Abstract
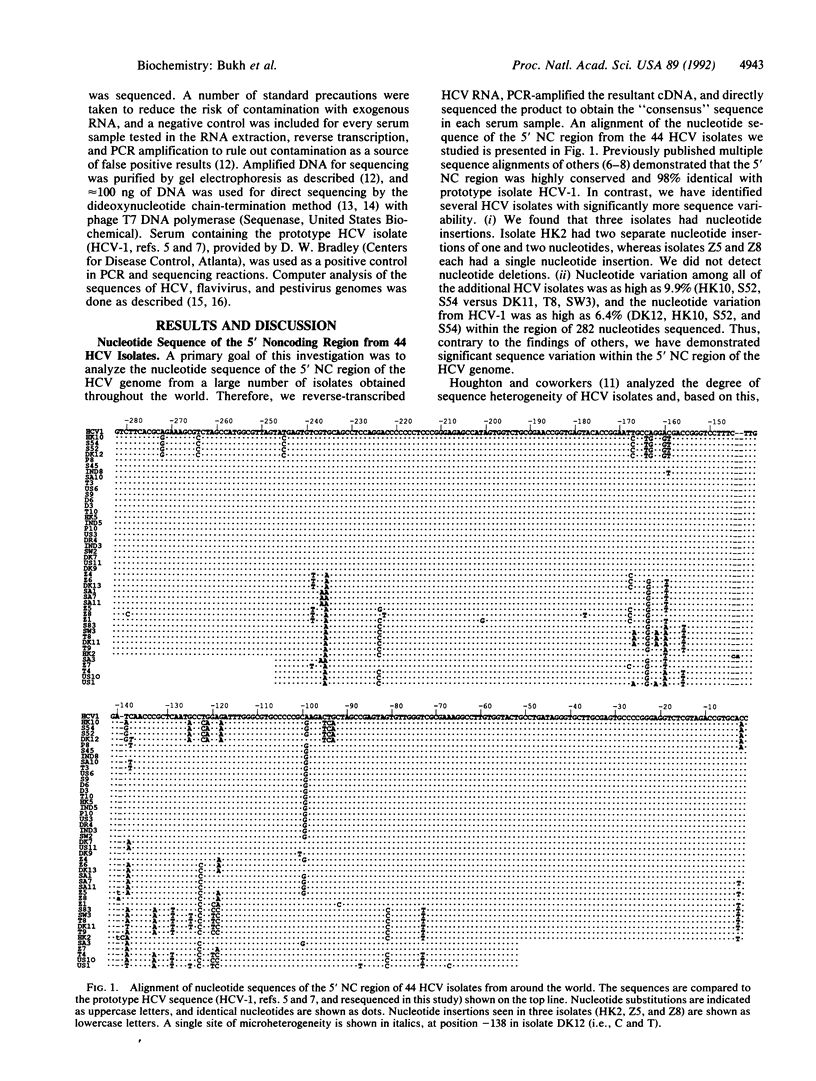
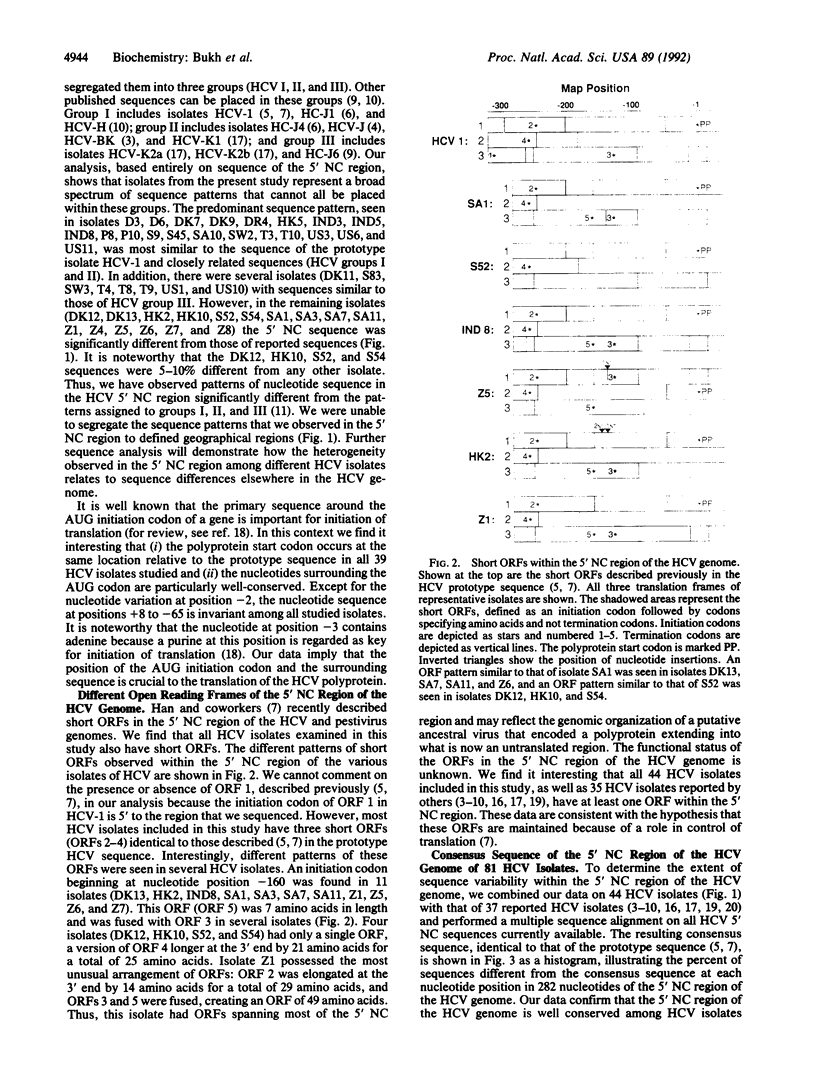
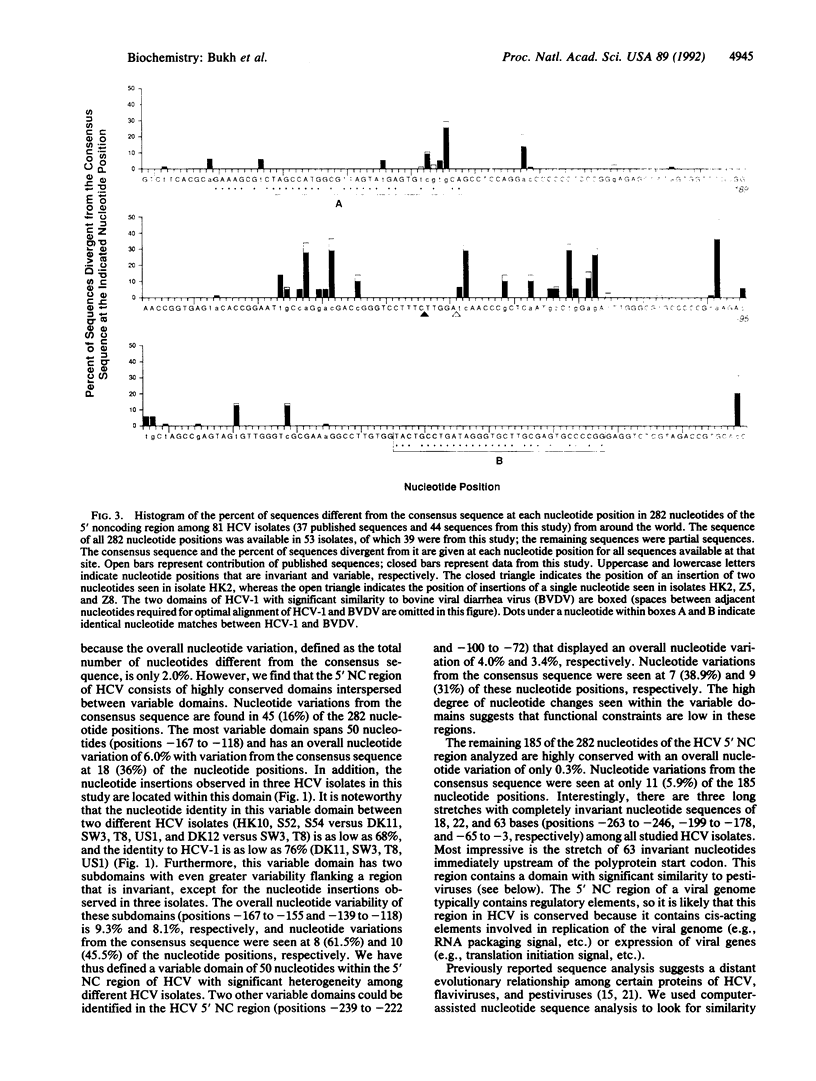
We have determined the nucleotide sequence of the 5' noncoding (NC) region of the hepatitis C virus (HCV) genome in 44 isolates from around the world. We have identified several HCV isolates with significantly greater sequence heterogeneity than reported previously within the 5' NC region. The most distantly related isolates were only 90.1% identical. Nucleotide insertions were seen in three isolates. Analysis of the nucleotide sequence from 44 HCV isolates in this study combined with that of 37 isolates reported in the literature reveals that the 5' NC region of HCV consists of highly conserved domains interspersed with variable domains. The consensus sequence was identical to the prototype HCV sequence. Nucleotide variations were found in 45 (16%) of the 282 nucleotide positions analyzed and were primarily located in three domains of significant heterogeneity (positions -239 to -222, -167 to -118, and -100 to -72). Conversely, there were three highly conserved domains consisting of 18, 22, and 63 completely invariant nucleotides (positions -263 to -246, -199 to -178, and -65 to -3, respectively). Two nucleotide domains within the 5' NC region, conserved among all HCV isolates studied to date, shared statistically significant similarity with pestivirus 5' NC sequences, providing further evidence for a close evolutionary relationship between these two groups of viruses. Additional analysis revealed the presence of short open reading frames in all HCV isolates. Our sequence analysis of the 5' NC region of the HCV genome provides additional information about conserved elements within this region and suggests a possible functional role for the region in viral replication or gene expression. These data also have implications for selection of optimal primer sequences for the detection of HCV RNA by the PCR assay.
Full text
PDF

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bachmann B., Lüke W., Hunsmann G. Improvement of PCR amplified DNA sequencing with the aid of detergents. Nucleic Acids Res. 1990 Mar 11;18(5):1309–1309. doi: 10.1093/nar/18.5.1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bukh J., Purcell R. H., Miller R. H. Importance of primer selection for the detection of hepatitis C virus RNA with the polymerase chain reaction assay. Proc Natl Acad Sci U S A. 1992 Jan 1;89(1):187–191. doi: 10.1073/pnas.89.1.187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cha T. A., Kolberg J., Irvine B., Stempien M., Beall E., Yano M., Choo Q. L., Houghton M., Kuo G., Han J. H. Use of a signature nucleotide sequence of hepatitis C virus for detection of viral RNA in human serum and plasma. J Clin Microbiol. 1991 Nov;29(11):2528–2534. doi: 10.1128/jcm.29.11.2528-2534.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choo Q. L., Kuo G., Weiner A. J., Overby L. R., Bradley D. W., Houghton M. Isolation of a cDNA clone derived from a blood-borne non-A, non-B viral hepatitis genome. Science. 1989 Apr 21;244(4902):359–362. doi: 10.1126/science.2523562. [DOI] [PubMed] [Google Scholar]
- Choo Q. L., Richman K. H., Han J. H., Berger K., Lee C., Dong C., Gallegos C., Coit D., Medina-Selby R., Barr P. J. Genetic organization and diversity of the hepatitis C virus. Proc Natl Acad Sci U S A. 1991 Mar 15;88(6):2451–2455. doi: 10.1073/pnas.88.6.2451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colett M. S., Larson R., Gold C., Strick D., Anderson D. K., Purchio A. F. Molecular cloning and nucleotide sequence of the pestivirus bovine viral diarrhea virus. Virology. 1988 Jul;165(1):191–199. doi: 10.1016/0042-6822(88)90672-1. [DOI] [PubMed] [Google Scholar]
- Fuchs K., Motz M., Schreier E., Zachoval R., Deinhardt F., Roggendorf M. Characterization of nucleotide sequences from European hepatitis C virus isolates. Gene. 1991 Jul 22;103(2):163–169. doi: 10.1016/0378-1119(91)90269-h. [DOI] [PubMed] [Google Scholar]
- Han J. H., Shyamala V., Richman K. H., Brauer M. J., Irvine B., Urdea M. S., Tekamp-Olson P., Kuo G., Choo Q. L., Houghton M. Characterization of the terminal regions of hepatitis C viral RNA: identification of conserved sequences in the 5' untranslated region and poly(A) tails at the 3' end. Proc Natl Acad Sci U S A. 1991 Mar 1;88(5):1711–1715. doi: 10.1073/pnas.88.5.1711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houghton M., Weiner A., Han J., Kuo G., Choo Q. L. Molecular biology of the hepatitis C viruses: implications for diagnosis, development and control of viral disease. Hepatology. 1991 Aug;14(2):381–388. [PubMed] [Google Scholar]
- Inchauspe G., Zebedee S., Lee D. H., Sugitani M., Nasoff M., Prince A. M. Genomic structure of the human prototype strain H of hepatitis C virus: comparison with American and Japanese isolates. Proc Natl Acad Sci U S A. 1991 Nov 15;88(22):10292–10296. doi: 10.1073/pnas.88.22.10292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato N., Hijikata M., Ootsuyama Y., Nakagawa M., Ohkoshi S., Sugimura T., Shimotohno K. Molecular cloning of the human hepatitis C virus genome from Japanese patients with non-A, non-B hepatitis. Proc Natl Acad Sci U S A. 1990 Dec;87(24):9524–9528. doi: 10.1073/pnas.87.24.9524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak M. An analysis of vertebrate mRNA sequences: intimations of translational control. J Cell Biol. 1991 Nov;115(4):887–903. doi: 10.1083/jcb.115.4.887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo G., Choo Q. L., Alter H. J., Gitnick G. L., Redeker A. G., Purcell R. H., Miyamura T., Dienstag J. L., Alter M. J., Stevens C. E. An assay for circulating antibodies to a major etiologic virus of human non-A, non-B hepatitis. Science. 1989 Apr 21;244(4902):362–364. doi: 10.1126/science.2496467. [DOI] [PubMed] [Google Scholar]
- Meyers G., Rümenapf T., Thiel H. J. Molecular cloning and nucleotide sequence of the genome of hog cholera virus. Virology. 1989 Aug;171(2):555–567. doi: 10.1016/0042-6822(89)90625-9. [DOI] [PubMed] [Google Scholar]
- Miller R. H., Purcell R. H. Hepatitis C virus shares amino acid sequence similarity with pestiviruses and flaviviruses as well as members of two plant virus supergroups. Proc Natl Acad Sci U S A. 1990 Mar;87(6):2057–2061. doi: 10.1073/pnas.87.6.2057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moormann R. J., Warmerdam P. A., van der Meer B., Schaaper W. M., Wensvoort G., Hulst M. M. Molecular cloning and nucleotide sequence of hog cholera virus strain Brescia and mapping of the genomic region encoding envelope protein E1. Virology. 1990 Jul;177(1):184–198. doi: 10.1016/0042-6822(90)90472-4. [DOI] [PubMed] [Google Scholar]
- Nakao T., Enomoto N., Takada N., Takada A., Date T. Typing of hepatitis C virus genomes by restriction fragment length polymorphism. J Gen Virol. 1991 Sep;72(Pt 9):2105–2112. doi: 10.1099/0022-1317-72-9-2105. [DOI] [PubMed] [Google Scholar]
- Ogata N., Alter H. J., Miller R. H., Purcell R. H. Nucleotide sequence and mutation rate of the H strain of hepatitis C virus. Proc Natl Acad Sci U S A. 1991 Apr 15;88(8):3392–3396. doi: 10.1073/pnas.88.8.3392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamoto H., Okada S., Sugiyama Y., Kurai K., Iizuka H., Machida A., Miyakawa Y., Mayumi M. Nucleotide sequence of the genomic RNA of hepatitis C virus isolated from a human carrier: comparison with reported isolates for conserved and divergent regions. J Gen Virol. 1991 Nov;72(Pt 11):2697–2704. doi: 10.1099/0022-1317-72-11-2697. [DOI] [PubMed] [Google Scholar]
- Okamoto H., Okada S., Sugiyama Y., Yotsumoto S., Tanaka T., Yoshizawa H., Tsuda F., Miyakawa Y., Mayumi M. The 5'-terminal sequence of the hepatitis C virus genome. Jpn J Exp Med. 1990 Jun;60(3):167–177. [PubMed] [Google Scholar]
- Takamizawa A., Mori C., Fuke I., Manabe S., Murakami S., Fujita J., Onishi E., Andoh T., Yoshida I., Okayama H. Structure and organization of the hepatitis C virus genome isolated from human carriers. J Virol. 1991 Mar;65(3):1105–1113. doi: 10.1128/jvi.65.3.1105-1113.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeuchi K., Kubo Y., Boonmar S., Watanabe Y., Katayama T., Choo Q. L., Kuo G., Houghton M., Saito I., Miyamura T. The putative nucleocapsid and envelope protein genes of hepatitis C virus determined by comparison of the nucleotide sequences of two isolates derived from an experimentally infected chimpanzee and healthy human carriers. J Gen Virol. 1990 Dec;71(Pt 12):3027–3033. doi: 10.1099/0022-1317-71-12-3027. [DOI] [PubMed] [Google Scholar]
- Winship P. R. An improved method for directly sequencing PCR amplified material using dimethyl sulphoxide. Nucleic Acids Res. 1989 Feb 11;17(3):1266–1266. doi: 10.1093/nar/17.3.1266. [DOI] [PMC free article] [PubMed] [Google Scholar]





