Abstract
Glucan phosphatases are a family of enzymes that are functionally conserved at the enzymatic level in animals and plants. These enzymes bind and dephosphorylate glycogen in animals and starch in plants. While the enzymatic function is conserved, the glucan phosphatases employ distinct mechanisms to bind and dephosphorylate glycogen or starch. The founding member of the family is a bimodular human protein called laforin that is comprised of a carbohydrate binding module 20 (CBM20) followed by a dual specificity phosphatase domain. Plants contain two glucan phosphatases: Starch EXcess4 (SEX4) and Like Sex Four2 (LSF2). SEX4 contains a chloroplast targeting peptide, dual specificity phosphatase (DSP) domain, a CBM45, and a carboxy-terminal motif. LSF2 is comprised of simply a chloroplast targeting peptide, DSP domain, and carboxy-terminal motif. SEX4 employs an integrated DSP-CBM glucan-binding platform to engage and dephosphorylate starch. LSF2 lacks a CBM and instead utilizes two surface binding sites to bind and dephosphorylate starch. Laforin is a dimeric protein in solution and it utilizes a tetramodular architecture and cooperativity to bind and dephosphorylate glycogen. This chapter describes the biological role of glucan phosphatases in glycogen and starch metabolism and compares and contrasts their ability to bind and dephosphorylate glucans.
Keywords: Glucan phosphatase, Starch, Glycogen, Reversible phosphorylation, Dual specificity phosphatase, Glucan interactions
Glycogen and starch
All life forms strive to maintain an internal cellular environment that is highly organized and more energy rich than the often harsh external conditions. Osmotic and energy balances are critical to this internal homeostasis, and their disruption can lead to catastrophic failure and death. To counter this threat, life forms have developed numerous coping mechanisms that include the ability to store and use energy. The vast majority of organisms, to one degree or another, use glucans as a repository of glucose to be stored in times of energy excess and to be used to generate energy in times of energy depletion.
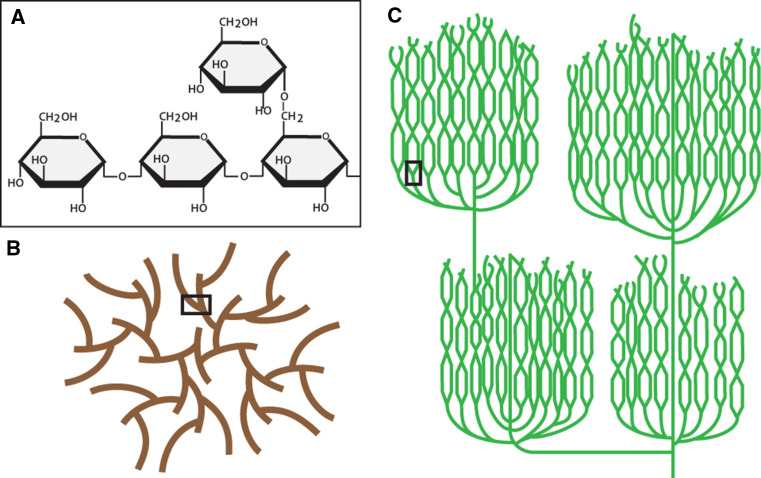
Glucans comprise two broad, major forms—water-soluble glycogen and water-insoluble, semi-crystalline starch. Glycogen is found in opisthokonts and other diverse eukaryotic lineages as well as in most prokaryotes, while the distribution of starch is largely restricted to the Archaeplastida—the green Chloroplastida, the red Rhodophyceae, and the blue–green Glaucophyta—and other eukaryotes with Archaeplastida-derived plastids via secondary endosymbiosis. Both macromolecules are composed of α-1,4 linked glucose residues with α-1,6 linked branch points (Fig. 1a). Both the chain length and the frequency of branching vary between glucan types and these two characteristics are typically associated with solubility, with longer and more scantly branched chains decreasing water solubility. Glucan polymerization is catalyzed by glycogen synthase or starch synthases that use either UDP-glucose or ADP-glucose to add successive glucose residues in an α-1,4 manner to the non-reducing end of the growing polysaccharide. The incorporation of branches is achieved by glycogen branching enzyme or starch branching enzymes that transfer a segment of an existing α-1,4 chain to form an α-1,6 linkage, resulting in an additional non-reducing end on which synthases can act.
Fig. 1.
The structure of glycogen and starch. a Starch and glycogen are both glucose polymers with α-1,4-linked linear chains and α-1,6-linked branch points. b Schematic diagram of a glycogen β-particle, 20–25 nm in diameter. Short chains of 12–14 glucose units per chain and continuous branching render glycogen water-soluble. c Schematic diagram of amylopectin, the major component of starch. Branching is discontinuous, with branches clustered within the amorphous layer. The long, linear chains contain 20+ glucose units per chain and form double helices producing the insoluble, crystalline lamellae. Starch granules can reach up to 100 μm in diameter. The black boxes in (b) and (c) delineate branch points as in (a)
Glycogen comprises α-1,4 linked, helical chains averaging 13 glucose residues in length with roughly two α-1,6 branch point per chain, approximately 7 % of all glycosidic bonds, yielding glycogen particles containing up to 55,000 glucose residues at a molecular mass of approximately 107 (Fig. 1b) [1–3]. These particles, termed β-particles, are 20–25 nm in diameter which, in some tissues such as liver, associate to form larger α-particles [4]. β-Particle size is limited by increasing molecular density at the periphery of growing granules that hinders the activity of biosynthetic enzymes towards glycogen [5, 6]. Glycogen breakdown is effected by the concerted action of glycogen phosphorylase, which releases glucose-1-phosphate from linear α-1,4 non-reducing ends, and glycogen debranching enzyme, which removes α-1,6 branch points via two distinct catalytic activities, thereby enabling glycogen phosphorylase to continue on the now-linear chain. Thus, the highly branched, soluble conformation of glycogen with its many enzyme-accessible non-reducing ends renders it an exceptional energy source for the rapid release of glucose in response to acute energy demands.
In contrast, both the structure and the metabolism of starch are more complex than for glycogen. Starch structure and metabolism are best described in members of the green lineage. Thus, this discussion will focus on that lineage, with differences in other lineages specifically stated. Starch in the green lineage is located within plastids whereas in the Glaucophyta, Rhodophyceae, and most other starch-accumulating lineages it is located in the cytosol. This difference in localization has consequences on the identity of the glucose donor used by starch synthases. Synthases operating in the cytosol utilize UDP-glucose, produced by the ancestral eukaryotic UDP-glucose pyrophosphorylase, whereas synthases producing starch in plastids utilize ADP-glucose, synthesized by ADP-glucose pyrophosphorylase of cyanobacterial origin [7].
Starch structure
A further complexity in starch is that it is composed of two different poly-glucans, amylopectin and amylose. The addition of glucose residues to the non-reducing ends of an amylopectin granule is, as with glycogen, accompanied by the action of branching enzymes introducing α-1,6 linked branches. However, specific debranching enzymes—isoamylases—acting during synthesis remove many of the α-1,6 branches incorporated by branching enzymes. This action results in the typical amylopectin architecture where a highly branched amorphous layer (similar to glycogen) alternates with a non-branched crystalline layer of elongated chains in a concentric manner (Fig. 1c). Only a small proportion of these crystalline, linear chains emerge to propagate the subsequent amorphous layer, resulting in a discontinuous branching pattern for amylopectin, unlike the continuous branching pattern observed for glycogen [8]. Approximately 5 % of glycosidic bonds are α-1,6 linked, with the chains of the crystalline lamellae typically containing 12–15 glucosyl residues [7]. Interestingly, deficiency in isoamylases during synthesis leads to the formation of phytoglycogen, soluble glucans that in most respects display similar properties to glycogen β-particles in both granule size and branching pattern [5, 8]. The second, minor component of starch, amylose, is polymerized by a specific starch synthase isoform called granule-bound starch synthase and amylose cannot form without amylopectin as a prerequisite. Amylose, unlike either amylopectin or glycogen, is seldom branched; it contributes both to the crystallinity of starch and serves to increase the efficiency—i.e., density—of glucose storage within the granule.
The glucan chains of the starch crystalline lamellae form left-handed double helices that exclude water from their center and come together in one of two (or a mixture of) major allomorphs, designated A-type and B-type [9]. The A-type allomorph, typically found in cereal endosperm starches, comprises densely packed crystalline helices that largely omit water. Conversely, the B-type allomorph, typically found in storage organs of dicots and as transitory starch in photosynthetic tissue, comprises helices packed in a hexagonal pattern that allows for water channels between helices but nevertheless retains crystallinity. As a result of the alternating crystalline and amorphous layers and the discontinuous branching pattern of amylopectin that prevents crowding, a starch granule, unlike glycogen and phytoglycogen, is essentially unlimited in size with granules becoming as large as 100 μm in diameter [7].
Starch functions
In the Chloroplastida, starch serves two broad biological functions that can affect its metabolism. In photosynthetic (or source) tissue, energy derived from the light-dependent reactions of photosynthesis during the day is used to assimilate carbon dioxide through the Calvin-Benson cycle; the assimilated carbon generally then has two main fates. The first is the synthesis of sucrose that serves the plant as an energy-transport molecule to deliver carbon to non-photosynthetic (or sink) tissue, in most multicellular green lineages. This sucrose may then be used as both an immediate energy supply and as a source of carbon for the production of starch in storage organs, such as tubers, roots, and seeds. The second main fate is the translocation of triose-phosphates into plastids that are used to generate ADP-glucose and, eventually, starch. Starch in source tissue is termed transitory starch since it is accumulated throughout the light period and then degraded during the dark period to provide energy for continued growth and metabolism when the production of ATP via photophosphorylation is halted. Thus, two broad pools of starch exist in most plants—transitory starch in source tissue and storage starch in sink tissue.
Starch catabolism
Of these two pools, the more metabolically active is the transitory starch of source tissue. Immediately following the onset of the dark period, transitory starch metabolism shifts from synthesis to degradation, the process of which has been best described in the model dicot angiosperm Arabidopsis thaliana. The major enzymes involved in starch degradation also differ from those involved in glycogenolysis. Firstly, hydrolysis primarily involves β-amylases—exo-amylases—that release maltose. β-Amylases, however, cannot degrade past α-1,6 branch points and thus isoamylase isoform 3—distinct from the isoamylase isoforms operating during granule synthesis—is required to hydrolyze the branches directly releasing short, linear oligosaccharides. This mechanism is unlike the two-step glycogen debranching enzyme action. Secondly, the crystallinity of starch prevents such access by the enzymes of starch degradation. This is in contrast to glycogen, in which glycogen phosphorylase and glycogen debranching enzyme can easily access the non-reducing ends of soluble glucan chains.
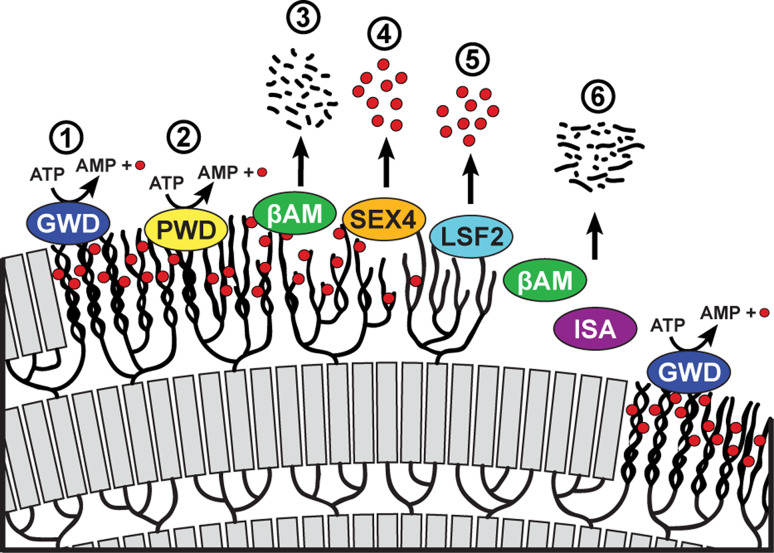
To overcome this crystallinity issue, Archaeplastida (and consequently the starch-accumulating lineages derived via secondary endosymbiosis involving an Archaeplastida endosymbiont) have developed a system of reversible glucan phosphorylation that serves to disrupt the crystalline lamellae. Enzymes termed glucan-water dikinases hydrolyze ATP, transferring the β-phosphate to a glucosyl residue within the glucan chain while releasing the γ-phosphate as orthophosphate to produce AMP (Fig. 2) [10]. Within the green lineage, the helices that form the crystalline lamellae are phosphorylated by glucan-water dikinase (GWD) on the hydroxyl groups at the C6-position of the glucosyl residue. A second dikinase, phosphoglucan-water dikinase (PWD), phosphorylates the C3-position but requires the prior C6 phosphorylation by GWD. Phosphorylation leads to disruption of the helices and increases the solubility of the glucan chains, thereby allowing β-amylases access to release maltose. β-Amylases, however, cannot hydrolyze glucan chains at or close to sites of phosphorylation and thus dephosphorylation is required to continue the degradative process. Dephosphorylation, in the green lineage, is effected by two phosphatases, known as SEX4 (s tarch-ex cess 4) and LSF2 (l ike-s ex 4 2). SEX4 preferentially dephosphorylates the C6-position of glucans while LSF2 specifically dephosphorylates the C3-position (Fig. 2) [11–13].
Fig. 2.
Starch degradation requires multi-step cycles of reversible phosphorylation. 1 Glucan, water dikinase (GWD) transfers the β-phosphate of ATP to the C6 position of glucosyl moieties within the starch lamellae. 2 Phosphoglucan, water dikinase (PWD) then phosphorylates the C3 position of glucosyl residues. 3 Phosphorylation leads to the disruption of the helices and solubilization of the starch surface, giving β-amylases (βAM) access to degrade the linear chains. 4 SEX4 then removes the C6- and C3-bound phosphate, with a preference for the former position, while 5 LSF2 releases C3-bound phosphate. 6 Dephosphorylation permits β-amylases to continue to degrade the linear chains. Isoamylase (ISA) is required for debranching within the amorphous layer. Another cycle begins with phosphorylation of the next crystalline layer by GWD. Red circles represent phosphate molecules
The centrality of each enzyme in the processive degradation of starch is demonstrated by the fact that mutant plants lacking (or with reduced) activity at any of these steps—β-amylase; isoamylase; glucan, water dikinase; or glucan phosphatase activities—invariably produces a starch-excess (or sex) phenotype [8]. Mutants lacking GWD or PWD display severe and mild sex phenotypes, respectively. The severity of GWD mutants resembles the sex phenotype of plants lacking β-amylase, indicating that GWD-mediated phosphorylation is most critical for starch degradation. Similarly, Arabidopsis mutants lacking SEX4 also display a severe sex phenotype with reduced growth [14, 15] as well as the accumulation during the dark period of phospho-oligosaccharides released via hydrolysis by α-amylase—an endo-amylase—and isoamylase [16]. Conversely, LSF2 mutants are largely indistinguishable from wild-type, though sex4-lsf2 double mutant plants display an exacerbated sex phenotype relative to sex4-only mutants [13].
The unique architecture of starch comprised of alternating layers of crystalline and amorphous lamellae requires the activity of isoamylase isoforms during amylopectin synthesis [17]. The alternating crystallinity of starch requires successive rounds of solubilization by phosphorylation of the crystalline layer, hydrolysis by β-amylases, and dephosphorylation, followed finally by further β-amylase digestion and isoamylase debranching of the amorphous layer (Fig. 2). Disruption at any point in this cycle leads to a starch-excess phenotype since each step in the process requires the action of the preceding activity.
Although reversible phosphorylation is a critical aspect of starch metabolism in green plants, the first glucan phosphatase described was in fact the human phosphatase laforin. It was identified as a gene mutated in a rare, progressive, and fatal neurodegenerative epilepsy known as Lafora disease, from which the enzyme’s name derives [18]. Interestingly, glycogen also contains small amounts of covalently attached phosphate—present at the C2-, C3-, and C6-positions [19, 20]—that are removed by laforin. Although neither the enzyme(s) responsible for phosphate incorporation into glycogen nor the role of phosphate in glycogen are known, laforin mutations in Lafora disease patients result in hyper-phosphorylated, aberrantly branched glucan with elongated chains that form insoluble, amylase-resistant bodies termed Lafora bodies.
Glucan phosphatase family
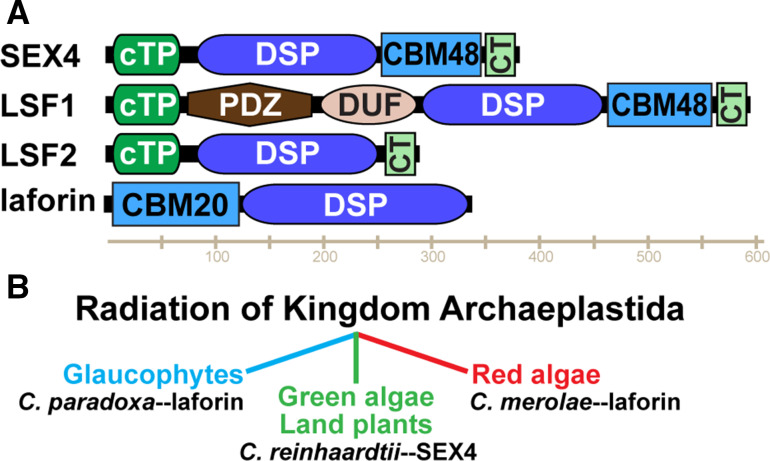
The discovery of glucan phosphatases began in the Gatehouse laboratory where a protein in Arabidopsis (gene locus At3g52180) was shown to be a phosphatase, but its endogenous substrate was unknown [21]. This finding was followed by the identification of a carbohydrate-binding module (CBM) in At3g52180 by the Moorhead laboratory as well as the Zeeman and Smith laboratories with each group demonstrating that At3g52180 binds starch and that it localizes to chloroplasts [14, 22]. The Zeeman and Smith laboratories demonstrated that mutation of At3g52180 produces a starch excess phenotype whereby plants synthesize starch but are unable to efficiently degrade it, and they designated the protein Starch EXcess4 (SEX4) [14]. Concurrently, we published that the human protein laforin possesses the unique ability to release phosphate from phospho-glucans, establishing laforin as the founding member of the glucan phosphatases [18]. Laforin is a bimodular protein that contains a CBM followed by a dual specificity phosphatase (DSP) domain (Fig. 3 a). SEX4 possesses a chloroplast targeting peptide (cTP), followed by a DSP domain, a CBM domain, and a carboxy-terminal (CT) motif that is integral for SEX4 enzymatic activity and structural stability (Fig. 3a). We subsequently demonstrated that SEX4 dephosphorylates amylopectin and defined a glucan phosphatase as a protein containing a CBM and DSP domain within the same polypeptide [18, 23]. Additionally, we demonstrated that human laforin partially complements the sex4 Arabidopsis mutant phenotype, demonstrating that laforin and SEX4 are functional equivalents at the enzymatic level [23]. Although laforin and SEX4 share a common enzymatic function and similar domains, the two proteins are not orthologs because: (1) their CBMs are of different families (discussed below) and (2) the DSP and CBM are in opposite orientations (Fig. 3a). A SEX4 ortholog contains a DSP domain followed by a CBM, while a laforin ortholog has a CBM followed by a DSP domain.
Fig. 3.
The domain structure and evolution of the glucan phosphatase family. a SEX4 contains a chloroplast-targeting peptide (cTP) at its N-terminus, followed by a dual-specificity phosphatase domain (DSP), a carbohydrate binding module (CBM) of the CBM48 family, and a C-terminal (CT) motif. LSF1 also contains a cTP at its N-terminus, a PDZ protein interaction domain, a domain of unknown function (DUF), a DSP domain, CBM48, and CT motif. LSF2 contains a cTP, DSP domain and CT motif but lacks a CBM. Laforin is composed of an N-terminal CBM of the CBM20 family and a C-terminal DSP domain. b Laforin is conserved in the glaucophyte lineage of Archaeplastida represented by Cyanophora paradoxa and in the red algal lineage represented by Cyanidioschyzon merolae. In contrast, green algae and land plants such as Chlamydomonas reinhardtii possess SEX4 orthologs
Bioinformatics searches for phosphatases similar to SEX4 and laforin uncovered Like SEX Four1 (LSF1) and Like SEX Four2 (LSF2) [13, 24]. LSF1 is comprised of a cTP, a PDZ protein interaction domain, domain of unknown function, DSP domain, CBM, and CT motif (Fig. 3a). Alternatively, LSF2 lacks a CBM and is comprised of only a cTP, DSP domain and CT motif. LSF1 and LSF2 each bind starch and are localized to chloroplasts [13, 24]. Mutations in LSF1 result in a starch excess phenotype though less extreme than the sex4 mutant phenotype. Mutations in LSF2 result in elevated levels of starch-bound phosphate, but no starch excess phenotype. Plants with mutations in either LSF1 or LSF2 in combination with SEX4 result in an exacerbated starch excess phenotype.
The phosphatase domains of laforin, SEX4, LSF1, and LSF2 are all members of the Protein Tyrosine Phosphatase (PTP) superfamily [25, 26]. The PTP superfamily includes a clade of sequence diverse phosphatases called the dual specificity phosphatases (DSPs) that dephosphorylate phospho-Ser, -Thr, and -Tyr residues of proteinaceous substrates, and other phosphatases that dephosphorylate phospho-lipids, phospho-nucleic acids, and phospho-glucans [25–27]. The glucan phosphatases fall within this diverse group of DSPs. The PTPs and DSPs all utilize a catalytic cysteine residue at the base of the active site CX5R motif to perform nucleophilic attack on the phosphorous atom of the substrate [28]. The human proteome contains ~65 DSPs and the plant proteome ~22 DSPs [28, 29].
Unlike DSP domains, CBMs are non-enzymatic domains that are appended to enzymatic domains. A CBM is a contiguous amino acid sequence with a conserved tertiary fold that possesses carbohydrate-binding ability. CBMs are curated within the carbohydrate active enzymes (CAZy) database (http://www.cazy.org) that defines CAZymes as a diverse collection of enzymes that synthesize and degrade a heterogeneous group of substrates that includes oligosaccharides, polysaccharides, and glycoconjugates [30]. The database defines nearly 300 families of catalytic and ancillary modules that include glycoside hydrolases, glycosyl transferases, polysaccharide lyases, carbohydrate esterases, and non-enzymatic carbohydrate binding modules [31]. CBMs are categorized based on amino acid sequence into families with each family sharing a common structural fold, but not always binding specificity. They are currently classified into 71 different families that bind to divergent carbohydrates such as starch, chitin, cellulose, xylan, and others [32].
The laforin CBM is of the CBM20 family while SEX4 and LSF1 both contain a CBM48. The CBM20 and CBM48 domains are typically 90–120 amino acids long and they are evolutionarily related [33, 34]. While there are no invariant residues shared by all CBM20 or CBM48 domains, they share a conserved β-strand-rich tertiary fold and there are highly conserved aromatic amino acids that typically directly interact with the glucan [33]. The CBM20 and CBM48 families are related based on primary amino acid sequence, conserved binding regions, and similar tertiary structure. Additionally, the CBM20 and CBM48 domains do not exclusively target starch or glycogen, and there are examples from each family that target either glucan [34, 35]. The evolutionary relatedness of these CBM families have been extensively analyzed and intermediates between the CBM48 and CBM20 families that contain conserved elements of both families have been identified [33, 34].
We initially defined a glucan phosphatase as a protein containing both a CBM and DSP domain [23]. However, LSF2 lacks a CBM domain yet still binds and dephosphorylates starch [13, 36]. LSF2 utilizes surface binding sites (SBSs; also referred to as secondary binding sites) instead of a CBM to bind starch [36]. While CBMs are defined domains within a contiguous polypeptide, SBSs are short stretches of non-contiguous amino acids that come together to impart glucan binding. SBSs are an emerging theme among glucan binding proteins, particularly among the glycoside hydrolases [37, 38]. SBSs are difficult to identify by sequence analysis and are typically identified after the X-ray or NMR structure is determined of the protein bound to a glucan. While there is not a strict SBS signature motif, aromatic residues often serve as the basis for the glucan interaction. Glycoside hydrolases have been identified that contain a CBM and SBSs, such as SusG [39] and others that possess only SBSs, such as barley α-amylase [40, 41] and yeast glucoamylase [42]. Thus, LSF2 does not contain a classical CBM and is not classified under the CAZy system, but it is a carbohydrate-modifying enzyme. While LSF2 utilizes two SBSs, no SBSs have been identified in laforin, SEX4, or LSF1 to date.
Conservation of glucan phosphatases
The glucan phosphatases have a varied and interesting evolutionary lineage that can be traced back to the radiation of Archaeplastida. First, a clear definition of what constitutes a SEX4 versus laforin orthologs must be defined. The SEX4 gene encodes a DSP domain followed by a CBM, while the laforin gene (EPM2A in vertebrates) encodes a CBM followed by a DSP domain [23, 43]. The green lineage (i.e. Chloroplastida) of glucan phosphatases is quite well conserved from land plants to single-cell green algae, highlighting the essential nature of reversible glucan phosphorylation in land plants and green algae (Fig. 3b) [43, 44]. LSF1 is conserved in vascular plants (tracheophyta) and mosses (bryophyta) with proteins being 30 % identical at the amino acid level, but LSF1 is not conserved in single-cell green algae (chlorophyta) [13]. Alternatively, SEX4 and LSF2 orthologs are present in vascular plants and mosses as well as unicellular green algae such as Chlamydomonas and Ostreococcus [13]. SEX4 proteins from land plants to unicellular green algae are 34 % identical and LSF2 orthologs are 36 % identical [13, 36, 45]. Thus, SEX4 and LSF2 orthologs are highly conserved throughout the green lineage, including the most ancient genomes.
Laforin exhibits a more unique and varied conservation. The gene encoding laforin is conserved in species as divergent as humans and the unicellular red alga Cyanidioschyzon merolae, but absent in the genomes of most non-vertebrate organisms. A laforin ortholog is conserved in all vertebrate genomes, but it has been lost in the vast majority of invertebrate genomes and lower organisms (including yeast, fly and worm genomes), and laforin-like orthologs are conserved in a subset of protists and invertebrates [43]. The vertebrate laforin orthologs are 66 % identical at the amino acid level, while laforin orthologs from humans to C. merolae are only 25 % identical [23, 43, 46]. The invertebrates and protists that contain laforin-like orthologs are organisms with genomes that are predicted to have undergone slower rates of molecular evolution and/or organisms that metabolize a carbohydrate similar to a Lafora body [43, 47]. Three criteria have emerged to define which genomes contain a laforin-like gene, the organism must: (1) be of red or blue-green algal descent, (2) possess a true mitochondrion, and (3) produce an insoluble glucan (e.g., floridean starch, amylopectin granules, etc.) [43]. The majority of organisms that fit these criteria are a group of unicellular protists classified as alveolates. The alveolates include apicomplexa and cilates as well as organisms from other phyla. We and others have identified laforin-like genes in the genomes of red lineage organisms back to the ancient unicellular red alga C. merolae, the green lineage traced to Chlamydomonas reinhardtii, and the blue-green lineage to Cyanophora paradoxa [23, 43, 44]. Thus, glucan phosphatases can be traced back to single-cell organisms representing the three main lineages that expanded from the Archaeplastida: red algae, green algae/land plants, glaucophytes, and other eukaryotes possessing an Archaeplastida-derived plastid (Fig. 3b).
Specificity of glucan phosphatases and consensus motif
To define the specificity of glucan phosphatases, we and others have employed a radiolabelling assay that utilizes the specific action of the glucan dikinases [11–13, 36, 45, 46]. Two independent paths are followed to generate radiolabelled starch at either the C6 or C3 position. To generate C6 labeled starch, starch from GWD-deficient Arabidopsis plants is first phosphorylated by GWD in the presence of [β-33P]ATP and then this radiolabelled starch is phosphorylated at the C3 position by PWD with cold ATP. To generate C3 labeled starch, starch is first phosphorylated by GWD with ATP and then phosphorylated by PWD with [β-33P]ATP. These two products are used as substrates for the glucan phosphatases to define their substrate preferences.
Using this assay, we recently demonstrated that laforin preferentially dephosphorylates hydroxyls at the C3 position and it has substantial activity against the C6 position as well [11]. Conversely, SEX4 preferentially dephosphorylates the C6 position while possessing activity against the C3 position [45]. LSF2 differs in that it exclusively dephosphorylates the C3 position [13, 36]. Insights from recent structures as well as structure-guided mutagenesis demonstrated that the DSP domain is responsible for engagement and orientation of the glucan to achieve position-specific specificity [11, 36, 45, 46]. These studies also allowed for the identification of a glucan phosphatase catalytic site (PTP-loop) consensus motif of Cys, hydrophilic (ζ), Ala, Gly, long chain aliphatic (Ψ), Gly, Arg (GζAGΨGR) [11]. This consensus motif is uniquely adapted for engaging glucan chains and directing them towards the active site for dephosphorylation. This consensus motif also explains the lack of glucan phosphatase activity for LSF1, as it possesses three residues within its catalytic site that are not in agreement with the consensus signature.
X-ray structures of glucan phosphatases
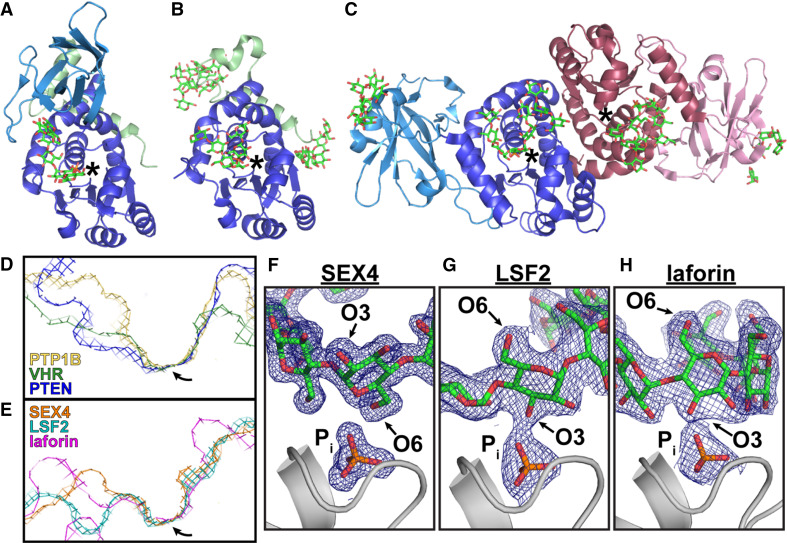
We recently determined the crystal structures of SEX4 (3NME, 4PHY), LSF2 (4KYQ, 4KYR), and laforin (4RKK) (Fig. 4a–c) [36, 45, 46, 48]. We determined a product bound structure for each glucan phosphatase and a ligand-free structure for SEX4 and LSF2. Each glucan phosphatase has a glucan bound at the active site and the glucan-protein interaction is coordinated by aromatic and hydrophilic residues. While SEX4, LSF2, and laforin each binds and dephosphorylates glucans, each of the three glucan phosphatases employs a different mechanism to engage their substrate.
Fig. 4.
The crystal structures of the glucan phosphatases reveal their unique structures and specificities. a–c X-ray crystal structures of a SEX4 (PDB: 4PYH), b LSF2 (PDB: 4KYR), and c laforin (PDB: 4RKK) bound to glucans and phosphate reveal their diverse domain arrangements and glucan-binding platforms. CBMs are shown in light blue, the DSP domains are shown in dark blue, and the cTPs are shown in green. The second subunit of the laforin dimer is shown in shades of pink. Active sites are marked with an asterisk. The active site topology among phosphatases varies based on substrate. (D) The tyrosine phosphatase PTP1B (PDB: 2HNP) displays a narrow and deep catalytic cleft for dephosphorylating pTyr but not pSer/pThr; VHR (PDB: 1VHR) is a DSP with a narrow and shallow cleft for dephosphorylating pTyr and pSer/pThr; and the lipid phosphatase PTEN (PDB: 1D5R) has a wide and shallow cleft suited for phospholipid substrates. e The glucan phosphatases also display wide and shallow active sites to accommodate phosphoglucans. Arrows mark the position of the active site cysteine. (F–H) 2Fo-Fc density maps (1.0σ) of bound glucans and phosphate (P i) at the active sites of SEX4 (f), LSF2 (g), and laforin (h). The glucans are oriented with either the O3 or O6 pointed toward the catalytic sites (in gray) corresponding to their respective dephosphorylation preferences
The active site architecture of glucan phosphatases is unique compared to other phosphatases (Fig. 4d, e). Tyrosine specific phosphatases utilize a narrow and deep catalytic cleft to accommodate pTyr while excluding the shorter pSer and pThr from the catalytic site [49]. Alternatively, the proteinaceous DSP catalytic cleft is narrow and shallow to dephosphorylate pTyr, pSer, and pThr [50]. The lipid phosphatases and glucan phosphatases both possess very wide and shallow active sites with each containing distinct characteristics to accommodate either phospholipids or phosphoglucans [36, 45, 46, 51].
SEX4 utilizes an integrated DSP-CBM glucan-binding platform
We determined the X-ray structures of catalytically inactive (C198S) Arabidopsis thaliana SEX4 lacking the cTP and bound to maltoheptaose and phosphate to 1.65 Å and bound to just phosphate to 2.4 Å [45, 48] (Fig. 4a). The SEX4 DSP domain and CBM directly interact with 457 Å2 of interfacial surface area via multiple highly conserved motifs found within each domain. Analysis of the structural data revealed a previously unidentified carboxy-terminal (CT) motif within SEX4 [48]. This CT motif was predicted to be unstructured, but instead it is comprised of two α-helices separated by a loop region with invariant residues conserved among all SEX4 orthologs. The CT motif helices intimately associate with the DSP domain residues, cradling the face of the DSP domain opposite the active site. Biochemical analysis revealed that the SEX4 CT motif is essential for maintaining the fold and stability of SEX4.
Maltoheptaose is a linear, seven-residue polymer of glucose with α-1,4-glycosidic linkages, representative of a glucan chain found within amylopectin. The maltoheptaose and phosphate bound at the active site represent the product of SEX4 glucan dephosphorylation. The maltopheptaose chain is integrated within a continuous binding pocket that spans both the CBM and the DSP domain (Fig. 4a). This extended binding surface is comprised of aromatic and hydrophilic residues that form a glucan-binding platform unique for SEX4 among the glucan phosphatases.
The first glucose unit from the non-reducing end of the maltoheptaose chain is located at the DSP and glucose 6 is at the CBM with glucose 2 directly above the phosphate bound at the DSP active site. The O6 hydroxyl of Glc2 interacts with phosphate in the DSP active site at a distance of 2.6 Å, while the O3 group is 7.1 Å away (Fig. 4F). These structural data are in agreement with the biochemical data demonstrating that SEX4 prefers to dephosphorylate the C6 position and suggests that the glucan chain must completely switch directionality in order for SEX4 to dephosphorylate the C3 position. The glucose residues adjacent to Glc2 (i.e. Glc1 and Glc3) are also oriented with the O6 groups positioned towards the active site, while the O6 group of Glc6 is solvent exposed. Thus, SEX4 accommodates the helical nature of the glucan and the glucan rotates from the CBM to the DSP. The maltoheptaose spans the ~33 Å DSP-CBM glucan binding pocket with 610 Å2 of total contact area. Within this contact area, 40 % of the glucan-SEX4 interactions occur via CBM residues and 60 % via DSP domain residues. The CBM residues are highly conserved in all SEX4 orthologs to form a glucan-binding site. Two tryptophan residues (W278 and W314) combine with H330 to interact with both faces of the glucan chain with additional hydrogen bonding with O3 groups and N332 and K307. This dual-tryptophan and lysine glucan-binding platform is a conserved motif of the CBM family 48 (CBM48) and the related CBM20 family [30, 34].
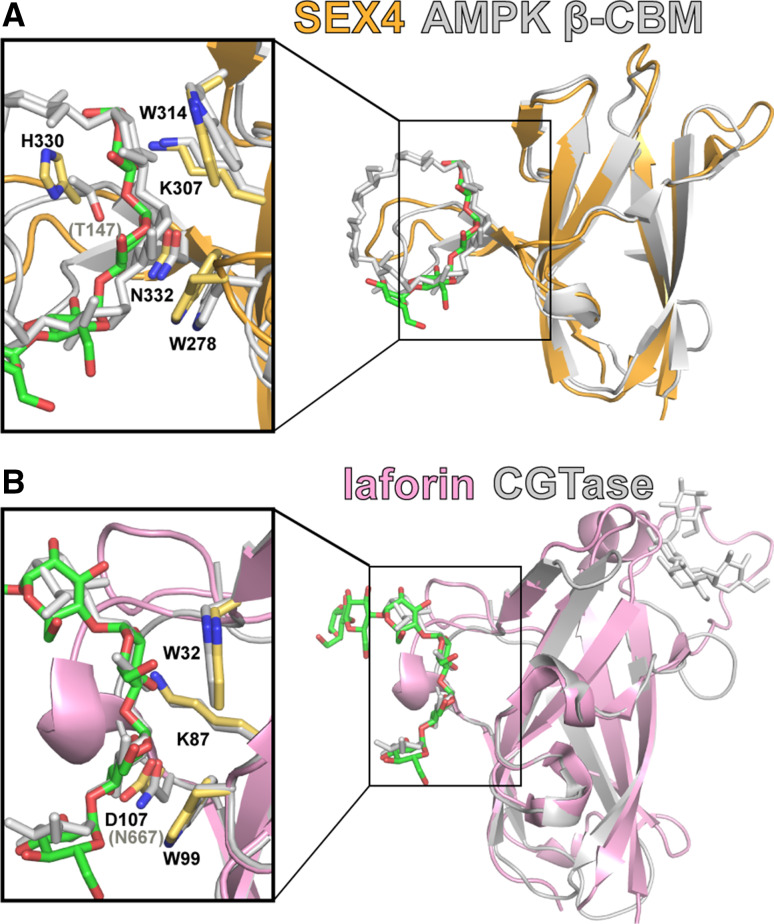
A query using the Dali server to define the most similar CBM to that of SEX4 identifies the CBM48 of the human AMP-activated protein kinase β-subunit (AMPK-β, PDB: 1Z0N) with a root mean square deviation (RMSD) of 1.4 Å compared to the SEX4 CBM (Fig. 5a) [45, 52–54]. AMPK-β shares all of the SEX4 CBM glucan- interacting residues except for SEX4 H330 where AMPK-β instead has a threonine residue (Fig. 5a). AMPK interacts with glycogen via the AMPK-β subunit bringing it in proximity to several enzyme targets such as glycogen synthase and also allowing it to sense glycogen levels that modulate kinase activity. The AMPK-β glucan-bound structure is co-crystallized with the circular, seven glucose oligosaccharide β-cyclodextrin [54]. However, the maltoheptaose bound to SEX4 possesses a similar curvature as the β-cyclodextrin bound to AMPK-β (Fig. 5a). Alignment of the maltoheptaose bound and ligand-free SEX4 structures reveals that H330 undergoes a conformational shift upon when bound to a glucan, positioning H330 in line with glucose 6 of the reducing end and opposite of W314 with maltoheptaose positioned between the two (Fig. 5a). Mutation of W314 or H330 to an alanine reduces SEX4 dephosphorylation of starch by 95 and 66 %, respectively. Additionally, alanine mutation of the key CBM SEX4 glucan-binding residues resulted in a dramatic decrease in glucan binding as defined via an amylopectin co-sedimentation assay. This decreased binding translated to an average decrease of 89 % in mutant CBM SEX4 to dephosphorylate starch even though the DSP active site is intact. Thus, the SEX4 CBM is essential for glucan binding and if SEX4 glucan binding is impacted so to is its ability to dephosphorylate starch.
Fig. 5.
The glucan phosphatases SEX4 and laforin utilize a conserved mode of glucan binding via CBMs. According to a DALI search, the CBM of the AMPK β-subunit (PDB: 1Z0N) is the most similar to the SEX4 CBM (a), and the laforin CBM is highly similar to the CBM of a bacterial cyclodextrin glycosyltransferase or CGTase bound to a glucan at two sites (PDB: 1CXK) (b). Interacting SEX4 and laforin residues are shown in yellow and labeled in black in (a) and (b), respectively; interacting AMPK β-CBM and CGTase residues are shown and labeled in gray in (a) and (b), respectively
While the SEX4 DSP domain contributes 60 % of the SEX4-maltoheptaose contact area, mutations of the DSP domain residues that interact with maltoheptaose only minimally decreased starch binding [45]. Similarly, these DSP domain mutations only decreased dephosphorylation by an average of 38 %, dramatically lower than the 89 % observed with CBM mutations. These data were surprising since the DSP contributes the majority of the SEX4-maltoheptaose contact area. While the DSP domain is important for glucan binding, binding is clearly not its major contribution. Utilizing the position-specific radiolabelling assay, we found that the glucan-binding residues within the DSP domain are essential to position the glucan for specific dephosphorylation of the C3 or C6 position. Two lines of evidence led to this conclusion: (1) mutation of the aromatic residues that form the boundary of the DSP active site architecture ablated the SEX4 C6 specificity and (2) while mutations of key CBM interacting residues dramatically decreased starch dephosphorylation, the mutant enzymes maintained the C6 position preference. Thus, SEX4 utilizes a coordinated binding platform comprised of its CBM and DSP domain whereby the CBM is largely responsible for glucan engagement and the DSP domain is responsible for site specificity.
LSF2 utilizes surface binding sites (SBSs)
The X-ray structures of catalytically inactive (C193S) A. thaliana LSF2 lacking the cTP (residues 79–292) either without ligand or with bound maltohexaose and phosphate was determined to 2.30 and 1.65 Å, respectively (Fig. 4b) [36]. While SEX4 was bound to a single maltoheptaose, LSF2 is bound to multiple maltohexaose oligosaccharides via three binding sites.
The LSF2 active site contains five highly conserved aromatic residues that integrate a glucan-binding channel within the DSP active site and coordinate binding to a single maltohexaose chain. The C3 position of Glc3 directly interacts with the phosphate at the LSF2 active site at a distance of 2.4 Å compared to 7.0 Å for the C6 position (Fig. 4g). These data support the biochemical data showing that LSF2 exclusively dephosphorylates the C3 position in vitro. Additionally, these data align with in planta data demonstrating that lsf2 mutant plants have an increase in C3 phosphorylated starch. The glucan binding at the LSF2 active site is coordinated by three Tyr residues and a Trp (Y83, Y85, Y135, and W136) that interact with the convex side of the maltohexaose chain. The glucose moieties of the concave surface form a helical structure around F162, interacting with both faces of the Phe ring. LSF2 containing mutated residues on both sides of the DSP glucan-binding channel to (W136A/F162A) resulted in a 99 % decrease in starch phosphorylation. However, LSF2-W136A/F162A maintained robust binding to starch. These data suggested that while the active site glucan-binding channel is necessary for glucan phosphatase activity, the other binding sites significantly contribute to glucan binding.
The two additional LSF2 glucan binding sites both intimately involve the CT motif and are referred to as Site-2 and Site-3 [36]. Site-2 is located ~21 Å away from the DSP active site and it is comprised of highly conserved residues from the DSP domain and CT motif that make extensive contacts with a single maltohexaose chain via hydrogen bonding and van der Waals contacts. Mutation of Site-2 residues to alanine decreased LSF2 starch phosphatase activity by 24–50 %. Additionally, mutation of R157 decreased amylopectin binding by 64 %, dramatically greater than observed with the active site W136A/F162A double mutant. Thus, Site-2 is necessary for the biological activity of starch dephosphorylation by LSF2.
The LSF2 Site-3 binding site is also located >20 Å away from the DSP active site, and it is comprised of highly conserved residues formed by the CT motif that form hydrogen bonding interactions and van der Waal’s interactions to bind two maltohexaose chains (Hex-1 and Hex-2). Mutation of Site-3 residues to alanine decreased LSF2 starch phosphatase activity by 35–87 %. Mutation of one Site-3 residue (F261A) decreased amylopectin binding by 74 %, showing the most dramatic decrease in binding. Combinatorial mutations of the active site, Site-2, and/or Site-3 showed additive effects, suggesting that the sites function synergistically. Intriguingly, the two maltohexaose chains at Site-3 possess a helical-like structure that is reminiscent of amylopectin glucan chain helices. Cumulatively, these data suggest a model whereby LSF2 may engage longer glucan chains or perhaps multiple chains simultaneously via the three glucan binding sites.
As discussed, glucan phosphatases were originally defined as proteins containing both a CBM and DSP domain [23]. However, LSF2 lacks a CBM and instead utilizes glucan binding sites at the active site and two that are >20 Å away, i.e. Site-2 and Site-3. These glucan binding sites away from the active site are referred to as surface binding sites (SBSs) and they are a common mechanism employed by enzymes within the glycoside hydrolase family [38]. There are numerous examples of glycoside hydrolases that possess one or more CBM, and recent structural studies have identified that some of these glycoside hydrolases also contain one or more SBS or they lack a CBM and only contain SBSs. Indeed, SusG is a cell membrane associated α-amylase from Bacteroides species that contains both a CBM58 and SBS [39]. Alternatively, barley α-amylase, human salivary α-amylase, pancreatic α-amylase, and yeast glucoamylase contain only SBSs [40–42, 55–57]. Similar to LSF2, Svensson and colleagues demonstrated that two SBSs that are >30 Å away from the barley α-amylase active site are directly involved in substrate binding and they act synergistically to engage the glucan [37, 55]. In each case, the SBSs are identified via structural studies and these sites prove difficult to identify via bioinformatics due to the absence of a strict, unique signature though they typically employ aromatic residues.
Laforin utilizes a tetramodular architecture and cooperativity
The structure of human laforin (residues 1-328 out of 331 total residues) bound to maltohexaose and phosphate was determined to 2.4 Å with two, superimposable molecules in the asymmetric unit (Fig. 4c) [46]. The laforin CBM and DSP domains are intimately associated with >1200 Å2 of interfacial surface area, forming an integrated tertiary structure. Additionally, the two laforin molecules form a tetramodular, antiparallel dimer that is mediated entirely by the DSP domain. This tetramodular architecture yields a CBM-DSP-DSP-CBM dimer that engages multiple maltohexaose chains at the CBM and DSP domain, along with a single phosphate at each DSP active site. The laforin DSP domain is structurally most similar to the plant glucan phosphatases LSF2 and SEX4 with a RMSD of 2.0 and 2.1 Å, respectively.
Similar to SEX4 and LSF2, there is a maltohexaose chain and phosphate tightly coordinated into the laforin DSP domain active site. The laforin DSP active site contains a number of highly conserved hydrophilic and aromatic residues that coordinate binding with the maltohexaose chain via hydrogen bonds and van der Walls interactions. Mutation of these residues did not decrease laforin binding to glycogen, but did decrease glycogen dephosphorylation by 69–95 %. These residues position the C2 and C3 groups of Glc3 3.8 Å and 2.7 Å from the phosphate, respectively, with the C6 position 7.7 Å away and pointed toward the solvent (Fig. 4h). As predicted by the structural data, laforin preferentially dephosphorylates the C3-position while maintaining substantial activity against the C6-position, opposite from the SEX4 specificity [45, 46].
While SEX4 and laforin each contain a CBM and DSP domain, their domain organization and utilization is quite different. SEX4 employs a continuous binding pocket spanning a tightly integrated CBM and DSP interface, while the laforin CBM and DSP domains are distant from each other with each binding a unique maltohexaose chain. The laforin CBM engages a maltohexaose chain via aromatic and hydrophilic residues. As with the SEX4 CBM48, the laforin CBM20 contains a dual-tryptophan and a lysine glucan binding platform that is a hallmark of the CBM20 and CBM48 family [30, 34]. Mutation of these residues decreased glycogen binding by 40–100 % and this decreased binding translated to a decrease in glycogen dephosphorylation of 57–96 %.
The laforin CBM is structurally most similar to a fungal glucoamylase (PDB: 2VN4 and 2VN7; RMSD = 2.0 Å), a bacterial cyclodextrin glycosyltransferase (CGTase, PDB: 1CXK; RMSD = 2.2 Å), and the CBM of the human glycerophosphocholine phosphodiesterase (GDE5/GPCPD1, PDB: 2z0b; RMSD = 2.5 Å) according to a Dali search [46, 58, 59]. Of these three CBMs, only the bacterial CGTase is bound to a glucan. The CBMs of these proteins all contain conserved tryptophan residues characteristic of the CBM20 family. These tryptophan residues are reported to support two separate glucan-binding sites in the CGTase [33]. The laforin CBM and CGTase CBM superimpose with good overlap at the first glucan binding site (Fig. 5b). The first site is composed of two tryptophan residues that stack with the glucan and hydrophilic residues that participate in hydrogen bonding. In the crystal structures of laforin and the CGTase, a glucan is bound to this first site, associating with W32, W99, K87 and D107 in laforin and W616, W662, K651 and N667 in the CGTase (Fig. 5b). The CGTase contains a second glucan binding site that is also occupied by a glucan in the CGTase structure (Fig. 5b). This site is in closer proximity to the CGTase catalytic domain, and it contains an additional aromatic residue (Y633) that guides the substrate toward the active site [60]. However, laforin lacks a tyrosine at this position and instead has a glutamate and there is no glucan bound at this position in the laforin crystal structure. Thus, the laforin CBM possesses just one glucan binding site.
The laforin crystal structure revealed that laforin is an anti-parallel dimer with the dimer interface mediated by the DSP domains, yielding a unique tetramodular architecture. The status of laforin dimerization has been a controversial topic with some data suggesting that laforin is a monomer in solution and other suggesting a dimer or multimer [61–65]. The structure of laforin in solution was defined using multiple biophysical techniques: analytical ultracentrifugation (AUC), size-exclusion chromatography–small angle X-ray scattering (SEC–SAXS), and deuterium exchange mass spectrometry (DXMS). AUC data demonstrated that laforin possess a molecular weight of 79 ± 6 kDa, consistent with the predicted dimer molecular weight of 78 kDa. Additionally, laforin eluted from SEC as a single species and ab initio reconstruction from SAXS data of the molecular envelope revealed an extended bilobal shape that matches the laforin crystal structure. DXMS data confirm that the laforin dimerization interface is via the DSP domain. In contrast, another study determined the structure of laforin using a hybrid approach [66]. These investigations determined the X-ray crystal structure of the DSP domain, generated a homology model of the CBM, and performed small angle X-ray scattering on the full-length protein. The models were manually docked inside the scattering envelope to produce a CBM–CBM dimerization interface. However, rigid-body modeling from SAXS data without complete structures can produce inaccuracies, and no further experimental evidence was presented to corroborate this quaternary arrangement [67, 68].
Laforin is structurally organized so that all four glucan binding sites in the tetramodular architecture align on one face of the dimer. Using a thermal shift assay, we found that the melting temperature (∆T m) of laforin increased minimally with shorter oligosaccharides (2 degrees to 8 degrees of polymerization, i.e. DP2-8), but there is a dramatic increase in ∆T m with longer oligosaccharides with DP24 yielding a 14 °C increase. A detailed analysis demonstrated that shorter oligosaccharides bind with two binding events without cooperativity while the longer DP24 oligosaccharide showed a single high-affinity binding event with cooperativity. Importantly, a mutation in the laforin DSP–DSP dimer interface that abolishes dimerization also abolished the cooperativity. Thus, laforin dimerization is the basis of laforin’s substrate binding cooperativity.
Binding soluble versus insoluble substrates
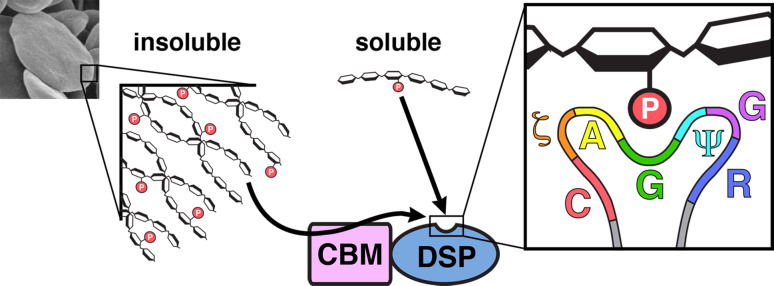
A major difference between glycogen, amylopectin, and starch is water solubility: glycogen is water-soluble, starch is water-insoluble, and amylopectin is moderately water-soluble. The insoluble and crystalline nature of starch presents a much different binding surface than that found in glycogen or amylopectin. Using structure guided mutagenesis to mutate residues and delete domains, we recently demonstrated that the glucan phosphatase DSP domain is sufficient for dephosphorylating water-soluble polyglucans (Fig. 6). Mutations in the laforin or SEX4 CBMs or the LSF2 SBSs that dramatically decreased both starch dephosphorylation and binding to water-soluble glycogen or amylopectin did not dramatically decrease dephosphorylation of amylopectin or phospho-oligosaccharides [11, 45, 46, 69]. Soluble glucans are thought to lack the higher-order helical structure found in crystalline, insoluble glucans. Thus, the glucan-interacting motifs within the glucan phosphatase DSP domains enable adequate engagement and dephosphorylation of the soluble, phospho-glucan. Conversely, the ancillary glucan-binding interfaces are critical for dephosphorylation of insoluble starch. Mutations in the ancillary glucan binding platforms of SEX4, LSF2, or laforin abolished the enzymes ability to dephosphorylate starch. The need for these ancillary binding platforms highlights the unique environment of glucans at the outer starch granule. The glucan surface possesses multiple microenvironments that glucan phosphatases will encounter. There are long oligosaccharide chains, phospho-oligosaccharides chains, branch points, residual helical structure, and non-phosphorylated oligosaccharide chains. In a water-soluble environment, the DSP domain alone can coordinate engagement and dephosphorylation of the phospho-glucan. However, these ancillary glucan-binding platforms are necessary for glucan binding and dephosphorylation within an insoluble microenvironment.
Fig. 6.
While the CBM of a glucan phosphatase is required for dephosphorylating insoluble glucan substrates, a DSP is sufficient for dephosphorylating water-soluble polyglucans. We have defined the active site consensus motif for a glucan phosphatase as: Cys, hydrophilic, Ala, Gly, long chain aliphatic, Gly, Arg (CζAGΨGR). Reproduced with permission from Meekins et al. [11]
Conclusions and future perspectives
Over the past decade, reversible glucan phosphorylation has emerged as a key regulatory mechanism governing starch and glycogen metabolism. The discovery, activity, and function of the glucan dikinases were first to be defined and more recently the glucan phosphatases have also emerged as essential enzymes in starch and glycogen metabolism. Despite an impressive amount of progress, there are still many outstanding questions regarding both starch- and glycogen-bound phosphate. GWD has a recently described role in starch synthesis, though the exact function still remains to be fully defined [70, 71]. Similarly, SEX4 and LSF2 could also function during starch synthesis. Since starch-bound phosphate affects surface properties and morphology of starch granules, these phosphate levels could be dynamic during synthesis involving interplay of starch phosphorylation by GWD and then removal of some phosphate by the glucan phosphatases. Since the loss of LSF1 also results in a starch excess phenotype, LSF1 is clearly intimately involved in starch metabolism, but its role remains entirely undefined. Additionally, whether LSF1 is an enzyme or scaffolding protein is unknown. Finally, many of the discoveries regarding reversible starch phosphorylation have focused on transitory leaf starch. The role of glucan phosphatases and reversible starch phosphorylation of storage starch is largely unknown.
An intriguing question regarding glycogen-bound phosphate centers on how phosphate is introduced into glycogen. While evidence suggests that at least some of this phosphate, present at the C2- and C3-positions of glucosyl moieties, may be introduced via an error of glycogen synthase [19, 72], the role of an as-yet-undiscovered glycogen-phosphorylating enzyme has also been proposed [20]. Further, whether phosphorylation and the action of laforin are involved in glycogen synthesis, degradation, or both has not yet been defined. Nevertheless, the X-ray crystal structures of the glucan phosphatases have defined a number of key issues regarding their mechanism of action and specificity.
Acknowledgments
This work was supported by National Institutes of Health Grants R01NS070899; Kentucky Science and Engineering Foundation Grants KSEF-2268RDE-014 and KSEF-2971-RDE-017; Mitzutani Foundation for Glycoscience Award; National Science Foundation Grants IIA-1355438 and MCB-1252345. M. S. G. is co-founder of OptiMol Enzymes LLC.
Abbreviations
- CBM
Carbohydrate binding module
- CGTase
Cyclodextrin glycosyltransferase
- DSP
Dual specificity phosphatase
- GWD
Glucan-water dikinase
- PWD
Phosphoglucan-water dikinase
- LSF2
Like Sex Four2
- SBS
Surface binding site
- SEX4
Starch EXcess4
Footnotes
S. Emanuelle and M. K. Brewer contributed equally to this work.
References
- 1.Shearer J, Graham TE. New perspectives on the storage and organization of muscle glycogen. Can J Appl Physiol. 2002;27:179–203. doi: 10.1139/h02-012. [DOI] [PubMed] [Google Scholar]
- 2.Roach PJ. Glycogen and its metabolism. Curr Mol Med. 2002;2:101–120. doi: 10.2174/1566524024605761. [DOI] [PubMed] [Google Scholar]
- 3.Roach PJ, Depaoli-Roach AA, Hurley TD, Tagliabracci VS. Glycogen and its metabolism: some new developments and old themes. Biochem J. 2012;441:763–787. doi: 10.1042/BJ20111416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ryu JH, Drain J, Kim JH, McGee S, Gray-Weale A, Waddington L, Parker GJ, Hargreaves M, Yoo SH, Stapleton D. Comparative structural analyses of purified glycogen particles from rat liver, human skeletal muscle and commercial preparations. Int J Biol Macromol. 2009;45:478–482. doi: 10.1016/j.ijbiomac.2009.08.006. [DOI] [PubMed] [Google Scholar]
- 5.Powell PO, Sullivan MA, Sheehy JJ, Schulz BL, Warren FJ, Gilbert RG. Acid hydrolysis and molecular density of phytoglycogen and liver glycogen helps understand the bonding in glycogen alpha (composite) particles. PLoS One. 2015;10:e0121337. doi: 10.1371/journal.pone.0121337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Goldsmith E, Sprang S, Fletterick R. Structure of maltoheptaose by difference Fourier methods and a model for glycogen. J Mol Biol. 1982;156:411–427. doi: 10.1016/0022-2836(82)90336-9. [DOI] [PubMed] [Google Scholar]
- 7.Ball S, Colleoni C, Cenci U, Raj JN, Tirtiaux C. The evolution of glycogen and starch metabolism in eukaryotes gives molecular clues to understand the establishment of plastid endosymbiosis. J Exp Bot. 2011;62:1775–1801. doi: 10.1093/jxb/erq411. [DOI] [PubMed] [Google Scholar]
- 8.Zeeman SC, Kossmann J, Smith AM. Starch: its metabolism, evolution, and biotechnological modification in plants. Ann Rev Plant Biol. 2010;61:209–234. doi: 10.1146/annurev-arplant-042809-112301. [DOI] [PubMed] [Google Scholar]
- 9.Hancock RD, Tarbet BJ. The other double helix—the fascinating chemistry of starch. J Chem Educ. 2000;77:988. doi: 10.1021/ed077p988. [DOI] [Google Scholar]
- 10.Ritte G, Heydenreich M, Mahlow S, Haebel S, Kotting O, Steup M. Phosphorylation of C6- and C3-positions of glucosyl residues in starch is catalysed by distinct dikinases. FEBS Lett. 2006;580:4872–4876. doi: 10.1016/j.febslet.2006.07.085. [DOI] [PubMed] [Google Scholar]
- 11.Meekins DA, Raththagala M, Auger KD, Turner BD, Santelia D, Kotting O, Gentry MS, Kooi CWV. Mechanistic insights into glucan phosphatase activity against polyglucan substrates. J Biol Chem. 2015;290:23361–23370. doi: 10.1074/jbc.M115.658203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hejazi M, Fettke J, Kotting O, Zeeman SC, Steup M. The Laforin-like dual-specificity phosphatase SEX4 from Arabidopsis hydrolyzes both C6- and C3-phosphate esters introduced by starch-related dikinases and thereby affects phase transition of alpha-glucans. Plant Physiol. 2010;152:711–722. doi: 10.1104/pp.109.149914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Santelia D, Kotting O, Seung D, Schubert M, Thalmann M, Bischof S, Meekins DA, Lutz A, Patron N, Gentry MS, Allain FH, Zeeman SC. The phosphoglucan phosphatase like sex Four2 dephosphorylates starch at the C3-position in Arabidopsis. Plant Cell. 2011;23:4096–4111. doi: 10.1105/tpc.111.092155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Niittyla T, Comparot-Moss S, Lue WL, Messerli G, Trevisan M, Seymour MDJ, Gatehouse JA, Villadsen D, Smith SM, Chen J, Zeeman SC, Smith AM. Similar protein phosphatases control starch metabolism in plants and glycogen metabolism in mammals. J Biol Chem. 2006;281:11815–11818. doi: 10.1074/jbc.M600519200. [DOI] [PubMed] [Google Scholar]
- 15.Zeeman SC, Northrop F, Smith AM, Rees T. A starch-accumulating mutant of Arabidopsis thaliana deficient in a chloroplastic starch-hydrolysing enzyme. Plant J. 1998;15:357–365. doi: 10.1046/j.1365-313X.1998.00213.x. [DOI] [PubMed] [Google Scholar]
- 16.Kotting O, Santelia D, Edner C, Eicke S, Marthaler T, Gentry MS, Comparot-Moss S, Chen J, Smith AM, Steup M, Ritte G, Zeeman SC. STARCH-EXCESS4 Is a laforin-like phosphoglucan phosphatase required for starch degradation in Arabidopsis thaliana . Plant Cell. 2009;21:334–346. doi: 10.1105/tpc.108.064360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ball SG, Morell MK. From bacterial glycogen to starch: understanding the biogenesis of the plant starch granule. Ann Rev Plant Biol. 2003;54:207–233. doi: 10.1146/annurev.arplant.54.031902.134927. [DOI] [PubMed] [Google Scholar]
- 18.Worby CA, Gentry MS, Dixon JE. Laforin: a dual specificity phosphatase that dephosphorylates complex carbohydrates. J Biol Chem. 2006;281:30412–30418. doi: 10.1074/jbc.M606117200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.DePaoli-Roach AA, Contreras CJ, Segvich DM, Heiss C, Ishihara M, Azadi P, Roach PJ. Glycogen phosphomonoester distribution in mouse models of the progressive myoclonic epilepsy, Lafora disease. J Biol Chem. 2015;290:841–850. doi: 10.1074/jbc.M114.607796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nitschke F, Wang P, Schmieder P, Girard JM, Awrey DE, Wang T, Israelian J, Zhao X, Turnbull J, Heydenreich M, Kleinpeter E, Steup M, Minassian BA. Hyperphosphorylation of glucosyl C6 carbons and altered structure of glycogen in the neurodegenerative epilepsy Lafora disease. Cell Metab. 2013;17:756–767. doi: 10.1016/j.cmet.2013.04.006. [DOI] [PubMed] [Google Scholar]
- 21.Fordham-Skelton AP, Chilley P, Lumbreras V, Reignoux S, Fenton TR, Dahm CC, Pages M, Gatehouse JA. A novel higher plant protein tyrosine phosphatase interacts with SNF1-related protein kinases via a KIS (kinase interaction sequence) domain. Plant J. 2002;29:705–715. doi: 10.1046/j.1365-313X.2002.01250.x. [DOI] [PubMed] [Google Scholar]
- 22.Kerk D, Conley TR, Rodriguez FA, Tran HT, Nimick M, Muench DG, Moorhead GB. A chloroplast-localized dual-specificity protein phosphatase in Arabidopsis contains a phylogenetically dispersed and ancient carbohydrate-binding domain, which binds the polysaccharide starch. Plant J. 2006;46:400–413. doi: 10.1111/j.1365-313X.2006.02704.x. [DOI] [PubMed] [Google Scholar]
- 23.Gentry MS, Dowen RH, 3rd, Worby CA, Mattoo S, Ecker JR, Dixon JE. The phosphatase laforin crosses evolutionary boundaries and links carbohydrate metabolism to neuronal disease. J Cell Biol. 2007;178:477–488. doi: 10.1083/jcb.200704094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Comparot-Moss S, Kotting O, Stettler M, Edner C, Graf A, Weise SE, Streb S, Lue WL, MacLean D, Mahlow S, Ritte G, Steup M, Chen J, Zeeman SC, Smith AM. A putative phosphatase, LSF1, is required for normal starch turnover in Arabidopsis leaves. Plant Physiol. 2010;152:685–697. doi: 10.1104/pp.109.148981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Moorhead GB, De Wever V, Templeton G, Kerk D. Evolution of protein phosphatases in plants and animals. Biochem J. 2009;417:401–409. doi: 10.1042/BJ20081986. [DOI] [PubMed] [Google Scholar]
- 26.Tonks NK. Protein tyrosine phosphatases–from housekeeping enzymes to master regulators of signal transduction. FEBS J. 2013;280:346–378. doi: 10.1111/febs.12077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Alonso A, Rojas A, Godzik A, Mustelin T. The dual-specific protein tyrosine phosphatase family. Berlin: Springer; 2003. [Google Scholar]
- 28.Tonks NK. Protein tyrosine phosphatases: from genes, to function, to disease. Nat Rev Mol Cell Biol. 2006;7:833–846. doi: 10.1038/nrm2039. [DOI] [PubMed] [Google Scholar]
- 29.Kerk D, Templeton G, Moorhead GB. Evolutionary radiation pattern of novel protein phosphatases revealed by analysis of protein data from the completely sequenced genomes of humans, green algae, and higher plants. Plant Physiol. 2008;146:351–367. doi: 10.1104/pp.107.111393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cantarel BL, Coutinho PM, Rancurel C, Bernard T, Lombard V, Henrissat B. The Carbohydrate-Active EnZymes database (CAZy): an expert resource for Glycogenomics. Nucleic Acids Res. 2009;37:D233–D238. doi: 10.1093/nar/gkn663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lombard V, Golaconda Ramulu H, Drula E, Coutinho PM, Henrissat B. The carbohydrate-active enzymes database (CAZy) in 2013. Nucleic Acids Res. 2014;42:D490–D495. doi: 10.1093/nar/gkt1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Coutinho PM, Henrissat B. Carbohydrate-active enzymes: an integrated database approach. In: Gilbert HJ, Henrissat GDB, Svensson B, editors. Recent advances in carbohydrate bioengineering. Cambridge: The Royal Society of Chemistry; 1999. pp. 3–12. [Google Scholar]
- 33.Christiansen C, Abou Hachem M, Janecek S, Vikso-Nielsen A, Blennow A, Svensson B. The carbohydrate-binding module family 20–diversity, structure, and function. FEBS J. 2009;276:5006–5029. doi: 10.1111/j.1742-4658.2009.07221.x. [DOI] [PubMed] [Google Scholar]
- 34.Janecek S, Svensson B, MacGregor EA. Structural and evolutionary aspects of two families of non-catalytic domains present in starch and glycogen binding proteins from microbes, plants and animals. Enzyme Microb Technol. 2011;49:429–440. doi: 10.1016/j.enzmictec.2011.07.002. [DOI] [PubMed] [Google Scholar]
- 35.Janecek S, Blesak K. Sequence-structural features and evolutionary relationships of family GH57 alpha-amylases and their putative alpha-amylase-like homologues. Protein J. 2011;30:429–435. doi: 10.1007/s10930-011-9348-7. [DOI] [PubMed] [Google Scholar]
- 36.Meekins DA, Guo HF, Husodo S, Paasch BC, Bridges TM, Santelia D, Kotting O, Vander Kooi CW, Gentry MS. Structure of the Arabidopsis glucan phosphatase like sex Four2 reveals a unique mechanism for starch dephosphorylation. Plant Cell. 2013;25:2302–2314. doi: 10.1105/tpc.113.112706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bozonnet S, Jensen MT, Nielsen MM, Aghajari N, Jensen MH, Kramhoft B, Willemoes M, Tranier S, Haser R, Svensson B. The ‘pair of sugar tongs’ site on the non-catalytic domain C of barley alpha-amylase participates in substrate binding and activity. FEBS J. 2007;274:5055–5067. doi: 10.1111/j.1742-4658.2007.06024.x. [DOI] [PubMed] [Google Scholar]
- 38.Cuyvers S, Dornez E, Delcour JA, Courtin CM. Occurrence and functional significance of secondary carbohydrate binding sites in glycoside hydrolases. Crit Rev Biotechnol. 2012;32:93–107. doi: 10.3109/07388551.2011.561537. [DOI] [PubMed] [Google Scholar]
- 39.Koropatkin NM, Smith TJ. SusG: a unique cell-membrane-associated alpha-amylase from a prominent human gut symbiont targets complex starch molecules. Structure. 2010;18:200–215. doi: 10.1016/j.str.2009.12.010. [DOI] [PubMed] [Google Scholar]
- 40.Kadziola A, Sogaard M, Svensson B, Haser R. Molecular structure of a barley alpha-amylase-inhibitor complex: implications for starch binding and catalysis. J Mol Biol. 1998;278:205–217. doi: 10.1006/jmbi.1998.1683. [DOI] [PubMed] [Google Scholar]
- 41.Robert X, Haser R, Mori H, Svensson B, Aghajari N. Oligosaccharide binding to barley alpha-amylase 1. J Biol Chem. 2005;280:32968–32978. doi: 10.1074/jbc.M505515200. [DOI] [PubMed] [Google Scholar]
- 42.Sevcik J, Hostinova E, Solovicova A, Gasperik J, Dauter Z, Wilson KS. Structure of the complex of a yeast glucoamylase with acarbose reveals the presence of a raw starch binding site on the catalytic domain. FEBS J. 2006;273:2161–2171. doi: 10.1111/j.1742-4658.2006.05230.x. [DOI] [PubMed] [Google Scholar]
- 43.Gentry MS, Pace RM. Conservation of the glucan phosphatase laforin is linked to rates of molecular evolution and the glycogen metabolism of the organism. BMC Evol Biol. 2009;9:138. doi: 10.1186/1471-2148-9-138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ball S, Colleoni C, Arias MC. The transition from glycogen to starch metabolism in cyanobacteria and eukaryotes. In: Nakamura Y, editor. Starch: metabolism and structure. Japan: Springer; 2015. pp. 93–158. [Google Scholar]
- 45.Meekins DA, Raththagala M, Husodo S, White CJ, Guo HF, Kotting O, Vander Kooi CW, Gentry MS. Phosphoglucan-bound structure of starch phosphatase Starch Excess4 reveals the mechanism for C6 specificity. Proc Natl Acad Sci USA. 2014;111:7272–7277. doi: 10.1073/pnas.1400757111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Raththagala M, Brewer MK, Parker MW, Sherwood AR, Wong BK, Hsu S, Bridges TM, Paasch BC, Hellman LM, Husodo S, Meekins DA, Taylor AO, Turner BD, Auger KD, Dukhande VV, Chakravarthy S, Sanz P, Woods VL, Li S, Vander Kooi CW, Gentry MS. Structural mechanism of laforin function in glycogen dephosphorylation and lafora disease. Mol Cell. 2015;57:261–272. doi: 10.1016/j.molcel.2014.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Putnam NH, Srivastava M, Hellsten U, Dirks B, Chapman J, Salamov A, Terry A, Shapiro H, Lindquist E, Kapitonov VV, Jurka J, Genikhovich G, Grigoriev IV, Lucas SM, Steele RE, Finnerty JR, Technau U, Martindale MQ, Rokhsar DS. Sea anemone genome reveals ancestral eumetazoan gene repertoire and genomic organization. Science. 2007;317:86–94. doi: 10.1126/science.1139158. [DOI] [PubMed] [Google Scholar]
- 48.Vander Kooi CW, Taylor AO, Pace RM, Meekins DA, Guo HF, Kim Y, Gentry MS. From the cover: structural basis for the glucan phosphatase activity of Starch Excess4. Proc Natl Acad Sci USA. 2010;107:15379–15384. doi: 10.1073/pnas.1009386107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Barford D, Flint AJ, Tonks NK. Crystal structure of human protein tyrosine phosphatase 1B. Science. 1994;263:1397–1404. doi: 10.1126/science.8128219. [DOI] [PubMed] [Google Scholar]
- 50.Yuvaniyama J, Denu JM, Dixon JE, Saper MA. Crystal structure of the dual specificity protein phosphatase VHR. Science. 1996;272:1328–1331. doi: 10.1126/science.272.5266.1328. [DOI] [PubMed] [Google Scholar]
- 51.Lee JO, Yang H, Georgescu MM, Di Cristofano A, Maehama T, Shi Y, Dixon JE, Pandolfi P, Pavletich NP. Crystal structure of the PTEN tumor suppressor: implications for its phosphoinositide phosphatase activity and membrane association. Cell. 1999;99:323–334. doi: 10.1016/S0092-8674(00)81663-3. [DOI] [PubMed] [Google Scholar]
- 52.Holm L, Sander C. Dali: a network tool for protein structure comparison. Trends Biochem Sci. 1995;20:478–480. doi: 10.1016/S0968-0004(00)89105-7. [DOI] [PubMed] [Google Scholar]
- 53.Mobbs JI, Koay A, Di Paolo A, Bieri M, Petrie EJ, Gorman MA, Doughty L, Parker MW, Stapleton DI, Griffin MD, Gooley PR. Determinants of oligosaccharide specificity of the carbohydrate-binding modules of AMP-activated protein kinase. Biochem J. 2015;468:245–257. doi: 10.1042/BJ20150270. [DOI] [PubMed] [Google Scholar]
- 54.Polekhina G, Gupta A, van Denderen BJ, Feil SC, Kemp BE, Stapleton D, Parker MW. Structural basis for glycogen recognition by AMP-activated protein kinase. Structure. 2005;13:1453–1462. doi: 10.1016/j.str.2005.07.008. [DOI] [PubMed] [Google Scholar]
- 55.Nielsen MM, Bozonnet S, Seo ES, Motyan JA, Andersen JM, Dilokpimol A, Abou Hachem M, Gyemant G, Naested H, Kandra L, Sigurskjold BW, Svensson B. Two secondary carbohydrate binding sites on the surface of barley alpha-amylase 1 have distinct functions and display synergy in hydrolysis of starch granules. Biochemistry. 2009;48:7686–7697. doi: 10.1021/bi900795a. [DOI] [PubMed] [Google Scholar]
- 56.Payan F, Qian M. Crystal structure of the pig pancreatic alpha-amylase complexed with malto-oligosaccharides. J Protein Chem. 2003;22:275–284. doi: 10.1023/A:1025072520607. [DOI] [PubMed] [Google Scholar]
- 57.Ragunath C, Manuel SG, Venkataraman V, Sait HB, Kasinathan C, Ramasubbu N. Probing the role of aromatic residues at the secondary saccharide-binding sites of human salivary alpha-amylase in substrate hydrolysis and bacterial binding. J Mol Biol. 2008;384:1232–1248. doi: 10.1016/j.jmb.2008.09.089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bott R, Saldajeno M, Cuevas W, Ward D, Scheffers M, Aehle W, Karkehabadi S, Sandgren M, Hansson H. Three-dimensional structure of an intact glycoside hydrolase family 15 glucoamylase from Hypocrea jecorina. Biochemistry. 2008;47:5746–5754. doi: 10.1021/bi702413k. [DOI] [PubMed] [Google Scholar]
- 59.Uitdehaag JC, Mosi R, Kalk KH, van der Veen BA, Dijkhuizen L, Withers SG, Dijkstra BW. X-ray structures along the reaction pathway of cyclodextrin glycosyltransferase elucidate catalysis in the alpha-amylase family. Nat Struct Biol. 1999;6:432–436. doi: 10.1038/8235. [DOI] [PubMed] [Google Scholar]
- 60.Penninga D, van der Veen BA, Knegtel RM, van Hijum SA, Rozeboom HJ, Kalk KH, Dijkstra BW, Dijkhuizen L. The raw starch binding domain of cyclodextrin glycosyltransferase from Bacillus circulans strain 251. J Biol Chem. 1996;271:32777–32784. doi: 10.1074/jbc.271.51.32777. [DOI] [PubMed] [Google Scholar]
- 61.Dukhande VV, Rogers DM, Roma-Mateo C, Donderis J, Marina A, Taylor AO, Sanz P, Gentry MS. Laforin, a dual specificity phosphatase involved in Lafora disease, is present mainly as monomeric form with full phosphatase activity. PLoS One. 2011;6:e24040. doi: 10.1371/journal.pone.0024040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Koksal AC, Cingolani G. Dimerization of vaccinia virus VH1 is essential for dephosphorylation of STAT1 at tyrosine 701. J Biol Chem. 2011;286:14373–14382. doi: 10.1074/jbc.M111.226357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Liu Y, Wang Y, Wu C, Liu Y, Zheng P. Dimerization of Laforin is required for its optimal phosphatase activity, regulation of GSK3beta phosphorylation, and Wnt signaling. J Biol Chem. 2006;281:34768–34774. doi: 10.1074/jbc.M607778200. [DOI] [PubMed] [Google Scholar]
- 64.Liu Y, Wang Y, Wu C, Zheng P. Deletions and missense mutations of EPM2A exacerbate unfolded protein response and apoptosis of neuronal cells induced by endoplasm reticulum stress. Hum Mol Genet. 2009;18:2622–2631. doi: 10.1093/hmg/ddp196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sanchez-Martin P, Raththagala M, Bridges TM, Husodo S, Gentry MS, Sanz P, Roma-Mateo C. Dimerization of the glucan phosphatase laforin requires the participation of cysteine 329. PLoS One. 2013;8:e69523. doi: 10.1371/journal.pone.0069523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sankhala RS, Koksal AC, Ho L, Nitschke F, Minassian BA, Cingolani G. Dimeric quaternary structure of human laforin. J Biol Chem. 2015;290:4552–4559. doi: 10.1074/jbc.M114.627406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Jacques DA, Trewhella J. Small-angle scattering for structural biology—expanding the frontier while avoiding the pitfalls. Protein Sci Publ Protein Soc. 2010;19:642–657. doi: 10.1002/pro.351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Schneidman-Duhovny D, Kim SJ, Sali A. Integrative structural modeling with small angle X-ray scattering profiles. BMC Struct Biol. 2012;12:17. doi: 10.1186/1472-6807-12-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wilkens C, Auger KD, Anderson NT, Meekins DA, Raththagala M, Abou Hachem M, Payne CM, Gentry MS, Svensson B. Plant alpha-glucan phosphatases SEX4 and LSF2 display different affinity for amylopectin and amylose. FEBS Lett. 2016;590:118–128. doi: 10.1002/1873-3468.12027. [DOI] [PubMed] [Google Scholar]
- 70.Mahlow S, Hejazi M, Kuhnert F, Garz A, Brust H, Baumann O, Fettke J. Phosphorylation of transitory starch by alpha-glucan, water dikinase during starch turnover affects the surface properties and morphology of starch granules. New Phytol. 2014;203:495–507. doi: 10.1111/nph.12801. [DOI] [PubMed] [Google Scholar]
- 71.Skeffington AW, Graf A, Duxbury Z, Gruissem W, Smith AM. Glucan, water dikinase exerts little control over starch degradation in arabidopsis leaves at night. Plant Physiol. 2014;165:866–879. doi: 10.1104/pp.114.237016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Tagliabracci VS, Heiss C, Karthik C, Contreras CJ, Glushka J, Ishihara M, Azadi P, Hurley TD, DePaoli-Roach AA, Roach PJ. Phosphate incorporation during glycogen synthesis and Lafora disease. Cell Metab. 2011;13:274–282. doi: 10.1016/j.cmet.2011.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]