with Miropeats. Alignment identities of collinear blocks between assemblies are shown above the corresponding GRCh37 sequence. Repeats internal to Susie3 are shown in red along the coordinates. Alignment identity across the entire locus is shown below the Susie3 contigs in 5-kbp windows (1 kbp sliding). Support for the proper organization of the Susie3 sequence is shown by the tiling path of concordant BAC end sequences from the Kamilah BAC library (CHORI-277). (D) A sequence-resolved complex gorilla genome structural variation orthologous to human chromosome 19:38,867,213–39,866,620 (GRCh38). The dot-matrix plot shows a 125,375-bp inversion flanked by a proximal 16-kbp deletion and 8-kbp insertion, and a 23-kbp distal deletion. The deletions remove the entire sequences of the SELV and CLC genes in gorilla when compared with human.
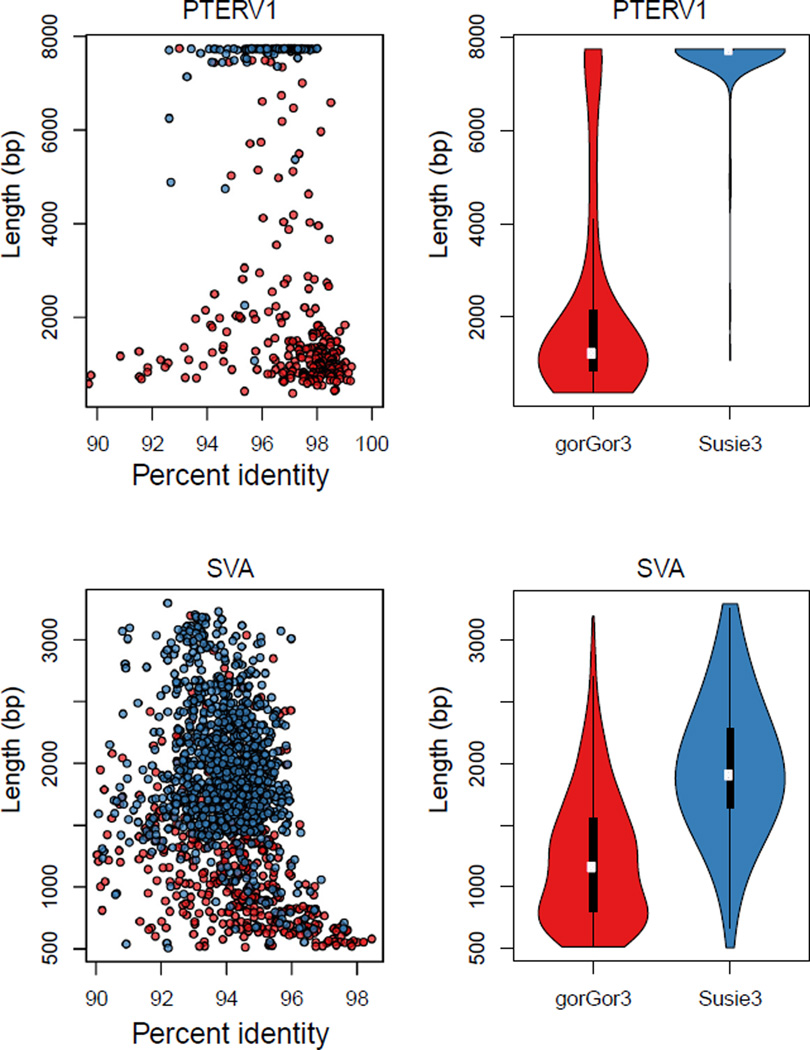
The greater sequence contiguity of the genome assembly provided an opportunity to comprehensively assess structural variation between gorilla and human genomes at a fine-scale resolution, down to ~50 bp. Comparison of the gorilla (Susie3) and human (GRCh38) assemblies revealed a total of 117,512 insertions and deletions (92 Mbp) and 697 inversion variants (Table 2 and table S20). More than 86% of these events were previously unidentified (fig. S47) (16, 17), and 72% of the events were determined to represent fixed differences specific to the gorilla lineage. Variants ranged in size from small structural variants within the coding sequence of genes to large, complex, gene-rich structural variant events spanning hundreds of kilobase pairs (Fig. 4, C and D). Our analysis also provided a comprehensive catalog of mobile element differences between human and gorilla (24.1% of all structural variation events). Compared with earlier gorilla genome assemblies, we found that the proportion of full-length retrotransposons is significantly greater for the longest [HERV Kolmogorov-Smirnov (KS) P = 0.0016, and PTERV1 KS P < 2.2 × 10−16] and most G+C–rich repeat elements (SVA KS P < 2.2 × 10−16) (Fig. 5). PTERV1 is particularly notable in this regard because its 9- to 10-kbp full-length insertions are specific to the gorilla lineage (18, 19); we found a 4.8-fold increase in full-length elements, with most elements in Susie3 being previously unidentified.
Table 2.
Gorilla genome structural variants. Contigs greater than 200 kbp were mapped to GRCh38 by using BLASR (30). Nonrepetitive sequences contained at most 70% of sequence annotated as repeat by RepeatMasker (3.3.0) (28) or Tandem Repeats Finder 4.07b (29). Mosaic repeats are defined as one or more different repeat annotations; however, mosaic structural variants composed solely of Alu are listed separately because of their frequency.
| Insertion | Deletion | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Repeat element |
Count | Fixed | Average length (bp) |
Standard deviation length (bp) |
Total bases | Count | Fixed | Average length (bp) |
Standard deviation length (bp) |
Total bases |
| Complex, not repetitive | 15,383 | 13,190 | 709.31 | 1807.29 | 10911284 | 13,749 | 10,250 | 763.17 | 1,954 | 10492765 |
| Complex, repetitive | 7,552 | 7,243 | 2716.96 | 2746.63 | 20,518,484 | 7,450 | 5,934 | 2757.39 | 2906.61 | 20,542,529 |
| AluY | 6,418 | 5,878 | 284.36 | 65.2 | 1,825,012 | 9,422 | 7,882 | 294.58 | 60.04 | 2,775,515 |
| AluS | 990 | 950 | 160.64 | 85.98 | 159,036 | 1,053 | 845 | 159.84 | 82.05 | 168,315 |
| L1Hs | 86 | 50 | 262.67 | 582.37 | 22,590 | 220 | 179 | 235.51 | 358.93 | 51,813 |
| L1 | 1,339 | 1,256 | 438.5 | 737.1 | 587,147 | 1,181 | 874 | 450.84 | 744.1 | 532,441 |
| L1P | 1,891 | 1,660 | 278.26 | 487.86 | 526,192 | 1,430 | 1,098 | 312.81 | 493.88 | 447,317 |
| SVA | 1,498 | 1,097 | 1561.2 | 1007.55 | 2,338,672 | 1,194 | 786 | 1279.32 | 914.18 | 1,527,511 |
| HERV | 324 | 285 | 241.06 | 498.09 | 78,104 | 260 | 194 | 362.79 | 797.4 | 94,325 |
| PTERV | 190 | 179 | 2973.22 | 3427 | 564,912 | 1 | 1 | 188 | 0 | 188 |
| Mosaic-Alu | 1,433 | 1,355 | 320.11 | 262.59 | 458,717 | 1,514 | 1,177 | 307.92 | 260.53 | 466,194 |
| STR | 11,049 | 4,152 | 228.73 | 313.35 | 2,527,257 | 10,564 | 4,573 | 156.93 | 175.96 | 1,657,802 |
| Tandem Repeats | 7,351 | 4,520 | 367.74 | 811.03 | 2,703,261 | 7,828 | 4,225 | 323.38 | 558.5 | 2,531,424 |
| Satellite | 1,757 | 1,613 | 437.46 | 1072.08 | 768,617 | 1,768 | 1,437 | 426.73 | 1047.71 | 754,455 |
| Other | 1,319 | 1,184 | 201.01 | 247.8 | 265,130 | 1,196 | 896 | 180.49 | 227.79 | 215,872 |
| Not base-pair resolved | 41 | N/A | 49,496 | 23817.46 | 2029323 | 61 | N/A | 55,971 | 43,496 | 3,414,209 |
| Total | 58,621 | 44,653 | 789.54 | 2183.43 | 46,283,738 | 58,891 | 40,482 | 775.55 | 2804.21 | 45,672,675 |
Fig. 5. Improved mobile element resolution.
(Left) PTERV1 and SVA insertion length and percent identity distributions in Susie3 (blue) and gorGor3 (red). The PTERV1 and SVA elements in gorGor3 are biased toward short but on average higher identity alignments to the consensus sequence because the more divergent long terminal repeat sequences are not resolved. (Right) The mean and median insertion lengths for gorGor3 and Susie3 are PTERV1, 2194.93, 7565.85 (median 1223 and 7725) and SVA, 1240.1, and 1965.63 (median 1162 and 1909).
A small fraction (0.3%) of the structural variants affect protein-coding genes, including those that appear to lead to a likely gene-disruptive event in gorilla when compared with human. Because such gene-disrupting events may represent polymorphisms or be enriched for errors in a reference, we restricted our analysis to previously undescribed events that were shared across an additional six western lowland gorillas (table S19). Using these criteria, we identified 145 structural variants (76 deletions and 69 insertions) that affect the coding sequence of 110 distinct RefSeq genes. Although no single functional gene category reached statistical significance for enrichment, several of the genic differences between the species were associated with sensory perception, keratin production, interleukin and cytokine secretion, reproduction, immunity, growth, transmembrane signaling, and nucleotide binding (table S19).
We also investigated structural variants that affected potential regulatory elements on the basis of recent annotations of the human genome (ENCODE). We annotated base pair–resolved variants that intersected with deoxyribonuclease I hypersensitive (DNase I HS) sites associated with open chromatin, histone H3 lysine 4 trimethylation (H3K4me3) marks associated with transcriptionally active regions/promoters, and histone H3 acetylation on lysine 27 (H3K27ac) signals associated with enhancers (12). Among the 10,466 insertions and deletions that intersected regulatory elements, 2151 represented fixed structural differences between humans and gorillas (GSVs). Similarly, we identified 133 GSVs that affect long intergenic noncoding RNA (lincRNA). As expected, we observed a significant (2- to 10-fold) depletion of structural variants intersecting these functional categories, with the exception of lincRNA.
We further quantified the spatial correlation between GSVs and the regulatory elements by comparing their proximity. The midpoints of the GSVs overlapped significantly less with the protein-coding genes (projection test P = 6.0 × 10−12) and with fetal DNase I HS sites (projection test P = 4.3 × 10−10), which is consistent with the action of purifying selection. We observed a modest spatial enrichment between GSVs that map within 50 to 200 bp of putative promoter and enhancer marks (projection test P = 0.041 and 4.1 × 10−15 for H3K4me3 and H3K27ac, respectively). This analysis identified 327 protein-coding genes within 10 kbp of an H3K4me3 mark that overlaps or maps near a GSV (<100 bp) and 672 genes that had the same spatial pattern for H3K27ac marks. We compared these fixed structural differences within putative regulatory DNA with 127 genes with differences in RNA expression and CTCF binding sites between human and gorilla (20). We identified nine genes in which a GSV intersected an H3K4me3 or HeK27Ac mark (ADAMTS10, ALDH1L1, CDH1, COL5A1, GRK5, IGF2BP1, INSR, IQGAP2, and SRC) and eight that intersected the 3′ untranslated region or a non-coding transcribed exon (AMOTL1, DUSP4, HNF1B, ITGB8, SGPP2, TCL1A, ZDHHC19, and ZNF607). None of these GSVs affected protein-coding regions of these genes, and thus, these fixed differences in noncoding regulatory DNA are strong candidates to explain the expression differences between the species.
Six inversions were identified that potentially disrupt an exon or break within a gene, in addition to 206 inversions and microinversions that map within introns. Among the insertion and deletion events, 15 of the structural variants occurred in genes that are largely intolerant to mutation in humans (residual variation intolerance score < 20 percentile) (21). This includes insertions within the insulin-degrading enzyme (IDE) and a negative regulator of WNT signaling (AXIN1), as well as deletions in a tubulin-specific chaperone (TBCD) and an extracellular sulfatase (SULF2) associated with cell signaling. Our analysis revealed a large number of in-frame deletions and insertions that result in the addition and loss of amino acids and protein domain differences between human and gorilla genes. Although the impact of these structural variants awaits further characterization, the greater resolution of structural variants provides a large number of likely high-impact species differences for future investigation.
Sequence and assembly of the major histocompatibility locus
As a test of the enhanced structural variation detection and its potential biomedical relevance, we compared the organization of the major histocompatibility complex (MHC) II locus between gorilla and human (GRCh37). MHC encodes genes that are critical for antigen presentation on immune cells, and its content and structure are known to differ radically between closely related primates (22). We found that segmental duplication has expanded the locus in gorilla relative to human so that in the assembly, there are 79,166 (~10%) duplicated base pairs, compared with 53,084 bp (~8%) in human. We identified three large gorilla insertions across the ~1 Mbp region (15, 48, and 52 kbp) that correspond to 14% of the locus (Fig. 4C), including several previously unidentified MHC genes. In terms of assembly statistics, the MHC region in gorGor3 has 168 gaps that are filled in Susie3, eight of which mapped within 100 bp of 27 distinct genes (human RefSeq).
To verify the organization of the HLA region in Susie3, we sequenced a tiling path of BACs derived from another gorilla, Kamilah, who previously contributed to the published gorGor3 assembly (table S6). We generated two high-quality, clone-based sequence haplotypes for the region (MHC haplotype 1, 863,324 bp; MHC haplotype 2, 289,560 bp). Both Kamilah MHC haplotypes mapped with higher sequence identity to Susie3 (99.6 and 99.9%) than gorGor3 (98.1 and 98.9%), which is consistent with assembly improvements for this complex region of the genome.
Population genetic inferences based on the new assembly
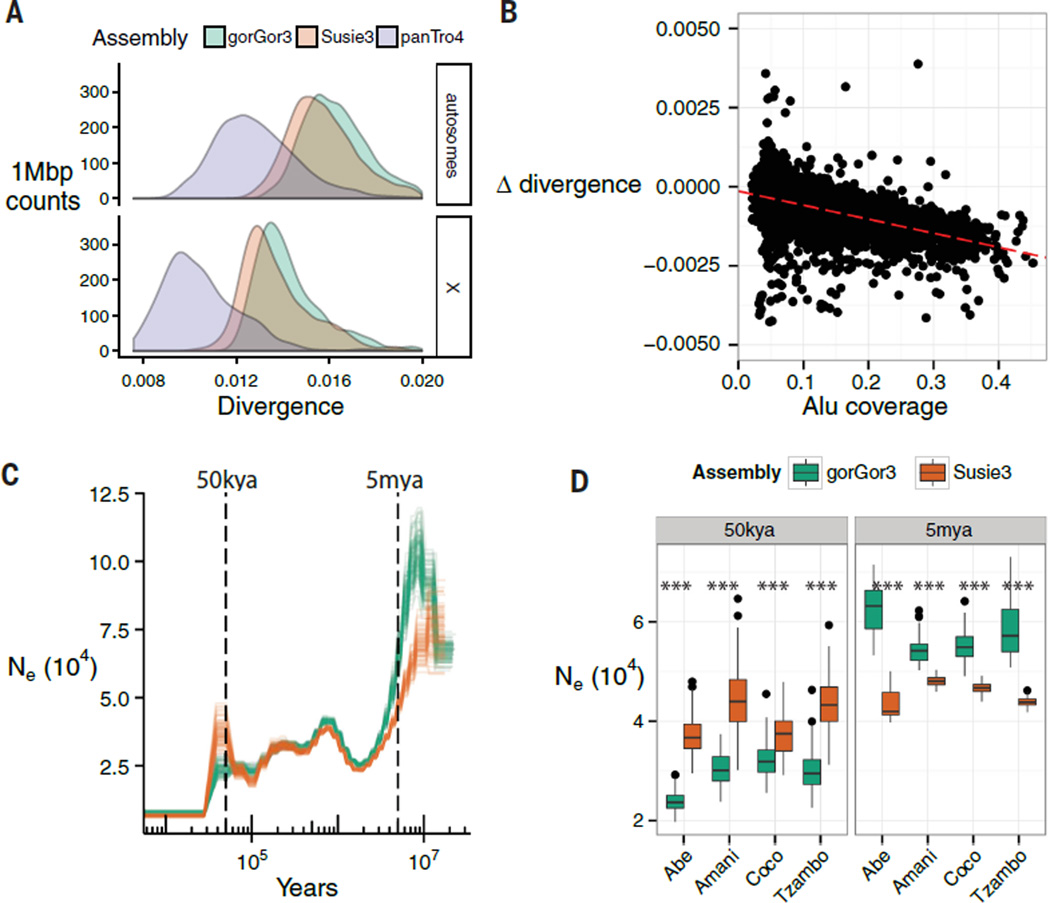
Last, we assessed whether the new gorilla genome assembly would have any influence on previous population genetic inferences as a result of potential changes in estimates of divergence and diversity. Although the difference was subtle, we found that human versus gorilla sequence alignments were significantly less divergent (P < 2.2 × 10−16; Welch two-sample t test) with Susie3 (1.60% divergent) when compared with the published gorilla assembly (1.65% divergent) (Fig. 6). Both regional and chromosomal analyses showed that the different estimates of gorilla-human divergence were nonrandomly distributed. In particular, we observed that specific gene-rich regions and chromosomes (such as chromosomes 19 and 22) showed the largest difference between the two assemblies (fig. S61B). We found a strong correlation with the difference in divergence and regions enriched for Alu and G+C content (r = −0.60; Pearson’s, P < 0.001) (Fig. 6B), suggesting that mismapping, collapse, or underrepresentation within these regions of the Illumina-based assemblies may be contributing to this excess of divergence (Fig. 6B).
Fig. 6. Population genetic analyses.
(A) Density of average divergence within 1-Mbp windows between human (GRCh38) and gorGor3, Susie3, or chimpanzee (panTro4) autosomes. (B) A comparison of human-gorGor3 and human-Susie3 divergence over 1-Mbp windows. The x axis is Alu coverage in each window, and the y axis is the difference in human-gorilla divergence between gorGor3 and Susie3. Positive y axis values indicate increased human–Susie3 divergence relative to human–gorGor3. The increased divergence of human–gorGor3 correlates with Alu content (slope, −0.0044094; intercept, 0.0001486; Pearson’s correlation, −0.60). (C) The effective population size (Ne) shown over time. A PSMC model was applied to the western lowland gorilla based on different genome assemblies. Illumina genome sequence data from western lowland gorillas (Abe, Amani, Coco, Tzambo) was mapped against gorGor3 (green) and Susie3 (orange), and PSMC was fit to the genome alignments (-N25 -t15 -r5 -b -p “4+25*2+4+6”; mutation rate = 1.25 × 10−8; generation time = 19 years). There are 100 bootstrap replicates for each gorilla and model. (D) The distribution of the bootstrap intervals that overlap 50 ka and 5 ma. At 50 ka, Susie3 estimates of the effective population size are significantly higher than that for gorGor3; the inverse pattern is true for 5 ma. All differences between Susie3 and gorGor3 are significant (***P ≤ 0.0001; Welch two-sample t test).
We also assessed the effect of the new assembly on estimates of diversity by comparing the pattern of single-nucleotide variants for six western lowland gorillas against Susie3 and gorGor3. Although a greater fraction of the Illumina paired-end reads mapped with higher mapping quality to Susie3 (table S8), we observed a higher ratio of heterozygous genotypes in gorGor3 (mean, 0.35) as compared with Susie3 (mean, 0.33). Next, we examined the average observed heterozygosity across both assemblies in 100-kbp windows (fig. S62). The largest difference in observed heterozygosity between the assemblies was on the X chromosome, in which the average gorGor3 heterozygosity was 0.27 versus 0.23 in Susie3.
It is likely that the increased gorGor3 heterozygosity is due to mapping errors owing to fewer reads successfully mapping (gorGor3 has more than 400,000 gaps, many of which contain repetitive sequences). Mapping software errs on the side of sensitivity, meaning reads derived from underrepresented regions may be incorrectly placed. To test this idea, we identified the reads (including mate-pairs) from Coco, a female gorilla, corresponding to heterozygous positions in gorGor3 and remapped them to both gorGor3 and Susie3. We found that only 87% of the heterozygous calls in gorGor3 resulted in heterozygous calls in Susie3 (table S33). The “lost” heterozygous genotypes correspond to regions with lower Illumina read depth as compared with that of gorGor3, supporting the notion that mapping error is likely inflating heterozygosity in the original assembly (fig. S63).
Accurate estimates of population histories are important for understanding how climate change, disease, and human activity shaped the genetic diversity within western lowland gorillas. Such estimates are critically dependent on genetic measures such as heterozygosity. We fit a pairwise sequentially Markovian coalescent (PSMC) model to Illumina data from four western lowland gorillas mapped against Susie3 and gorGor3 (Fig. 6). Comparing the two assemblies, we observed two statistically significant differences in the predicted effective population size of the gorilla species at ~50 thousand years ago (ka) and ~5 million years ago (ma) (Fig. 6). The effects occur in opposite directions. We observed a significant decrease in the effective population size at more ancient time periods (P < 0.0001; Welch two-sample t test), possibly because of a greater fraction of repetitive sequence and segmental duplications being resolved in Susie3. In contrast, use of Susie3 as a reference predicts a significantly greater effective population size at 50 ka (P < 0.0001; Welch two-sample t test). Our results suggest that the most recent bottleneck of the western lowland gorilla may have been underestimated by a factor of ~1.5, highlighting the importance of using higher-quality assemblies when fitting demographic models.
Our results demonstrate the utility of long-read sequence technology to generate high-quality working draft genomes of complex vertebrate genomes without guidance from preexisting reference genomes. Comparisons against recent published human genomes that used alternate assemblers indicate that Falcon provides superior performance with respect to assembled contig length, repeat resolution, and computational time (6, 7). The genome assembly that results from using the long-read data provides a more complete picture of gene content, structural variation, and repeat biology as well as allows us to refine population genetic and evolutionary inferences. Notwithstanding the dramatic improvements to the sequence contiguity of euchromatin, gaps still remain. Our analysis suggests that both heterochromatin and large, high-identity segmental duplications remain largely unresolved because read lengths are insufficiently long to traverse these repetitive structures (Fig. 1C). Although all long-read assembly algorithms should still be considered work in progress, it is clear that both sequencing and computational technologies have now advanced to a stage that allows individual laboratories to generate high-quality mammalian genomes with vastly higher contiguity of the euchromatin. This capability promises to revolutionize our understanding of genome evolution and species biology.
Materials and methods
High-quality DNA was extracted (Qiagen Puregene kit, Valencia, CA) from peripheral blood drawn from a captive female western lowland gorilla (Susie) from the Lincoln Park Zoo (Chicago, IL). We generated SMRTbell genomic libraries (>20 kbp in length) and SMRT sequence data (P6C4 chemistry, RSII platform) with average subread lengths of 12.9 kbp. A gorilla genome assembly, Susie3, was generated by using Falcon (v0.3.0) (https://github.com/PacificBiosciences/FALCON-integrate). The Falcon assembly method operates in two phases: First, overlapping sequence reads are compared [DALIGNER (23)] to generate 97 to 99% accurate consensus sequences for reads with lengths in the top percentile (>15 kbp). Next, overlaps between the corrected longer reads are used to generate a string graph (24). The graph is reduced so that multiple edges formed by heterozygous structural variation are replaced to represent a single haplotype. Contigs are formed by using the sequences of nonbranching paths. Two supplemental graph cleanup operations are defined so as to improve assembly quality by removing spurious edges from the string graph: tip removal (25) and chimeric duplication edge removal. Tip removal discards sequences with errors that prevent 5′ or 3′ overlaps. Chimeric duplication edges may result from the raw sequence information or during the first sequence cleanup step and artificially increase the copy number of a duplication. Indel errors in the assembly were corrected by aligning Illumina (HiSEq. 2500 and NextSeq) sequence data generated from six additional western lowland gorillas (10), including 14-fold sequence data generated from the new gorilla reference genome (Susie). Consensus gene models were built with Augustus (13) by use of TransMap (14, 15) alignments of GENCODE transcripts to Susie3 and previously published RNA-seq data (26, 27).
Supplementary Material
Acknowledgments
We are grateful to A. Scally and Z. Ning for early access to the upgraded Kamilah gorilla assembly (gorGor4) and for discussion regarding its assembly. We thank M. Duyzend, L. Harshman, and C. Lee for technical assistance and quality control in generating sequencing data and H. Li for helpful suggestions for the PSMC analysis. The authors thank M. Heget, K. Gillespie, and M. Shender from the Lincoln Park Zoo for providing gorilla peripheral blood and T. Brown for assistance in editing this manuscript. This work was supported, in part, by grants from the U.S. National Institutes of Health (NIH grant HG002385 to E.E.E. and HG007635 to R.K.W. and E.E.E.; HG003079 to R.K.W.; HG007990 to D.H. and B.P.; and HG007234 to B.P.). E.E.E., J.S., and D.H. are investigators of the Howard Hughes Medical Institute. E.E.E. is on the scientific advisory board (SAB) of DNAnexus and was a SAB member of Pacific Biosciences. (2009–2013); E.E.E. is a consultant for Kunming University of Science and Technology (KUST) as part of the 1000 China Talent Program. M.J.P.C. is a former employee of (2009–2012) and owns shares in Pacific Biosciences. On 24 February 2011, Pacific Biosciences filed a patent entitled “Sequence assembly and consensus sequence determination” (U.S. patent no. US20120330566, issued 27 December 2012); M.J.P.C. is identified as inventor of this patent. Pacific Biosciences has filed two patents related to the Falcon assembler algorithm entitled “String graph assembly for polyploid genomes” (U.S. patent no. US2015/0169823 A1 filed 18 December 2014, and U.S. patent no. US2015/0286775 A1 filed 18 June 2015); C.C. is identified as inventor for both patents.
Footnotes
SUPPLEMENTARY MATERIALS
www.sciencemag.org/content/352/6281/aae0344/suppl/DC1
Supplementary Text
Figs. S1 to S63
Tables S1 to S36
References (32–67)
The Susie3 assembly, PacBio and Illumina sequencing data for Susie, and clone sequences have been deposited in the European Nucleotide Archive under the project accession PRJEB10880. E.E.E., D.G., J.H., M.J.P.C., C.M.H., and Z.N.K. designed experiments; K.M.M., M.M., and C.B. prepared libraries and generated sequencing data; D.G., J.H., M.J.P.C., C.M.H., Z.N.K., L.W.H., and A.R. performed bioinformatics analyses; I.F., J.A., M.D., B.P., R.K.W., and D.H. analyzed gene accuracy. J.S. helped in the evaluation of Hi-C data. C.D. and C.-S.C. aided in Falcon assembler modifications. J.H. deposited SMRT sequencing data into SRA. E.E.E., D.G., J.H., M.J.P.C., C.M.H., and Z.N.K. wrote the manuscript.
REFERENCES AND NOTES
- 1.Lam HYK, et al. Performance comparison of whole-genome sequencing platforms. Nat. Biotechnol. 2012;30:78–82. doi: 10.1038/nbt.2065. pmid: 22178993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rogers J, Gibbs RA. Comparative primate genomics: Emerging patterns of genome content and dynamics. Nat. Rev. Genet. 2014;15:347–359. doi: 10.1038/nrg3707. pmid: 24709753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chaisson MJP, Wilson RK, Eichler EE. Genetic variation and the de novo assembly of human genomes. Nat. Rev. Genet. 2015;16:627–640. doi: 10.1038/nrg3933. pmid: 26442640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Scally A, et al. Insights into hominid evolution from the gorilla genome sequence. Nature. 2012;483:169–175. doi: 10.1038/nature10842. pmid: 22398555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Carbone L, et al. Gibbon genome and the fast karyotype evolution of small apes. Nature. 2014;513:195–201. doi: 10.1038/nature13679. pmid: 25209798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Berlin K, et al. Assembling large genomes with single-molecule sequencing and locality-sensitive hashing. Nat. Biotechnol. 2015;33:623–630. doi: 10.1038/nbt.3238. pmid: 26006009. [DOI] [PubMed] [Google Scholar]
- 7.Pendleton M, et al. Assembly and diploid architecture of an individual human genome via single-molecule technologies. Nat. Methods. 2015;12:780–786. doi: 10.1038/nmeth.3454. pmid: 26121404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chin C-S, et al. Nonhybrid, finished microbial genome assemblies from long-read SMRT sequencing data. Nat. Methods. 2013;10:563–569. doi: 10.1038/nmeth.2474. pmid: 23644548. [DOI] [PubMed] [Google Scholar]
- 9.Royle NJ, Baird DM, Jeffreys AJ. A subterminal satellite located adjacent to telomeres in chimpanzees is absent from the human genome. Nat. Genet. 1994;6:52–56. doi: 10.1038/ng0194-52. pmid: 8136835. [DOI] [PubMed] [Google Scholar]
- 10.Prado-Martinez J, et al. Great ape genetic diversity and population history. Nature. 2013;499:471–475. doi: 10.1038/nature12228. pmid: 23823723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.O’Leary NA, et al. Reference sequence (RefSeq) database at NCBI: Current status, taxonomic expansion, and functional annotation. Nucleic Acids Res. 2016;44(D1):D733–D745. doi: 10.1093/nar/gkv1189. pmid: 26553804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Harrow J, et al. GENCODE: The reference human genome annotation for The ENCODE Project. Genome Res. 2012;22:1760–1774. doi: 10.1101/gr.135350.111. pmid: 22955987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stanke M, Diekhans M, Baertsch R, Haussler D. Using native and syntenically mapped cDNA alignments to improve de novo gene finding. Bioinformatics. 2008;24:637–644. doi: 10.1093/bioinformatics/btn013. pmid: 18218656. [DOI] [PubMed] [Google Scholar]
- 14.Siepel A, et al. Targeted discovery of novel human exons by comparative genomics. Genome Res. 2007;17:1763–1773. doi: 10.1101/gr.7128207. pmid: 17989246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhu J, et al. Comparative genomics search for losses of long-established genes on the human lineage. PLOS Comput. Biol. 2007;3:e247. doi: 10.1371/journal.pcbi.0030247. pmid: 18085818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ventura M, et al. Gorilla genome structural variation reveals evolutionary parallelisms with chimpanzee. Genome Res. 2011;21:1640–1649. doi: 10.1101/gr.124461.111. pmid: 21685127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sudmant PH, et al. Evolution and diversity of copy number variation in the great ape lineage. Genome Res. 2013;23:1373–1382. doi: 10.1101/gr.158543.113. pmid: 23825009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yohn CT, et al. Lineage-specific expansions of retroviral insertions within the genomes of African great apes but not humans and orangutans. PLOS Biol. 2005;3:e110. doi: 10.1371/journal.pbio.0030110. pmid: 15737067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Polavarapu N, Bowen NJ, McDonald JF. Identification, characterization and comparative genomics of chimpanzee endogenous retroviruses. Genome Biol. 2006;7:R51. doi: 10.1186/gb-2006-7-6-r51. pmid: 16805923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xue Y, et al. Mountain gorilla genomes reveal the impact of long-term population decline and inbreeding. Science. 2015;348:242–245. doi: 10.1126/science.aaa3952. pmid: 25859046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Petrovski S, Wang Q, Heinzen EL, Allen AS, Goldstein DB. Genic intolerance to functional variation and the interpretation of personal genomes. PLOS Genet. 2013;9:e1003709. doi: 10.1371/journal.pgen.1003709. pmid: 23990802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Little AM, Parham P. Polymorphism and evolution of HLA class I and II genes and molecules. Rev. Immunogenet. 1999;1:105–123. pmid: 11256568. [PubMed] [Google Scholar]
- 23.Myers EW. Efficient local alignment discovery amongst noisy long reads. Lect. Notes Comput. Sci. 2014;8701:52–67. [Google Scholar]
- 24.Myers EW. The fragment assembly string graph. Bioinformatics. 2005;21(suppl. 2):ii79–ii85. doi: 10.1093/bioinformatics/bti1114. pmid: 16204131. [DOI] [PubMed] [Google Scholar]
- 25.Pevzner PA, Tang H, Tesler G. De novo repeat classification and fragment assembly. Genome Res. 2004;14:1786–1796. doi: 10.1101/gr.2395204. pmid: 15342561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Brawand D, et al. The evolution of gene expression levels in mammalian organs. Nature. 2011;478:343–348. doi: 10.1038/nature10532. pmid: 22012392. [DOI] [PubMed] [Google Scholar]
- 27.Necsulea A, et al. The evolution of lncRNA repertoires and expression patterns in tetrapods. Nature. 2014;505:635–640. doi: 10.1038/nature12943. pmid: 24463510. [DOI] [PubMed] [Google Scholar]
- 28.Smit A, Hubley R, Green P. RepeatMasker Open-3.0. 1996 available at www.repeatmasker.org. [Google Scholar]
- 29.Benson G. Tandem repeats finder: A program to analyze DNA sequences. Nucleic Acids Res. 1999;27:573–580. doi: 10.1093/nar/27.2.573. pmid: 9862982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chaisson MJ, Tesler G. Mapping single molecule sequencing reads using basic local alignment with successive refinement (BLASR): Application and theory. BMC Bioinformatics. 2012;13:238. doi: 10.1186/1471-2105-13-238. pmid: 22988817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Parsons JD. Miropeats: Graphical DNA sequence comparisons. Comput. Appl. Biosci. 1995;11:615–619. doi: 10.1093/bioinformatics/11.6.615. pmid: 8808577. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.