Abstract
Purpose
To examine the predictions of alternative models for the stochastic shut-off of activated rhodopsin (R*) and their implications for the interpretation of experimentally recorded single-photon responses (SPRs) in mammalian rods.
Theory
We analyze the transitions that an activated R* molecule undergoes as a result of successive phosphorylation steps and arrestin binding. We consider certain simplifying cases for the relative magnitudes of the reaction rate constants and derive the probability distributions for the time to arrestin binding. In addition to the conventional model in which R* catalytic activity declines in a graded manner with successive phosphorylations, we analyze two cases in which the activity is assumed to occur not via multiple small steps upon each phosphorylation but via a single large step. We refer to these latter two cases as the binary R* shut-off and three-state R* shut-off models.
Methods
We simulate R*’s stochastic reactions numerically for the three models. In the simplifying cases for the ratio of rate constants in the binary and three-state models, we show that the probability distribution of the time to arrestin binding is accurately predicted. To simulate SPRs, we then integrate the differential equations for the downstream reactions using a standard model of the rod outer segment that includes longitudinal diffusion of cGMP and Ca2+.
Results
Our simulations of SPRs in the conventional model of graded shut-off of R* conform closely to the simulations in a recent study. However, the gain factor required to account for the observed mean SPR amplitude is higher than can be accounted for from biochemical experiments. In addition, a substantial minority of the simulated SPRs exhibit features that have not been reported in published experiments. Our simulations of SPRs using the model of binary R* shut-off appear to conform closely to experimental results for wild type (WT) mouse rods, and the required gain factor conforms to biochemical expectations. However, for the arrestin knockout (Arr−/−) phenotype, the predictions deviated from experimental findings and led us to invoke a low-activity state that R* enters before arrestin binding. Our simulations of this three-state R* shut-off model are very similar to those of the binary model in the WT case but are preferred because they appear to accurately predict the mean SPRs for four mutant phenotypes, Arr+/−, Arr−/−, GRK1+/−, and GRK1−/−, in addition to the WT phenotype. When we additionally treated the formation and shut-off of activated phosphodiesterase (E*) as stochastic, the simulated SPRs appeared even more similar to real SPRs, and there was very little change in the ensemble mean and standard deviation or in the amplitude distribution.
Conclusions
We conclude that the conventional model of graded reduction in R* activity through successive phosphorylation steps appears to be inconsistent with experimental results. Instead, we find that two variants of a model in which R* activity initially remains high and then declines abruptly after several phosphorylation steps appears capable of providing a better description of experimentally measured SPRs.
Introduction
Vertebrate phototransduction represents an extremely well-studied example of sensory signaling at a molecular, biochemical, electrophysiological, and modeling level. The activation steps are particularly well understood and have been modeled quantitatively [1] in a manner that has been shown to accurately describe the onset of the electrical response of rods in many studies. The shut-off and light adaptation of the phototransduction cascade have also been modeled in several studies, but there has not yet been acceptance of a single comprehensive description.
Intense research interest has centered on the nature of the shut-off of a single activated molecule of rhodopsin (R*) and how this may account for the observed properties of the single-photon response (SPR) of rods. Ever since the discovery that the amplitude distribution of SPRs exhibits a coefficient of variation (standard deviation [SD]/mean) of cv ≈ 0.2 [2], far lower than the value of unity expected for a single stochastic event, there has been speculation about the underlying mechanisms. Models have been put forward that involve a postulated long series of unknown steps in the shut-off of R* [3,4] or alternatively a single stochastic shut-off step under feedback regulation by Ca2+ [5].
A molecular model of the SPR was formulated by Hamer et al. [6,7], who developed a stochastic “front end” description of R* shut-off that fed into a deterministic downstream description of the remainder of the cascade. In conformity with the earlier biochemical findings of Gibson et al. [8], the stochastic description postulated that successive phosphorylation steps (mediated by rhodopsin kinase, encoded by GRK1) led to successive reductions in R* catalytic activity; subsequently, arrestin binding completely terminated the activity. The downstream part of the model resembled other fully deterministic models of phototransduction, and in particular it incorporated Ca2+ feedback (via guanylate cyclase-activating proteins, GCAPs) onto guanylyl cyclase activity. The resulting simulations provided a good description of the amplitude distribution and kinetics of SPRs [6,7]. That stochastic model has been modified and extended in a variety of ways in subsequent studies [9-12], some of which have explicitly included longitudinal diffusion of the cytoplasmic messengers, cGMP and Ca2+ [9,11]. In addition, various refinements to the deterministic downstream reactions have also been proposed [13-15].
Each of these descriptions of stochastic R* activity [6,7,9-12] has modeled R* activity as declining in a graded manner with successive phosphorylation events, as suggested in a biochemical study [8]. However, there has been no direct evidence for multiple partial reductions in R* activity by successive phosphorylation steps. In this paper, we investigate alternative possibilities.
Two troublesome issues become apparent when simulations are undertaken using the model of graded shut-off in R* activity. First, the required catalytic activity of the phosphodiesterase (PDE) is approximately double the magnitude that has been determined through biochemical measurements. Second, a minority of the simulated SPRs exhibit a late plateau-like region that is not obviously consistent with the individual SPRs that one observes experimentally.
Because of these considerations, we decided to examine the consequences of dispensing with the assumption that the sequential phosphorylation steps gradually reduce R* activity. We discovered that when R* activity does not decline in this way it is possible to provide a parsimonious account of the properties of the SPR while avoiding the shortcomings alluded to above.
Theory
In this section, we examine several models of stochastic shut-off of R* activity involving sequential phosphorylation of activated R* followed by arrestin binding. In accord with previous analysis, we assume that the SPR is not significantly affected by altered Ca2+ concentration acting via recoverin [6,9,11]. More generally, we assume that the rate constants of transition are independent of downstream events in the phototransduction cascade so that, importantly, we are able to restrict consideration to cases where the molecular transition rates depend only upon the state in which the molecule currently exists.
Sequential phosphorylation of equivalent residues, followed by arrestin binding
As in previous studies, we assume that all the phosphorylation sites, whether Ser or Thr, are equivalent to each other. In the general case illustrated in Figure 1A, the R* molecule passes sequentially through a series of states in which one extra phosphate is added at each step (green arrows). Because the sites are assumed equivalent, it is only necessary to monitor the number of phosphates attached, rather than keep track of which sites they have been attached to. Arrestin can bind to any of these states (blue arrows), thereby terminating R* activity.
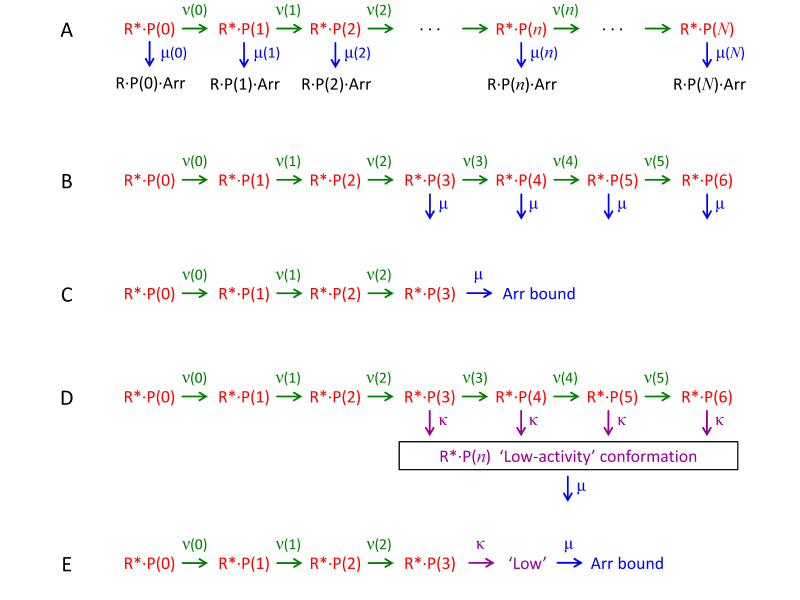
Figure 1.
Models for the progression of an R* molecule through successive states of phosphorylation and arrestin binding. Photoisomerization generates the fully active state R*·P(0), representing non-phosphorylated metarhodopsin II. The C-terminus is successively phosphorylated by rhodopsin kinase, and the number n of attached phosphate groups is indicated as R*·P(n). At each step, there is a possibility that arrestin can bind to create the form R*·P(n)·Arr, which is inactive. A: General case for phosphorylation and arrestin binding. The rate constants are unconstrained, and for state R*·P(n) the rate constants of phosphorylation and arrestin binding are denoted as ν(n) and μ(n), respectively; however, the single molecule can undergo only one of these two reactions. B: Arrestin binding is assumed not to occur until M = 3 phosphates have attached and then to occur with the same rate constant irrespective of how many phosphates have been attached. For simplicity, the arrestin-bound states are not shown, as they are inactive. C: The truncated linear sequence to which the scheme in panel B is equivalent (see Theory). D: Alternative “three-state” scheme, in which the catalytic activity of phosphorylated R* drops to a lower level after M = 3 phosphates have attached and before arrestin binding. The rate constant of transition to this low-activity state is κ, and the subsequent rate constant of arrestin binding is again μ. E: The truncated linear sequence to which the scheme in panel D is equivalent.
The active states, R*·P(n), are shown in red, and the inactivated states, R*·P(n)·Arr, are shown in black. The integer n indicates the number of phosphates attached and runs from zero up to the number of residues that can be phosphorylated, that is, n = 0 ... N, where typically N = 6, as shown explicitly in Figure 1B. The initial activate state, R*·P(0), represents non-phosphorylated metarhodopsin II, which is formed rapidly after photoisomerization. Each state R*·P(n) may undergo one of two reactions: it may be phosphorylated to become R*·P(n+1), or it may bind arrestin to become R*·P(n)·Arr. We denote the rate constant at which R*·P(n) undergoes phosphorylation as ν(n), in green, and the rate constant at which it undergoes arrestin binding as μ(n), in blue. The mean rate at which the molecule exits state R*·P(n) is ν(n)+μ(n), while the probability that the exit reaction corresponds to phosphorylation rather than to arrestin binding is ν(n)/{ν(n)+μ(n)}. Because each of the arrestin-bound states R*·P(n)·Arr has zero ability to activate transducin, these states can all be ignored, and there is no need to indicate any reactions involving them.
Note that we have not yet needed to specify the catalytic activity of R*·P(n). That activity only needs to be known when we solve the downstream deterministic equations.
Analysis of arrestin binding times
We now examine the time to arrestin binding, tArr, and its probability distribution, p(tArr). For the arrestin binding reaction, we follow recent studies by assuming that some number, M, of phosphates need to have been attached before arrestin can bind and that subsequent states bind arrestin with a fixed rate constant. This assumption is based on the biochemical experiments of Vishnivetskiy et al. [16]. The rate constant μ(n) of arrestin binding to state R*·P(n) is
For example, with M = 3, as shown in Figure 1B, the states with zero, one, or two phosphates cannot bind arrestin, whereas each of the subsequent states with three or more phosphates binds arrestin with the fixed rate constant μ. At this stage, we leave the values for the phosphorylation rate constants completely general and simply denote them as ν(0), ν(1) ... ν(N−1). Note that ν(N) = 0 because the fully phosphorylated state cannot be further phosphorylated. The parameters N and M and the required rate constants are described in Table 1, together with their default values.
Table 1. Stochastic R* parameters.
|
Symbol |
Description |
Value |
Units |
|---|---|---|---|
| Model 1: Conventional graded R* shut-off | |||
|
N |
Number of sites for phosphorylation |
6 |
|
|
M |
Minimum phosphates for Arr binding |
3 |
|
| νmax |
Maximal rate constant of phosphorylation |
80 |
s−1 |
| μ |
Rate constant of Arr binding |
20 |
s−1 |
| ωP |
Exponential factor for phosphorylation |
1 |
|
| ωG | Exponential factor for R* activity | 1 |
| Model 2: Binary R* shut-off Case 1: All rate constants equal | |||
|---|---|---|---|
|
N |
Number of sites for phosphorylation |
Any, ≥3 |
|
|
M |
Minimum phosphates for Arr binding |
3 |
|
| ν, μ | Rate constants for all reactions | 60 | s−1 |
| Model 3: Three-state R* shut-off Case 1: All rate constants equal | |||
|---|---|---|---|
|
N |
Number of sites for phosphorylation |
Any, ≥3 |
|
|
M |
Minimum phosphates before Arr binding |
3 |
|
| ν, κ, μ |
Rate constants for all reactions |
60 |
s−1 |
| ρlow | Fractional R* activity in low-activity state | 0.1 |
Amazingly, the assumption embodied in Equation (0.1) leads to a major simplification. Because each of the states with M or more phosphates inactivates by binding arrestin at the common rate, μ, it is irrelevant whether any additional phosphates are added once M have been attached. Therefore, we reach the astonishing prediction that the cascade can be truncated at any stage after state M, and yet the predicted kinetics of arrestin binding will be unaltered. In particular, the cascade can be truncated right at stage M, as illustrated for the case of M = 3 in Figure 1C. What this means is that the only phosphorylation rate constants that affect the kinetics of arrestin binding are those for the first M steps leading up to the formation of R*·P(M). This prediction came as such a surprise that we checked it numerically, by running simulations using different numbers N of available sites for phosphorylation, and for each N ≥ M that we tested, we obtained probability distributions (not shown) that were identical, within the noise.
For reaction schemes of the type illustrated in Figure 1C, the macroscopic solution for the impulse response was analyzed by Baylor, Hodgkin and Lamb [17]. For several simplifying cases of the rate constants, they derived the time course of state M, and in our formulation this represents the probability density function for the time to arrestin binding, tArr. From their equations we can readily obtain analytical solutions for the predicted probability density p(tArr) in three straightforward cases. The expressions that we derive have utility in two distinct ways. First, these equations provide us with multiple checks on the accuracy of the stochastic simulations that we perform. Second, in our “binary R* shut-off” model, they provide conveniently simple analytical expressions for the distribution of R* lifetimes, that can be used in conjunction with the deterministic set of reactions for the downstream phototransduction cascade to provide analytical expressions for the predicted distribution of SPR amplitudes.
Case 1: All rate constants equal
The simplest case occurs when all the non-zero rate constants are equal, in other words, when all the phosphorylation rate constants (at least those up to M−1) are the same as the common arrestin binding rate constant, μ, defined above in Equation (0.1), so that
In this case of “all rate constants equal”, the probability density function for the time to arrestin binding, p(tArr), is obtained from Equation (44) in [17] as
Note that, as we discovered above, this solution is independent of the total number of sites, N, and instead is a function of the minimum number of sites, M, required for arrestin binding. Note also that Equation (0.3) is the same “Poisson equation” that was derived for equal rate constants by Fuortes and Hodgkin [18] in their classic study of Limulus photoreceptor responses.
In this Poisson case, it is straightforward to integrate Equation (0.3) to obtain the cumulative probability density for arrestin binding, P(tArr), as
One can also obtain the mean time to arrestin binding and the coefficient of variation (cv = SD/mean) of the time to arrestin binding as
Case 2: Common arrestin binding rate constants and common phosphorylation rate constants
When each of the phosphorylation rate constants (at least up to M−1) is common, ν, but different from the common non-zero arrestin binding rate constant, μ, we have
with the arrestin binding rate constants still defined by Equation (0.1). This case was termed “Final rate constant different” in [17], where the solution was given as their Equation (45). That solution may be rewritten here as
In this case, the mean and cv are more complicated and will not be presented.
Case 3: Rate constants arranged arithmetically
A third tractable simplification arises when the rate constants of phosphorylation and arrestin binding happen to be arranged arithmetically; that is, when
with the common non-zero arrestin binding rate constant μ still defined by Equation (0.1). In this model, with M = 3, the four non-zero rate constants would be 4 μ, 3 μ, 2 μ, and μ. This situation was termed “Independent activation” in [17], where the solution was given as their Equation (37), and may be written as
In this case, one can apply integration to obtain the mean time to arrestin binding as
which, for M = 3, reduces to mean(tArr) = 25/12 μ-1. It is noteworthy that both Equation (0.5) and Equation (0.11) conform to the expectation for a stochastic cascade that the mean time taken to reach a given state is equal to the sum of the mean dwell times for the individual transitions leading up to that state.
Next, we describe three models for the way in which the R* molecule becomes inactivated as it progresses through the reaction steps illustrated in Figure 1 and analyzed above.
Model 1: Conventional case of graded reductions in R* activity
In the conventional model of graded reductions in R* catalytic activity, we follow the formulation of previous studies by specifying the transition rate constants for state R*·P(n) as
This equation indicates that the strength of the phosphorylation reactions declines monotonically as the number of attached phosphates increases, as reported in biochemical experiments [8]. We are not aware of any way of obtaining an analytical solution for the probability distributions of the states in this case. Together, Equations (0.1) and (1.1) contain three parameters (νmax, ωP, and μ) that need to be specified before stochastic simulation can be undertaken.
For subsequent substitution into the downstream transduction cascade, we need the catalytic activity of each state, R*·P(n). It is convenient to express this in fractional terms, ρ(n), where ρ(0) = 1 for the initial state R*·P(0). As in previous studies, this fractional catalytic activity is presumed to decline exponentially with n:
Here, we allow for the possibility that the steepness parameter, ωG, may be different from the steepness parameter ωP for phosphorylation in Equation (1.1); this was also permitted in [9], whereas in [6] and [11] it was assumed that ωG = ωP.
Model 2: “Binary” R* activity
In the second model, we dispense with the assumption that the catalytic activity of R*·P(n) declines with the increasing number of attached phosphates, and instead we assume that the activity ρ(n) is constant. We refer to this as “binary” R* activity, because the R* remains fully active until it binds arrestin.
To determine the time course of R* activity in this model, we only need the time tArr until arrestin binds, and it was the realization of this fact that provided the main impetus for deriving analytical expressions for the probability distributions of tArr in the preceding section.
Of particular importance is the cumulative probability distribution, P(tArr), because in the binary activity model the activity of R* is unity until arrestin binds, whereupon it drops to zero. Therefore, the mean R* activity is simply the complement of P(tArr), that is,
Therefore, for the case of “all rate constants equal”, Equation (0.4) gives the mean time course of R* activity as
At very early times, this expression approximates to
Therefore, in the binary R* model, the initial time course of R* activity does not approximate an exponential decay but is instead “flat-topped”, as shown in the Results section.
The expected form for the probability distribution of SPR amplitudes can be readily determined in the binary R* model, because the solution of the downstream reaction cascade involves only a single stochastic parameter, tArr, for which we know the probability distribution in certain simplifying cases. Therefore, if we solve the downstream cascade of reactions for a range of values of tArr, we can obtain any desired feature of the SPR as a function of tArr; for example, we can obtain the peak amplitude, SPRpeak, as a function SPRpeak(tArr) of tArr. Then, for any case where we know the probability density p(tArr), we can evaluate the probability density of SPRpeak as
In practice, the function SPRpeak(tArr) can be evaluated at a small number of values of tArr and then fit using a spline function and differentiated. This approach is illustrated subsequently in the Results section.
Model 3: Three-state model with low R* activity before arrestin binding
One drawback of the binary R* model turns out to be its inability to predict a sensible SPR waveform in the arrestin knockout phenotype (see Results). This occurs because in that case the R* activity never shuts off. To deal with this issue, we have investigated an alternative model in which the R* activity drops to a lower level after M phosphates have been attached. As illustrated in Figure 1D, we envisage that once M phosphates have attached, the R* molecule can enter a new state according to a rate constant κ. We further envisage that, following this change, arrestin can now bind, again with rate constant µ (Figure 1D). Therefore, instead of Equation (0.1), we have
Therefore, simply by replacing μ with κ, the analytical expressions that we derived above for arrestin binding time now apply instead to the time tlow to reach the conformation with low activity. We additionally need to specify the arrestin binding rate constant as
Finally, we need to specify the R* activity, which we assume drops to a low value, ρlow, and then drops to zero upon arrestin binding, which we formulate as
To derive the mean R* activity deterministically in this three-state case, it is convenient to define the following three variables: PF(t), the probability that the molecule remains in the fully activated state; PFL(t), the probability that it is either in the fully activated or in the low-activity state (i.e., the probability that it is not arrestin-bound); and the difference PL(t) = PFL(t) − PF(t), the probability that it is in the low-activity state. In the three-state case, PF(t) is analogous to R*bin(t), except with μ replaced by κ, while PFL(t) is a corresponding expression with the additional rate constant μ. Accordingly, we can express the mean R* activity in the three-state case as PF(t) plus a scaled-down version of PL(t) to give
In the general case, with arbitrary rate constants, it is necessary to evaluate the terms PF(t) and PFL(t) in Equation (3.4) by numerical integration. However, for the simplest case, with all rate constants equal (ν = μ = κ), Equation (3.4) reduces to
As in the case of the binary R* activity model, we can also predict the form of the distribution of SPR peak amplitudes, although in this case there is an approximation involved. Thus, when we obtain the relationship between SPR peak amplitude and the time tlow to the low-activity state, our approximation is to take the subsequent time to arrestin binding as fixed at 1/μ rather than being stochastic. Our subsequent results show that this provides a remarkably accurate prediction. By analogy with Equation (2.5), the predicted distribution in the three-state model is
Downstream phototransduction cascade
For any of the models above, we can simulate the time course of R* activity, or, in the special cases of simple ratios of rate constants, we know the probability density of arrestin binding times. We can then solve the deterministic reactions for the downstream phototransduction cascade to generate the predicted individual SPRs. To do this, we employed a well-established model of phototransduction, with Ca2+-mediated feedback onto cyclic GMP concentration via GCAPs and guanylyl cyclase. We used a distributed model with longitudinal diffusion of cGMP and Ca2+ within the outer segment, and as a check we also used a reduced set of these equations for the isotropic case of a well-stirred outer segment.
Photoisomerizations are deemed to occur at discrete spatial locations, that is, at disks, and PDE activity is likewise deemed to be restricted to those disks so that its activity is specified by an ordinary differential equation (o.d.e.). For both cGMP and Ca2+, a partial differential equation (p.d.e.) is required to describe the longitudinal diffusion. The three differential equations and their ancillary equations are as follows. Most variables include the spatial (x) coordinate as well as time. The parameters in these equations are defined in Table 2.
Table 2. Downstream phototransduction cascade parameters.
| Symbol | Description | Default value a | Model 1 value b | Units |
|---|---|---|---|---|
| βDark |
Dark rate constant of cGMP hydrolysis |
3.2 |
4 |
s−1 |
| αmax |
Maximal rate of cGMP synthesis by GC |
120 |
150 |
µM/s |
|
fCa |
Fraction of CNGC current carried by Ca2+ |
0.12 |
||
|
KGCAP |
Ca2+ concentration parameter of GCAP |
80 |
nM |
|
|
mGCAP |
Ca2+ cooperativity of GCAP |
1.5 |
||
|
JcG, max |
Maximal CNGC current for the OS |
2000 |
pA |
|
|
KcG |
cG concentration parameter of CNGC |
20 |
µM |
|
|
Jex, max |
Maximal exchange current for the OS |
4.6 |
pA |
|
| Kex | Ca2+ concentration parameter of exchanger | 1100 | nM |
| Calculated resting dark state | ||||
|---|---|---|---|---|
|
cGDark |
Dark cGMP concentration |
4.12 |
µM |
|
|
CaDark |
Dark Ca2+ concentration |
322 |
nM |
|
| JDark | Dark current | 18.4 | pA |
| Other kinetic parameters | ||||
|---|---|---|---|---|
| νRE |
Rate of G*-E* activation by an R* |
300 c |
s−1 |
|
|
kE |
Rate of G*-E* shut-off |
5 c |
s−1 |
|
| βsub |
Rate constant of cGMP hydrolysis by an E* |
0.024 d |
0.063 d |
s−1 |
|
BCa |
Buffering power of cytoplasm for Ca2+ |
50 |
||
| Vcyto | Available cytoplasmic volume of OS | 0.02 | pL |
| Longitudinal diffusion parameters | ||||
|---|---|---|---|---|
|
DcG |
Longitudinal diffusion coefficient for cG |
40 |
µm2/s |
|
|
DCa |
Longitudinal diffusion coefficient for Ca2+ |
2 |
µm2/s |
|
| L | Length of outer segment | 22 | µm |
a. All values apply to the entire outer segment; however, suction pipette experiments record only a fraction of the total current. b. The default values applied in all cases, except for the three values listed under Model 1. c. The formation (νRE) and shut-off (kE) of E* are modeled as either deterministic or stochastic. d. For comparison with the values used in Gross et al. [11], note that they employed a different parameter for hydrolysis by E*, that they referred to as βidv, which is shown in Appendix 1 to be related to βsub by βidv = Ndisks βsub, where Ndisks = 710 is the number of disks in the outer segment.
The membrane currents above are expressed as current density per unit length of outer segment, jcG(x,t) and jex(x,t). However, to simplify comparison with the conventional lumped formulation, they have been written in the form L jcG(x,t), with units of current over the entire outer segment length, L. Consequently, when these equations are coded numerically, we can conveniently use the same symbols in both the spatial and lumped cases and then use either the integral Equation (4.9) in the spatial case or a simple sum in the lumped case.
Boundary conditions at ends
The boundary conditions at each end of the outer segment are of the reflective (zero flux) kind. They apply for both cGMP and Ca2+, and can be written as:
Boundary condition at location of photoisomerization
For these partial differential equations, Equation (4.4) above does not incorporate a term for light-induced PDE activity because the spatial extent of R* activity at each location where a photoisomerization has occurred is deemed to be infinitesimally narrow. Therefore, at each such location we need to apply a boundary condition for cGMP hydrolysis. In the symmetric case of a single photoisomerization delivered at the middle of the outer segment, defined as x = x0, this is straightforward. Equating the flux in the positive x direction with the rate at which cGMP is hydrolyzed by half of the activated PDE on that disk, we obtain the boundary condition as
The full derivation of this equation is given in Appendix 1, and it is identical to Equation (B 1) derived by Lamb and Pugh [1]. However, we note that the corresponding Equation (6) in Gross et al. [11] differs in having a factor of 4 in its denominator, but in our view that formulation was incorrect. Gross et al. [11] invoked a second factor of 2, on account of the fraction of the outer segment volume that is cytoplasmic. However, analysis of the equations shows that this is not appropriate. One way of rationalizing this is to note that the volume factor has already been accounted for in the definition of the effective longitudinal diffusion coefficient. In comparing the equations between the two studies, one also needs to note that they used a different parameter for the PDE hydrolytic efficacy that they termed βidv, which is Ndisks times our βsub, that is, βidv = Ndisks βsub, where Ndisks is the number of disks in the outer segment.
Although this analytical form of the boundary condition is important in estimating the hydrolytic activity underlying the rogue responses, it is not required for numerical simulation. For numerical simulation, we instead used spatial elements of finite width δx, and we took the PDE activity to be distributed over the width of the element where the photoisomerization occurred. Therefore, in any element in which a photoisomerization occured, Equation (4.4) was replaced with the following modified form of the conventional lumped case equation:
Ensemble variance for dim-flash responses
One can consider an idealized experiment, where the number of photoisomerizations per trial is Poisson-distributed with a mean of m, and where the individual SPRs have a mean time-course a(t) and variance time-course σ12(t). In this idealized case, the expected ensemble variance for a very long series of identical flashes is
Typically, one would expect the ratio σ1(t)/a(t) to be quite small (around 0.3; see Results section) so that its square would be around 0.1. As a result, the experimentally recorded variance is dominated by the Poisson contribution of the first term in Equation (5.1), and the “between SPR” variability elevates the predicted variance by only about 10%. In practice, the situation is more complicated because of the finite number of flashes that can be delivered, nonlinear summation (especially when the light stimulus is spatially restricted), noise, and lack of recording stability.
Methods
All numerical computations were performed using custom Matlab (The MathWorks, Inc., Natick, MA) scripts, which are available upon request.
Numerical simulation of stochastic reactions for R*
The stochastic reactions for R*, described in the panels in Figure 1, were simulated using Gillespie’s method [19]. We will begin by describing the simplest cases (Figure 1A–C) where just two fates are possible for each state: either phosphorylation or arrestin binding. First, the mean transition rate constants μ(n) and ν(n) were assigned. Then, for each states, two pseudo-random numbers, r1(n) and r2(n), uniformly distributed between zero and unity, were used. The lifetime that the molecule remained in state R*·P(n) was simulated as −ln(r1(n))/{ν(n)+μ(n)}, and the state to which the molecule transitioned was simulated as phosphorylation if r2(n) < ν(n)/{ν(n)+μ(n)} and as arrestin binding otherwise. If a finite flash width was required, the time of photoisomerization was simulated as uniformly distributed over that flash width. These simulations were very fast, and 106 iterations could be performed in ~1 s on a laptop PC. For the three-state R* activity model, simulation of the full version (Figure 1D) would have required more complicated code, so we chose instead to simulate only the truncated linear configuration (Figure 1E), and in this case 106 iterations again took only ~1 s.
Numerical simulation of stochastic reactions for E*
For most of the simulations described here, the time course of SPR-induced PDE activity, E*(t), was taken to be deterministic, as described by the o.d.e. Equation (4.1). In one case, however, we additionally simulated the generation and decay of E* as a stochastic variable. At the single location of the photoisomerization, E* molecules were generated stochastically at a mean rate given by νRE times the instantaneous R* activity (ρ = 1 or ρ = ρlow), and each generated E* was assigned a stochastic lifetime with a mean of kE−1.
Numerical integration of equations for downstream phototransduction cascade
The differential equations for the deterministic reactions of the downstream phototransduction cascade were integrated numerically using a Matlab o.d.e. solver, typically “ode15s”. For the case of longitudinal diffusion in the outer segment, the method of partial discretization was employed [20] to represent the system as a set of spatially coupled o.d.e.s, with the outer segment divided into a finite number of spatial elements (50 in the illustrated Figures).
The parameters we chose are specified in Table 2. The values in the “Default” column were used for each model, with the exception that for Model 1 the three values in the column “Model 1” were used. We began by evaluating the dark resting state as follows. For Equations (4.1)–(4.9), we set all the spatial and temporal derivatives to zero and expressed the steady cGMP in terms of the steady Ca using Equation (4.4) with (4.6)–(4.8). We then determined the value of Ca that rendered the net influx of calcium zero by numerically finding the zero for the numerator of Equation (4.5). The resulting dark resting values are listed in Table 2. Note that the resting state was identical in each model because we constrained the ratio αmax/βDark to be the same in each case.
The numerical integration of the downstream reactions was orders of magnitude slower than the stochastic simulations, and 105 repetitions of the spatial diffusion case typically took around 5 h on a laptop PC. In practice, as few as 1000 repetitions were sufficient to provide extremely good estimates of the ensemble mean and variance of the SPR. However, to obtain a good histogram of the distribution of SPR peak amplitudes for the illustrated Figures we ran 105 repetitions.
Results
Model 1: Conventional model of R* graded shut-off
We began by verifying that our implementation of the stochastic model of graded R* shut-off in conjunction with our formulation of the downstream reactions predicted SPRs with properties conforming to those recently reported by Gross et al. [11]. To this end, we adopted parameter values that were nearly identical to theirs, as listed in Table 1. One minor difference was that for consistency with our subsequent simulations of Models 2 and 3, we chose the “abrupt” arrestin-binding cut-in described by Equation (0.1), as used recently in [10], rather than the graded approximation to this relation employed by Gross et al. [11] in their Equation (10).
Figure 2 collects the results of our simulations for the graded R* shut-off model. The top pair of panels summarize the behavior of individual R* molecules in 106 repetitions. The remaining panels show SPRs predicted by integrating the downstream cascade reactions: the middle pair of panels each show a handful of SPRs, while the bottom pair plot the ensemble behavior for 105 simulations.
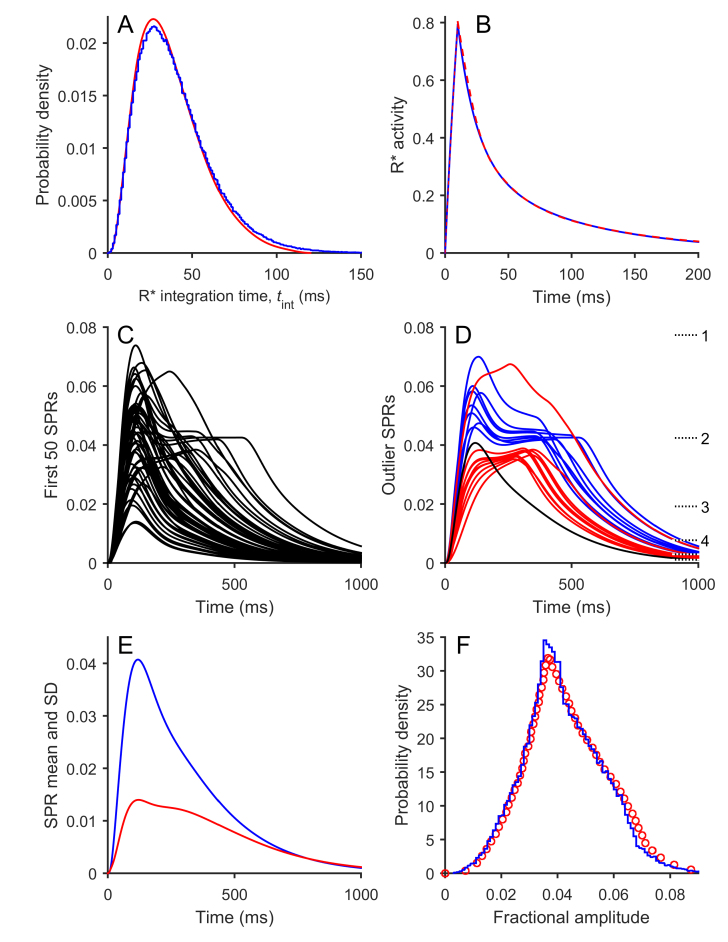
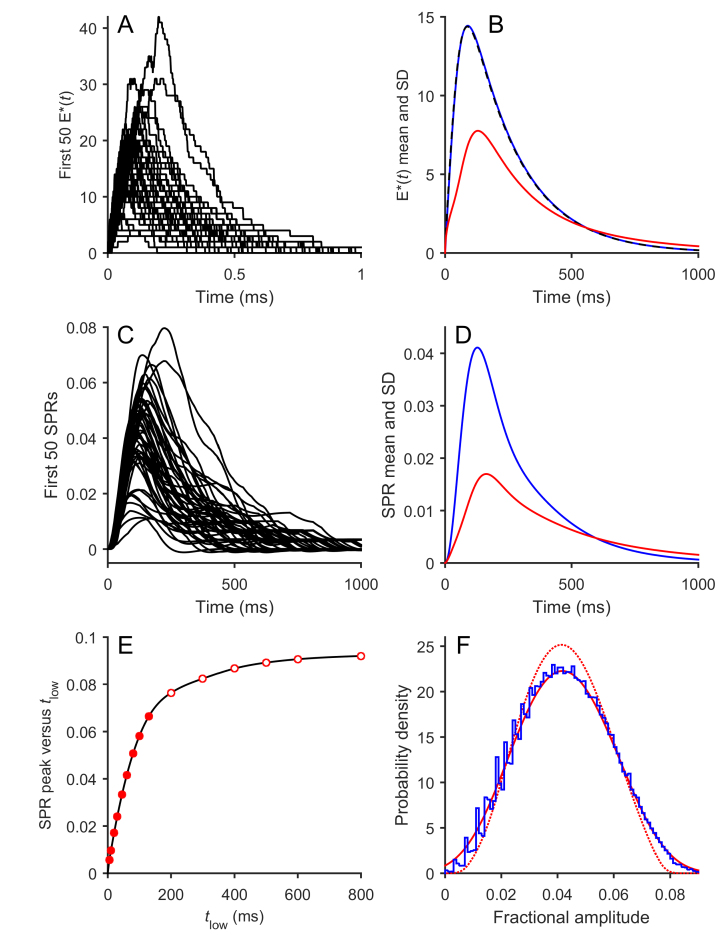
Figure 2.
Results of simulation of Model 1 (Graded R* shut-off model). The equations for Model 1 were simulated numerically using stochastic parameters n = 6, M = 3, νmax = 80 s−1, μ = 20 s−1, and ωP = ωG = 1 (Table 1); the flash duration was set to 10 ms. The downstream parameters are given in Table 2, with αmax = 150 µM/s, βDark = 4 s−1, and βsub = 0.063 s−1. The number of simulations of R* activity in panels A and B was 106, while for the subsequent panels the downstream reactions were integrated using the first 50 R* simulations (C), the first 200 R* simulations (D), or the first 105 R* simulations (E and F). A: Distribution of times to Arr binding for the 106 simulations (blue trace). The red trace is the corresponding curve from the simulations of Gross et al. [11] plotted in their illustration 6E. B: Mean R*(t) time course for the 106 simulations (blue trace). The red trace is the corresponding curve from the simulations of Gross et al. [11] plotted in their Supplementary illustration S2C. C: The first 50 simulated SPRs. D: Sample “outlier” SPR responses, selected as described in the text from the first 200 R* simulations. Blue: The 10 SPRs with the longest times in state R*·P(2). Red: The 10 SPRs with the longest time-to-peak. The horizontal dotted lines at the right indicate the steady-state levels reached with very long dwell times in each of the states R*·P(n) . These levels have been numbered for n = 1 – 4, but are not numbered for 5 or 6. The level for state R*·P(0) is above the top of the vertical scale, at 0.114. E: Time course of the mean (blue) and SD (red) for the ensemble of 105 simulated SPRs. F: Probability distribution of the peak amplitudes for the 105 simulated SPRs (blue). The red circles show the corresponding histogram plotted by Gross et al. [11] for their simulations in their illustration 6E.
Figure 2A compares the probability distribution for R* integration times, p(τR*), that we obtained (blue) with the corresponding curve from [11] (red) that was plotted as the inset in their illustration 6E. The similarity is very close, although our simulations gave marginally fewer short integration times and a marginally greater number of long integration times. Figure 2B compares the time-course of the mean of the simulated R* activity for the 106 repetitions and shows that the traces were barely distinguishable between the two studies. Figure 2C plots the first 50 SPRs simulated and gives a flavor of the individual waveforms; Figure 2D selects some outlier responses and will be considered shortly. Figure 2E plots the ensemble mean and SD for 105 SPRs integrated using the downstream reactions, and the traces are extremely similar to the corresponding curves illustrated in [11]. Figure 2F plots the probability distribution for the peak amplitudes of these 105 SPRs (in blue), and the close resemblance to the distribution obtained in [11] (red circles) is striking.
These simulations generated the following values of interest, which are compared with the corresponding values from [11] in brackets, where available. For the dark steady-state: cGDark, 4.1 µM (4.1 µM); CaDark, 322 nM (320 nM); and total outer segment current, 18.4 pA (18.7 pA). For the stochastic responses: R* integration time, mean 39.7 ms (40 ms), and cv 0.547 (0.52); cv of SPR area, 0.459; fractional amplitude of SPR at peak, mean 0.0420, and cv 0.332; and mean time-to-peak of SPR: 119 ms. Based on the closeness of these values and the similarity of the four comparisons in panels A, B, E, and F, we conclude that our SPR simulations conformed very closely to those of Gross et al. [11].
Despite this close similarity, we observed a disturbing feature of a minority of the simulated SPRs that does not accord with our experience of the form of real SPRs. As exemplified by Figure 2C,D, a minority of the simulated responses tended to exhibit a plateau after the initial peak. In Figure 2C, it is apparent that a sub-set of the first 50 simulated responses exhibited a fairly flat region at around half the amplitude of the peak; qualitatively the same phenomenon was apparent in any group of 50 consecutive simulations that we selected (not shown). Investigation of this phenomenon revealed that the plateau resulted from trials that happened to exhibit relatively long durations in the doubly phosphorylated state, R*·P(2). Therefore, to better illustrate these events, we selected outlier responses representing 5% of the total trials based on two criteria, and we present these in Figure 2D. From the first 200 trials we selected the 10 responses that had the longest times to exit from state R*·P(2), and these are plotted in blue. Each of these traces has the appearance of heading toward a fractional amplitude of ~0.042 at times of 400–500 ms. Next, from the same set of 200 trials we selected the 10 responses that exhibited the longest time-to-peak, and these are plotted in red. Most of these red traces reach peak later than 300 ms at a level of roughly 0.037. We observed qualitatively the same kind of behavior when we selected the outliers from any group of 200 consecutive trials (not shown).
We suggest that this tendency for a sizable fraction of trials to exhibit a plateau is an inevitable feature of the graded shut-off model and arises (at least for the parameter values chosen) from trials that happened to exhibit a long dwell time in state R*·P(2). Although each state is broadly analogous to every other state, there are some factors that combine to generate “unusual” events for the doubly phosphorylated state. First, the mean lifetime in this state is moderately long, at 1/(80 s−1 e−2) = 92 ms, so that 5% of trials are expected to display dwell times in this state of longer than ~270 ms. Second, the amplitude elicited by a long dwell time in this state happens to asymptote to a level close to the peak amplitude of the mean SPR (see dotted horizontal lines on the right in Figure 2D). As a result, the inevitable events that exhibit long dwell times in state R*·P(2) generate responses that are noticeably unusual. For the other states, the effects of comparatively long dwell times are visually less noticeable, either because the mean lifetime is quite short (states n = 0 or 1) or because the asymptotic response level for that state is considerably smaller than the peak (states n ≥ 3).
We think it very likely that it is the existence of a moderate number of SPRs exhibiting such a plateau that causes the rather “flat-topped” shape for the time-course of the ensemble standard deviation, shown by the red trace in Figure 2E. In addition, we think that it is the existence of responses such as those shown in Figure 2D that gives rise to the sharp peak in the amplitude histogram (between 0.035 and 0.042) in Figure 2F. We are not aware of published experiments that have reported SPRs exhibiting kinetics of this kind, and nor have we observed responses that obviously resemble these events in our own recordings of dim flash responses. Nevertheless, we cannot completely rule out the possibility that SPRs with this shape do occur in real rods. However, our firm inclination is to suggest that this feature predicted by the model of graded reduction in R* activity is inconsistent with the properties of observed rod responses.
In addition to this issue with plateau-like events, another major problem was that the required amplification of the downstream reactions was excessively high, being greater than can be accounted for by the measured biochemical parameters. Thus, the product νRE βsub that was needed both in Model 1 (for νRE = 300 s−1 and βsub = 0.063 s−1) and in [11] was νRE βsub ≈ 19 s−2. This corresponds to an amplification constant for phototransduction of A = νRE βsub ncG ≈ 57 s−2, far higher than the values reported in experiments on mammalian rods (see Discussion).
For comparison with biochemical estimates, βsub is given by (½ kCAT/Km) / (½ πr2L BP NAv) [1], where kCAT/Km is the catalytic activity of the fully activated PDE dimer, BP is the buffering power of the cytoplasm for cGMP, NAv is Avogadro’s number, and r = 0.75 µm and L = 22 µm are the radius and length of the outer segment. The factor of 1/2 in the numerator reflects the fact that two G* molecules are required to fully activate the dimeric PDE, while the factor of 1/2 in the denominator corresponds to the fraction of the envelope volume that is cytoplasmic. For amphibian rods at room temperature, kCAT/Km has been measured as 4.4 × 108 M−1 s−1 [21]. Substitution of these values gives βsub = 0.019 s−1 / BP. If the cytoplasm exhibited finite buffering power for cGMP (i.e., BP > 1), then the value would be lower than 0.019 s−1. Conversely, if the mammalian enzyme exhibited higher activity at body temperature, then the value could be higher (see Discussion). However, taking these two factors into account, it is difficult to envisage that the biochemical measurements could correspond to a hydrolytic rate constant of more than ~50% above 0.019 s−1, which would set a realistic constraint that βsub ≤ 0.03 s−1. On this basis, the value of βsub = 0.063 s−1 required in Model 1 again appears excessive.
An independent estimate of βsub for mouse rods was recently reported by Gross et al. [13], based on measurements of “rogue” responses representing the failure of activated R* to shut off. However, in our view their estimate needs to be halved for the following reason. As derived in Appendix 1 and noted following Equation (4.11), their analysis incorrectly introduced a second factor of 2 in the denominator of their Equation (6). Accordingly, their analysis of rogue responses actually predicts a value of βsub = 0.03 s−1 in GCAPs−/− rods. Again, this is approximately half the value of βsub required to achieve responses of appropriate amplitude in Model 1.
An additional problem with the graded R* shut-off model is that its prediction of the arrestin-knockout phenotype is not very realistic. When the rate constant for arrestin binding was set to zero (μ = 0), the predicted mean SPR exhibited a tail considerably smaller than observed in experiments, decaying to <4% of the peak after 5 s (not shown). This occurred because in the graded shut-off model the activity of state R*·P(6) is extremely small, at e−6 ≈ 0.0025, with the consequence that the activity of R* is substantially quenched even in the absence of arrestin binding.
The combination of these problems leads us to conclude that the graded R* shut-off model, in the form described both here and in [11], does not provide a plausible description of the observed SPRs in mammalian rods.
Model 2: “Binary” R* activity
In light of these difficulties with the model of a graded decline in R* activity, we chose to investigate a model in which the R* activity does not decline in a graded manner with successive phosphorylations and instead remains maximal until arrestin binds. We refer to this situation as “binary” R* activity. Such a model has the convenience that the probability distribution of R* lifetimes can be readily predicted in several simplifying cases, as derived in the Theory section. Not only does this provide a means of checking the accuracy of our stochastic simulations, but it also allows us to predict the distribution of SPR amplitudes.
The results of our simulations for the binary R* shut-off model are collected in Figure 3, where the organization of panels is broadly similar to that in Figure 2. The top pair of panels summarize the simulated activity of R* molecules in 106 repetitions; the middle pair show a selection of SPRs; and the bottom pair plot the ensemble behavior for 105 simulations.
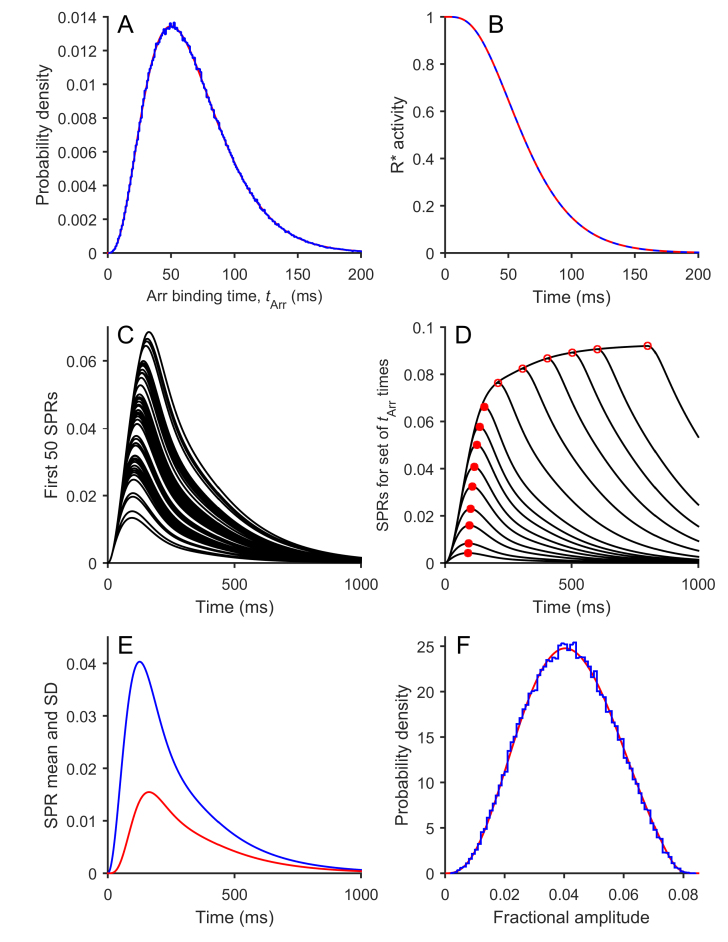
Figure 3.
Results of simulation of Model 2 (Binary model). The equations for Model 2 were simulated numerically using M = 3 and ν = μ = 60 s−1 for the stochastic parameters (Table 1) and with the downstream parameters from Table 2; the flash duration was set to zero. The number of simulations of R* activity in panels A and B was 106, while for the subsequent panels the downstream reactions were integrated using the first 50 R* simulations (panel C) or the first 105 R* simulations (panels E and F). A: Distribution of times to Arr binding for the 106 simulations (blue trace). The red trace is the theoretical prediction of Equation (0.3). B: Mean R*(t) time course for the 106 simulations (dashed blue trace). The red trace is the theoretical prediction obtained using Equation (2.3). The blue trace is drawn dashed, as otherwise it would have completely obscured the theoretical prediction. C: The first 50 simulated SPRs. D: SPRs for a set of defined times to arrestin binding. Rather than being simulated stochastically, the time to arrestin binding was instead set to 5, 10, 20, 30, 45, 60, 80, 100, 130, 200, 300, 400, 500, 600, or 800 ms. Red circles indicate the peaks. These measurements were fit by a spline function and used to predict the expected distribution in panel F. E: Time course of the mean (blue) and SD (red) for the ensemble of 105 simulated SPRs. F: Probability distribution of the peak amplitudes for the 105 simulated SPRs (blue). The smooth red trace plots the predicted distribution obtained using the approach described in the Theory section, using Equation (2.5) and the measurements of peak amplitude from panel D.
For the stochastic shut-off parameters, we chose to set all the rate constants equal, both for simplicity and also so that we could examine whether the results were described by the theory developed in Equations (0.3)–(0.6) and (2.3)–(2.5); we used ν = μ = 60 s−1 (Table 1), and we set the flash duration to zero. For the downstream reactions, we used the default parameters in Table 2. These differ in the following respects from those used in the graded R* shut-off model in [11] and duplicated in Figure 2. First, we reduced the maximum cyclase rate to the value used by Koch and Dell’Orco [22], αmax = 120 µM s−1. Next, to keep the dark resting state unchanged, we held the ratio αmax/βDark constant by reducing βDark in the same ratio to βDark = 3.2 s−1. Finally, we adjusted the magnitude of the PDE hydrolytic activity to generate SPRs of the expected amplitude, ~ 4% of the dark current. The resulting value of βsub = 0.024 s−1 (Table 2) turned out to be very close to that predicted both by biochemical measurements and by the analysis of “rogue” R* responses [13].
For 106 simulations of the R* stochastic reactions, Figure 3A plots the probability distribution for arrestin binding times p(tArr) as the blue histogram. For comparison, the smooth red curve is the prediction of Equation (0.3), and the fit is extremely good. For M = 3 and ν = μ = 60 s−1, its peak is predicted to occur at 3/(60 s−1) = 50 ms, as observed. The mean and the cv of the arrestin binding time (and thus of R* activity) are predicted by Equations (0.5) and (0.6) to be 66.67 ms and 0.5, respectively, and the values we obtained in the simulations were 66.66 ms and 0.4995. In Figure 3B, the blue curve plots the ensemble mean time course of R*(t) for the 106 binary R* activity events, while the dashed red trace plots the prediction of Equation (2.3) and is seen to completely overlie the blue curve for the simulations. The closeness of the simulated activity to the predictions gives us confidence that our simulation algorithm provided an accurate rendition of the stochastic reactions.
Figure 3C superimposes the first 50 SPRs simulated, and two features are apparent. First, in this case with zero flash duration, the traces begin rising along a common curve. Second, there is no hint of any plateau-like responses of the kind seen in Figure 2. Figure 3D plots the SPRs obtained for a series of specified values of tArr, ranging from 5 ms to 800 ms, and confirms the common early rising phase. The red circles mark the peaks of these individual traces, with filled red used for tArr up to 130 ms and open symbols used for tArr equal to or greater than 200 ms. The magnitudes of these peaks are subsequently used to generate the relationship between SPRpeak and tArr that is required in Equation (2.5) for predicting the distribution of amplitudes. Note that the probability of tArr > 150 ms was 2.2%.
For 105 simulated SPRs, Figure 3E plots the time-course of the ensemble mean and SD and shows a characteristic feature of experimentally measured SPRs, which is that the ensemble SD reaches peak almost 40 ms later than the ensemble mean at 163 ms compared to 126 ms. Figure 3F plots the probability distribution for the peak amplitudes of the 105 SPRs as the blue histogram and compares this with the prediction of Equation (2.5) in red, demonstrating that the prediction is very accurate. The peak amplitudes of the individual SPRs (measured as a fraction of the dark current) had a mean of 0.0413, with a cv of 0.348, considerably reduced from the cv for arrestin binding time of 1/2.
Our interpretation of these results is as follows. First, the good fit of the predictions to the simulated results supports the view that our algorithms accurately simulated the stochastic reactions. Second, the binary R* activity model with the parameters we have chosen generates simulated responses that account for many of the features observed for mammalian rod SPR experiments. Third, the binary R* activity model does not exhibit the first two problems we noted for the graded R* activity model; that is, it does not generate occasional SPRs that exhibit a plateau-like late phase and it only requires a value for βsub that is consistent with predictions from biochemical measurements. We suggest that adoption of a realistic estimate for the gain product νRE βsub (of approximately 7 s−2) necessitates the use of an integration time for R* activity of the order of 65 ms or more to generate SPRs of realistic amplitude. In our view, the proposal of an R* integration time of 40 ms in previous work seems unrealistic and appears to have arisen as a consequence of assuming that R* activity shuts off in a graded manner.
Despite the good description of WT SPRs obtained in Figure 3, there is a problem with the binary R* model when the arrestin knockout phenotype is modeled (by setting μ = 0). As the R* activity then never shuts off, the simulated response simply rises to a maximum and remains there. This behavior is not illustrated but is essentially the same as shown subsequently for the GRK1 knockout (obtained by setting ν = 0). We therefore investigated an extension of the binary R* model in which the catalytic activity declined to a low level after a certain number of phosphates had been attached.
Model 3: Three-state model, with low R* activity before arrestin binding
To deal with the failure to account for the arrestin knockout phenotype, we modified the binary R* activity model to include a penultimate state with lower activity, as shown schematically in Figure 1D. In this “three-state” model, we hypothesize that the R* catalytic activity remains maximal until a change is triggered by the attachment of some threshold number of phosphates. When this change occurs, the catalytic activity drops to a low level, and a new conformation amenable to arrestin binding is formed. As indicated in Figure 1D, we assume for simplicity that the rate constant of entering the low-activity conformation is fixed (at κ) for all states with M or more phosphates so that the scheme simplifies to the linear sequence illustrated in Figure 1E. It is straightforward to simulate this linear scheme stochastically and likewise to obtain analytical solutions in the simplifying cases of ratios of rate constants, as set out in the Theory section.
The results of our simulations for this three-state model are collected in Figure 4, where the organization of panels follows that in Figure 3. The top pair of panels summarize the simulated activity of the R* molecule in 106 repetitions; the middle pair show a selection of responses; and the bottom pair plot the ensemble behavior for SPRs in 105 simulations.
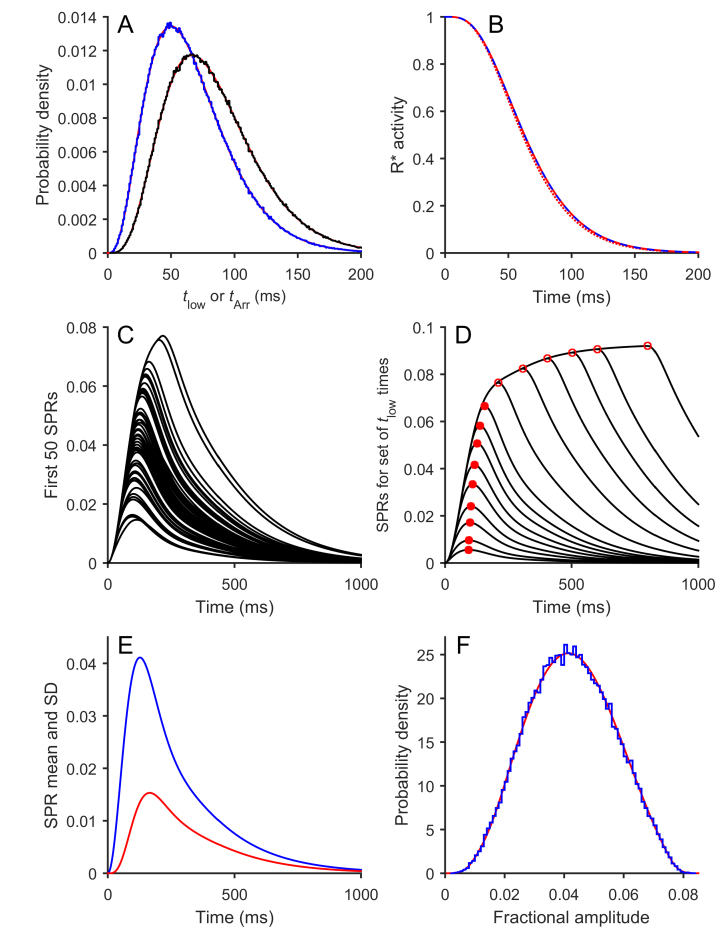
Figure 4.
Results of simulation of Model 3 (Three-state model). The equations for Model 3 were simulated numerically using M = 3 and ν = κ = μ = 60 s−1 for the stochastic parameters (Table 1) and with the downstream parameters from Table 2; the flash duration was set to zero. The number of simulations of R* activity in panels A and B was 106, while for the subsequent panels the downstream reactions were integrated using the first 50 R* simulations (panel C) or the first 105 R* simulations (panels E and F). A: Distribution of times to the low-activity state (left) and to Arr binding (right) for the 106 simulations. Blue traces are simulations, and red curves are theoretical predictions. B: Mean R*(t) time-course, for the 106 simulations (dashed blue trace). The overlying dashed red trace is the prediction of Equation (3.5). For comparison, the dotted red trace shows the predicted response for the binary model using the same values of ν and μ; thus it plots the dashed red trace from Figure 3B. The similarity of these traces shows that inclusion of the low-activity state made little difference to the mean time-course of the simulated and predicted R*(t) responses. C: The first 50 simulated SPRs. D: SPRs for a set of defined times to the low-activity state. Rather than being simulated stochastically, the interval until low activity was instead set to 5, 10, 20, 30, 45, 60, 80, 100, 130, 200, 300, 400, 500, 600, or 800 ms. The subsequent delay to arrestin binding was fixed at its mean value of 1/μ. Red circles indicate the peaks. These measurements were fit by a spline function and used to predict the expected distribution in panel F. E: Time course of the mean (blue) and SD (red) for the ensemble of 105 simulated SPRs. F: Probability distribution of the peak amplitudes for the 105 simulated SPRs (blue). The smooth red trace plots the predicted distribution obtained using the approach described in the Theory section using Equation (3.6) and the measurements of peak amplitude from panel D.
For the downstream reactions, we retained the parameters employed for the binary R* model. For the stochastic shut-off parameters, we retained M = 3, and we set the rate constant κ for the change to the low-activity conformation equal to the other rate constants at κ = ν = μ = 60 s−1. To account for the late phase of the arrestin knockout phenotype, we chose the low-state activity as ρlow = 0.1; see Equation (3.3).
For 106 simulations of the R* stochastic reactions, Figure 4A plots the probability distributions for both the time to entering the low-activity state (tlow, blue histogram) and the time to arrestin binding (tArr, black histogram). For comparison, the smooth red curves plot the predictions of the Poisson expression, Equation (0.3), and in both cases the fit is excellent. In Figure 4B, the dashed blue curve plots the mean time course of R*(t) for the ensemble of 106 R* events in this three-state case. The overlying dashed red curve shows the prediction of Equation (3.5), while the dotted red curve shows the prediction of the binary R* activity model, Equation (2.3). The mean of the R* activity (simulated or predicted) barely differs from the prediction of the binary activity model, indicating that the inclusion of the third state, with low activity ρlow = 0.1, causes little change in the mean R* activity in this WT case.
Figure 4C plots the first 50 simulated SPRs, and the traces are qualitatively similar to the corresponding simulations in Figure 3C, with no hint of outliers exhibiting unusual kinetics. Figure 4D superimposes the SPRs obtained for a series of specified values of tlow, ranging from 5 ms to 800 ms, with a fixed subsequent delay to arrestin binding; see the Figure caption. The red circles mark the peaks of these individual traces, with filled symbols used for tlow from 5 ms to 130 ms and open symbols used for tlow from 200 ms to 800 ms. The magnitudes of these peaks were subsequently used to generate the relationship between SPRpeak and tlow that is required for predicting the probability distribution of amplitudes. Note that the probability of tlow > 150 ms was 2.2%, and the probability of tArr > 150 ms was 5.6%.
For 105 simulated SPRs, Figure 4E plots the time course of the ensemble mean and SD. Just as was the case for the binary model, the ensemble SD reaches peak around 40 ms later than the ensemble mean, at 167 ms compared to 127 ms. Figure 4F plots the probability distribution for the peak amplitudes of the 105 SPRs as the blue histogram and compares this with the prediction in red, obtained using Equation (3.6) with the measurements from Figure 4D. Again, the prediction is very accurate. With all three rate constants set to 60 s−1, the SPR peak amplitudes had a mean of 0.0423 and a cv of 0.339. We therefore conclude that the three-state model generates responses that are barely distinguishable from those of the binary activity model for these parameters applicable to WT mammalian rods.
Stochastic generation and decay of E*
For the simulations above, we have approximated the time course of PDE activity, E*(t), as being deterministic, according to Equation (4.1) and as used in recent modeling [11]. However, because of the modest number of E* molecules activated during the SPR, this approach has its limitations. For example, for a mean R* lifetime of τR* ≈ 70 ms and a mean rate of E* activation of νRE ≈ 300 s−1, the mean number of total E* subunits activated throughout the duration of the SPR would be nE* = τR* νRE ≈ 21, and the number active at any instant would be smaller than this. We therefore now follow Hamer et al. [6] and Reingruber and Holcman [9], who considered stochastic activation and shut-off of E* in their analyses of the graded R* shut-off model (i.e., Model 1), and we simulate the stochastic E* case applied to the three-state Model 3.
Figure 5 shows the results obtained when stochastic activation and shut-off of E* were added to the stochastic R* activity for Model 3. We used the same seed for the random number generator and thereby ensured that the sequence of R*(t) trials was identical to that obtained in Figure 4. Panel A illustrates the simulated E*(t) time course for the first 50 SPR trials, corresponding to the same R* events that were used to generate Figure 4C above, and shows substantial fluctuations in the number of activated E* subunits. Panel B plots the mean (blue) and SD (red) of E*(t) for the entire set of 105 trials. In addition, the dashed black trace plots the deterministic time course (see Equation (4.1)) predicted by convolving the mean R*(t) time course with an exponential decay, exp(−kE t), and it provides an excellent fit. Panel C plots the photocurrent responses for the first 50 SPR trials and therefore corresponds to Figure 4C but with the addition of stochastic generation and decay of E*. Note that at late time in the simulations (>400 ms), discrete levels of response are visible, corresponding to the presence of the last few E* subunits in each trial. Panel D plots the mean (blue) and SD (red) of the SPRs for the entire set of simulations. For the parameter values listed in Table 2 (and in particular for this value of βDark), the waveforms in panels C and D are quite similar to those in panels A and B, indicating that in this case the downstream reactions introduce relatively minor filtering of the E*(t) kinetics. The ensemble of SPRs had a mean peak amplitude of 0.0427 and a cv of 0.382. Panel E plots the relationship between peak SPR amplitude and the time to the low-activity state of R* for the sample traces shown previously in Figure 4D.
Figure 5.
Stochastic generation and shut-off of E* combined with the three-state R* model. The three-state model has been extended by simulating E* generation and shut-off as being stochastic, with the same rate constants (νRE and kE) as in the deterministic E* case in Figure 4. All other features of the simulation were identical to those in Figure 4, including the seed for the random number generator, so that the sequence of R*(t) simulations was the same. A: The first 50 simulations of E*(t). B: Mean (blue) and SD (red) of the E*(t) time-course for the 105 simulations. The dashed black trace is the predicted mean E*(t) time-course, obtained by convolving the mean R*(t) time-course with the deterministic E* impulse response, νRE exp(−kE t). C: The first 50 simulations of SPRs. D: Mean (blue) and SD (red) SPR time course for the 105 simulations. E: Relationship between SPR amplitude and tlow from Figure 4D. Symbols plot the measured SPR peak amplitudes, and the curve is a spline fit. F: Probability distribution of the peak amplitudes for the 105 simulated SPRs (blue). Minor peaks are visually resolvable for what we presume to be the first eight E*s. The dotted red trace is redrawn from the red trace in Figure 4F, while the continuous red trace is the convolution of the dotted trace with a Gaussian function representing variability of the number of E*s; see Text.
Figure 5F plots the probability distribution for the peak amplitudes of the 105 SPRs as the blue histogram. Note that this trace exhibits multiple minor peaks that we interpret as corresponding to discrete numbers of E* molecules being present at around the time of the peak; the first eight such peaks are visually apparent with this bin width. With fewer than 5 × 104 trials or with broader sampling bins, multiple peaks like this were not readily resolvable (not shown). To predict an expected distribution in this more complicated case with stochastic E* time-course, we began with the same approach as in Figure 4 and plotted the predicted histogram from Figure 4F as the dotted red trace in Figure 5F; this curve had its peak at 0.0415. Next, we assumed that stochastic fluctuations in E* numbers would contribute variability with a coefficient of variation 1/√nE*. Therefore, we convolved the dotted histogram with a Gaussian distribution having an SD of 0.0415/√21 to generate the continuous red trace in Figure 5F. Although we cannot provide a rigorous justification for this prediction, we note that it provides a reasonable description of the histogram obtained by simulation, apart from the absence of the multiple minor peaks. We conclude that the inclusion of stochastic steps for the generation and decay of E* leads on the one hand to SPR responses (Figure 5C) that more nearly resemble the shape of observed dim flash responses but on the other hand leads to only minor changes in the form of the ensemble mean, SD, and amplitude distribution.
Simulation of responses for mutant phenotypes
To investigate the wider applicability of our model, we repeated our simulations of the three-state R* activity case (Model 3) after adjusting the parameters to correspond to those expected for four mutations in the phototransduction cascade: Arr+/−, Arr−/−, GRK1+/−, and GRK1−/−. For the arrestin hemizygote, we halved μ and for the arrestin knockout, we set μ to zero. For the rhodopsin kinase hemizygote, we halved ν and for the rhodopsin kinase knockout, we set ν to zero. In the case of the three-state model, Figure 6 presents the ensemble mean responses for the WT case and these four mutants. As a check, we employed a deterministic description for R*tri(t), corresponding to Equations (3.4)–(3.5), as driving functions for numerical integration of the downstream reactions, and the computation time was then extremely rapid. This generated traces that were very similar (not shown), although the simulated responses were marginally smaller at around the peak of the mean SPR, presumably because of nonlinearity caused by greater response compression for larger SPRs than for smaller ones.
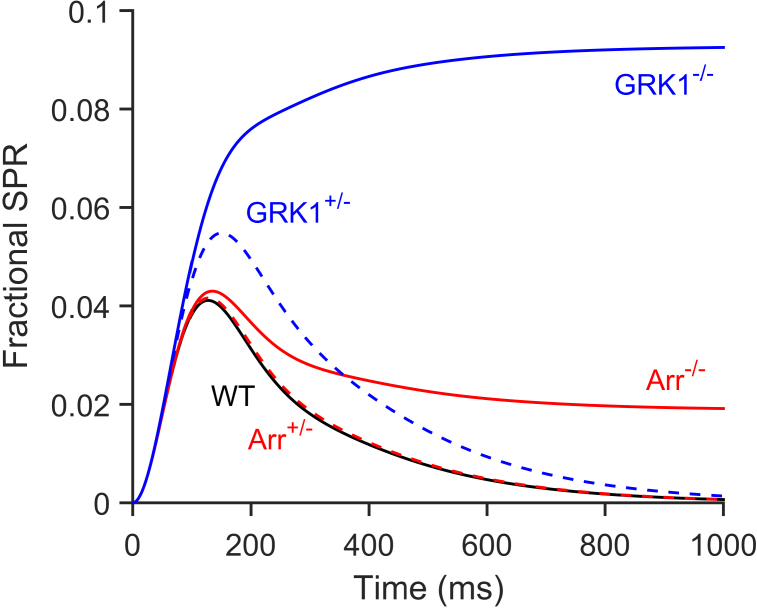
Figure 6.
Simulation of SPRs for mutant phenotypes using the three-state model. Simulated mean SPRs predicted by the three-state model are shown. The wild-type (WT) response is the same as in Figure 4E and was averaged from 105 simulations. The remaining traces were each averaged from 104 simulations. The hemizygote responses (dashed traces) for Arr+/− (red) and GRK1+/− (blue) were obtained by halving the respective rate parameter, μ or ν, from the value shown in Table 1. The knockout responses (solid traces) for Arr−/− (red) and GRK1−/− (blue) were obtained by reducing the respective rate parameter, μ or ν, to zero.
The ensemble mean response for the Arr+/− case (dashed red trace) differs only marginally from the WT response (black), whereas the Arr−/− response exhibits a final steady level of roughly half the peak amplitude of the WT SPR. The magnitude of this final level in the Arr−/− simulation depended on the choice of catalytic activity in the “low-activity” R* state, and a magnitude of roughly half the WT peak amplitude was obtained by setting ρlow = 0.1. The GRK1+/− mean response is about 30% larger than the WT response, whereas the GRK1−/− response rises to a plateau at a little over double the peak amplitude of the WT SPR. In qualitative terms, each of these simulated mean responses bears a remarkable similarity to the respective SPR responses reported in experiments on mouse rods; see, for example illustration 8 in [23] with data from [24,25] and illustration 1 in [26], with data from Dr. M.E. Burns. Responses very similar to those recorded from GRK1−/− rods are also obtained for rods in which rhodopsin has a truncated C-terminus [27,28].
Discussion
The combination of theoretical analysis and numerical simulation of the stochastic steps in three models of R* inactivation leads us to several conclusions. We shall deal first with the inadequacy of the conventional model of graded R* shut-off and then consider the implications of our results from the simulations using our new models of abrupt shut-off of R* activity.
Inadequacy of graded R* shut-off model
In our view, the conventional model, of a sequence of graded steps in the reduction of R* catalytic activity as recently simulated in [11] and duplicated here in Model 1, fails to provide an adequate description of the SPRs. Although the ensemble mean and SD resemble those reported in the literature, there are serious problems with two other aspects of the predictions.
First, the graded R* shut-off model is characterized by a small proportion (5%–10%) of simulated SPRs that exhibit either a plateau-like region or a very late peak (Figure 2D). This result appears to be an inevitable consequence of the assumption of an exponential decline in both the catalytic activity and the rate constant of phosphorylation. These occasional events do not correspond to published reports of mammalian rod SPRs. Their occurrence appears to generate the rather sharp peak in the amplitude distribution histogram of the simulated responses (Figure 2F), a feature that again has not been reported in published experiments.
Second, the required “gain parameter” needs to be set to roughly double the value predicted from other experiments. This gain parameter can be represented either by the product νRE βsub or equivalently by the “amplification constant” A = νRE βsub ncG [1], where ncG is the cooperativity of channel opening by cGMP, which is widely accepted to be ncG = 3. To obtain a peak amplitude for the WT SPRs of ~4% of the dark current, the gain product needed to be set to νRE βsub ≈ 19 s−2 both in the present work and in [11], corresponding to an amplification constant of A ≈ 57 s−2. We now compare the magnitude of this gain factor required for the graded R* shut-off simulations with three estimates derived from experiments, two using electrophysiological approaches and one using biochemical measurements.
Electrophysiological measurements of the amplification constant can be obtained by fitting the rising phase of responses to families of flash responses [1], and reported values in the literature range from A = 5–23 s−2 in experiments on rodent rods [29-33]. It is likely that some of the lower values resulted from excessive low-pass filtering [32], and our best guess for the true value of A in WT mouse rods is A ≈ 24 s−2, corresponding to νRE βsub ≈ 8 s−2. Recently, a second completely independent estimate for this gain parameter was obtained by Gross et al. [13] in their experiments on “rogue” SPRs in mouse GCAPs−/− rods, where they obtained a value that we have corrected to νRE βsub ≈ 9 s−2 (see Results section for Model 1). Biochemical experiments on amphibian rods [21] can be extrapolated (with caution) from room temperature to mammalian body temperature. For the rate of E* activation per R*, we anticipate an upper limit of νRE ≤ 350 s−1, but there are some uncertainties in estimating βsub. Although it is often thought that the Q10 for biochemical reactions may be around 2, the relevant Q10 for the PDE activity is likely to be smaller than this because the reaction rate is already quite near the limit for aqueous diffusion, and the increase in temperature of ~15 °C (from 22 °C to 37 °C) is unlikely to cause as much as a twofold increase in rate. Accordingly, we think that the amphibian value of kCAT/Km ≈ 4.4 × 108 M−1 s−1 [21] will convert to a mammalian value of <8 × 108 M−1 s−1. Substitution into the expression for βsub (see Appendix 1, Equation A.10) then gives βsub <0.033 s−1 / BP, where BP ≥ 1 is the cytoplasmic buffering power for cGMP. As a result we think that the biochemistry points to a realistic upper limit for the gain parameter of mouse rods of νRE βsub ≈ 12 s−2, provided that BP ≈ 1, but it would be lower than this if the cytoplasm exhibits any buffering power for cGMP. In summary, the three experimental estimates for νRE βsub are 8, 9, and <12 s−2, and these are all considerably smaller than the value of 19 s−2 that is required in the graded R* shut-off model to fit the observed SPR amplitude.
An alternative possibility that we tested, to try to avoid an excessive magnitude for the gain parameter in the graded shut-off model, was to slow the R* shut-off reactions from the values proposed by Gross et al. [11,13] to increase the mean R* lifetime. When we halved the parameters νmax and μ (thereby doubling the mean R* lifetime), and reduced νRE βsub to 12 s−2, the simulated SPRs had approximately the correct mean amplitude, but several other features were unusual and did not resemble physiological responses. The shape of the mean response had an unrealistic, considerably more “triangular” shape, with its width at half-height increased to ~400 ms, the tail phase was too slow, the time-to-peak was slightly increased to 132 ms, and the outlier events became much more prominent, appearing to be double the duration of those illustrated in Figure 2D. Accordingly, we rejected the possibility that the use of slower R* shut-off reactions could avoid the need for an excessive gain parameter in the graded R* shut-off model.
Binary and three-state R* shut-off models
Because of these deficiencies in the graded R* shut-off model, we investigated two variants of a scenario in which R* retains maximal activity until at least three phosphates have attached (referred to here as M = 3). In the first variant, the “binary R* shut-off” model, the R* activity remains at unity until it drops abruptly to zero when arrestin binds. In the second variant, the “three-state R* shut-off” model, we invoked an additional state in which the R* catalytic activity dropped to a low level, ρlow, before the binding of arrestin. These two variants of abrupt shut-off predicted nearly identical behavior to each other and one that conformed closely to the behavior reported in the literature (Figure 3 and Figure 4) in the case of parameters appropriate for WT mouse rods. However, the binary model was not able to account for the kinetics of the responses recorded from the rods of Arr−/− mice, and we required the additional low activity state. As shown in Figure 6, the simulated responses of the three-state model provided a satisfactory description of the ensemble mean SPRs simulated for the rods of four genotypes of mouse: Arr+/−, Arr−/−, GRK1+/−, and GRK1−/−. The height of the late plateau level in the Arr−/− simulations provided our basis for choosing the low-state activity as ρlow = 0.1.
Inclusion of stochastic reactions for E* generation and decay
The sample simulated SPR responses for both versions of the abrupt R* shut-off model, illustrated in Figure 3C and Figure 4C, exhibit a smooth time course for each response because all the steps other than R* formation and decay have been assumed to be deterministic. To approximate more closely to reality, we added stochastic simulation of the generation and decay of E* to the downstream reactions to generate the responses illustrated in Figure 5. With the inclusion of E* fluctuations, the sample SPR responses (Figure 5C) more nearly resembled the shape of observed dim flash responses. Incorporation of this stochastic description caused only minor change in the form of the ensemble mean, SD, and amplitude distribution of the SPRs.
R* time-course and integration time
Our modeling and simulations provide some new perspectives on the time course of R* activity and on the R* “integration time”, τR*, defined as the area under the time course of fractional R* activity, averaged across the simulated trials. In particular, we find that to achieve a sufficiently large SPR amplitude (~4% of the dark current) with a realistic magnitude of the gain product νRE βsub ≈ 7 s−2, the R* integration time τR* needs to be of the order of 60–70 ms, considerably larger than the value of 40 ms recently reported [11,34] for WT mouse rods. We think that our estimate applies irrespective of the shape of the decline in the ensemble mean R* activity, that is, that the estimate τR* ≈ 60–70 ms is dependent primarily on the downstream model and in particular on its “gain” parameters rather than on the stochastic model chosen to describe R* activity.
Ensemble mean R* time course
In the binary and three-state models, the time course of the ensemble mean R* activity does not approximate an exponential decay but instead exhibits an initial flat top as illustrated in Figure 3B and Figure 4B, with the early time course given by 1 – k tM+1; see Equation (2.4). To the extent that our models are realistic in this regard, this predicted time course provides some noteworthy insights.
First, there is an issue with the extraction of estimates of R* lifetime from “Pepperberg plot” experiments conducted on mutant mice with different expected rates of R* shut-off. In such experiments, the shift of the relationship between saturation time and log intensity has been used to provide estimates of R* lifetime [34]. However, the theoretical basis for that approach is predicated upon the assumption that R*’s mean activity declines exponentially; see [34]. Accordingly, we urge caution in the interpretation of these estimates of R* lifetime, unless one obtains clear evidence to support the assumption of an exponential decline in R* activity.
Another issue relates to the accuracy (or otherwise) of the “delayed Gaussian” prediction of the Lamb and Pugh [1] model for the onset phase of the rod’s response to bright flashes of light. That model ignored shut-off reactions and was therefore explicitly restricted to times shorter than the fastest shut-off reaction. Nevertheless, experiments on electroretinogram (ERG) scotopic a-wave responses in both human subjects and anesthetized mammals demonstrate that the predictions are quite accurate until at least 20–30 ms; see, for example illustrations 4C, D in [35]. If it were the case that the mean R* activity in the rods declined exponentially with a time constant of 40 ms, then the activity would have declined to ~60% after 20 ms, and one might expect the recorded photocurrents to deviate substantially from the Gaussian approximation. In contrast, if the mean R* activity in rods follows a time course similar to that illustrated in Figure 3B or Figure 4B and remains above 98% at 20 ms, then one might indeed expect the recorded photocurrents to conform reasonably closely to the Gaussian approximation until at least 20 ms.
These concepts are examined in Figure 7, where families of bright-flash responses have been predicted under five different assumptions concerning the shut-off reactions. When neither R* nor G*/PDE* shuts off, the predictions are shown in black, with the dashed traces giving the Gaussian approximation and the continuous traces giving the solution of the differential equations, with the R* shut-off rate constant kR* = 0 and with kE = 0. The green traces present the solution when G*/PDE* shuts off normally (kE specified in Table 2) but R* does not shut off (kR* = 0). Next, the red traces plot the predictions for the three-state model with parameters as in the Tables and in Figure 4. The blue traces plot the predictions for the case of exponential shut-off of R* with a time constant of kR*−1 = 40 ms.
Figure 7.
Predicted bright flash responses. Bright flash responses are predicted under five sets of conditions, indicated by the different colors. Black traces: No shut-off of either R* or G*/PDE*. The dashed black trace plots the Gaussian analytical expression of Lamb and Pugh [1]; the continuous black trace plots the solution of the differential equations (with kR* = 0 and kE = 0). Green traces: With normal shut-off of G*/PDE* but no shut-off of R*, achieved by setting kR* = 0 but leaving kE as in Table 2. Red traces: Three-state model of R* shut-off, with parameters as in the Tables and Figure 4. Here, the mean R* time course was calculated by numerical integration of the differential equations representing the reactions in Figure 1E. Blue traces: Exponential decay of mean R* activity, with a time constant kR*−1 = 40 ms. At higher intensities, some traces are obscured by overlying traces, which were plotted in the sequence black, green, red, blue. For all traces except the Gaussian analytical model, the downstream reactions were integrated using the lumped model versions of Equations (4.1) – (4.9), with the default parameters in Table 2 (except where kE was set to zero). In all cases, the gain factor νRE βsub was set to 7.2 s−2 (giving an amplification constant of A = 21.6 s−2), the flash duration was zero, and the flash intensities were 10, 30, 100, 300, 1000, 3000, and 10,000 photoisomerizations.
These calculations show that the Gaussian approximation fails to provide a perfect fit to the “no shut-off” case. They also show very close agreement between the green and red traces, for both of which G*/PDE* shuts of normally and where the only difference is whether R* fails to shut off (green) or whether it shuts off according to the three-state model (red). Importantly, the blue traces for an R* exponential lifetime of 40 ms are seen to deviate markedly from the other traces beyond about 20 ms after the flash. In other words, the observed good fit of the Lamb and Pugh theory to the ensemble of responses is more difficult to account for if the R* activity is assumed to drop exponentially with a time constant as short as 40 ms than is the case where the R* activity is assumed to be “flat-topped”.
Finally, as far as the R* time course is concerned, the previously assumed rapid exponential decline in R* activity has always been difficult to rationalize from a teleological perspective. Given that it took hundreds of millions of years for rod phototransduction to evolve the exquisite sensitivity needed for single-photon detection, it seems difficult to envisage what advantage there could be in causing that very high R* activity to begin declining (on average) from the instant that the R* is formed. Instead, one might envisage that it would be advantageous to maintain that hard-won catalytic activity for as long as possible and then to shut it off rapidly. This, in fact, is approximately what the binary and three-state models achieve.
Physical nature of the postulated low-activity state
To account for SPRs in the arrestin-knockout phenotype, we invoked a low-activity state that the R* molecule enters once several phosphates have been attached, but we did not specify the nature of this state. One possibility would be that once several phosphates have bound, the R* molecule undergoes a conformation change, with this altered conformation exhibiting not only greatly increased affinity for arrestin but also lowered activity toward transducin. Another possibility would be that once several phosphates have bound, the kinase (GRK1) binds more tightly than previously, making access for transducin more difficult. However, we emphasize that our simulations provide no prospect for distinguishing the precise nature of such a postulated low-activity state.
Comparison with biochemical estimates of R* affinity for transducin and for arrestin
The biochemical measurements of transducin affinity (and hence of assumed R* activity) reported by Gibson et al. [8] were obtained using macroscopic experiments in which the mean number of bound phosphates across different preparations was measured. Any interpretation about the probability distribution of rhodopsin molecules having specific numbers of attached phosphates is model-dependent, involving, for example, the rate constants of phosphorylation under the particular experimental conditions. In particular, one cannot assume that a graded relationship between measured affinity and mean number of bound phosphates in any way reflects a graded relationship between R* activity and actual numbers of bound phosphates. Future experiments will be needed to measure the catalytic activity of the molecule in its different phosphorylation states. As far as we can see, our postulated abrupt drop in R* activity is in no way inconsistent with the observed graded decline in biochemical affinity measured as a function of the mean number of phosphates. For the binding of arrestin, Vishnivetskiy et al. [16] measured the abundance of rhodopsins bearing different numbers of phosphates in their different samples, and they then measured the degree of arrestin binding to these samples. They found that high-affinity binding of arrestin required at least two bound phosphates and was maximal with three. Despite that finding, recent stochastic models of R* activity (including ours) have assumed that the binding of just two phosphates is insufficient to cause rapid binding of arrestin and that instead three are required.
Plateau-like SPRs
Plateau-like SPR events have been routinely observed experimentally when phosphorylation of R* is disrupted, as, for example, in GRK1−/− rods [25], in rhodopsin C-terminus mutants [27,28] and in certain cases of substitution of the Ser and Thr residues in the carboxy terminus [36,37]. In addition, they are also observed in WT rods in a very small proportion (~0.3%) of flash presentations, and as such they have been termed “aberrant” or “rogue” responses [13,38]. Typically, these aberrant SPRs exhibit a plateau level around 50% higher than the normal peak, followed by an abrupt recovery to baseline. To explain these experimentally observed aberrant responses in the conventional model of graded reduction in R* activity, it has been necessary to assume that phosphorylation fails to occur for a small minority of R* events. However, in terms of our three-state stochastic model, an alternative explanation would be that phosphorylation occurs normally but that the subsequent transition to the low-activity state is delayed for the order of seconds. It is difficult to speculate on a possible molecular mechanism, but it might involve either a change to or a degradation of a tiny fraction of the rhodopsin molecules or alternatively a disruption of some component with which R* interacts.
In conclusion, we think that our simulations rule out the conventional graded shut-off model as providing a realistic description of experimentally recorded SPRs. And while we emphasize that our simulations do not provide any direct evidence that the activity of R*·P(n) remains unattenuated up to n = 3, we think they demonstrate that a model of this kind is capable of accounting for many of the properties of experimentally recorded SPRs.
Our model opens up some exciting possibilities for the future. The analysis of additional experimental data may permit refinement of the model and provide improved estimates of the underlying physical parameters. In particular, we think that such future refinements may readily account for the different classes of SPRs recorded by Azevedo et al. [37] using mice with Ser-only and Thr-only C-terminus mutations of rhodopsin. Furthermore, now that we have a deterministic expression (Equations (3.4)–(3.5)) for the mean R*(t) time course in the three-state shut-off model, it should be straightforward to extend the model to account for more complicated aspects of light adaptation, including modulation by recoverin, modulation of channel activity, and so on.
Acknowledgments
The authors declare no conflict of interest. This work was supported by award number R01EY023603 from the US National Eye Institute.
Appendix 1.
To access the data, click or select the words “Appendix 1.”
References
- 1.Lamb TD, Pugh EN., Jr A quantitative account of the activation steps involved in phototransduction in amphibian photoreceptors. J Physiol. 1992;449:719–57. doi: 10.1113/jphysiol.1992.sp019111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baylor DA, Lamb TD, Yau KW. Responses of retinal rods to single photons. J Physiol. 1979;288:613–34. [PMC free article] [PubMed] [Google Scholar]
- 3.Rieke F, Baylor DA. Origin of reproducibility in the responses of retinal rods to single photons. Biophys J. 1998;75:1836–57. doi: 10.1016/S0006-3495(98)77625-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Field GD, Rieke F. Mechanisms regulating variability of the single photon responses of mammalian rod photoreceptors. Neuron. 2002;35:733–47. doi: 10.1016/s0896-6273(02)00822-x. [DOI] [PubMed] [Google Scholar]
- 5.Whitlock GG, Lamb TD. Variability in the time course of single photon responses from toad rods: termination of rhodopsin’s activity. Neuron. 1999;23:337–51. doi: 10.1016/s0896-6273(00)80784-9. [DOI] [PubMed] [Google Scholar]
- 6.Hamer RD, Nicholas SC, Tranchina D, Liebman PA, Lamb TD. Multiple steps of phosphorylation of activated rhodopsin can account for the reproducibility of vertebrate rod single-photon responses. J Gen Physiol. 2003;122:419–44. doi: 10.1085/jgp.200308832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hamer RD, Nicholas SC, Tranchina D, Lamb TD, Jarvinen JL. Toward a unified model of vertebrate rod phototransduction. Vis Neurosci. 2005;22:417–36. doi: 10.1017/S0952523805224045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gibson SK, Parkes JH, Liebman PA. Phosphorylation modulates the affinity of light-activated rhodopsin for G protein and arrestin. Biochemistry. 2000;39:5738–49. doi: 10.1021/bi991857f. [DOI] [PubMed] [Google Scholar]
- 9.Reingruber J, Holcman D. The dynamics of phosphodiesterase activation in rods and cones. Biophys J. 2008;94:1954–70. doi: 10.1529/biophysj.107.116202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Caruso G, Bisegna P, Lenoci L, Andreucci D, Gurevich VV, Hamm HE, DiBenedetto E. Kinetics of rhodopsin deactivation and its role in regulating recovery and reproducibility of rod photoresponse. PLOS Comput Biol. 2010;6:e1001031. doi: 10.1371/journal.pcbi.1001031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gross OP, Pugh EN, Jr, Burns ME. Calcium feedback to cGMP synthesis strongly attenuates single-photon responses driven by long rhodopsin lifetimes. Neuron. 2012;76:370–82. doi: 10.1016/j.neuron.2012.07.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Reingruber J, Pahlberg J, Woodruff ML, Sampath AP, Fain GL, Holcman D. Detection of single photons by toad and mouse rods. Proc Natl Acad Sci USA. 2013;110:19378–83. doi: 10.1073/pnas.1314030110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gross OP, Pugh EN, Jr, Burns ME. Spatiotemporal cGMP dynamics in living mouse rods. Biophys J. 2012;102:1775–84. doi: 10.1016/j.bpj.2012.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Invergo BM, Dell’Orco D, Montanucci L, Koch K-W, Bertranpetit J. A comprehensive model of the phototransduction cascade in mouse rod cells. Mol Biosyst. 2014;10:1481–9. doi: 10.1039/c3mb70584f. [DOI] [PubMed] [Google Scholar]
- 15.Astakhova L, Firsov M, Govardovskii V. Activation and quenching of the phototransduction cascade in retinal cones as inferred from electrophysiology and mathematical modeling. Mol Vis. 2015;21:244–63. [PMC free article] [PubMed] [Google Scholar]
- 16.Vishnivetskiy SA, Raman D, Wei J, Kennedy MJ, Hurley JB, Gurevich VV. Regulation of arrestin binding by rhodopsin phosphorylation level. J Biol Chem. 2007;282:32075–83. doi: 10.1074/jbc.M706057200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Baylor DA, Hodgkin AL, Lamb TD. The electrical response of turtle cones to flashes and steps of light. J Physiol. 1974;242:685–727. doi: 10.1113/jphysiol.1974.sp010731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fuortes MG, Hodgkin AL. Changes in time scale and sensitivity in the ommatidia of Limulus. J Physiol. 1964;172:239–63. doi: 10.1113/jphysiol.1964.sp007415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gillespie DT. Exact stochastic simulation of coupled chemical reactions. J Phys Chem. 1977;81:2340–61. [Google Scholar]
- 20.Iserles A. A First Course in the Numerical Analysis of Differential Equations. Cambridge University Press, 1996. [Google Scholar]
- 21.Leskov IB, Klenchin VA, Handy JW, Whitlock GG, Govardovskii VI, Bownds MD, Lamb TD, Pugh EN, Jr, Arshavsky VY. The gain of rod phototransduction: Reconciliation of biochemical and electrophysiological measurements. Neuron. 2000;27:525–37. doi: 10.1016/s0896-6273(00)00063-5. [DOI] [PubMed] [Google Scholar]
- 22.Koch K-W, Dell’Orco D. A calcium-relay mechanism in vertebrate phototransduction. ACS Chem Neurosci. 2013;4:909–17. doi: 10.1021/cn400027z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pugh EN Jr, Lamb TD. Phototransduction in vertebrate rods and cones: molecular mechanisms of amplification, recovery and light adaptation. In: Stavenga DG, de Grip WJ, Pugh EN Jr, editors. Handbook of Biological Physics. Vol. 3, Molecular Mechanisms of Visual Transduction. Amsterdam: Elsevier; 2000. p. 183–255. [Google Scholar]
- 24.Xu J, Dodd RL, Makino CL, Simon MI, Baylor DA, Chen J. Prolonged photoresponses in transgenic mouse rods lacking arrestin. Nature. 1997;389:505–9. doi: 10.1038/39068. [DOI] [PubMed] [Google Scholar]
- 25.Chen C-K, Burns ME, Spencer M, Niemi GA, Chen J, Hurley JB, Baylor DA, Simon MI. Abnormal photoresponses and light-induced apoptosis in rods lacking rhodopsin kinase. Proc Natl Acad Sci USA. 1999;96:3718–22. doi: 10.1073/pnas.96.7.3718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fu Y, Yau K-W. Phototransduction in mouse rods and cones. Eur J Phys. 2007;454:805–19. doi: 10.1007/s00424-006-0194-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen J, Makino CL, Peachey NS, Baylor DA, Simon MI. Mechanisms of rhodopsin inactivation in vivo as revealed by a COOH-terminal truncation mutant. Science. 1995;267:374–7. doi: 10.1126/science.7824934. [DOI] [PubMed] [Google Scholar]
- 28.Kraft TW, Allen DE, Petters RM, Peng Y-W, Wong F. Altered light responses of single rod photoreceptors in transgenic pigs expressing P347L or P347S rhodopsin. Mol Vis. 2005;11:1246–56. [PubMed] [Google Scholar]
- 29.Zhang X, Wensel TG, Kraft TW. GTPase regulators and photoresponses in cones of the eastern chipmunk. J Neurosci. 2003;23:1287–97. doi: 10.1523/JNEUROSCI.23-04-01287.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nikonov SS, Kholodenko R, Lem J, Pugh EN., Jr Physiological features of the S- and M-cone photoreceptors of wild-type mice from single-cell recordings. J Gen Physiol. 2006;127:359–74. doi: 10.1085/jgp.200609490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen CK, Woodruff ML, Chen FS, Cillufo MC, Fain GL. Replacing the rod with the cone transducin α subunit decreases sensitivity and accelerates response decay. J Physiol. 2010;588:3231–41. doi: 10.1113/jphysiol.2010.191221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Woodruff ML, Rajala A, Fain GL, Rajala RVS. Modulation of mouse rod photoreceptor responses by Grb14 protein. J Biol Chem. 2014;289:358–64. doi: 10.1074/jbc.M113.517045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vinberg F, Turunen TT, Heikkinen H, Pitkänen M, Koskelainen A. A novel Ca2+-feedback mechanism extends the operating range of mammalian rods to brighter light. J Gen Physiol. 2015;146:307–21. doi: 10.1085/jgp.201511412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gross OP, Burns ME. Control of rhodopsin’s active lifetime by arrestin-1 expression in mammalian rods. J Neurosci. 2010;30:3450–7. doi: 10.1523/JNEUROSCI.5391-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lamb TD, Pugh EN., Jr Phototransduction, dark adaptation, and rhodopsin regeneration. The Proctor Lecture. Invest Ophthalmol Vis Sci. 2006;47:5137–52. doi: 10.1167/iovs.06-0849. [DOI] [PubMed] [Google Scholar]
- 36.Mendez A, Burns ME, Roca A, Lem J, Wu LW, Simon MI, Baylor DA, Chen J. Rapid and reproducible deactivation of rhodopsin requires multiple phosphorylation sites. Neuron. 2000;28:153–64. doi: 10.1016/s0896-6273(00)00093-3. [DOI] [PubMed] [Google Scholar]
- 37.Azevedo AW, Doan T, Moaven H, Sokal I, Baameur F, Vishnivetskiy SA, Homan KT, Tesmer JJG, Gurevich VV, Chen J, Rieke F. C-terminal threonines and serines play distinct roles in the desensitization of rhodopsin, a G protein-coupled receptor. eLife. 2015;4:e05981. doi: 10.7554/eLife.05981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kraft TW, Schnapf JL. Aberrant photon responses in rods of the macaque monkey. Vis Neurosci. 1998;15:153–9. doi: 10.1017/s0952523898151131. [DOI] [PubMed] [Google Scholar]