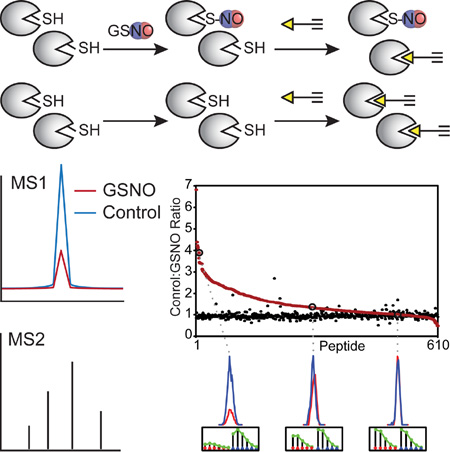
Summary
S-nitrosoglutathione (GSNO) is an endogenous transnitrosation donor involved in S-nitrosation of a variety of cellular proteins, thereby regulating diverse protein functions. Quantitative proteomic methods are necessary to establish which cysteine residues are most sensitive to GSNO-mediated transnitrosation. Here, a competitive cysteine-reactivity profiling strategy was implemented to quantitatively measure the sensitivity of >600 cysteine residues to transnitrosation by GSNO. This platform identified a subset of cysteine residues with a high propensity for GSNO-mediated transnitrosation. Functional characterization of previously unannotated S-nitrosation sites revealed that S-nitrosation of a cysteine residue distal to the 3-hydroxyacyl-CoA dehydrogenase type-2 (HADH2) active site impaired catalytic activity. Similarly, S-nitrosation of a non-catalytic cysteine residue in the lysosomal aspartyl protease cathepsin D (CTSD) inhibited proteolytic activation. Together, these studies revealed two previously uncharacterized cysteine residues that regulate protein function and established a chemical-proteomic platform with capabilities to determine substrate specificity of other cellular transnitrosation agents.
Graphical Abstract
eTOC Blurb
Zhou et al. apply a reactive-cysteine profiling strategy to rank ~600 cysteine residues by sensitivity to S-nitrosoglutathione (GSNO). Previously uncharacterized GSNO-sensitive cysteines on HADH2 and CTSD were shown to be important for catalytic activity and proteolytic processing, respectively.
Introduction
Nitric oxide (NO) plays critical roles in mammalian physiology. In the immune response, cytotoxic NO concentrations are generated by inducible NOS (iNOS) at infection sites (Forstermann and Sessa, 2012). In cardiovascular and neuronal signaling, endothelial nitric oxide synthase (eNOS) and neuronal NOS (nNOS) (Forstermann and Sessa, 2012) produce local bursts of NO that activate soluble guanylate cyclase (Derbyshire and Marletta, 2012) and mediate intercellular communication. Recently, an alternate NO signaling pathway has emerged involving modification of cysteines to form S-nitrosothiols (S-nitrosation) (Broniowska and Hogg, 2012; Smith and Marletta, 2012). Once formed, S-nitrosothiols are transferred via transnitrosation, whereby specific protein-small molecule or protein-protein interactions direct nucleophilic attack of a thiolate at the nitrogen of the S-nitrosothiol (Perissinotti et al., 2005; Wang et al., 2001). Cellular transnitrosation is predominately mediated by abundant thiol containing small molecules or proteins, such as glutathione (GSH) and thioredoxin, which form the corresponding S-nitrosated species GSNO and Trx-SNO, respectively (Broniowska et al., 2013; Smith and Marletta, 2012). GSNO is the primary small molecule S-nitrosothiol in eukaryotes, and cellular GSNO concentrations range from high nanomolar levels in cells with eNOS or nNOS activity to micromolar levels in cells where iNOS has been induced and activated (Seneviratne et al., 2013).
Over 3000 sites of protein S-nitrosation have been identified including cysteines within two NOS isoforms (eNOS and iNOS), glyceraldehyde-3-phosphate dehydrogenase (GAPDH), and caspase-3 (Casp3) (Doulias et al., 2013; Erwin et al., 2006; Gould et al., 2015; Lee et al., 2012; Mitchell et al., 2005; Mitchell and Marletta, 2005; Tristan et al., 2011). Transnitrosation of the active-site cysteine of Casp3 by Trx-SNO inhibits protease activity and governs the initiation of apoptosis (Mitchell et al., 2007). Similarly, S-nitrosation of GAPDH at Cys152 triggers GAPDH translocation to the nucleus and subsequent transnitrosation of nuclear proteins and initiation of apoptosis (Hara et al., 2005; Kornberg et al., 2010). Dysregulation of S-nitrosation is implicated in a variety of diseases including cystic fibrosis, asthma, hypertension, cancer, and neurodegeneration (Anand and Stamler, 2012). Importantly, small-molecule inhibitors of GSNO reductase, the primary enzyme that degrades cellular GSNO, are currently in clinical development for treatment of diseases that exhibit decreased endogenous S-nitrosothiol levels (Green et al., 2012).
Although the endogenous regulation of protein function through S-nitrosation is well established, the properties of an individual cysteine that govern sensitivity to S-nitrosation are poorly characterized (Doulias et al., 2010; Gould et al., 2015; Greco et al., 2006; Marino and Gladyshev, 2010). A potential consensus sequence for S-nitrosation has been identified for the protein complex comprised of iNOS, S100A8 and S100A9 (Jia et al., 2014), but no sequence or structural motif exists for the majority of physiological transnitrosation donors. The most widely utilized platform for identifying S-nitrosated proteins in complex proteomes is the Biotin-Switch Method (BSM) (Jaffrey and Snyder, 2001), which is comprised of three main steps: (1) blocking free cysteines with a thiol-reactive agent such as iodoacetamide, N-ethylmaleimide, or methyl methanethiosulfonate; (2) selective reduction of S-nitrosothiols with ascorbate; and (3) labeling the newly reduced cysteines with a biotinylated thiol-reactive agent. Since the initial development of the BSM, several modifications to this platform have been developed. These include the SNO-RAC and d-Switch methodologies, which introduce resin-assisted capture and isotopic labeling, to identify and quantify S-nitrosated species from proteomes (Fares et al., 2014; Forrester et al., 2009b; Sinha et al., 2010). Furthermore, protein S-nitrosothiols can be enriched with mercury resin (Doulias et al., 2013; Gould et al., 2015), phosphine reagents that undergo a Staudinger-type reaction (Bechtold et al., 2010; Seneviratne et al., 2013; Seneviratne et al., 2016; Wang and Xian, 2008), and biotinylated sulfinic acids that form stable thiosulfonates (Majmudar et al., 2016).
A significant limitation of existing methods to monitor S-nitrosation is the inability to quantitatively determine the extent of S-nitrosation for each individual cysteine, and thereby evaluate the sensitivity of a particular cysteine to a given transnitrosation donor. In this study, this limitation is addressed by implementing a competitive cysteine-labeling strategy to concurrently and quantitatively monitor the sensitivity to transnitrosation of hundreds of cysteines within the human proteome. Specifically, 609 cysteines in the human proteome were screened in parallel to determine the stoichiometry of S-nitrosation upon treatment with GSNO. To evaluate the functional consequences of S-nitrosation, two proteins that demonstrated high sensitivity to GSNO transnitrosation, 3-hydroxyacyl-CoA dehydrogenase type-2 (HADH2) and cathepsin D (CTSD), were selected for further biochemical evaluation. In both cases, S-nitrosation regulates protein function through a unique mechanism, thereby revealing previously uncharacterized functional roles for these cysteines.
Results
A competitive cysteine-labeling strategy to quantitatively measure sensitivity to transnitrosating agents
The extent of cysteine S-nitrosation was quantitatively monitored using a mass spectrometry (MS)-based proteomic platform that exploits the loss in nucleophilicity of a cysteine thiol upon conversion to a S-nitrosothiol, which is reflected in blocked reactivity with a cysteine-reactive chemical probe. This platform is derived from the previously reported isotopic tandem orthogonal proteolysis-ABPP (isoTOP-ABPP) method (Qian et al., 2013; Weerapana et al., 2010), which reliably and quantitatively monitors the intrinsic reactivity of 500–1000 cysteines concurrently within cell and tissue proteomes generated without protein denaturation. To date, the isoTOP-ABPP method and derivatives thereof have been successfully applied to quantitatively measure the loss in cysteine reactivity upon oxidation (Deng et al., 2013), reaction with lipid-derived electrophiles (Wang et al., 2014), and metal binding (Pace and Weerapana, 2014). In this approach (Figure 1), a cell lysate is exposed to GSNO or a buffer control. Both proteomes are then treated with a cysteine-reactive iodoacetamide-alkyne (IA) probe (Figure 1 inset), which covalently modifies hundreds of reactive and functional cysteines within the proteome. Following alkylation by the iodoacetamide of the IA probe, the terminal alkyne of the IA probe can be further functionalized through bioorthogonal copper(I)-catalyzed azide-alkyne cycloaddition (CuAAC) reactions (Rostovtsev et al., 2002; Weerapana et al., 2010). The IA probe-labeled proteins are covalently linked to light (GSNO-treated sample) or heavy (buffer control sample) biotin-azide tags (Azo-L and Azo-H) that contain an azobenzene linker for reductive cleavage by sodium dithionite (Qian et al., 2013). The light and heavy samples are then combined and subjected to streptavidin enrichment, on-bead trypsin digestion, and sodium dithionite cleavage to afford a mixture of light and heavy-labeled peptides for MS analysis. These cysteine-containing isotopically labeled peptides are identified using the fragmentation (MS/MS or MS2) spectra and the relative abundance between the GSNO and control samples is measured by the heavy:light ratio (R) that reflects the intensity of these species in the parent ion (MS1) spectra. An R-value of ~1 signifies a cysteine that was unaffected by GSNO treatment, whereas R-values > 1 indicate modification of that cysteine by GSNO, and a larger R-value reflects a greater percentage of cysteine modification.
Figure 1. A competitive cysteine-profiling strategy to quantify sensitivity to transnitrosation.
Cell lysates are treated with a transnitrosation donor (e.g. GSNO) or a buffer control, labeled with the IA probe, and subjected to CuAAC to append either the Azo-L (GSNO) or Azo-H (buffer) biotin-azide tags. The resulting isotopically tagged proteomes are mixed together and subjected to streptavidin enrichment and on-bead trypsin digestion. The probe-labeled peptides are eluted by reductive cleavage of the azobenzene with sodium dithionite and analyzed by high resolution LC/LC-MS/MS to identify (MS2) and quantify (MS1) the IA-probe reacted cysteines within each sample. A loss in cysteine reactivity is indicative of transnitrosation donor-mediated S-nitrosation or other cysteine modification.
Quantitatively measuring cysteine sensitivity to GSNO
MCF-7 breast cancer-cell lysate proteomes were subjected to the following treatments; (1) one hour treatment with 500 µM GSNO (light) versus no GSNO added (heavy) as the experimental sample (n=5); and (2) both the light and heavy samples left untreated by GSNO as a control (n=2) to verify the quantitative accuracy of the MS approach. In total, 610 cysteines were identified with heavy:light ratios in all of the experimental and control samples (Figures 2A and S1; Table S1). As expected, the control samples showed a limited distribution of R-values, closely centered on R = 1. In contrast, the experimental samples showed a wider distribution of R-values, where the majority of cysteines exhibited R-values ≥ 1 (Figure 2B). The majority of cysteines (583 out of 609; 96%) were minimally affected by GSNO treatment with R-values < 3. However, a small subset of cysteines (27 out of 609) showed high sensitivity (R ≥ 3) to GSNO treatment. The proteins containing these GSNO-sensitive cysteines serve diverse cellular functions, with the majority located on enzymes or structural proteins according to gene ontology (GO) analysis (Figure S2B).
Figure 2. Identifying cysteines sensitive to modification by GSNO.
(A) Heavy:light ratios (R) are shown for 610 cysteines identified in GSNO-treated (in red; light: 500 µM GSNO; heavy: buffer;) and control (in black; light: buffer; heavy: buffer) MS analyses (Table S1). R-values from the control sample are centered around R ~ 1, whereas R-values from the experimental sample show a large proportion of cysteines with R >> 1, indicating a loss in the free reduced form of individual cysteines upon GSNO treatment. A greater R-value is indicative of a higher extent of cysteine modification. Representative extracted ion chromatograms and isotopic envelopes (light: red; heavy: blue) are shown for three cysteines with high, moderate, and low sensitivity to GSNO. Ratio values are averaged across the different charge states identified, but displayed chromatograms are for a single representative charge state. The data shown here are for a representative dataset of GSNO and control proteomes. The averaged ratio values from 5 GSNO-treated and 2 buffer-treated analyses are shown with error bars in Figure S1. A complete list of all identified cysteines are displayed in Table S1 and gene ontology (GO) analyses for cysteines sensitive and insensitive to GSNO are shown in Figure S2. (B) Distribution of R-values for the control and GSNO-treated (500 µM) samples. (C) Of the four cysteines identified in PCBP1, Cys194 demonstrated high sensitivity to GSNO transnitrosation (R = 3.4), whereas Cys54, Cys109, and Cys158 were insensitive (R ~ 1). (D) All three cysteines identified in CLIC1 were insensitive to GSNO transnitrosation compared to the control (0 µM GSNO) sample (R ~ 1).
Of the GSNO-sensitive cysteines, one of the highest R-values of 4.9 was observed for Cys152 in GAPDH, which is indicative of ~80% modification of this cysteine when exposed to 500 µM GSNO. Cys152 is the active-site nucleophile of GAPDH and a well-characterized biologically relevant site of S-nitrosation (Hara et al., 2005; Kornberg et al., 2010), thereby validating the ability of this platform to detect functionally important sites of S-nitrosation. To evaluate the data obtained here within the context of previously identified sites of S-nitrosation, a comparison to the dbSNO database (Lee et al., 2012) was performed, which integrates data from 219 research articles to generate a list of >3000 S-nitrosation sites obtained through global proteomic analyses. Of the 27 cysteines observed to be highly sensitive to GSNO transnitrosation (R ≥ 3 at 500 µM GSNO), 14 were previously identified in the dbSNO database (Table S1). This comparison highlights the ability of this cysteine-profiling platform to identify both known and previously uncharacterized S-nitrosation sites. Furthermore, since this platform monitors S-nitrosation indirectly by the loss in cysteine reactivity to IA, it also enables detection of other cysteine modifications that decrease reactivity (e.g. glutathionylation) upon GSNO treatment.
Importantly, since this platform measures the extent of modification at the level of individual cysteines, the sensitivity of multiple cysteines within the same protein can be measured in a single experiment. For example, Cys194 in the poly(rC)-binding protein 1 (PCBP1), is highly sensitive to GSNO (R = 3.4) whereas three other observed cysteines, Cys54 (R = 1.2), Cys109 (R = 1.2), and Cys158 (R = 1.0), are insensitive to GSNO (Figure 2C). In contrast, the three cysteines observed for the chloride intracellular channel protein 1 (CLIC1), Cys24 (R = 1.1), Cys178 (R = 0.8), and Cys223 (R = 1.0), are all insensitive to GSNO (Figure 2D).
To further quantify cysteine sensitivity to GSNO, proteomes were treated with varying GSNO concentrations (0, 50, 100, 200, 500 µM) followed by labeling with the IA probe and Azo tags and subsequent MS analysis. R-values were quantified at each GSNO concentration for 576 cysteines (Table S2). Cysteines that were highly sensitive to transnitrosation at 500 µM GSNO, such as Cys329 in cathepsin D (CTSD) and Cys58 in HADH2, showed a concentration-dependent increase in modification at increasing GSNO concentrations (Figure 3A). In contrast, Cys32 in glutathione-S-transferase omega 1 (GSTO1) showed a R-value of ~1 at all GSNO concentrations (Figure 3A), confirming GSNO insensitivity. A wide array of GSNO transnitrosation sensitivities were observed as exemplified by plotting the R-values at increasing GSNO concentrations for cysteines from six different proteins (GAPDH, CTSD, HADH2, DCXR, GSTO1 and CLIC4) (Figure 3B).
Figure 3. Concentration-dependent analysis of cysteine-reactivity upon GSNO treatment.
(A) The light-tagged proteomes were treated with 0, 50, 100, 200, or 500 µM GSNO for 1 hr and compared to a buffer-treated heavy-tagged sample. Heavy:light ratio values (R) obtained for each identified cysteine at every GSNO concentration are shown in Table S2. Representative extracted ion chromatograms (light:red; heavy:blue) are shown for a cysteine that is insensitive to GSNO (GSTO1 Cys32) and two cysteines with high GSNO sensitivity (CTSD Cys329 and HADH2 Cys58). Cysteines with high sensitivity to GSNO show a clear concentration-dependent increase in R-values. (B) R-values are plotted with increasing GSNO concentration for six cysteines (from CLIC4, GSTO1, DCXR, HADH2, CTSD, and GAPDH) with varying GSNO sensitivities. The quantitative nature of the analysis allows ranking of cysteines by their GSNO sensitivity. (C) Corroboration of the MS-based identification of cysteine sensitivity and insensitivity using a gel-based assay for two proteins. Myc/His-tagged GSTO1 or CTSD overexpressing cell lysates were treated with varying GSNO concentrations followed by IA labeling. IA-labeled proteins were tagged with biotin using CuAAC and subjected to streptavidin enrichment, SDS-PAGE analysis, and immunoblotting with an anti-Myc antibody. A decrease in CTSD enrichment was observed at increasing GSNO concentrations, indicative of decreased cysteine reactivity and therefore IA labeling upon GSNO treatment. In contrast, no change in IA labeling and GSTO1 enrichment was observed at increasing GSNO concentrations, indicating insensitivity of the GSTO1 cysteines to GSNO.
In addition to the concentration-dependent labeling experiment, a time-dependent analysis was performed to determine how the abundance of individual cysteine modifications is altered over the course of 60 min upon treatment with 500 µM GSNO (Table S3). In this analysis, 191 cysteine-containing peptides were identified with ratios at 5 different time points (5, 10, 20, 30 and 60 min). Based on the kinetics and lifetime of modification, these cysteines fell into three classes: (1) a gradual increase in cysteine modification with time; (2) an initial increase in modification at 5 min that then stays constant with time; and (3) an increase in modification at an early time point that then decreases with time. The active-site cysteine of GAPDH (Cys152) falls in the first category, with a steady increase in modification over time. Interestingly, Cys73 of thioredoxin falls into the third category, where maximal modification is observed at 10 min (R = 4.5) followed by a decrease in modification at 60 min (R = 1.6). This observation suggests that S-nitrosated Cys73 in thioredoxin (Trx-C73-SNO) is short lived in cell lysates relative to other S-nitrosated species, consistent with the previously characterized role of Trx-C73-SNO as a transnitrosating agent that targets a variety of cellular proteins (Mitchell et al., 2007).
One highly sensitive cysteine to GSNO (CTSD) and one completely insensitive cysteine (GSTO1) were selected for verification using a secondary detection method. According to the MS data (Tables S1 and S2), both CTSD and GSTO1 were selectively labeled by the IA probe at a single cysteine. CTSD and GSTO1 were overexpressed by transient transfection in HEK293T cells with a C-terminal Myc/His tag. CTSD and GSTO1 overexpressing lysates were treated with increasing GSNO concentrations, followed by IA labeling, CuAAC and enrichment steps identical to that used for the MS analysis. Proteins were eluted from the streptavidin beads with sodium dithionite, separated by SDS-PAGE, and analyzed by immunoblotting with an anti-Myc antibody. Protein cysteine modification by GSNO is reflected by reduced protein detection in the immunoblot. In agreement with the MS data, GSTO1 enrichment was not affected by GSNO treatment (Figure 3C), confirming the insensitivity of GSTO1 to GSNO. In contrast, CTSD enrichment showed a concentration-dependent decrease upon GSNO treatment, mirroring the increasing R-values observed in the MS studies for CTSD Cys329 at increasing GSNO concentrations (Figure 3C).
Functional characterization of Cys58 in HADH2
To identify previously uncharacterized cysteines on enzymes within the human proteome that are posttranslationally regulated by S-nitrosation, the GSNO proteomics data was mined for cysteines that satisfied the following criteria: (1) demonstrated high sensitivity and clear concentration-dependence to GSNO transnitrosation (R > 1.3 at 50 µM GSNO, R > 1.5 at 100 µM GSNO, and R > 3 at 200 µM GSNO); (2) were not active-site or known regulatory cysteines; (3) were located distal to the active site of the enzyme; and (4) were found on enzymes implicated in human disease. Only two cysteines satisfied these four criteria–HADH2 Cys58 and CTSD Cys329–and were characterized further.
HADH2 (a.k.a. HSD17B10, HCD2, ERAB, MRPP2, SCHAD, and XH98G2) (Figure 4A) is a mitochondrial protein associated with numerous catalytic functions, including the third step in fatty acid beta-oxidation as well as oxidation of androgens and estrogens (Pu and Yang, 2000; Shafqat et al., 2003). HADH2 also has several non-catalytic functions, including forming a complex with mitochondrial ribonuclease P and amyloid-beta (Holzmann et al., 2008; Yan et al., 1997). Aberrant HADH2 activity is implicated in Alzheimer’s disease and other neurological disorders (Holzmann et al., 2008; Yan et al., 1997; Yang et al., 2011). Furthermore, inherited HADH2 mutations lead to 2-methyl-3-hydroxybutyryl-CoA dehydrogenase deficiency, which is characterized by neurological abnormalities (Ofman et al., 2003).
Figure 4. Functional characterization of Cys58 of HADH2.
(A) Structure of HADH2 (PDB: 2O23) with NAD+ bound in the active site. The GSNO transnitrosation-sensitive cysteine, Cys58, is located distal to the active site within a hydrophobic pocket flanked by Leu39 and Leu54 (inset). (B) S-nitrosation of recombinantly expressed and purified HADH2 WT treated with varying concentrations of GSNO or DEA NONOate. (C) S-nitrosation of recombinantly expressed and purified HADH2 WT or C58A treated with 100 µM GSNO or varying concentrations of Cys-NO. (D) HADH2 S-nitrosation in lysates from HEK293T cells overexpressing mock (empty vector), HADH2 WT, or HADH2 C58S treated with 100 µM GSNO. (E) HADH2 S-nitrosation in HEK293T cells overexpressing mock (empty vector), HADH2 WT, or HADH2 C58S were treated with DMSO (control), L-NAME (pan-NOS inhibitor), or 1400W (iNOS-selective inhibitor). (F) HADH2 S-nitrosation in A549 cells overexpressing HADH2 WT or HADH2 C58A were treated with cytokines (IFNγ, TNF-α, IL-1β) and/or L-NAME. In panels B-F, S-nitrosation was monitored using the biotin-switch method as detailed under Experimental Procedures. (G) HADH2-catalyzed reduction of acetoacetyl-CoA by NADH monitored in vitro by a decrease in absorbance at 340 nm. (H) Catalytic efficiency (kcat/KM) for WT and C58A-mutant HADH2 with varying acetoacetyl-CoA concentrations in the presence of 100 µM GSNO or GSH. (I) Catalytic efficiency (kcat/KM) for WT and C58S mutant HADH2 with varying acetoacetyl-CoA concentrations. (J) Effect of cysteine oxidants on HADH2 activity. HADH2 WT and C58A activity was monitored as described in Experimental Procedures in the presence of increasing concentrations of GSNO, DEA NONOate, SIN-1, GSSG, or H2O2. Error bars represent standard error from the mean.
Since the cysteine-profiling platform indirectly measures cysteine modification through loss in reactivity and since GSNO can result in S-nitrosation or glutathionylation, the ability of GSNO to S-nitrosate and glutathionylate HADH2 was directly compared. Recombinantly expressed and purified HADH2 (Figure S3A) was treated in vitro with increasing GSNO concentrations and analyzed for glutathionylation and S-nitrosation. While HADH2 glutathionylation was not observed under these conditions, a clear concentration-dependent increase in HADH2 S-nitrosation was observed (Figure S3B). The relative sensitivity of HADH2 cysteines towards GSNO, NO (released from DEA NONOate), and the alternative transnitrosating agent S-nitrosocysteine (Cys-NO) was determined. Purified HADH2 showed increasing levels of S-nitrosation up to 30 µM GSNO without further increases in S-nitrosation at up to 100 µM GSNO (Figure 4B and S3B). Conversely, DEA NONOate treatment did not result in HADH2 S-nitrosation (Figure 4B), which can be attributed to the differences in reaction mechanism between transnitrosation and S-nitrosation by NO generated from DEA NONOate. Interestingly, treatment with the alternative transnitrosation donor, Cys-NO, resulted in only low levels of S-nitrosation near the detection limit of the BSM (Figure 4C) indicating HADH2 is uniquely sensitive to GSNO transnitrosation. The observed HADH2 S-nitrosation signal was ascorbate-dependent, further confirming that GSNO treatment results in S-nitrosation (Figure 4B and 4C). To determine if HADH2 possesses a GSNO binding site that is necessary for transnitrosation, S-methylglutathione was used as a competitive inhibitor. Increasing S-methylglutathione concentrations partially diminished GSNO transnitrosation of HADH2, consistent with GSNO transnitrosation of HADH2 preceding through a GSNO-binding step (Figure S3C). The inability of S-methylglutathione to fully compete against GSNO transnitrosation may be due to the irreversibility of S-nitrosation on the timescale of the treatment or weak binding of S-methylglutathione to HADH2.
Although HADH2 contains four cysteines, the concentration-dependent cysteine-profiling studies identified Cys58 as the only cysteine in HADH2 sensitive to increasing GSNO concentrations (Table S2). Cys58 is located distal to the HADH2 active site within a hydrophobic pocket flanked by two leucines (Leu39 and Leu54) (Figure 4A). HADH2 transnitrosation by GSNO was dependent on the presence of Cys58 (Figure 4C and S3D) confirming that GSNO selectively modifies HADH2 only at Cys58. To confirm HADH2 is selectively S-nitrosated at Cys58 in the context of a proteome, HEK293T cell lysates overexpressing HADH2 with a C-terminal Myc/His tag were treated with GSNO, and as seen with recombinant HADH2, HADH2 WT but not HADH2 C58S was S-nitrosated under these conditions (Figure 4D). Overexpressed Myc/His tagged HADH2 exhibited immunofluorescence staining consistent with the mitochondrial localization of endogenous HADH2 (Figure S3E). To determine if HADH2 is selectively S-nitrosated in intact cells and if this S-nitrosation requires endogenous nitric oxide synthase (NOS) activity, HEK293T cells overexpressing HADH2 WT or C58S were treated with a pan-NOS inhibitor (L-N-nitroarginine methyl ester; L-NAME), an iNOS specific inhibitor (1400W), or left untreated. S-nitrosation of HADH2 WT but not HADH2 C58S was observed in cells without exogenous NO or S-nitrosothiol addition (Figure 4E). Although the exact cellular GSNO concentrations under these conditions are unknown, previous studies showed that endogenous GSNO concentrations in a variety of cell lines ranges from ~1–4 µM, and macrophage activation results in a ~2-fold increase in cellular GSNO concentrations (Seneviratne et al., 2013). Furthermore, NOS inhibition resulted in decreased HADH2 WT S-nitrosation for the pan-NOS inhibitor (L-NAME) but not the iNOS specific inhibitor (1400W) (Figure 4E), indicating that HADH2 is endogenously S-nitrosated at Cys58 by eNOS or nNOS activity and that either iNOS activity does not lead to HADH2 S-nitrosation or iNOS is not expressed under these conditions. Gene expression profiling studies show the presence of eNOS, nNOS and iNOS transcripts in HEK293T cells (Wu et al., 2009), but the relative protein levels of each isoform is unknown. To determine if HADH2 can be S-nitrosated by NO endogenously generated by iNOS, HADH2 S-nitrosation was monitored in A549 cells, a human adenocarcinoma line in which iNOS can be induced with cytokines. A549 cells overexpressing HADH2 WT or C58A were treated with cytokines (IFNγ, TNF-α, and IL-1β) with and without the pan-NOS inhibitor L-NAME. Cytokine-treated HADH2 WT was S-nitrosated under these conditions; S-nitrosation was diminished in the presence of L-NAME, in the absence of cytokines, by omitting ascorbate in the BSM in cells transfected with HADH2 WT, and by removal of Cys58 in the HADH2 C58A mutant (Figure 4F). These results suggest that NO synthesized by NOS, while not capable of directly S-nitrosating HADH2 (Figure 4B), generates transnitrosating agents (e.g. GSNO) that then transnitrosate Cys58 of HADH2.
Upon confirming that Cys58 in HADH2 is targeted by S-nitrosation, the effect of S-nitrosation on HADH2 enzymatic activity was investigated (Figure 4G). Initially, the activities of recombinant HADH2 WT and C58A (Figure 4H) were compared. The catalytic efficiency (kcat/KM) at varying acetoacetyl-CoA concentrations for HADH2 C58A (923 ± 90 M−1s−1) was indistinguishable from that of HADH2 WT (888 ± 37 M−1s−1). Treatment with GSNO led to a significant ~50% decrease (p = 0.005) in catalytic efficiency for the HADH2 WT protein (456 ± 27 M−1s−1), whereas HADH2 C58A activity was unaffected by GSNO treatment (906 ± 149 M−1s−1). Therefore, HADH2 S-nitrosation disrupts active-site binding or positioning of the acetoacetyl-CoA substrate to form a catalytically productive complex, likely through long-range structural perturbations. Interestingly, a ~75% decrease in catalytic efficiency (kcat/KM) at varying acetoacetyl-CoA concentrations was observed for HADH2 C58S (196 ± 41 M−1s−1) relative to HADH2 WT, even in the absence of GSNO (Figure 4I). Replacement of Cys58 by the more polar serine likely disrupts the hydrophobic pocket defined by Leu39 and Leu54, in which Cys58 is located (Figure 4A). Similarly, S-nitrosated Cys58 is likely poorly accommodated within this hydrophobic pocket, resulting in structural alterations that are transmitted to the active site. To determine if HADH2 activity is uniquely sensitive to GSNO compared to other cysteine-modifying agents, HADH2 WT was treated with varying concentrations of GSNO, NO (released from DEA NONOate), peroxynitrite (generated from from SIN-1), oxidized glutathione (GSSG), and hydrogen peroxide. Kinetic analyses at sub-saturating acetoacetyl-CoA concentrations demonstrated clear concentration-dependent inhibition of HADH2 WT by GSNO, but inhibition was not observed for HADH2 WT treated with the other oxidants or HADH2 C58A treated with GSNO (Figure 4J). Taken together, these studies identify a previously uncharacterized site of GSNO transnitrosation and demonstrate that HADH2 activity is allosterically regulated through this posttranslational modification.
Functional characterization of Cys329 in CTSD
The cysteine corresponding to Cys329 in the mouse CTSD ortholog was previously identified as S-nitrosated in a proteomic analysis of mouse heart tissue after myocardial ischemic preconditioning (Kohr et al., 2012; Lee et al., 2012), however, the functional consequences were not determined. CTSD levels are elevated in primary breast cancers and correlate with greater incidence of clinical metastasis, and high CTSD concentrations are an effective marker of cancer aggressiveness (Joyce et al., 2004; Liaudet-Coopman et al., 2006; Masson et al., 2010). CTSD is a lysosomal aspartyl protease that is biosynthesized as an inactive pre-pro-enzyme (Zaidi et al., 2008). To generate the active enzyme, CTSD undergoes several glycosylation and proteolytic cleavage steps during translocation from the site of synthesis in the endoplasmic reticulum (ER), through the Golgi compartment, to the lysosomes (Figure 5A) (Masa et al., 2006). The final catalytically active form of CTSD is comprised of a glycosylated and proteolytically cleaved light and heavy-chain that interact non-covalently. The uncleaved pro, intermediate, and pre-pro forms of CTSD are catalytically inactive. In crystal structures of the cleaved, active form of human CTSD, Cys329 forms a disulfide with Cys366 located distal to both the active site of the enzyme as well as the site of light and heavy chain-forming proteolytic cleavage (Figure 5B). The annotation of Cys329 as a disulfide was surprising, because in this and a previous study (Weerapana et al., 2010) Cys329 was found highly reactive towards the IA probe. A typical structural disulfide is inert and cannot react with the IA probe without prior reduction, suggesting the observed disulfide between Cys329 and Cys366 is either dynamic and redox modulated or a crystallization artifact.
Figure 5. Functional characterization of Cys329 of CTSD.
(A) CTSD is expressed in the endoplasmic reticulum as catalytically inactive pre-pro-CTSD, which undergoes a series of glycosylation and proteolytic cleavage steps during lysosomal translocation. The fully processed light and heavy-chain forms of CTSD interact non-covalently to form the active CTSD protease. (B) Structure of CTSD (PDB: 1LYB) showing the protease active site, the site of proteolytic cleavage into the active form, and the S-nitrosation-sensitive Cys329, which is found in a structural disulfide with Cys366. (C) CTSD S-nitrosation in HEK293T cell lysates overexpressing mock (empty vector), CTSD WT, C329A, and C366A treated with GSNO (100 µM). (D) CTSD S-nitrosation in HEK293T cells overexpressing CTSD WT or C329A pre-treated with Cys-SNO (100 µM). (E) CTSD S-nitrosation in lysates from HEK293T cells overexpressing CTSD WT or mock (empty vector). The lysates were split and analyzed for S-nitrosation with and without the ascorbate S-nitrosothiol reduction step of the BSM. In panels C-E S-nitrosation was monitored using the biotin-switch method as detailed under Experimental Procedures. (F) CTSD proteolytic activity of HEK293T cell lysates overexpressing CTSD WT, C329A, or C366A. The background activity from mock (empty plasmid)-treated lysates was subtracted. (G) Immunoblot with anti-CTSD antibody to monitor processing of pre-pro-CTSD (overexpressing CTSD WT, C329A, C366A, and C91A) into the active heavy-chain form. Lysates were normalized by GAPDH levels. Gel data from a second biological replicate are displayed in Figure S4A. (H) Processed heavy-chain CTSD was monitored in HEK293T cells overexpressing mock (empty vector) or CTSD WT upon treatment with Cys-SNO (0, 50, or 100 µM), by immunoblot with an anti-CTSD antibody. To control for protein loading, GAPDH levels in these same samples were measured by immunoblot with an anti-GAPDH antibody. The full gel image, as well as a gel image from a second biological replicate are shown in Figure S4B/C. (I) Quantification of the band intensities for CTSD and GAPDH at each Cys-SNO concentration in panel H.
Due to the numerous glycosylation and processing steps CTSD undergoes, recombinant expression and purification in a properly folded and active form was not feasible; therefore, to confirm the observed selective modification of CTSD at Cys329, HEK293T cells were transfected with plasmids containing CTSD WT, C329A, or C366A with a C-terminal Myc/His tag. CTSD-overexpressing cell lysates were treated with GSNO; S-nitrosation of CTSD WT was observed only upon GSNO treatment, and the observed S-nitrosation was abolished in the CTSD C329A mutant (Figure 5C). Conversely, mutation of Cys366, the disulfide partner of Cys329, to alanine did not eliminate the observed S-nitrosation (Figure 5C). Therefore, CTSD is selectively transnitrosated by GSNO at Cys329, despite the presence of nine other cysteines. In addition to treating cell lysates with GSNO, CTSD overexpressing cells were treated with S-nitrosocysteine (Cys-SNO), a cell-permeable transnitrosation donor (Broniowska et al., 2006; Zhang and Hogg, 2004). Analysis of lysates from Cys-SNO treated cells demonstrated S-nitrosation of CTSD WT, but not CTSD C329A, indicating CTSD is selectively S-nitrosated at Cys329 in intact cells (Figure 5D). To confirm the observed signal in the BSM was due to S-nitrosation and not other potential cysteine oxidations (e.g. glutathionylation), the BSM was repeated with and without ascorbate, which selectively reduces S-nitrosothiols over disulfides (Wang et al., 2008). The signal observed for CTSD WT S-nitrosation was dependent on the use of ascorbate in the BSM confirming that CTSD is modified intracellularly by S-nitrosation (Figure 5E). However, it is unclear in these cases if GSNO is the cellular transnitrosation donor.
To determine the role of Cys329 and S-nitrosation in CTSD function, cell lysates overexpressing CTSD WT or CTSD C329A and C366A mutants were assayed for CTSD catalyzed proteolysis. Interestingly, significant proteolysis activity above background was observed only for CTSD WT but not C329A and C366A lysates (Figure 5F). Further evaluation of CTSD expression by immunoblotting with an anti-CTSD antibody demonstrated that CTSD WT is proteolytically processed to generate the active heavy-chain form of the enzyme, whereas no heavy-chain form was observed for CTSD C329A or C366A (Figure 5G and S4A). The lack of active heavy-chain form correlated with the lack of CTSD activity in the CTSD C329A and C366A-overexpressing lysates. Therefore, Cys329 and the corresponding disulfide partner, Cys366, are critical to maintain the structural or specificity determinants necessary for efficient proteolytic processing into the active form of CTSD. Importantly, mutation of another CTSD cysteine, Cys91, to alanine did not affect CTSD processing into the active heavy chain form (Figure 5G and S4A). Cys91 is annotated to form a separate structural disulfide with Cys160 in CTSD. Successful proteolytic processing of CTSD C91A indicates the observed effect of cysteine mutation on CTSD processing is unique to the Cys329/Cys366 dynamic disulfide and is not common to all CTSD disulfides.
To determine if Cys329 S-nitrosation affects proteolytic processing in a manner similar to the C329A mutation, CTSD WT overexpressing cells were treated with Cys-SNO. Addition of increasing Cys-SNO concentrations to the media resulted in a reproducible decrease in the fully proteolytically processed, active heavy-chain form of CTSD (Figure 5H/I and S4B/C) consistent with the hypothesis that Cys329 S-nitrosation inhibits CTSD processing, resulting in decreased cellular CTSD activity. Treatment of CTSD WT overexpressing HEK293T cell lysates that contain fully processed CTSD with increasing GSNO concentrations did not inhibit proteolytic activity (Figure S5), further indicating that S-nitrosation does not affect the proteolytic activity of fully processed CTSD but instead inhibits cellular processing of the pre-pro-enzyme into the active heavy-chain form when S-nitrosated in intact cells.
Discussion
With the advent of proteomic methods to study S-nitrosation within complex proteomes ~3,000 cysteines targeted by S-nitrosation have been identified, which testifies to the ubiquity of this modification as a potential regulatory signaling mechanism. However, the majority of these proteomic methods are qualitative and provide minimal information on the stoichiometry of S-nitrosation. To discover functionally relevant S-nitrosation sites, a competitive cysteine-profiling strategy was utilized to concurrently and quantitatively rank 610 cysteines by their sensitivity to GSNO (Figure 2A and S1). In validation of this approach, one of the most sensitive cysteines to GSNO was the active-site cysteine of GAPDH, which is a well-established site of S-nitrosation that leads to GAPDH nuclear translocation, transnitrosation of nuclear proteins, and induction of apoptosis (Hara et al., 2005; Kornberg et al., 2010; Tristan et al., 2011). The cysteine-profiling platform described herein affords several advantages over the BSM, primarily the ability to quantify the extent of modification of individual cysteine residues. However, several limitations currently exist that may be improved in subsequent iterations of this platform. The platform is optimized for analysis of cell lysates treated with exogenous GSNO at largely supraphysiological concentrations. The data obtained provide important insight into the sensitivity of hundreds of cysteines to GSNO treatment, and for HADH2 in particular, the in vitro data correlated with cellular data (Figure 4 and S3). However, some targets may not be S-nitrosated to the extent predicted in vitro either due to lower cellular GSNO levels or instability in different subcellular reducing environments. Furthermore, the sensitivity of the current platform does not allow accurate quantification where the stoichiometry of modification is less than 5%.
This cysteine-profiling platform enabled identification and functional annotation of two uncharacterized modes of regulation through S-nitrosation. The dehydrogenase HADH2 was shown to be S-nitrosated at Cys58 and S-nitrosation of this distal cysteine allosterically inhibits HADH2 enzymatic activity (Figure 4). S-nitrosation of Cys58 is hypothesized to disrupt the hydrophobic interactions surrounding this cysteine (Figure 4A) resulting in long-range structural perturbations that reduce the ability of the acetoacetyl-CoA substrate to bind or form a productive catalytic complex in the HADH2 active site. Further studies are needed to determine if increased HADH2 S-nitrosation contributes to Alzheimer’s disease and other neurological disorders (Holzmann et al., 2008; Yan et al., 1997; Yang et al., 2011) that exhibit decreased HADH2 activity. Importantly, even though the initial proteomic studies that indicated HADH2 is highly sensitive to GSNO were performed at supraphysiological GSNO concentrations, follow-up characterization demonstrated Cys58 of HADH2 is S-nitrosated under physiologically relevant and endogenously generated nitrosative stress. This observation further underscores the promise of translating GSNO sensitivity data acquired through cysteine-reactivity profiling in cell lysates to (patho)physiologically relevant sites of S-nitrosation.
S-nitrosation of Cys329 on CTSD does not directly inhibit CTSD enzymatic activity (Figure S5) but indirectly lowers active cellular levels of CTSD by inhibiting proteolytic processing of the enzyme into its active light and heavy-chain form (Figure 5H/I and S4B/C). The inhibition of CTSD proteolytic processing is a novel mechanism for protein regulation through S-nitrosation. Interestingly, many aggressive breast cancers are characterized by high concentrations of the catalytically inactive, pre-pro-form of CTSD in the extracellular milieu (Fusek and Vetvicka, 2005; Joyce et al., 2004; Liaudet-Coopman et al., 2006). This secreted, unprocessed, and inactive CTSD increases breast cancer-cell proliferation, suggesting the role of CTSD in breast cancer expands beyond the proteolytic activity of this enzyme and may involve protein-protein interactions that lead to increased tumorigenesis (Glondu et al., 2001). The trigger for misprocessing and secretion of the pre-pro-form of CTSD in breast cancer is unknown, but these studies suggest that increased NOS activity results in CTSD S-nitrosation, misprocessing, and accumulation of the active pre-pro-form of CTSD for subsequent secretion from the cell. Indeed, increased iNOS expression and activity is associated with decreased survival in breast cancer (Glynn et al., 2010).
In conclusion, the application of a competitive cysteine reactivity profiling strategy to investigate transnitrosation unearthed two previously uncharacterized and mechanistically distinct modes of allosteric protein regulation through S-nitrosation. Future studies will focus on further functional characterization of unannotated S-nitrosation-sensitive cysteines identified in these global MS studies, as well as expansion of this platform to other biologically relevant transnitrosation donors.
Significance
S-nitrosation has recently emerged as a ubiquitous endogenous protein posttranslational modification that significantly impacts cellular protein function through a variety of mechanisms. Despite the advent of chemical and proteomic methods to study S-nitrosation, the subset of cellular cysteines that show uniquely high reactivity to endogenous transnitrosation donors is poorly characterized. To further these existing global studies, a cysteine-reactivity profiling strategy is applied herein to rank ~600 cysteines by sensitivity to GSNO, an abundant cellular transnitrosation donor. These proteomic studies revealed several previously uncharacterized sites of S-nitrosation, including Cys58 in HADH2 and Cys329 in CTSD. Further characterization of these protein targets revealed HADH2 catalytic activity is allosterically regulated by S-nitrosation, and importantly this modification occurs in cells at (patho)physiological levels of nitrosative stress. A novel mode of protein regulation by S-nitrosation was revealed for CTSD, whereby CTSD processing to the active protease was inhibited by S-nitrosation. Together, these studies lay the foundation for evaluating the substrate specificity of other physiologically relevant transnitrosation donors and enable the identification of previously uncharacterized, functionally relevant sites of S-nitrosation.
Experimental Procedures
Mass-spectrometry analysis of GSNO-treated MCF7 lysates
S-nitrosoglutathione (GSNO) was dissolved in freshly-made HEN buffer (125 mM HEPES pH 7.7, 1 mM EDTA, 0.1 mM neocuproine). The GSNO concentration was determined by UV absorbance at 336 nm based on an extinction coefficient of 0.9 mM−1cm−1. MCF7 cell lysates (4 mg/mL) were aliquoted to 500 µL and incubated with 5 µL HEN buffer or 5 µL of GSNO (50 µM, 100 µM, 200 µM, or 500 µM final concentration) in HEN buffer at 37 °C for 1 h r. Excess GSNO was removed by filtration through a Nap-5 column. GSNO and buffer-treated samples were treated with the IA probe (100 µM), subjected to CuAAC with light or heavy Azo-tags, and LC/LC-MS/MS analysis on an Orbitrap Discovery Mass Spectrometer (ThermoFisher) as previously described (Qian et al., 2013). Tandem MS data was searched using the SEQUEST algorithm against the human UniProt database and quantification of heavy:light ratios (R) was performed using the CIMAGE quantification package as detailed in the Supplemental Information.
Biotin switch method
All biotin switch assays were conducted similar to previously described (Forrester et al., 2009a) with modifications as detailed in the Supplemental Information. Briefly, free thiols were alkylated with blocking buffer (HEN buffer with 1% w/v SDS, 6 M Urea and 200 mM iodoacetamide) for 1 hr at 37 °C. Proteins were precipitated with 1.2 mL ice-cold high-purity acetone and incubated at -80 °C for 15 min. Precipi tated protein was pelleted, washed with acetone, air dried and resuspended into label buffer (PBS containing 1% w/v SDS, 30 mM ascorbate, 200 µM EZ-link biotin iodoacetamide). After 1 hr at 37 °C, the reactions were quenched with 6× Laemmli Sample Buffer (LSB) containing 100 mM DTT and analyzed by immunoblotting for biotin.
HADH2 activity assays
The purified HADH2 enzyme activity was measured spectrophotometrically using the decrease in absorbance at 340 nm due to NADH oxidation as described previously (Binstock and Schulz, 1981). Briefly, HADH2 WT and C58A (100 µL of 25 µM HADH2 in HEN buffer) were reduced with 100 µL of a 50% slurry of TCEP immobilized resin (Pierce) for 20–25 min at room temperature. The TCEP resin was removed by centrifugation, and the reduced HADH2 supernatant was diluted to 2.5 µM and incubated with 100 µM GSNO or GSH in HEN buffer at room temperature. After 30 min, HADH2 activity (0.5 µM final concentration) was assayed by monitoring the absorbance at 340 nm in 150 µL reactions containing 400 µM NADH, 100 mM sodium phosphate pH 7.5, and varying acetoacetyl-CoA concentrations from 50 µM to 1 mM as detailed in the Supplemental Information.
CTSD activity assays
CTSD activity was monitored using the Cathepsin D Activity Assay Kit (BioVision Incorporated, Milpitas, CA) as detailed in the Supplemental Information.
Monitoring processing of WT and mutant CTSD
The Myc-tagged pcDNA3.1(+) vectors containing CTSD WT, C91A, C329A or C366A were transfected into HEK293T cells as described in the Supplemental Information. The cells were harvested and the soluble lysates were isolated by removal of the supernatant after ultracentrifugation (45 min, 45,000 rpm, 4 °C). The cell lysates were normalized to 4.0 mg/mL before immunoblotting with an anti-CTSD antibody (1:1000 dilution) followed by an anti-Rabbit HRP-linked antibody (1:3333 dilution) to evaluate the processing of CTSD. The protocol for monitoring CTSD processing upon Cys-SNO treatment of cells is detailed in the Supplemental Information.
Supplementary Material
Highlights.
Over 600 cysteine residues are ranked by sensitivity to S-nitrosoglutathione (GSNO)
GSNO-sensitive cysteines include several previously uncharacterized residues
S-nitrosation of Cys58 on HADH2 allosterically regulates catalytic activity
Cys329 on CTSD is important for proteolytic processing to the active enzyme
Acknowledgments
We thank Dr. Megan C. Harwig for assistance with immunofluorescence staining. This work was financially supported by the Smith Family Foundation (E.W), the Damon Runyon Cancer Research Foundation DRR-18-12 (E.W.), Boston College (E.W.), NIH grant 1R01GM118431-01A1 (E.W.), The Scripps Research Institute (M.A.M.), an NIH postdoctoral fellowship 5F32GM095023 (B.C.S.), an American Heart Association Scientist Development Grant 15SDG25830057 (B.C.S.), an Institutional Research Grant # 14-247-29-IRG and # 86-004-26-IRG from the American Cancer Society (B.C.S.), and the Advancing a Healthier Wisconsin Endowment (B.C.S.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Supplemental Information includes detailed protocols for cell culture, mass-spectrometry analysis, biotin switch method to detect protein S-nitrosation, HADH2 and CTSD activity assays as well as five figures and three supplemental tables. These materials can be found with this article online.
Author Contributions
Conceptualization, B.C.S., M.A.M., and E.W.; Formal Analysis, E.W., B.C.S., Y.Z., S.M.C., S.L.W-S, and K.S.K.; Investigation, Y.Z., S.M.C, S.L.W-S., K.S.K., and B.C.S.; Resources, B.C.S., M.A.M., and E.W.; Writing – Original Draft, B.C.S. and E.W.; Writing – Review & Editing, B.C.S., M.A.M., and E.W.; Visualization, E.W., B.C.S., Y.Z., S.M.C, S.L.W-S, and K.S.K.; Supervision, B.C.S., M.A.M., and E.W.; Project Administration, B.C.S., M.A.M., and E.W.; Funding Acquisition, B.C.S., M.A.M., and E.W.
References
- Anand P, Stamler JS. Enzymatic mechanisms regulating protein S-nitrosylation: implications in health and disease. J Mol Med (Berl) 2012;90:233–244. doi: 10.1007/s00109-012-0878-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bechtold E, Reisz JA, Klomsiri C, Tsang AW, Wright MW, Poole LB, Furdui CM, King SB. Water-soluble triarylphosphines as biomarkers for protein S-nitrosation. ACS Chem Biol. 2010;5:405–414. doi: 10.1021/cb900302u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binstock JF, Schulz H. Fatty acid oxidation complex from Escherichia coli. Methods Enzymol. 1981;71 Pt C:403–411. doi: 10.1016/0076-6879(81)71051-6. [DOI] [PubMed] [Google Scholar]
- Broniowska KA, Diers AR, Hogg N. S-nitrosoglutathione. Biochim Biophys Acta. 2013;1830:3173–3181. doi: 10.1016/j.bbagen.2013.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broniowska KA, Hogg N. The chemical biology of S-nitrosothiols. Antioxid Redox Signal. 2012;17:969–980. doi: 10.1089/ars.2012.4590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broniowska KA, Zhang Y, Hogg N. Requirement of transmembrane transport for S-nitrosocysteine-dependent modification of intracellular thiols. J Biol Chem. 2006;281:33835–33841. doi: 10.1074/jbc.M603248200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng X, Weerapana E, Ulanovskaya O, Sun F, Liang H, Ji Q, Ye Y, Fu Y, Zhou L, Li J, et al. Proteome-wide quantification and characterization of oxidation-sensitive cysteines in pathogenic bacteria. Cell Host Microbe. 2013;13:358–370. doi: 10.1016/j.chom.2013.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derbyshire ER, Marletta MA. Structure and regulation of soluble guanylate cyclase. Annual Review of Biochemistry. 2012;81:533–559. doi: 10.1146/annurev-biochem-050410-100030. [DOI] [PubMed] [Google Scholar]
- Doulias PT, Greene JL, Greco TM, Tenopoulou M, Seeholzer SH, Dunbrack RL, Ischiropoulos H. Structural profiling of endogenous S-nitrosocysteine residues reveals unique features that accommodate diverse mechanisms for protein S-nitrosylation. Proc Natl Acad Sci U S A. 2010;107:16958–16963. doi: 10.1073/pnas.1008036107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doulias PT, Tenopoulou M, Greene JL, Raju K, Ischiropoulos H. Nitric oxide regulates mitochondrial fatty acid metabolism through reversible protein S-nitrosylation. Sci Signal. 2013;6:rs1. doi: 10.1126/scisignal.2003252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erwin PA, Mitchell DA, Sartoretto J, Marletta MA, Michel T. Subcellular targeting and differential S-nitrosylation of endothelial nitric-oxide synthase. J Biol Chem. 2006;281:151–157. doi: 10.1074/jbc.M510421200. [DOI] [PubMed] [Google Scholar]
- Fares A, Nespoulous C, Rossignol M, Peltier JB. Simultaneous identification and quantification of nitrosylation sites by combination of biotin switch and ICAT labeling. Methods Mol Biol. 2014;1072:609–620. doi: 10.1007/978-1-62703-631-3_41. [DOI] [PubMed] [Google Scholar]
- Forrester MT, Foster MW, Benhar M, Stamler JS. Detection of protein S-nitrosylation with the biotin-switch technique. Free Radic Biol Med. 2009a;46:119–126. doi: 10.1016/j.freeradbiomed.2008.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forrester MT, Thompson JW, Foster MW, Nogueira L, Moseley MA, Stamler JS. Proteomic analysis of S-nitrosylation and denitrosylation by resin-assisted capture. Nat Biotechnol. 2009b;27:557–559. doi: 10.1038/nbt.1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forstermann U, Sessa WC. Nitric oxide synthases: regulation and function. Eur Heart J. 2012;33:829–837. 837a–837d. doi: 10.1093/eurheartj/ehr304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fusek M, Vetvicka V. Dual role of cathepsin D: ligand and protease. Biomed Pap Med Fac Univ Palacky Olomouc Czech Repub. 2005;149:43–50. doi: 10.5507/bp.2005.003. [DOI] [PubMed] [Google Scholar]
- Glondu M, Coopman P, Laurent-Matha V, Garcia M, Rochefort H, Liaudet-Coopman E. A mutated cathepsin-D devoid of its catalytic activity stimulates the growth of cancer cells. Oncogene. 2001;20:6920–6929. doi: 10.1038/sj.onc.1204843. [DOI] [PubMed] [Google Scholar]
- Glynn SA, Boersma BJ, Dorsey TH, Yi M, Yfantis HG, Ridnour LA, Martin DN, Switzer CH, Hudson RS, Wink DA, et al. Increased NOS2 predicts poor survival in estrogen receptor-negative breast cancer patients. J Clin Invest. 2010;120:3843–3854. doi: 10.1172/JCI42059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gould NS, Evans P, Martinez-Acedo P, Marino SM, Gladyshev VN, Carroll KS, Ischiropoulos H. Site-Specific Proteomic Mapping Identifies Selectively Modified Regulatory Cysteine Residues in Functionally Distinct Protein Networks. Chem Biol. 2015;22:965–975. doi: 10.1016/j.chembiol.2015.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greco TM, Hodara R, Parastatidis I, Heijnen HF, Dennehy MK, Liebler DC, Ischiropoulos H. Identification of S-nitrosylation motifs by site-specific mapping of the S-nitrosocysteine proteome in human vascular smooth muscle cells. Proc Natl Acad Sci U S A. 2006;103:7420–7425. doi: 10.1073/pnas.0600729103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green LS, Chun LE, Patton AK, Sun X, Rosenthal GJ, Richards JP. Mechanism of inhibition for N6022, a first-in-class drug targeting S-nitrosoglutathione reductase. Biochemistry. 2012;51:2157–2168. doi: 10.1021/bi201785u. [DOI] [PubMed] [Google Scholar]
- Hara MR, Agrawal N, Kim SF, Cascio MB, Fujimuro M, Ozeki Y, Takahashi M, Cheah JH, Tankou SK, Hester LD, et al. S-nitrosylated GAPDH initiates apoptotic cell death by nuclear translocation following Siah1 binding. Nat Cell Biol. 2005;7:665–674. doi: 10.1038/ncb1268. [DOI] [PubMed] [Google Scholar]
- Holzmann J, Frank P, Loffler E, Bennett KL, Gerner C, Rossmanith W. RNase P without RNA: identification and functional reconstitution of the human mitochondrial tRNA processing enzyme. Cell. 2008;135:462–474. doi: 10.1016/j.cell.2008.09.013. [DOI] [PubMed] [Google Scholar]
- Jaffrey SR, Snyder SH. The biotin switch method for the detection of S-nitrosylated proteins. Sci STKE. 2001;2001:l1. doi: 10.1126/stke.2001.86.pl1. [DOI] [PubMed] [Google Scholar]
- Jia J, Arif A, Terenzi F, Willard B, Plow EF, Hazen SL, Fox PL. Target-selective protein S-nitrosylation by sequence motif recognition. Cell. 2014;159:623–634. doi: 10.1016/j.cell.2014.09.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joyce JA, Baruch A, Chehade K, Meyer-Morse N, Giraudo E, Tsai FY, Greenbaum DC, Hager JH, Bogyo M, Hanahan D. Cathepsin cysteine proteases are effectors of invasive growth and angiogenesis during multistage tumorigenesis. Cancer Cell. 2004;5:443–453. doi: 10.1016/s1535-6108(04)00111-4. [DOI] [PubMed] [Google Scholar]
- Kohr MJ, Aponte A, Sun J, Gucek M, Steenbergen C, Murphy E. Measurement of S-nitrosylation occupancy in the myocardium with cysteine-reactive tandem mass tags: short communication. Circ Res. 2012;111:1308–1312. doi: 10.1161/CIRCRESAHA.112.271320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornberg MD, Sen N, Hara MR, Juluri KR, Nguyen JV, Snowman AM, Law L, Hester LD, Snyder SH. GAPDH mediates nitrosylation of nuclear proteins. Nat Cell Biol. 2010;12:1094–1100. doi: 10.1038/ncb2114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee TY, Chen YJ, Lu CT, Ching WC, Teng YC, Huang HD. dbSNO: a database of cysteine S-nitrosylation. Bioinformatics. 2012;28:2293–2295. doi: 10.1093/bioinformatics/bts436. [DOI] [PubMed] [Google Scholar]
- Liaudet-Coopman E, Beaujouin M, Derocq D, Garcia M, Glondu-Lassis M, Laurent-Matha V, Prebois C, Rochefort H, Vignon F. Cathepsin D: newly discovered functions of a long-standing aspartic protease in cancer and apoptosis. Cancer Lett. 2006;237:167–179. doi: 10.1016/j.canlet.2005.06.007. [DOI] [PubMed] [Google Scholar]
- Majmudar JD, Konopko AM, Labby KJ, Tom CT, Crellin JE, Prakash A, Martin BR. Harnessing Redox Cross-Reactivity To Profile Distinct Cysteine Modifications. J Am Chem Soc. 2016;138:1852–1859. doi: 10.1021/jacs.5b06806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marino SM, Gladyshev VN. Structural analysis of cysteine S-nitrosylation: a modified acid-based motif and the emerging role of trans-nitrosylation. J Mol Biol. 2010;395:844–859. doi: 10.1016/j.jmb.2009.10.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masa M, Maresova L, Vondrasek J, Horn M, Jezek J, Mares M. Cathepsin D propeptide: mechanism and regulation of its interaction with the catalytic core. Biochemistry. 2006;45:15474–15482. doi: 10.1021/bi0614986. [DOI] [PubMed] [Google Scholar]
- Masson O, Bach AS, Derocq D, Prebois C, Laurent-Matha V, Pattingre S, Liaudet-Coopman E. Pathophysiological functions of cathepsin D: Targeting its catalytic activity versus its protein binding activity? Biochimie. 2010;92:1635–1643. doi: 10.1016/j.biochi.2010.05.009. [DOI] [PubMed] [Google Scholar]
- Mitchell DA, Erwin PA, Michel T, Marletta MA. S-Nitrosation and regulation of inducible nitric oxide synthase. Biochemistry. 2005;44:4636–4647. doi: 10.1021/bi0474463. [DOI] [PubMed] [Google Scholar]
- Mitchell DA, Marletta MA. Thioredoxin catalyzes the S-nitrosation of the caspase-3 active site cysteine. Nat Chem Biol. 2005;1:154–158. doi: 10.1038/nchembio720. [DOI] [PubMed] [Google Scholar]
- Mitchell DA, Morton SU, Fernhoff NB, Marletta MA. Thioredoxin is required for S-nitrosation of procaspase-3 and the inhibition of apoptosis in Jurkat cells. Proc Natl Acad Sci U S A. 2007;104:11609–11614. doi: 10.1073/pnas.0704898104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ofman R, Ruiter JP, Feenstra M, Duran M, Poll-The BT, Zschocke J, Ensenauer R, Lehnert W, Sass JO, Sperl W, et al. 2-Methyl-3-hydroxybutyryl-CoA dehydrogenase deficiency is caused by mutations in the HADH2 gene. Am J Hum Genet. 2003;72:1300–1307. doi: 10.1086/375116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pace NJ, Weerapana E. A competitive chemical-proteomic platform to identify zinc-binding cysteines. ACS Chem Biol. 2014;9:258–265. doi: 10.1021/cb400622q. [DOI] [PubMed] [Google Scholar]
- Perissinotti LL, Turjanski AG, Estrin DA, Doctorovich F. Transnitrosation of nitrosothiols: characterization of an elusive intermediate. Journal of the American Chemical Society. 2005;127:486–487. doi: 10.1021/ja044056v. [DOI] [PubMed] [Google Scholar]
- Pu X, Yang K. Guinea pig 11beta-hydroxysteroid dehydrogenase type 1: primary structure and catalytic properties. Steroids. 2000;65:148–156. doi: 10.1016/s0039-128x(99)00098-7. [DOI] [PubMed] [Google Scholar]
- Qian Y, Martell J, Pace NJ, Ballard TE, Johnson DS, Weerapana E. An isotopically tagged azobenzene-based cleavable linker for quantitative proteomics. Chembiochem. 2013;14:1410–1414. doi: 10.1002/cbic.201300396. [DOI] [PubMed] [Google Scholar]
- Rostovtsev VV, Green LG, Fokin VV, Sharpless KB. A stepwise huisgen cycloaddition process: coppeRI)-catalyzed regioselective "ligation" of azides and terminal alkynes. Angew Chem Int Ed Engl. 2002;41:2596–2599. doi: 10.1002/1521-3773(20020715)41:14<2596::AID-ANIE2596>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- Seneviratne U, Godoy LC, Wishnok JS, Wogan GN, Tannenbaum SR. Mechanism-based triarylphosphine-ester probes for capture of endogenous RSNOs. Journal of the American Chemical Society. 2013;135:7693–7704. doi: 10.1021/ja401565w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seneviratne U, Nott A, Bhat VB, Ravindra KC, Wishnok JS, Tsai LH, Tannenbaum SR. S-nitrosation of proteins relevant to Alzheimer's disease during early stages of neurodegeneration. Proc Natl Acad Sci U S A. 2016 doi: 10.1073/pnas.1521318113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shafqat N, Marschall HU, Filling C, Nordling E, Wu XQ, Bjork L, Thyberg J, Martensson E, Salim S, Jornvall H, et al. Expanded substrate screenings of human and Drosophila type 10 17beta-hydroxysteroid dehydrogenases (HSDs) reveal multiple specificities in bile acid and steroid hormone metabolism: characterization of multifunctional 3alpha/7alpha/7beta/17beta/20beta/21-HSD. Biochem J. 2003;376:49–60. doi: 10.1042/BJ20030877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinha V, Wijewickrama GT, Chandrasena RE, Xu H, Edirisinghe PD, Schiefer IT, Thatcher GR. Proteomic and mass spectroscopic quantitation of protein S-nitrosation differentiates NO-donors. ACS Chem Biol. 2010;5:667–680. doi: 10.1021/cb100054m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith BC, Marletta MA. Mechanisms of S-nitrosothiol formation and selectivity in nitric oxide signaling. Curr Opin Chem Biol. 2012;16:498–506. doi: 10.1016/j.cbpa.2012.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tristan C, Shahani N, Sedlak TW, Sawa A. The diverse functions of GAPDH: views from different subcellular compartments. Cell Signal. 2011;23:317–323. doi: 10.1016/j.cellsig.2010.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C, Weerapana E, Blewett MM, Cravatt BF. A chemoproteomic platform to quantitatively map targets of lipid-derived electrophiles. Nat Methods. 2014;11:79–85. doi: 10.1038/nmeth.2759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Xian M. Fast reductive ligation of S-nitrosothiols. Angew Chem Int Ed Engl. 2008;47:6598–6601. doi: 10.1002/anie.200801654. [DOI] [PubMed] [Google Scholar]
- Wang K, Wen Z, Zhang W, Xian M, Cheng JP, Wang PG. Equilibrium and kinetics studies of transnitrosation between S-nitrosothiols and thiols. Bioorg Med Chem Lett. 2001;11:433–436. doi: 10.1016/s0960-894x(00)00688-0. [DOI] [PubMed] [Google Scholar]
- Wang X, Kettenhofen NJ, Shiva S, Hogg N, Gladwin MT. Copper dependence of the biotin switch assay: modified assay for measuring cellular and blood nitrosated proteins. Free Radic Biol Med. 2008;44:1362–1372. doi: 10.1016/j.freeradbiomed.2007.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weerapana E, Wang C, Simon GM, Richter F, Khare S, Dillon MB, Bachovchin DA, Mowen K, Baker D, Cravatt BF. Quantitative reactivity profiling predicts functional cysteines in proteomes. Nature. 2010;468:790–795. doi: 10.1038/nature09472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C, Orozco C, Boyer J, Leglise M, Goodale J, Batalov S, Hodge CL, Haase J, Janes J, Huss JW, 3rd, et al. BioGPS: an extensible and customizable portal for querying and organizing gene annotation resources. Genome Biol. 2009;10:R130. doi: 10.1186/gb-2009-10-11-r130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan SD, Fu J, Soto C, Chen X, Zhu H, Al-Mohanna F, Collison K, Zhu A, Stern E, Saido T, et al. An intracellular protein that binds amyloid-beta peptide and mediates neurotoxicity in Alzheimer's disease. Nature. 1997;389:689–695. doi: 10.1038/39522. [DOI] [PubMed] [Google Scholar]
- Yang SY, He XY, Miller D. Hydroxysteroid (17beta) dehydrogenase X in human health and disease. Mol Cell Endocrinol. 2011;343:1–6. doi: 10.1016/j.mce.2011.06.011. [DOI] [PubMed] [Google Scholar]
- Zaidi N, Maurer A, Nieke S, Kalbacher H. Cathepsin D: a cellular roadmap. Biochem Biophys Res Commun. 2008;376:5–9. doi: 10.1016/j.bbrc.2008.08.099. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Hogg N. The mechanism of transmembrane S-nitrosothiol transport. Proc Natl Acad Sci U S A. 2004;101:7891–7896. doi: 10.1073/pnas.0401167101. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.