Abstract
The study of the composition of the intestinal flora is important to the health of the host, playing a key role in maintaining intestinal homeostasis and the evolution of the immune system. For these studies, various universal primers of the 16S rDNA gene are used in microbial taxonomy. Here, we report an evaluation of 5 universal primers to explore the presence of microbial DNA in colon biopsies preserved in RNAlater solution. The DNA extracted was used for the amplification of PCR products containing the variable (V) regions of the microbial 16S rDNA gene. The PCR products were studied by restriction fragment length polymorphism (RFLP) analysis and DNA sequence, whose percent of homology with microbial sequences reported in GenBank was verified using bioinformatics tools. The presence of microbes in the colon of rats was quantified by the quantitative PCR (qPCR) technique. We obtained microbial DNA from rat, useful for PCR analysis with the universal primers for the bacteria 16S rDNA. The sequences of PCR products obtained from a colon biopsy of the animal showed homology with the classes bacilli (Lactobacillus spp) and proteobacteria, normally represented in the colon of rats. The proposed methodology allowed the attainment of DNA of bacteria with the quality and integrity for use in qPCR, sequencing, and PCR-RFLP analysis. The selected universal primers provided knowledge of the abundance of microorganisms and the formation of a preliminary test of bacterial diversity in rat colon biopsies.
Keywords: biopsy, bacterial, RFLP, rDNA gene
INTRODUCTION
The human colon is inhabited by a large number of bacteria, which have a symbiotic relationship with the host. The composition of intestinal flora is important for the health status of the host, as gut microbiota plays important roles in the maintenance of gut homeostasis by direct bactericidal effects and its participation in the evolution of innate and adaptive immune systems.1 Approximately 1010–1012 bacteria colony-forming units (CFUs)/ml have been described in the colon.2 Bacteria are involved in the processing of complex molecules to simple molecules; assist in host functions, such as food processing; regulate intestinal function, immune system development, and synthesis of essential vitamins; and provide protection against pathogens. These bacteria also are involved in the reduction of allergies and prevent the formation of tumors. Some of these bacteria produce lactic acid, thus promoting an acidic environment that prevents the growth of bacteria harmful to the health. Some species may become pathogenic when a microbial dysbiosis (microbial imbalance) occurs, producing a disorganization of interactions among different groups of taxa.3, 4
For the molecular analysis of microbial species that colonize colonic mucosa, several universal primers for the hypervariable region of 16S rDNA have been reported in the literature. Wang et al.5 performed the partial sequence of amplicons of 2 of the 9 variable regions of the 16S rDNA. The amplicons were 350 base pairs (bp) long, and these were produced by using eubacterial primers of V1–V2 hypervariable regions. The most frequently represented clones in humans using this pair of primers were from the Clostridium (Firmicutes) and Bacteroidetes. Furthermore, to explore the V4 hypervariable region of the 16S rDNA gene, Santiago et al.6 amplified in human stools Gram-positive bacteria, such as blautia (phylum Firmicutes) and Bifidobacterium (Actinobacteria phylum), commonly detected in theses samples. Other universal primers were used by Anderson and Lebepe-Mazur7 with the aim of exploring the pig gastrointestinal microflora. The primers S-D-Bact [Small subunit rDNA (S), domain level (D), and domain Bacteria (Bact)], whose nomenclature was suggested by Alm et al.,8 explore the V7–V8 regions of 16S rDNA of Bacteria fragilis, Bacteroides uniformis, Eubacterium eligens rectale, Lactobacillus reuteri, and Selenomonas ruminantium. On the other hand, the universal primers lac1 and SG-Lab-0677-aA-17 have been designed to quantify the lactobacilli. The presence of probiotics or prebiotics in the gut microflora, in particular, the enrichment of lactobacilli and bifidobacteria, is considered a relevant parameter to evaluate the health of the intestinal ecosystem and resistance against pathogens, as well as to promote a beneficial immune response.5–11
The aim of our study was to develop a simple and inexpensive method to compare the number and diversity of bacteria present in the colon of rats. The methodology presented here can be used for the following: to obtain bacterial DNA present in the colon biopsies stored in RNAlater solution (Ambion; Thermo Fisher Scientific, Waltham, MA, USA) at −20°C, to quantify by real-time PCR the approximate number of microorganisms, and to study the colon bacterial diversity.
MATERIALS AND METHODS
Biopsies' Collection and Storage
To know the intestinal microbiota, 17 transverse colon biopsies (0.10 ± 0.03 g) were obtained from male rats (n = 17) weighing 300–400 g. Biopsies were conserved in 0.7 ml RNAlater solution at −20°C until use.
Bacteria DNA Isolation from Colon Biopsies Conserved in RNAlater
To isolate the bacteria preserved in colonic biopsies stored in RNAlater solution at −20°C, the protocol Wizard Genomic DNA Purification Kit (Promega, Madison, WI, USA), described for the isolation of DNA from Gram-positive and -negative bacteria, was used following the manufacturer's instructions. In brief, colonic biopsies, weighing 0.10 ± 0.03 g, were thawed at room temperature and transferred to a clean tube containing 750 µl 50 mM EDTA/200 mg tissue. Then, it was stirred by vortex per 5 min to help loosen from the tissue the colonized bacteria, 60 µl lysozyme (10 mg/ml) was added, and the sample was incubated at 37°C during 60 min. After this time, we follow the protocol recommended by the supplier. The DNA concentration was analyzed by NanoDrop (Thermo Fisher Scientific, Waltham, MA, USA) and then conserved at −20°C until use.
PCR Primers for the 16S rDNA Gene
Five universal primer pairs for the phylogenetic marker 16S rDNA gene of bacteria were selected from literature, following the criteria that they matched for 5 of the 9 variable regions of this gene. These primers were synthesized by the Oligonucleotides Synthesis Department of the Center for Genetic Engineering and Biotechnology (CIGB) in Havana, Cuba. The sequences of these primers cover the DNA amplification from the variables regions V1, V2, V4, V7, and V8 (Table 1).
TABLE 1.
Universal PCR Primers for Bacteria 16S rDNA Gene
| Primers' name, variable (V) genome region | Primers' sequences of conserved regions, 5′–3′ | Escherichia coli nucleotide position | mer | Ta (°C) | Product (pb) |
|---|---|---|---|---|---|
| 27f and 342r | GAAGAGTTTGATCATGGCTCAG | 8–27 | 22 | 65 | 350 |
| General V1–V25 | CTGCTGCCTCCCGTAG | 342–358 | 16 | ||
| V4F-517 and V4R-805 | GCCAGCAGCCGCGGTAA | 517–532 | 17 | 60 | 289 |
| General V46 | GACTACCAGGGTATCTAAT | 787–805 | 19 | ||
| Total bacterial 16S | GTGGTGCACGGCTGTCGTCA | 1047–1067 | 20 | 65 | 142 |
| V77,12,13 | ACGTCATCCACACCTTCCTC | 1175–1189 | 20 | ||
| Consensus sequence of 7 strains; S-D-Bact 1030 and 1519; V7–V87 | TGCATGCCTGTCGTCAGCTC | 1051–1071 | 20 | 65 | 338 |
| GACCCGAGAACGTATTCACC | 1370–1389 | 20 | |||
| Lactobacillus spp. | AGCAGTAGGGAATCTTCCA | 7–27 | 19 | 65 | 340 |
| lac1 and SG-Lab-0677-aA-179–11 | CACCGCTACACATGGAG | 331–347 | 17 |
Ta, Annealing temperature. Complete genome of the E. coli DH1Ec169 strain was obtained from the Sequence ID: CP012127.1; Lactobacillus plantarum strain S-C2L 16S ribosomal RNA gene was obtained from the Accession Sequence KP056259. Database, National Center for Biotechnology Information, U.S. National Library of Medicine (Bethesda MD, USA).
The best annealing temperature was determined in E. coli DH5a strain and in colonic microbial DNA by performing gradient-endpoint PCR in a Mastercycler (Eppendorf, Nijmegen, The Netherlands) using the GoTaq DNA Polymerase Kit (Promega), according to manufacturer’s instructions. The PCR products were analyzed in 1% Tris-borate-EDTA (TBE) agarose gel via electrophoresis with 0.5 µg/ml ethidium bromide at 16 V/cm during 30 min.
Total DNA obtained from the E. coli DH5a strain, cultured overnight at 37°C in Luria Broth (LB) media, was used as control DNA. Biopsies from colon transverse, conserved during 11 mo at −20°C, were used for the standardization of the bacteria DNA isolation protocol and PCR conditions analysis.
PCR products were purified by the PCR Clean-Up Kit from Promega and were sent to a sequencing service (Macrogen, Seoul, South Korea). To obtain the most homologous alignment, the sequence of nucleotide obtained for each primer was analyzed using the Basic Local Alignment Search Tool (BLAST; http://blast.ncbi.nlm.nih.gov/Blast.cgi).
Standard Curve Method of Absolute Quantification
DNA from the E. coli DH5a strain was used as standard for determining the number of bacteria by real-time PCR. For this, 200 µl E. coli DH5a conserved in glycerol was grown in 5 ml LB overnight (∼16 h) at 37°C, with agitation of 200 rpm/s. The next day, the original culture was used to make serial dilution 1:10 (10−1–10−5). Then, 1 ml each dilution was grown in LB agar plates during ∼16 h at 37°C. Furthermore, to relate the amount of bacteria in CFUs with the amount of DNA in nanograms and the cycle threshold (Ct) for the primers described in Table 1, 1 ml each previous serial dilution was used for bacteria DNA purification and 16S rDNA gene quantification.
Design of Real-Time PCR for Absolute Quantification
All qPCR assays were performed on a CapitalBio RT-Cycler 636 (Capitalbio, Beijing, China). Amplification and detection were carried out in a final concentration of 75 nM each primer, using different dilutions of DNA sample, from 100 to 10−5, in a final volume of 20 µl/reaction. The variable regions of 16S rDNA gene were amplified by qPCR technique using ABsolute qPCR Mix, SYBR Green (Thermo Fisher Scientific), with an initial hold of 95°C during 900 s to activate the DNA polymerase, followed by 40 cycles of 95°C for 15 s, 60°C for 30 s, and 72°C for 30 s, and a melting step with 95°C during 30 s and 40°C during 30 s.
To obtain an standard curve (SC) of the E.coli DH5a strain, different concentrations of DNA were analyzed by qPCR, starting from 100 (128.8 ng), 10−1 (12.88 ng), 10−2 (1.28 ng), 10−3 (0.12 ng), 10−4 (0.012 ng), and 10−5 (0.0012 ng). The amplification efficiency (E) was measured calculating the slope of a DH5a SC for 4 primer pairs. The E calculation was obtained using the web application (see http://www.genomics.agilent.com/biocalculators/calcSlopeEfficiency), which is based in the formula calculation
| (1) |
To known the number of bacteria present in the rat colon, DNA microbiota was analyzed by qPCR with the pairs of primers mentioned in the Table 1.
Construction of Power Curve
The SC data of the E.coli DH5a were used to construct a power curve to have a tool for predicting the CFU from new values of Ct. The prediction curve allows the knowledge of the value of a dependent variable based on the calculation of intermediate independent variables that have not been measured. The graphic used to represent the power curve is a scatter plot. With the use of this curve, we were able to determine the intermediate Ct values corresponding to the microbial DNA obtained from the colon of rats.
DNA Library Construction and RFLP Analysis
Colonic bacterial DNA was amplified by PCR with the primers for 16S rDNA named General V1–V2. A total of 75 ng each purified amplicons was cloned in a pGEM–T Easy Vector System (Promega) and transformed into E.coli DH10B chemo-competent cells, according to the standard procedure for KCM cells, using KCl (0.5 M), CaCl2 (0.15 M), and MgCl2 (0.25 M).14 Recombinant bacteria were detected using the screening of blue-white colonies.15
Fifteen plasmids were purified using the QIAprep Spin Miniprep Kit (Qiagen, Germantown, MD, USA) and then sent to a sequencing service Macrogen. The sequence of nucleotide obtained for the plasmid was analyzed in BLAST. Furthermore, with the aim to perform a PCR-RFLP analysis, the recombinant plasmids (2 ng) were used to amplify the insert sequence using the GoTaq DNA Polymerase Kit, following the supplier's instructions.
The PCR products were analyzed in 1% TBE agarose gel via electrophoresis with 0.5 µg/ml ethidium bromide at 16 V/cm during 30 min. PCR was performed in a Mastercycler.
Finally, to perform the PCR-RFLP, 3 μl PCR products was digested with HaeIII restriction enzyme (Promega) in a total volume of 30 μl at 37°C during 2 h. The restriction enzyme digestion was mixed with 3 μl orange loading dye 6×, and the length of the restriction fragments was determined using 3.5% agarose gel electrophoresis in 0.5× TBE buffer, run at 16 V/cm during 30 min.
RESULTS
DNA Isolation from Colon Biopsies Conserved in RNAlater
Seventeen colon biopsies were processed for the bacteria DNA purification. The DNA obtained had a concentration average of 16.01 µg ± 13.84 in a total volume of 50 μl, with an average ratio of 260/280 of 1.64 ± 0.12. All of these results suggest a good DNA quality in purity and concentration for PCR analysis.
Standardization of PCR Primers
The standardization of PCR primer using E.coli DH5a DNA consisted of the determination of the best annealing temperature to obtain a PCR product with high specificity. Optimal annealing temperatures ranged between 50 and 65°C, with 60°C selected as the optimal value for all primer combinations, except for Lactobacillus primers, which as expected, do not produce PCR products in this bacterium.
Sequence Analysis Results
The PCR products obtained from E.coli DH5a were sequenced, and the results were analyzed to know the most significant alignments using the default settings for the standard nucleotide BLAST.
In this case, all DH5a sequences were highly homologous (97–100%) to E. coli, as expected.
Furthermore, to check the functioning of the primers in the colonic microbiota, a sample of microbial DNA was used to be sequenced in both directions (forward and reverse) using the 5 universal primer pairs under study. The results showed that the isolated DNA had the quality to be sequenced, and their nucleotide composition belongs to microorganisms that normally inhabit the colon, such as bacteria from the phylum Firmicutes [Gram-positive (G+)], class bacilli, genus Lactobacillus, and bacteria from phylum proteobacteria [Gram-negative (G−)], class Alpha proteobacteria (Table 2).
TABLE 2.
Summary of the Sequences with Highest Homology for Each PCR Product Obtained from the Amplification of a DNA Sample of Colonic Bacteria, with 5 Pairs of Primers
| PCR products obtained from colonic microbiota DNA | Primer direction | Phylum/class | Genus | Identity % | Accession | E value |
|---|---|---|---|---|---|---|
| Regions V1–V2 | F | Bacilli | Lactobacillus | 75 | HQ717267.1 | 8E-08 |
| R | Phylum: Firmicutes | 96 | LN800877.1 | 6E-35 | ||
| Region V4 | F | Bacilli | Lactobacillus | 90 | KC561107.1 | 2E-78 |
| R | Lactobacillus | 88 | HQ851026.1 | 1E-70 | ||
| Region V7 | F | Phylum proteo bacteria | Desulfobacca acetoxidans | 94 | CP002629.1 | 5E-12 |
| R | Unculture A. proteobacteria | 85 | EF219728.1 | 3E-04 | ||
| Regions V7–V8 | F | Bacilli | Lactobacillus | 85 | GU454851.1 | 1E-66 |
| R | 84 | JX079468.1 | 1E-52 | |||
| Lactobacillus spp | F | Not sequenced | ||||
| R | Bacilli | Lactobacillus | 93 | KM250395.1 | 4E-106 | |
F, forward; R, reverse. Identity %, The extent to which 2 sequences have the same residues at the same positions in an alignment, expressed as a percentage.
By taking into account all possible DNA homologies obtained from the BLAST analysis of a sample of colonic bacteria DNA, amplified for the 5 primer pairs, we conclude that 26% of the total homologies correspond to phylum Firmicutes, 0.67% to phylum proteobacteria, and 73% to uncultured bacterium. In Fig. 1, there appears a more detailed description of the number of sequences with homology for each primer pairs. As is observed in our conditions, all primers, except the total bacterial 16S V7, were able to identify bacteria from the phylum Firmicutes.
Figure 1.
Description for each PCR product of the total number of sequences identified with homology to the phylum Firmicutes, proteobacteria, and uncultured bacterium. This analysis was performed using BLAST.
SC Method of Absolute Quantification
To determine the value of an unknown quantity of the different bacteria colonizing the colon, firstly, we determine the relation between DNA concentration and CFU/ml through the construction of an SC with E. coli DH5a DNA. Then, we evaluated the relation between CFU/ml and Ct values. In Table 3, the results of the SC are shown for DH5a, which was made with 4 different pairs of primers. In the equations below, the SC functions (y) describe the relation between Ct and the log of the starting template concentration
| (2) |
| (3) |
| (4) |
| (5) |
The average of efficiency calculation = 75% ± 8.
TABLE 3.
Data Obtained from the SC for E. coli DH5a
| E. coli DH5a DNA concentration (ng) | CFU/ml | Ct general V1–V2 | Ct general V4 | Ct total bacteria 16S V7 | Ct consensus sequence of 7 strains V7–V8 |
|---|---|---|---|---|---|
| 128.8 | 6.3 × 108 | 10.96 ± 0.48 | 13.9 ± 0.72 | 12.23 ± 0.38 | 13.05 ± 0.34 |
| 12.88 | 1.1 × 107 | 14.19 ± 0.36 | 17.54 ± 0.12 | 15.65 ± 0.05 | 16.21 ± 0.17 |
| 1.28 | 1.5 × 106 | 18.35 ± 0.09 | 21.81 ± 0.57 | 19.27 ± 0.02 | 19.9 ± 0.07 |
| 0.12 | 3.5 × 105 | 23.41 ± 0.11 | 28.155 ± 1.71 | 23.00 ± 0.07 | 24.04 ± 0.33 |
| 0.012 | 5.3 × 104 | 27.60 ± 0.07 | 31.71 ± 0.16 | 26.80 ± 0.06 | 27.68 ± 0.00 |
| 0.0012 | 4 × 103 | 32.15 ± 0.47 | >40 | 30.81 ± 0.035 | 32.27 ± 0.04 |
| qPCR negative control | – | >40 | >40 | 34.15 ± 0.60 | >40 |
The table shows the Ct values for PCR products of 4 pairs of primers and their relation with the bacteria DNA concentration and the number of CFU/ml. Ct is defined as the number of cycles required for the fluorescent signal to cross the threshold.
The construction of the power regression curve using the Ct and CFU values from the SC data of E.coli DH5a allowed the attainment of a model for nonlinear curve for each pairs of primers. The relationship between Ct and CFU was represented by creating a scatter plot in Excel and then selecting the Power Trendline option. The model described below was used to predict the values of the y-axis (CFU), which correspond to known values of the x-axis (Ct values). The scatter plots for the primers general V1–V2 is shown in Fig. 2. The same analysis was performed for the rest of the primers (results not shown). As we do not have a well-characterized strain of Lactobacillus, we used the power regression curve obtained for V1–V2 primers to know approximately the CFU values obtained with the primers “Lactobacillus spp”.
Figure 2.
Power regression curve obtained from the SC data of the E.coli DH5a strain for the pairs of primers named general V1–V2; power function, y = 1E + 19 x −10.06, R2 = 0.9758.
Once the power regression curves were constructed, we proceeded to perform the analysis by qPCR of bacterial DNA in rat colon. Only 10−1 (12.88 ng) and 10−2 (1.28 ng) dilutions of bacterial DNA showed acceptable results by qPCR (Ct ≤ 30), obtaining an efficiency of PCR efficiency of 1.8 ± 0.11. Ct values obtained from the 5 primer pairs were converted to CFU, and the results are shown in Fig. 3. Below the results of descriptive statistics are shown by calculating the mean value and the sd for each pair of primer used. This analysis allows the examination of the CFU dispersion in the sample.
Figure 3.
Total bacterial load (CFU) in 17 colon samples. The figure shows the mean values ± sd.
According to these results, the pair of primers named general V1–V2 could be the most useful for detecting bacterial variability in the colon, as it is able to detect the greater amount of microorganisms in the sample. On the other hand, the “consensus sequences 7 strains primers” detects the least amount of microorganisms.
DNA Library Construction and RFLP Analysis
With the aim of obtaining an approach to study the diversity of species in the colonic biopsies, a library of 16S rDNA genes of bacteria was constructed from colon biopsies amplified with the primer pair general V1–V2. A total of 1723 colonies was obtained. The average amount of plasmids obtained from 15 white colonies was 6.2 ± 2.9 μg. PCR analysis of purified plasmids determined that all selected colonies/group were recombinant (data not shown).
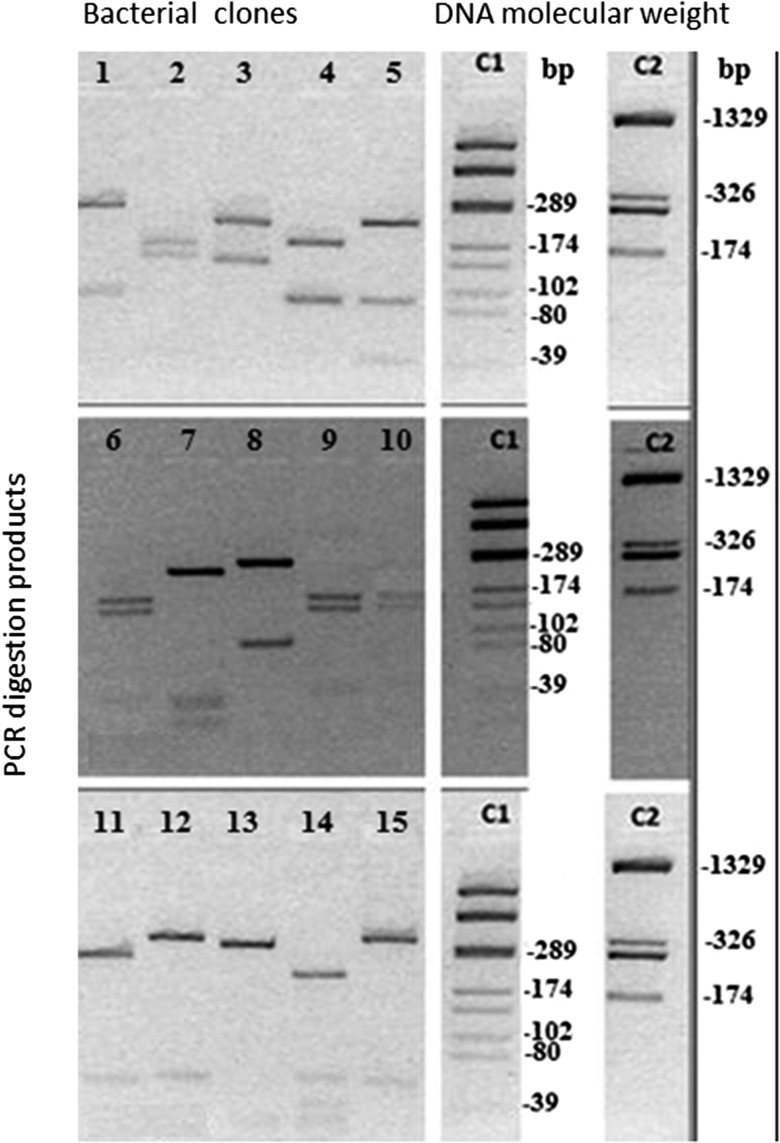
The PCR-RFLP analysis showed the structural diversity of the cloned DNA, which may reflect the bacterial diversity in rat colon samples (Fig. 4). This diversity was verified by the sequencing of PCR products obtained of the 15 recombinant bacterial clones. The sequencing showed that the most represented bacteria in rat colon were from phylum Firmicutes, followed by phyla Bacteroidetes and Fusobacteria. The analysis of the library clones by DNA sequencing allowed the knowledge of the diversity of bacteria within each phylum. For example, within the phylum Firmicutes bacteria species were found, such as Prevotella marshii, Prevotella bivia, Paenibacillus polymyxa, Turicibacter sanguinis, Romboutsia ilealis, Anaerostipes butyraticus, and Lactobacillus hominis. Furthermore, Bacteroidetes and Fusobacterium varium were found.
Figure 4.
Agarose gel electrophoresis (3.5%) in 0.5× TBE buffer, run at 16 V/cm during 30 min. MW marker. C1 and C2 positions are digestion of pGEM–T vector with HaeIII and BslI, respectively. Numbers from 1 to 5 represent PCR products of 15 recombinant plasmids/group, digested with HaeIII.
The percentage of each phylum of bacteria found in the analyzed clones is shown in Fig. 5. The phylum most represented in the transverse colon of rats was Firmicutes, followed by Bacteroidetes and Fusobacteria.
Figure 5.
Results are given in percent of microorganisms found in the sequencing of PCR products (general V1–V2) from 15 recombinant colonies.
DISCUSSION
Rat colon biopsies provides an opportunity to identify microbes involved in diseases and host-microbiota responses to therapies, which can be difficult to discern in humans, given their genetic diversity and variability in environmental and treatment exposures. Colonic biopsies taken from rats were preserved in RNAlater solution at −20°C for the standardization of an inexpensive methodology for assessing the amount of microorganisms inhabiting the colon. Our work was focused on techniques, such as DNA isolation from bacteria, quantitative and semiquantitative PCR using regions of the 16S rDNA gene, plasmid library construction, analysis of microbial diversity through PCR-RFLP technique, and sequencing by the Sanger method. The processing of colonic biopsies stored at −20°C yielded sufficient bacteria DNA, with the quality and integrity required for PCR analysis, showing that although RNAlater solution is a bacteriostatic and a suitable storage method for conserving colonic bacteria and obtaining nondegraded DNA.
Firmicutes and proteobacteria are normally represented in the colon of rats,16, 17 and the primers' difference in the detection of these bacteria may be a result of the different specificity of the primers and the different size of the amplicons generated, allowing a more or less specific homology search in GenBank. With the use of the primer pair of Lactobacillus, the results of the sequence were as expected, determining high homology with this microorganism.
PCR-RFLP analysis is a molecular technique that provides rapid comparison of community structures and diversity of complex bacterial flora in gut microflora. The 16S rDNA cloned library analysis is a powerful tool for bacteria species determination. As is known, there is bacterial diversity in the colonic mucosa; some of this diversity can be observed in our results. As a few recombinant bacterial clones were studied through PCR-RFLP analysis, it was not possible to determine whole bacterial diversity present in the samples. Because of this, bacteria that normally inhabit the colon, such as bacteria from the phyla proteobacteria and actinomycetes and Clostridium difficile from phylum Firmicutes, were not detected during this test. High-density technologies, such as deep sequencing-based metagenomic evaluation, are today the best option to know more precisely the microbial composition of the samples.
The microorganisms detected by the 5 primer sets used resemble those reported by other researchers,5,7,9,11 where the colonic microflora studied in humans is dominated primarily by microorganisms of the phyla Firmicutes and Bacteroidetes.
In this work, we have been adjusting the technical basis for the development of future research that could help us to know which bacteria are responsible for the abundance of microorganisms during the different stages of inflammatory bowel diseases, what functional significance would have this abundance in a group of animals with different colonic damage, and what the relationship is among inflammation, the species of bacteria resident in the colon, and the gene-expression signature of oxidative stress. These questions and others should be addressed in the future.
Conclusions
The methodology proposed allowed us to obtain bacterial DNA from colon biopsies stored in RNAlater solution at −20°C. The obtained DNA had the required quality and integrity for their use in qPCR studies, sequencing, and PCR-RFLP analysis. Of the 5 pairs of primers studied, the general V1–V2 offers the higher information about the colonic microflora diversity.
ACKNOWLEDGMENTS
The authors thank Dr. Julio Raul Fernandez from CIGB for his helpful discussions and recommendations. The authors declare no conflicts of interest.
REFERENCES
- 1.Lee BJ, Bak YT.. Irritable bowel syndrome, gut microbiota and probiotics. J Neurogastroenterol Motil 2011;17: 252–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Holzapfel WH, Haberer P, Snel J, Schillinger U, Huis in’t Veld JH. Overview of gut flora and probiotics. Int J Food Microbiol 1998;41:85–101. [DOI] [PubMed] [Google Scholar]
- 3.Canny GO, McCormick BA. Bacteria in the intestine, helpful residents or enemies from within? Infect Immun 2008;76:3360–3373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cummings JH, Macfarlane GT, Macfarlane S. Intestinal bacteria and ulcerative colitis. Curr Issues Intest Microbiol 2003;4:9–20. [PubMed] [Google Scholar]
- 5.Wang X, Heazlewood SP, Krause DO, Florin THJ Molecular characterization of the microbial species that colonize human ileal and colonic mucosa by using 16S rDNA sequence analysis. J Appl Microbiol 2003;95:508–520. [DOI] [PubMed] [Google Scholar]
- 6.Santiago A, Panda S, Mengels G, et al Processing faecal samples: a step forward for standards in microbial community analysis. BMC Microbiol 2014;14:112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Anderson KL, Lebepe-Mazur S. Comparison of rapid methods for the extraction of bacterial DNA from colonic and caecal lumen contents of the pig. J Appl Microbiol 2003;94:988–993. [DOI] [PubMed] [Google Scholar]
- 8.Alm EW, Oerther DB, Larsen N, Stahl DA, Raskin L. The oligonucleotide probe database. Appl Environ Microbiol 1996;62:3557–3559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Heilig HG, Zoetendal EG, Vaughan EE, Marteau P, Akkermans AD, de Vos WM. Molecular diversity of Lactobacillus spp. and other lactic acid bacteria in the human intestine as determined by specific amplification of 16S ribosomal DNA. Appl Environ Microbiol 2002;68:114–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Walter J, Hertel C, Tannock GW, Lis CM, Munro K, Hammes WP. Detection of Lactobacillus, Pediococcus, Leuconostoc, and Weissella species in human feces by using group-specific PCR primers and denaturing gradient gel electrophoresis. Appl Environ Microbiol 2001;67:2578–2585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Verdenelli MC, Silvi S, Cecchini C, Orpianesi C. Influence of a combination of two potential probiotic strains, Lactobacillus rhamnosus IMC 501 and Lactobacillus paracasei IMC 502 on bowel habits of healthy adults. Lett Appl Microbiol 2011;52:596–602. [DOI] [PubMed] [Google Scholar]
- 12.Fredricks DN, Relman DA. Improved amplification of microbial DNA from blood cultures by removal of the PCR inhibitor sodium polyanetholesulfonate. J Clin Microbiol 1998;36:2810–2816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Maiwald M. Broad-range PCR for detection and identification of bacteria. In Persing DH., Tenover FC, Tang YW, Nolte FS, Hayden RT, van Belkum A (eds). Molecular Microbiology: Diagnostic Principles and Practice, 2nd ed. Washington, DC: American Society for Microbiology, 2011. [Google Scholar]
- 14.Chung CT, Miller RH. A rapid and convenient method for the preparation and storage of competent bacterial cells. Nucleic Acids Res 1988;16:3580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sambrook J, Russell D. Plasmids and their usefulness in molecular cloning. Molecular Cloning. A Laboratory Manual, 3rd ed. Cold Spring Harbor, NY: Cold Spring Laboratory, 2011;1.27. [Google Scholar]
- 16.Lagier JC, Million M, Hugon P, Armougom F, Raoult D. Human gut microbiota: repertoire and variations. Front Cell Infect Microbiol 2012;2:136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Eckburg PB, Bik EM, Bernstein CN, et al Diversity of the human intestinal microbial flora. Science 2005;308:1635–1638. [DOI] [PMC free article] [PubMed] [Google Scholar]