Summary
TGF-β signaling regulates a variety of cellular processes, including proliferation, apoptosis, differentiation, immune responses, and fibrogenesis. Here, we describe a lysine methylation-mediated mechanism that controls the pro-fibrogenic activity of TGF-β. We find that the methyltransferase Set9 potentiates TGF-β signaling by targeting Smad7, an inhibitory downstream effector. Smad7 methylation promotes interaction with the E3 ligase Arkadia and, thus, ubiquitination-dependent degradation. Depletion or pharmacological inhibition of Set9 results in elevated Smad7 protein levels and inhibits TGF-β-dependent expression of genes encoding extracellular matrix components. The inhibitory effect of Set9 on TGF-β-mediated extracellular matrix production is further demonstrated in mouse models of pulmonary fibrosis. Lung fibrosis induced by bleomycin or Ad-TGF-β treatment was highly compromised in Set9-deficient mice. These results uncover a complex regulatory interplay among multiple Smad7 modifications and highlight the possibility that protein methyltransferases may represent promising therapeutic targets for treating lung fibrosis.
Graphical Abstract

Highlights
-
•
Set9 (Setd7) methylates Smad7 at lysine-70
-
•
Methylated Smad7 interacts with Arkadia and is rapidly degraded
-
•
Set9 function is required for TGF-β-mediated activation of ECM genes
-
•
Set9 function is required for bleomycin- or Ad-TGF-β-induced pulmonary fibrosis
Elkouris et al. find that Set9 regulates the TGF-β-signaling pathway and is required for the development of pulmonary fibrosis in mice. The authors find that Set9-mediated methylation of inhibitory Smad7 leads to its degradation, thus affecting TGF-β-dependent activation of extracellular matrix genes.
Introduction
TGF-β is a pleiotropic cytokine involved in the regulation of cell fate determination, extracellular matrix (ECM) production, cell growth, motility, differentiation, and the immune response (Massagué, 2012, Travis and Sheppard, 2014, Lamouille et al., 2014, ten Dijke and Hill, 2004, Wakefield and Hill, 2013). TGF-β cytokines bind to type I and type II serine-threonine kinase receptors and transmit their signals through Smad proteins. Receptor-regulated Smads (R-Smads; Smad2 and Smad3) are directly phosphorylated by type I receptor kinases to form a heteromeric complex with the Co-Smad protein Smad4, which can translocate to the nucleus and regulate target gene transcription (Massagué, 2012, ten Dijke and Hill, 2004, Wakefield and Hill, 2013). The pathway is under control by the inhibitory Smad7, which escorts E3-ubiquitin ligases (Smurf1 and Smurf2) to the plasma membrane and facilitates the proteasomal destruction of TGF-β receptors and also itself (Ebisawa et al., 2001, Kavsak et al., 2000, Yan et al., 2009, Grönroos et al., 2002, Ogunjimi et al., 2005, Simonsson et al., 2005).
Dysfunction of TGF-β signaling has been implicated in the pathogenesis of fibrotic diseases, rheumatoid arthritis, and cancer (Blobe et al., 2000). In particular, TGF-β signaling plays a central role in the development of pulmonary fibrosis by promoting the proliferation and transdifferentiation of lung fibroblasts and epithelial cells to collagen-producing myofibroblasts (Wynn, 2011). Although the link between TGF-β signaling and pulmonary fibrosis was identified two decades ago (Border and Noble, 1994), little progress has been made in pharmacological intervention strategies for this fatal disease. Effective therapy could be achieved by targeting the profibrogenic activity of TGF-β.
A potential strategy toward the above goal would involve targeting posttranslational modifications of proteins involved in the regulation of the pathway. Apart from the phosphorylation and ubiquitination described above, Smad proteins are also subject to regulation by other modifications. For example, Smad7 can be acetylated by the histone acetyltransferase p300 at lysine-64 and lysine-70, which antagonizes its ubiquitination and degradation (Grönroos et al., 2002, Simonsson et al., 2005). The BMP-signaling effector molecule Smad6 interacts with the protein arginine methyltransferase PRMT and is di-methylated at arginine-74 and -81 residues (Inamitsu et al., 2006, Xu et al., 2013). Arginine-74-methylated Smad6 is released from the type I receptor and facilitates the initial steps of the BMP-signaling pathway via de-repression of Smad1/5 (Xu et al., 2013).
Here we show that Set9 methylates Smad7, the inhibitory Smad of the TGF-β-signaling pathway, and influences the profibrogenic effects of TGF-β by modulating the expression of ECM genes. Set9 (SETD7; KMT7) is the founding member of the methyltransferase enzymes, which can methylate non-histone protein substrates, including the transcription factors TAF10, p53, Stat3, E2F1, Rb, ERα, NF-κB, and DNMT1 (Kouskouti et al., 2004, Chuikov et al., 2004, Yang et al., 2010, Kontaki and Talianidis, 2010a, Carr et al., 2011, Munro et al., 2010, Subramanian et al., 2008, Pradhan et al., 2009, Sarris et al., 2014, Hamamoto et al., 2015). Methylation can regulate transcription factor function in many different ways: it can modulate protein stability, the deposition of other modifications, nuclear retention and recruitment to genomic targets, transactivation potential, and interaction with other proteins (Sarris et al., 2014). Depending on the substrate, Set9 regulates various signaling and developmental pathways. Despite the plethora of studies in different cell lines, the in vivo function of Set9 in animals remains unexplored.
The results presented here suggest that Set9-dependent methylation and consequent modulation of Smad7 protein stability plays a central role in TGF-β-dependent activation of ECM genes and the development of pulmonary fibrosis. Inhibition of Set9 has a protective effect against cellular damage- or cytokine-induced pulmonary fibrosis.
Results
Set9 Is Required for TGF-β-Mediated Activation of ECM Genes
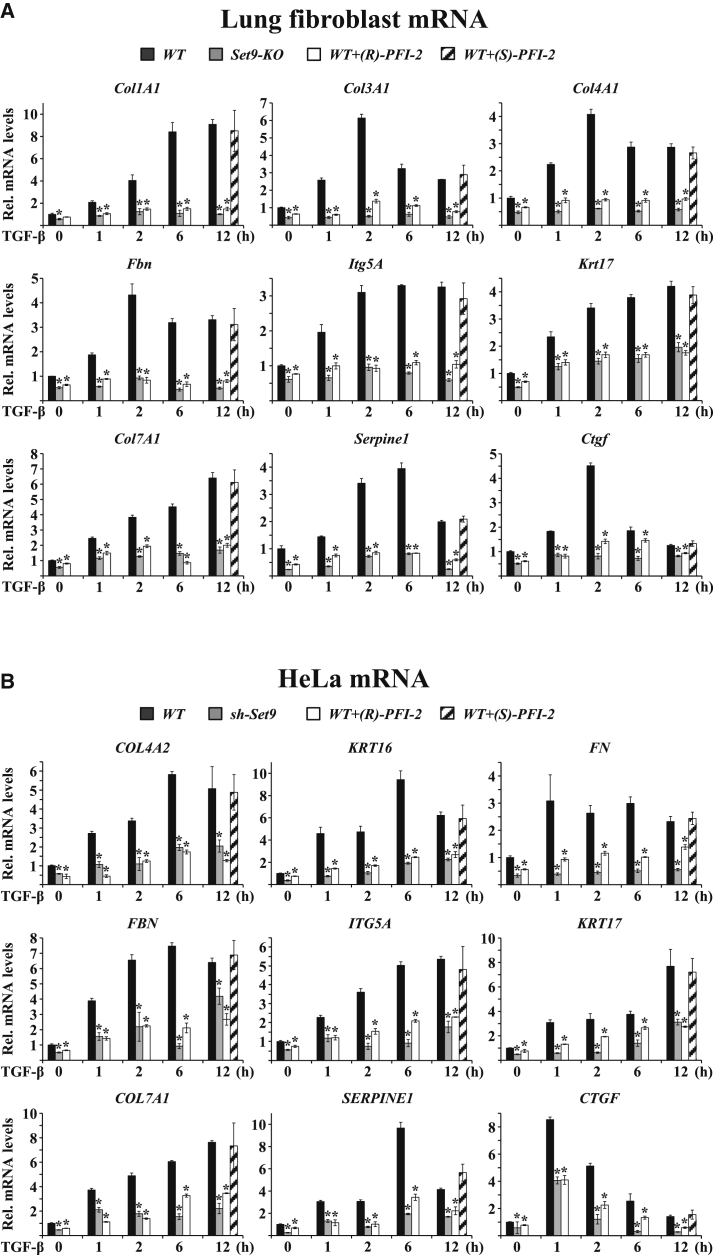
To gain insight into the in vivo function of Set9, we generated a knockout (KO) mouse model by deleting exon-4 of the Setd7 gene (Figures S1A–S1C). Mice with homozygous deletions (Set9-KO) develop normally and are fertile. Examination of 2-month-old Set9-KO mice did not reveal visible phenotypes. To expose potential hidden phenotypes, we isolated primary lung fibroblasts from wild-type (WT) and Set9-KO mice and subjected them to different induction conditions. The initial data indicated that the steady-state mRNA levels of several ECM genes were decreased in Set9-deficient cells. Since TGF-β-signaling pathway plays a key role in the regulation of ECM gene expression (Wynn, 2011), we tested the effect of Set9 inactivation in TGF-β-dependent gene activation. As expected, the mRNA levels of several ECM genes, including Col1A1, Col3A1, Col4A1, Fbn, Itg5A, Krt17, Col7A1, Serpine1, and Ctgf, were highly increased in TGF-β-treated WT cells, reaching a maximum at different time periods after treatment (Figure 1A). However, TGF-β-dependent stimulation of all of the studied ECM genes was greatly compromised in fibroblasts isolated from Set9-KO mice (Figure 1A).
Figure 1.
Set9-Dependent Regulation of TGF-β-Induced Expression of Extracellular Matrix Genes
(A) qPCR analysis of the mRNA levels of the indicated panel of extracellular matrix (ECM) genes in isolated primary lung fibroblasts that were treated in vitro with 5 ng/ml TGF-β for the indicated time periods (h, hours). Lung fibroblasts were from wild-type mice (WT, black bars) or Set9-KO mice (gray bars). WT mice were treated with 1 μM (R)-PFI-2 Set9 inhibitor (white bars) or with 1 μM (S)-PFI-2 inactive enantiomer (stripped bars). Bars represent mean values of mRNA levels normalized to Gapdh mRNA ± SEM from four experiments performed in triplicates (∗p < 0.05, two-tailed Student’s t test).
(B) qPCR analysis of mRNA from WT HeLa cells and HeLa cells stably expressing sh-Set9 (sh-Set9). Treatments and analyses were performed as in (A).
See also Figure S1.
Co-treatment of WT cells with a newly characterized, cell-permeable, highly specific Set9 inhibitor, (R)-PFI-2 (Barsyte-Lovejoy et al., 2014), also prevented TGF-β-dependent activation of ECM genes (Figure 1A). The specificity of the effect was confirmed by the lack of inhibition in cells treated with the inactive enantiomer (S)-PFI-2 (Figure 1A). Similar to primary fibroblasts, TGF-β-mediated stimulation of a panel of ECM genes was diminished in the human cervical carcinoma HeLa cell line following small hairpin RNA (shRNA)-mediated knockdown of Set9 or upon inhibition of Set9 enzymatic activity by (R)-PFI-2 treatment (Figure 1B). Since TGF-β or (R)-PFI-2 inhibitor treatment did not influence the protein levels of Set9 (Figures S1D and S1E), the above results demonstrate that Set9 enzymatic activity is required for TGF-β-dependent activation of ECM genes. The effect of Set9 inactivation also was observed on other classes of TGF-β-regulated genes, including HmgA2, Has2, Twist, and Snai1, which regulate epithelial-mesenchymal transition (EMT) (Valcourt et al., 2005, Moustakas and Heldin, 2014) (Figure S1F). The high number of genes affected indicates that Set9 may not function in a gene-specific manner but at a more upstream level of the signaling pathway.
Set9 Methylates Smad7 at Lysine-70 Residue and Regulates Its Stability
To investigate the molecular mechanism, we first tested the role of p53, a known substrate of Set9 (Chuikov et al., 2004), which can cooperate with Smad proteins to mediate TGF-β responses (Cordenonsi et al., 2003). Loss of Set9 in lung fibroblasts did not affect TGF-β-mediated stabilization of p53 (Figure S1G). Furthermore, in p53-deficient H1299 lung carcinoma cells, Set9 depletion greatly compromised TGF-β-dependent activation of COL7A1, SERPINE1, and CTGF expression (Figure S1H), demonstrating that p53 is not required for Set9-dependent modulation of ECM gene expression.
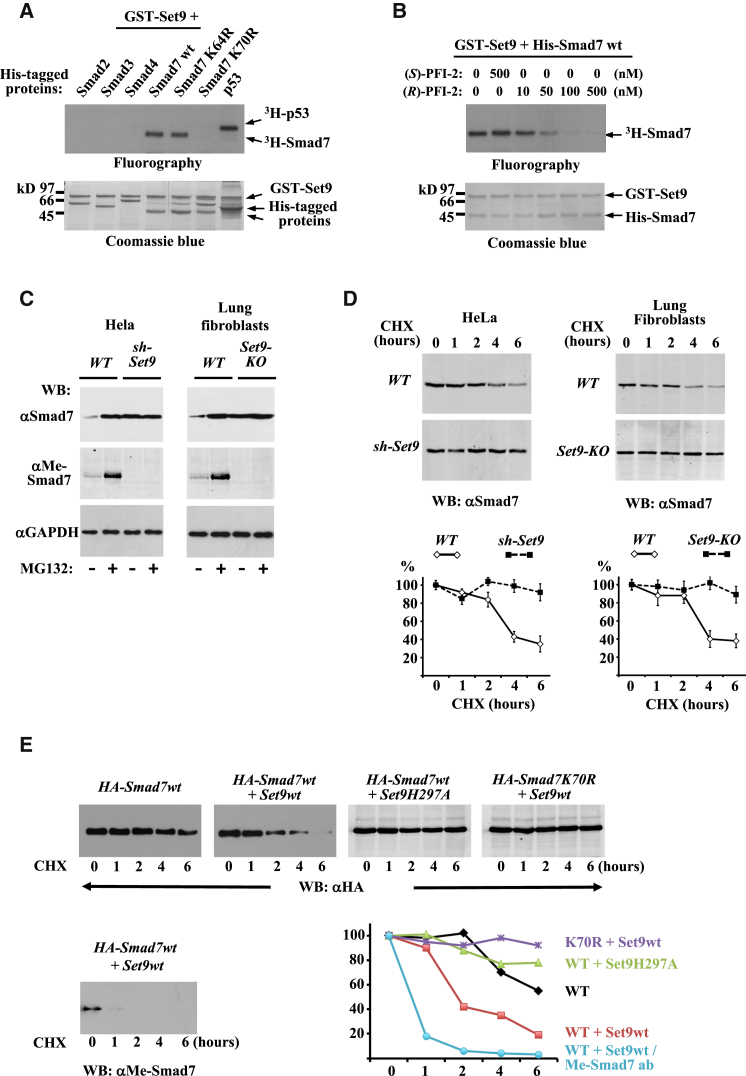
Next we investigated whether Smad proteins, the main downstream effectors of the pathway, are potential enzymatic substrates of Set9. In vitro methylation assays performed with Smad2, Smad3, Smad4, and Smad7 recombinant proteins revealed that only Smad7 could be methylated by Set9 (Figure 2A). Methylation of Smad7 in vitro was sensitive to nanomolar concentrations of (R)-PFI-2, but not to (S)-PFI-2 (Figure 2B). Mass spectrometry (MS) analysis was used to identify sites of methylation on in-vitro-methylated Smad7 protein. We detected 28 tryptic and four AspN-specific peptides in total, covering 57% of the Smad7 sequence. Accurate mass measurement revealed one mono-methylated peptide of 26 amino acids, and tandem MS identified lysine-70 as the site of methylation (Figure S2). No corresponding non-methylated peptide was identified, as the non-methylated lysine would be subject to tryptic digestion in contrast to the methylated one. Indeed, we identified the 23-amino acid-long peptide starting at the glycine following lysine-70. To verify the MS results, we constructed a mutant Smad7 replacing the identified lysine with an arginine. The K70R mutant was not methylated, suggesting that K70 is the main methylation site of Smad7 (Figure 2A).
Figure 2.
Set9 Methylates Smad7 and Modulates Smad7 Stability
(A and B) Set9 methylates Smad7. Fluorographs and Coomassie blue-stained gels of in vitro methylation reactions carried out with the indicated recombinant Smad proteins and p53 as positive control are shown. Where indicated in (B), (R)-PFI-2 and (S)-PFI-2 also were included in the reactions.
(C) Western blot analysis, with the indicated antibodies recognizing all forms of Smad7 proteins (αSmad7) or K70-methylated Smad7 (αMe-Smad7) and Gapdh, was performed in whole-cell extracts from WT and Set9-depleted (sh-Set9) HeLa cells or primary lung fibroblasts from WT and Set9-KO mice. The cells were treated with 5 ng/ml TGF-β and, where indicated (+), with 5 μM MG132 for 5 hr.
(D) WT and sh-Set9 HeLa cells or WT and Set9-KO lung fibroblasts were treated with 100 μg/ml cycloheximide (CHX). Representative western blots of whole-cell extracts prepared at the indicated time points following CHX addition with αSmad7 antibody are shown. Graphs show average band intensities, measured by ImageJ, ±SEM relative to untreated controls from three experiments.
(E) HeLa cells were transfected with the indicated combinations of HA-tagged WT (HA-Smad7wt) or K70R non-methylatable mutant form of Smad7 and myc-tagged WT (Set9wt) or enzymatically inactive H297A mutant form of Set9 expression vectors. Then 2 days after transfection, the cells were treated with 50 μg/ml CHX for the indicated times. Whole-cell extracts were analyzed in western blots using anti-HA (α-HA) antibodies (upper panels) or anti-Me-Smad7 antibody (bottom panel) to estimate the amounts of total Smad7 and methylated Smad7 proteins. One representative of two experiments is shown. The graph shows band intensities, measured by ImageJ software, relative to untreated controls.
See also Figure S2.
These data indicated that Smad7 is a primary candidate target through which Set9 exerts its modulatory function on ECM gene regulation. This was confirmed by the data showing rescue of TGF-β-mediated inducibility of gene expression in Set9-deficient HeLa cells when Smad7 also was depleted by small interfering RNA (siRNA). As expected, knockdown of Smad7 resulted in elevated steady-state- and TGF-β-induced mRNA levels of COL4A2, FN, FBN, ITG5A, KRT16, and SERPINE1 genes (Figures S2B and S2C). Importantly, increased expression of these genes was also observed in Set9-depleted cells after Smad7 knockdown (Figure S2B), suggesting that Set9-mediated regulation of ECM genes requires Smad7.
To examine the effect of methylation of Smad7 in vivo, we generated a methyl-specific Smad7 antibody, whose specificity was verified using in-vitro-methylated proteins (Figure S3). Methylated endogenous Smad7 was readily detected in extracts of lung fibroblasts or HeLa cells that were treated with the proteasome inhibitor MG132 (Figure 2C). In untreated cells, however, the methyl-Smad7 signal could hardly be detected, and it was completely absent in extracts from Set9-KO lung fibroblasts or sh-Set9-expressing HeLa cells (Figure 2C). The above results suggest that methylation leads to destabilization of Smad7. This scenario was further supported by the detection of highly elevated total Smad7 protein levels in sh-Set9 HeLa cells and in Set9-KO lung fibroblasts (Figure 2C). In fact, the amount of total Smad7 in untreated Set9-deficient cells was similar to that detected in MG132-treated WT cells and was not increased further by MG132 treatment (Figure 2C). In agreement with the above, the half-life of endogenous Smad7 significantly increased in Set9-deficient lung fibroblasts and Set-9-depleted HeLa cells (Figure 2D). In transfection experiments, overexpression of WT Set9, but not its enzymatically inactive mutant form (Set9H297A), resulted in decreased half-life of Smad7 protein (Figure 2E). When anti-MeSmad7 antibody was used in the assays, the signal corresponding to methylated Smad7 protein was largely lost already 1 hr after protein synthesis inhibition, suggesting that the methylated form of Smad7 is predisposed to degradation. In agreement with this, the protein levels of Smad7K70R mutant remained more stable during the 6-hr time frame of the assay (Figure 2E).
Previous studies have established that Smad7 stability is regulated by interplay between acetylation and ubiquitination at lysine residues K64 and K70 (Grönroos et al., 2002, Ogunjimi et al., 2005, Simonsson et al., 2005). Acetylation stabilizes Smad7 by competing with Smurf-dependent ubiquitination and subsequent proteasomal degradation. To understand the mechanism of methylation-mediated destabilization of Smad7, we investigated the possibility of a crosstalk among methylation, acetylation, and ubiquitination. Since lysine-70 is a common target for Set9-mediated methylation and p300-mediated acetylation, these modifications should be mutually exclusive. In agreement with this, an acetylation-specific antibody did not recognize methylated endogenous Smad7 in western blot assays, even when the protein was stabilized by MG132 treatment (Figure 3A). Consistent with the increased stability of Smad7 in Set9-deficient primary lung fibroblasts and in Set9-depleted HeLa cells, these cells contained reduced amounts of endogenous ubiquitinated protein after MG132 treatment, suggesting that Set9-mediated methylation may stimulate Smad7 ubiquitination (Figure 3B). This notion was further supported by the results of in vivo ubiquitination assays using mutant forms of Smad7. Overexpression of WT Set9, but not its enzymatically inactive mutant form (Set9H297A), increased the amount of ubiquitinated Smad7, while the Smad7K70R mutant was poorly ubiquitinated (Figure 3C). Interestingly, mutation of lysine-64 also impaired Set9-mediated induction of ubiquitination, suggesting that methylation at lysine-70 promotes ubiquitination mainly at the neighboring lysine-64 residue (Figure 3C).
Figure 3.
Methylation Prevents Acetylation and Promotes Smad7 Ubiquitination
(A) Whole-cell extracts from HeLa cells treated with 5 ng/ml TGF-β and 5 μM MG132 for 5 hr were used for IPs with αSmad7 (rabbit polyclonal) and αMe-Smad7 antibodies. Immunoprecipitated proteins were analyzed in western blots with α-panacetyl-lysine (upper panel) or αSmad7 (lower panel) antibodies.
(B) Set9 depletion reduces Smad7 polyubiquitination. Extracts from WT and sh-Set9 HeLa cells or WT and Set9-KO lung fibroblasts that were treated with 5 ng/ml TGF-β and 5 μM MG132 for 5 hr were subjected to IP with αSmad7 (rabbit polyclonal) antibody. Immunoprecipitated proteins were analyzed in western blots with αUbiquitin or αSmad7 antibodies.
(C) Methylation of Smad7 promotes polyubiquitination. HeLa cells were transfected with the indicated expression vectors and treated with 5 μM MG132 for 5 hr. Whole-cell extracts were immunoprecipitated with αHA antibody and analyzed in western blots with αUbiquitin or αSmad7 antibodies.
(D–F) Methylated Smad7 interacts with Arkadia. Extracts from WT and sh-Set9 HeLa cells or WT and Set9-KO lung fibroblasts that were treated with 5 ng/ml TGF-β and 5 μM MG132 for 5 hr were used for IPs with αSmad7 or αMe-Smad7 antibodies, and they were analyzed in western blots using α-Arkadia (D), α-Smurf1 (E), or α-Smurf2 (F) antibodies. Before IPs, the extracts were diluted to obtain input materials with equal amounts of Smad7. Equal loadings of total Smad7 proteins in the immunoprecipitates were verified in separate gels using αSmad7 antibody (bottom panels).
See also Figure S3.
To understand how methylation leads to increased ubiquitination and degradation of Smad7, we examined the physical association of the methylated protein with Smurf1, Smurf2, and Arkadia (RNF111), three known E3 ligases promoting Smad7 degradation (Grönroos et al., 2002, Ogunjimi et al., 2005, Simonsson et al., 2005, Koinuma et al., 2003, Liu et al., 2006). Co-immunoprecipitation (coIP) assays with methylation-specific Smad7 antibody revealed that methylated Smad7 efficiently interacts with Arkadia, but not with Smurf1 or Smurf2 (Figures 3D–3F). A much fainter Arkadia signal was detected in immunoprecipitates recognizing total Smad7, which disappeared in Set9-depleted primary lung fibroblasts or HeLa cells (Figure 3D). On the contrary, immunoprecipitates with total Smad7 contained similar amounts of Smurf1 or Smurf2 in extracts from both WT and Set9-depleted cells (Figures 3E and 3F), corroborating the conclusion that methylated Smad7 selectively interacts with Arkadia.
Additional evidence for the selective interaction between Arkadia and methylated Smad7 was provided by in vitro pull-down assays using unmethylated or premethylated recombinant Smad7 proteins and cellular extracts overexpressing Arkadia, Smurf1, or Smurf2. As shown in Figure 4A, premethylation of Smad7 by Set9 greatly increased interaction with Arkadia, while it essentially diminished the interaction with Smurf1 or Smurf2 proteins. The preferential association of Arkadia with methylated Smad7 was surprising, since methylation occurs at the N-terminal region of the protein while Arkadia interacts only with the C-terminal domain (Koinuma et al., 2003, Liu et al., 2006). To clarify the binding mechanism, we performed the pull-down experiments using in-vitro-translated Arkadia and Axin2 proteins. Axin2 is the partner of Arkadia, which facilitates the formation of a multimeric complex with Smad7 (Liu et al., 2006). We detected only a weak direct interaction between Arkadia and unmethylated Smad7, which was marginally increased after methylation (Figure 4B, panel at left). In contrast, Axin2 was strongly bound to both unmethylated and methylated Smad7. Interestingly, however, inclusion of Axin2 protein in the binding reaction resulted in a dramatic increase of Arkadia-Smad7 interaction, which was absolutely dependent on methylation (Figure 4B, panel at right). These results suggest that selective interaction of Arkadia with methylated Smad7 requires Axin2.
Figure 4.
Arkadia Preferentially Interacts with Methylated Smad7 and Promotes Its Ubiquitination
(A) In vitro interactions of Smad7 with Arkadia, Smurf1, and Smurf2 proteins. Recombinant His6-tagged Smad7 proteins immobilized on Talon™ Ni-affinity column were preincubated with GST-Set9 in the absence (His-Smad7) or presence (His-Smad7-Me) of 1 μM S-adenosyl-methionine. The beads were then incubated with extracts from HeLa cells that were transfected with CMV-Arkadia (left panel), CMV-Smurf1 (middle panel), and CMV-Smurf2 (right panel). The presence of Arkadia, Smurf1, and Smurf2 proteins in the Smad7-containing beads was evaluated by western blot analysis.
(B) Non-methylated and methylated His-Smad7 proteins immobilized in Talon™ beads as in (A) were incubated with in-vitro-translated 35S-methionine-labeled full-length Arkadia (Ark) or Axin2 proteins (corresponding to 10 ng of translated protein), as indicated. In the right panel, the beads were incubated with a mix of unlabeled Axin2 translation product (30 ng Axin2 protein) and 35S-Arkadia (10 ng protein) under the same conditions. After washings, bound 35S-Arkadia or 35S-Axin2 proteins were detected by SDS-PAGE followed by autoradiography.
(C) HeLa cells were transfected with Arkadia-specific smart pool siRNAs or control siRNA. Then 3 days after transfection, endogenous levels of Arkadia (first panel), methylated Smad7 (third panel), total Smad7 (fourth panel), and Gapdh (fifth panel) were evaluated by western blot analysis. In the experiment shown in the second panel, cells that were transfected with Arkadia-specific siRNA were co-transfected 36 hr before harvest with CMV-Arkadia plasmid, containing the mouse ORF of the gene.
(D) HeLa cells were transfected with the indicated expression vectors and treated with 5 μM MG132 for 5 hr. Transfections of siRNAs and the mammalian expression vectors (CMV-HA-Smad7wt, CMV-Myc-Set9wt, and CMV-Arkadia) were performed 72 and 36 hr before harvest, respectively. Whole-cell extracts were immunoprecipitated with αHA antibody and analyzed in western blots with αUbiquitin or αSmad7 antibodies.
(E) Control siRNA or Arkadia-specific siRNA-transfected HeLa cells were re-transfected with pCMV-HA-Smad7 (HA-Smad7wt) and pCMV-myc-Set9 (Set9wt) as in (C). Before harvesting, the cells were treated with 50 μg/ml CHX for the indicated times. Whole-cell extracts were analyzed in western blots using anti-Me-Smad7 or anti-HA (α-HA) antibodies as indicated. One representative of two experiments is shown. The graph shows band intensities, measured by ImageJ software, relative to untreated controls.
See also Figure S4.
To provide direct functional evidence for the role of Arkadia in destabilizing methylated Smad7, we depleted Arkadia in HeLa cells by siRNA-mediated silencing. The endogenous levels of Smad7 were increased about 2-fold in Arkadia-depleted cells (Figure 4C). Importantly, the endogenous amounts of methylated Smad7 increased more substantially, reaching levels that could be easily and reproducibly detected by western blots using Me-Smad7 antibody (Figure 4C). Set9 overexpression-mediated increased ubiquitination could not be detected in Arkadia-depleted HeLa cells, while complementation of Arkadia, by overexpressing the mouse counterpart that is not targeted by the siRNAs, resulted in increased levels of Smad7 ubiquitination (Figure 4D). In agreement with this, the half-life of total Smad7 and, importantly, that of methylated Smad7 was greatly increased in Arkadia-depleted HeLa cells (Figure 4E).
The above results demonstrate that Set9 regulates TGF-β-signaling pathway via methylation-dependent destabilization of Smad7, which is elicited primarily by the enhanced interaction with the E3-ubiquitin ligase Arkadia. As a result of this effect, elevated levels of Smad7 protein in Set9-deficient cells inhibit TGF-β-dependent activation of ECM genes. Smad7 inhibits TGF-β signaling by transfering Smurf1 and Smurf2 to the plasma membrane, which triggers the proteasomal destruction of TGF-β receptors (Ebisawa et al., 2001, Kavsak et al., 2000, Yan et al., 2009, Grönroos et al., 2002, Ogunjimi et al., 2005, Simonsson et al., 2005). In agreement with this, the endogenous levels of TGF-β receptor I protein (TGF-β-RI) were highly decreased in Set9-deficient lung fibroblasts and HeLa cells (Figure S4).
Loss of Set9 Protects against Bleomycin- or Ad-TGF-β-Mediated Pulmonary Fibrosis in Mice
The in vivo relevance of these findings was assessed in the pathological condition of lung fibrosis, as uncontrolled expression of ECM genes is a hallmark feature of this disease (Wynn, 2011). We tested bleomycin-induced pulmonary fibrosis, the best-characterized and most widely used model for the disease (Moore and Hogaboam, 2008, Della Latta et al., 2015). Bleomycin treatment of WT mice resulted in a characteristic lung histology phenotype of interstitial thickening, alveolar collapse, and the presence of cystic air spaces (Figure 5A). These pathological features were highly reduced in Set9-deficient mice (Figure 5A). The protective effect of Set9 depletion was confirmed by Sirius red and Masson’s trichrome staining, which revealed decreased cross-linked collagen fiber deposition in the lungs of Set9-KO mice (Figures 5A, 5C, and S5A).
Figure 5.
Set9 Deficiency Confers Resistance to Mice against Bleomycin- or Ad-TGF-β-Induced Pulmonary Fibrosis
(A) Representative H&E and Sirius red staining of lung sections of WT and Set9-KO mice after bleomycin treatment (n = 5). Control mice were treated with PBS (n = 5).
(B) Representative H&E and Sirius red staining of lung sections of WT and Set9-KO mice after Ad-TGF-β (n = 7) treatment. Control mice were treated with empty Adenoviral vector (Ad-0) (n = 6).
(C) Quantitation of Sirius red-positive areas in 20 different high-power fields in lung sections of each untreated, bleomycin-, or Ad-TGF-β-treated WT and Set9-KO mice. Data are presented as mean ± SEM (∗p value < 0.0001, two-tailed Student’s t test).
(D) Effect of Set9 deficiency on bleomycin-induced changes of lung mechanics. Static (Cst) and dynamic (Cdyn) compliance percentage differences (bleomycin- versus PBS-treated mice) in response to bleomycin-induced injury are shown. Bars represent mean values ± SEM from n = 5 PBS-treated WT mice, n = 5 bleomycin-treated WT mice, n = 3 PBS-treated Set9-KO mice, and n = 4 bleomycin-treated Set9-KO (∗p < 0.05, two-tailed Student’s t test).
(E) Effect of Set9 deficiency on Ad-TGF-β-induced changes of lung mechanics. Static (Cst) and dynamic (Cdyn) compliance percentage differences (Ad-TGF-β versus Ad-0-treated mice) in response to Ad-TGF-β-induced injury are shown. Bars represent mean values ± SEM from n = 5 Ad0-treated WT mice, n = 5 Ad-TGF-β-treated WT mice, n = 4 Ad-0-treated Set9-KO mice, and n = 7 Ad-TGF-β-treated Set9-KO mice (∗p < 0.05, two-tailed Student’s t test).
See also Figure S5.
The pathogenesis of pulmonary fibrosis in the bleomycin model involves severe tissue damage, followed by the accumulation of inflammatory cells and pro-fibrogenic cytokine secretion, which in turn induce collagen production by resident fibroblasts or by myofibroblasts derived from transdifferentiation of epithelial cells (Wynn, 2011, Moore and Hogaboam, 2008, Liu et al., 2010). Impairment of one of these early events could explain the reduced fibrotic response in Set9-KO mice. Our immunostaining experiments, with the pan-leukocyte marker CD45 and the myofibroblast marker αSma, demonstrate that Set9 inactivation does not affect bleomycin-mediated accumulation of these cell types (Figure S5B). These findings suggest that Set9 is specifically required for later stages of the fibrotic process, such as the production or deposition of ECM components. The possibility that bleomycin-mediated damage may influence Set9 expression was excluded by western blot analysis, which showed similar amounts of Set9 protein in untreated and bleomycin-treated lungs (Figure S6A).
Although TGF-β is recognized as the main mediator of bleomycin-induced pulmonary fibrosis, intrinsic changes in the activation of epithelial cells or fibroblasts, such as activation of Wnt-β-catenin signaling or defects in PTEN-dependent suppression of PI3K-Akt-S6K1 pathway, may promote growth factor-independent fibrotic responses (Königshoff et al., 2009, Xia et al., 2008). Therefore, we analyzed the effect of Set9 deficiency on the pulmonary fibrosis phenotype in another experimental model, involving intratracheal delivery of TGF-β-producing adenovirus (Ad-TGF-β) (Koltsida et al., 2011). Similar to bleomycin treatment, Set9-KO mice developed highly reduced fibrotic lesions following Ad-TGF-β instillation compared to WT counterparts (Figures 5B and 5C).
In agreement with the morphological data, significant differences between WT and Set9-KO mice were detected in lung mechanic parameters following bleomycin or Ad-TGF-β challenge: the static compliance (Cst), which reflects the elastic recoil of the lung at a given pressure, and the dynamic compliance (Cdyn), which captures the ease with which the lungs can be extended, were decreased to a much lesser extent in Set9-KO mice than in WT littermates (Figures 5D and 5E).
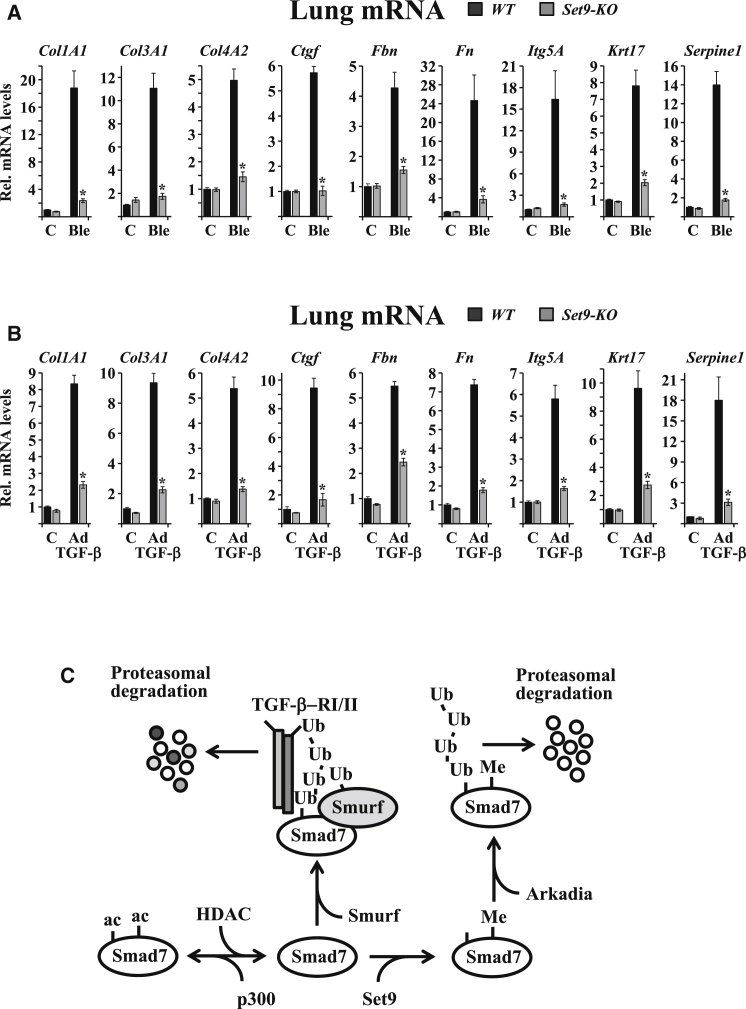
The mechanistic basis of the above histological and physiological phenotypes was revealed by the analysis of ECM gene expression profiles. The activation of a large panel of ECM genes, including collagens (Col1A1, Col3A1, and Col4A2), connective tissue growth factor (Ctgf), fibrillin (Fbn), fibronectin (Fn), integrin-5A (Itg5A), keratin-17 (Krt17), and Serpine1, was greatly diminished in the lungs of bleomycin-treated Set9-KO mice (Figure 6A). Similar to the observations in the bleomycin model, Ad-TGF-β-induced activation of ECM genes was highly compromised in Set9-KO mice (Figure 6B).
Figure 6.
Set9 Is Required for Bleomycin- and Ad-TGF-β-Dependent Activation of ECMGenes
(A) qPCR analysis of the mRNA levels of the indicated panel of ECM genes in reverse-transcribed total lung RNAs from WT and Set9-KO mice after treatment with PBS (C) or bleomycin (Ble). Bars represent mean values of mRNA levels normalized to Gapdh mRNA ± SEM. Data are from triplicate measurements of total RNAs from five individual mice (∗p < 0.05, two-tailed Student’s t test).
(B) qPCR analysis of mRNA levels of the indicated ECM genes in reverse-transcribed total lung RNAs from WT and Set9-KO mice after treatment with empty adenovirus vector (C) or Ad-TGF-β. Bars represent mean values of mRNA levels normalized to Gapdh mRNA ± SEM. Data are from triplicate measurements of total RNAs from five individual mice (∗p < 0.05, two-tailed Student’s t test).
(C) Schematic presentation shows methylation-ubiquitination-acetylation interplay in balancing intracellular Smad7 levels.
See also Figure S6.
In the lungs of both bleomycin- and Ad-TGF-β-treated mice, the mRNA levels of several well-characterized regulators of the EMT process (HmgA2, Has, Twist, and Snai1) were significantly increased, while those of E-cadherin (Cdh1) decreased (Figures S6B and S6C). These effects were largely prevented in the lungs of Set9-KO mice (Figures S6B and S6C). The above data were obtained with RNAs from total lung tissue containing different cell populations. The particular cell type(s) where EMT takes place remains to be determined. Notwithstanding this limitation, the impaired activation of another group of TGF-β target genes in Set9-KO mice provides additional support for the in vivo role of Set9 function in TGF-β signaling.
Discussion
The results of this study demonstrate that the methyltransferase Set9 is required for the development of pulmonary fibrosis, and they identify Set9 as a potential candidate therapeutic target for treatment of the disease.
Complete loss of Set9 in mice did not lead to macroscopically visible lung phenotypes under normal conditions. This was surprising, since Set9 is known to methylate several regulatory proteins, which control diverse biological processes such as cell growth, apoptosis, metabolism, and hormonal or cytokine signaling (Chuikov et al., 2004, Subramanian et al., 2008, Pradhan et al., 2009, Yang et al., 2010, Kontaki and Talianidis, 2010a, Kontaki and Talianidis, 2010b, Munro et al., 2010, Carr et al., 2011, Sarris et al., 2014). Our failure to detect major phenotypic changes in Set9-KO mice indicates that Set9 is redundant to normal lung development and function, either due to the activation of factor(s) and/or alternative pathways, which compensate its function, or because Set9-mediated methylation of the target proteins may have only minor, fine-tuning effects. In pathological situations, however, when specific biological processes are hyperactivated or repressed, the modulatory effects of posttranslational modifications may be manifested in a more dominant manner. This is likely the case with Set9 function, since under cellular damage conditions triggering lung fibrosis, Set9-KO mice exhibited a robust and clinically relevant phenotype. The strong protective effect of Set9 deficiency on the development of pulmonary fibrosis in two different mouse models warrants further studies in human subjects, as Set9 inhibitors could be considered in treating the disease at the early, developing stages. The essentially normal phenotype of Set9-KO mice in physiological conditions provides the expectation that pharmacological inhibition of Set9 may have negligible side effects.
Excessive production and accumulation of ECM proteins is the hallmark event of fibrotic diseases, including pulmonary fibrosis (Wynn, 2011). Our findings suggest that Set9 function is required for TGF-β-dependent activation of ECM genes. We identified Smad7, the inhibitory molecule of the TGF-β-signaling pathway, as a bona fide enzymatic target of Set9, and we provide evidence for the role of methylation-mediated destabilization of Smad7 in the regulation of ECM gene expression.
Previous studies have established that Smad7 is subject to reversible acetylation, which prevents Smurf1-dependent ubiquitination and degradation (Grönroos et al., 2002, Ogunjimi et al., 2005, Simonsson et al., 2005). Non-acetylated Smad7 molecules can exit the nucleus, interact with Smurf1/2 E3 ligases, and escort them to the plasma membrane, where they ubiquitinate TGF-β receptor (TβRI and TβRII), parallel to the ubiquitination of Smad7 proteins. Subsequent proteasomal degradation of both leads to feedback inhibition of TGF-β signal transduction. Our results reveal an additional level of complexity in the interplay of posttranslational modifications that regulate Smad7 stability (see scheme in Figure 6C). According to this model, the size of the non-acetylated pool of Smad7 is controlled by Set9-mediated methylation. Methylated Smad7 selectively interacts with the E3 ligase Arkadia, which catalyzes ubiquitination and facilitates its proteasomal degradation. When Set9 activity is inhibited, Arkadia-mediated clearance of Smad7 is blocked, leading to the expansion of the pool of non-modified Smad7 molecules available for interaction with Smurf1/2 E3 ligases and Smurf-mediated destruction of the TGF-β receptor complex. In this way, Set9 deficiency or pharmacological inhibition of its activity prevents activation of TGF-β-induced genes, including those encoding for ECM components.
Previous studies have established that Arkadia interacts only with the C-terminal domain of Smad7 and that this interaction is stimulated by Axin (Koinuma et al., 2003, Liu et al., 2006). On the other hand, Axin can interact with both the N-terminal and C-terminal parts of Smad7 (Liu et al., 2006). Set9-mediated methylation of the K70 lysine residue at the N-terminal region of Smad7 did not affect direct interaction with Axin2. However, selective interaction of Arkadia with methylated Smad7 was absolutely dependent on Axin2, suggesting that the latter may influence the overall structure of the non-methylated and methylated protein in distinct manners. Axin2 binding to methylated Smad7 may generate a configuration optimal for interaction with Arkadia. Future studies will be required to obtain structural insights into this interaction, which may facilitate the development of novel pharmaceutical intervention strategies aimed at the modulation of Smad7 stability, in addition to the abovementioned Set9 enzyme inhibitors.
Experimental Procedures
Animals and Treatments
The generation of Set9-KO mice by gene targeting is described in Figure S1. Mice that were used in this study had a mixed C57Bl6/CBA genetic background. Mice were maintained in grouped cages in a temperature-controlled, pathogen-free facility on a 12-hr light/dark cycle, and they were fed a standard chow diet (Altromin 1324; 19% protein and 5% fat) and water ad libitum.
Anesthetized, 12-week-old male Set9-KO and WT mice (weight 28–34 g) were treated intratracheally with 0.033 U/mouse bleomycin (Nippon Kayaku) diluted in endotoxin-free PBS, or PBS alone (vehicle), in 30 μl total volume. Animals were analyzed on days 5 and 15 post-bleomycin treatment.
For intratracheal instillation of recombinant adenovirus, a total of 2 × 108 infectious particles of adenoviruses expressing porcine, biologically active TGF-β1 (Ad-TGF-β1223/225), or a control virus (Ad-0) were used in 40 μl total volume as described (Koltsida et al., 2011). Animals were analyzed on day 14 post-adenovirus instillation.
All animal experiments were approved by the Prefecture of Attica and the Institutional Review Board of Biomedical Sciences Research Center Alexander Fleming, and they were performed in accordance with the respective national and European Union regulations. All the experiments were performed in randomly chosen age-matched male mice. No blinding was used in this study.
Histological Analysis
Histological assays were performed in paraffin-embedded or frozen tissue sections as described previously (Apostolou et al., 2012, Nikolaou et al., 2012, Nikolaou et al., 2015). Briefly, lungs were dissected, fixed in 4% paraformaldehyde, and embedded in paraffin. Lung sections (5–6 μm thick) were boiled in 10 mM Na-citrate for 20 min, and, after washings with PBS, the sections were blocked with normal goat serum and used for staining with the αSMA and αCD45 antibodies using AlexaFluor 568 and 488 (Molecular Probes) secondary antibodies. After counterstaining with DAPI, fluorescence images were observed using a Zeiss Axioscope 2 Plus microscope. Paraffin-embedded tissue sections were used for staining with H&E or with 0.1% Sirius red dissolved in saturated picric acid, as described previously (Nikolaou et al., 2012, Nikolaou et al., 2015). Sirius red-positive areas were quantitated using NIH ImageJ software.
Cell Culture and Transfections
For the isolation of primary lung fibroblasts, lung tissue from WT and Set9-KO mice was cut into small pieces in cold PBS, and it was incubated in a buffer containing 0.1% collagenase (Roche), 2.4 U/ml Dispase, 2 mM CaCl2, 150 mM NaCl, and 10 mM HEPES (pH 7.9) for 1.5 hr at 37°C with constant shaking. Liberated cells were drained through a 70-μm mesh filter and collected by centrifugation at 1,500 rpm for 5 min. The cells were washed twice with DMEM-F12 and resuspended in DMEM-F12/10% fetal calf serum (FCS), 100 U/ml penicillin/streptomycin-containing medium. Then 4 hr after transferring to plates, floating cells were removed by changing the medium. The culture medium was changed again after 24 and 48 hr.
HeLa cells (ATCC) were maintained in DMEM supplemented with 10% FCS and 100 U/ml penicillin/streptomycin. Sh-Set9-expressing HeLa cell line was generated by transfecting pRS-Set9 vector (OriGene) harboring the following short hairpin sequence: 5′ GAT CGA TGA CTG GAG AGA AGA TAG CCT ATG TGT TCA AGA GAC ACA TAG GCT ATC TTC TCT CCA GTC ATC TTT TT. Cells stably expressing sh-Set-9 were obtained by selection with 0.5 μg/ml puromycin. Tranfections of HeLa cells were performed using Jet-PEI reagent (Polyplus).
Vectors expressing HA-tagged WT and mutant forms of Smad7 were constructed by cloning the open reading frame (ORF) of human Smad7 into pCMV-HA vector (Kontaki and Talianidis, 2010a). pCMV-Myc-Set9wt and pCMV-myc-Set9 H297A have been described (Kontaki and Talianidis, 2010a). Mammalian expression vectors for Arkadia (pCMV6-RNF111), Smurf1 (pCMV6-Smurf1), and Smurf2 (pCMV6-Smurf2) were obtained from OriGene. SiRNA-mediated knockdown of Arkadia or Smad7 in HeLa cells was achieved by transfection of predesigned ON-TARGET plus SMART pool human RNF111 siRNAs or Smad7 siRNAs (Dharmacon) using DharmaFECT transfection reagent. The cells were used for analyses 72 hr following transfection. In reconstitution experiments, 36 hr before harvesting, the cells were transiently transfected with CMV-RNF111 mouse ORF clone using Jet-PEI reagent. Mutagenesis was performed with GeneEditor kit (Promega).
IPs and Western Blot Analysis
Cells were lysed by resuspension in 10 vol modified radio-IP assay (RIPA) buffer containing 50 mM Tris (pH 7.5), 1% NP40, 0.25% Na-Deoxycholate, 150 mM NaCl, 1 mM EDTA, 10% Glycerol, 1 mM NaF, and protease inhibitors cocktail (Roche). After 20-min incubation by shaking at 4°C, extracted proteins were recovered by centrifugation at 14,000 rpm for 10 min. In IP assays, cellular lysates were incubated with 2–5 μg antibodies and 30-μl bed volume of Protein-G-Sepharose beads (Millipore) for 5 hr at 4°C, followed by four successive washes with modified RIPA buffer. For IP-western blot assays analyzing ubiquitinated proteins, the RIPA extraction buffer contained 0.5% SDS and the lysates were diluted five times with RIPA buffer without SDS before IP. Extracts and immunoprecipitated proteins were resolved by 10%, 12%, or 6% SDS-PAGE, transferred to nitrocellulose membrane, and analyzed by western blot as described (Hatzis et al., 2006).
The antibodies used in this study are described in the Supplemental Experimental Procedures.
In Vitro Methylation Assays
The ORFs of human Smad2, Smad3, Smad4, and Smad7 cDNAs were subcloned into PRSET-A vector, in frame of the 6xHis-tag sequence, and used for transformation of BL21(DE3)-competent cells. Recombinant proteins from the extracts of IPTG-induced bacteria were affinity purified using TALON beads (Clontech Laboratories). GST-Set9 protein was purified by glutathione Sepharose affinity chromatography, as described previously (Kontaki and Talianidis, 2010a).
In vitro methylation assays were performed by incubation of GST-Set9 with purified recombinant Smad proteins with 1 μCi 3H-SAM (S-adenosyl-methionine) in a buffer containing 50 mM Tris-HCl (pH 9.1), 0.5 mM DTT, and 4 mM EDTA for 1 hr at 30°C. The reactions were stopped by adding SDS-PAGE loading buffer and analyzed by SDS-PAGE and fluorography.
For the analysis of protein-protein interactions with unmethylated or premethylated Smad7, Talon™ Ni-affinity beads containing His6-Smad7 were incubated with GST-Set9 for 2 hr at 30°C, in the presence or absence of 1 μM cold SAM in a buffer containing 50 mM Tris-HCl (pH 9.1) and 10 mM NaCl. The beads were washed with modified RIPA buffer lacking EDTA, and they were incubated with extracts of HeLa cells that were transfected with Arkadia, Smurf1, or Smurf2 expression vectors for 5 hr at 4°C with constant rotation. For the analyses of direct protein interactions in vitro, 35S-methionine labeled Arkadia and Axin2 were synthesized by the TnT Quick Coupled Transcription/Translation System (Promega), according to the manufacturer’s instructions. The translation products were diluted in modified RIPA buffer lacking EDTA and incubated with the beads as above. After extensive washing with modified RIPA buffer lacking EDTA, interacting proteins were detected by western blot analysis (in assays with cellular extracts) or by autoradiography (in assays with in-vitro-synthesized proteins).
RNA Analysis
Total RNA was prepared by the Trizol extraction protocol (Tatarakis et al., 2008). After digestion with DNase I, 1 μg total RNA was used for first-strand cDNA synthesis using Moloney murine leukemia virus (MMLV) reverse transcriptase. The qPCR analyses were carried out in STEP1 Real-time PCR detection system by using SYBR Green reagent as described before (Tatarakis et al., 2008, Nikolaou et al., 2015). The nucleotide sequences of primer sets used are described in the Supplemental Experimental Procedures.
Statistical Analysis
Data are reported as mean values from the number of replicates indicated in the legends, with error bars denoting ±SEM. Statistical significance was evaluated by two-tailed Student’s t test.
Author Contributions
Conceptualization, H.K. and I.T.; Investigation, M.E., H.K., A.S., A.A., M.S., E.S., D.S., and I.T.; Resources, K.C.N. and P.J.B.; Supervision, P.S., G.P., and I.T.; Writing, I.T.
Acknowledgments
We thank Dr. P. Hatzis for comments and critical reading of the manuscript. This work was supported by the European Union (European Research Council [ERC] Advanced Investigator Grant, ERC-2011-AdG294464), FP7 Marie Curie ITN (PITN-GA-2013-606806), and the European Regional Development Fund (ERDF)-GSRT grant (ARISTEIA II-3451).
Published: June 9, 2016
Footnotes
Supplemental Information includes Supplemental Experimental Procedures and six figures and can be found with this article online at http://dx.doi.org/10.1016/j.celrep.2016.05.051.
Supplemental Information
References
- Apostolou E., Stavropoulos A., Sountoulidis A., Xirakia C., Giaglis S., Protopapadakis E., Ritis K., Mentzelopoulos S., Pasternack A., Foster M. Activin-A overexpression in the murine lung causes pathology that simulates acute respiratory distress syndrome. Am. J. Respir. Crit. Care Med. 2012;185:382–391. doi: 10.1164/rccm.201105-0784OC. [DOI] [PubMed] [Google Scholar]
- Barsyte-Lovejoy D., Li F., Oudhoff M.J., Tatlock J.H., Dong A., Zeng H., Wu H., Freeman S.A., Schapira M., Senisterra G.A. (R)-PFI-2 is a potent and selective inhibitor of SETD7 methyltransferase activity in cells. Proc. Natl. Acad. Sci. USA. 2014;111:12853–12858. doi: 10.1073/pnas.1407358111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blobe G.C., Schiemann W.P., Lodish H.F. Role of transforming growth factor beta in human disease. N. Engl. J. Med. 2000;342:1350–1358. doi: 10.1056/NEJM200005043421807. [DOI] [PubMed] [Google Scholar]
- Border W.A., Noble N.A. Transforming growth factor beta in tissue fibrosis. N. Engl. J. Med. 1994;331:1286–1292. doi: 10.1056/NEJM199411103311907. [DOI] [PubMed] [Google Scholar]
- Carr S.M., Munro S., Kessler B., Oppermann U., La Thangue N.B. Interplay between lysine methylation and Cdk phosphorylation in growth control by the retinoblastoma protein. EMBO J. 2011;30:317–327. doi: 10.1038/emboj.2010.311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuikov S., Kurash J.K., Wilson J.R., Xiao B., Justin N., Ivanov G.S., McKinney K., Tempst P., Prives C., Gamblin S.J. Regulation of p53 activity through lysine methylation. Nature. 2004;432:353–360. doi: 10.1038/nature03117. [DOI] [PubMed] [Google Scholar]
- Cordenonsi M., Dupont S., Maretto S., Insinga A., Imbriano C., Piccolo S. Links between tumor suppressors: p53 is required for TGF-beta gene responses by cooperating with Smads. Cell. 2003;113:301–314. doi: 10.1016/s0092-8674(03)00308-8. [DOI] [PubMed] [Google Scholar]
- Della Latta V., Cecchettini A., Del Ry S., Morales M.A. Bleomycin in the setting of lung fibrosis induction: From biological mechanisms to counteractions. Pharmacol. Res. 2015;97:122–130. doi: 10.1016/j.phrs.2015.04.012. [DOI] [PubMed] [Google Scholar]
- Ebisawa T., Fukuchi M., Murakami G., Chiba T., Tanaka K., Imamura T., Miyazono K. Smurf1 interacts with transforming growth factor-beta type I receptor through Smad7 and induces receptor degradation. J. Biol. Chem. 2001;276:12477–12480. doi: 10.1074/jbc.C100008200. [DOI] [PubMed] [Google Scholar]
- Grönroos E., Hellman U., Heldin C.H., Ericsson J. Control of Smad7 stability by competition between acetylation and ubiquitination. Mol. Cell. 2002;10:483–493. doi: 10.1016/s1097-2765(02)00639-1. [DOI] [PubMed] [Google Scholar]
- Hamamoto R., Saloura V., Nakamura Y. Critical roles of non-histone protein lysine methylation in human tumorigenesis. Nat. Rev. Cancer. 2015;15:110–124. doi: 10.1038/nrc3884. [DOI] [PubMed] [Google Scholar]
- Hatzis P., Kyrmizi I., Talianidis I. Mitogen-activated protein kinase-mediated disruption of enhancer-promoter communication inhibits hepatocyte nuclear factor 4alpha expression. Mol. Cell. Biol. 2006;26:7017–7029. doi: 10.1128/MCB.00297-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inamitsu M., Itoh S., Hellman U., Ten Dijke P., Kato M. Methylation of Smad6 by protein arginine N-methyltransferase 1. FEBS Lett. 2006;580:6603–6611. doi: 10.1016/j.febslet.2006.11.008. [DOI] [PubMed] [Google Scholar]
- Kavsak P., Rasmussen R.K., Causing C.G., Bonni S., Zhu H., Thomsen G.H., Wrana J.L. Smad7 binds to Smurf2 to form an E3 ubiquitin ligase that targets the TGF beta receptor for degradation. Mol. Cell. 2000;6:1365–1375. doi: 10.1016/s1097-2765(00)00134-9. [DOI] [PubMed] [Google Scholar]
- Koinuma D., Shinozaki M., Komuro A., Goto K., Saitoh M., Hanyu A., Ebina M., Nukiwa T., Miyazawa K., Imamura T., Miyazono K. Arkadia amplifies TGF-beta superfamily signalling through degradation of Smad7. EMBO J. 2003;22:6458–6470. doi: 10.1093/emboj/cdg632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koltsida O., Hausding M., Stavropoulos A., Koch S., Tzelepis G., Ubel C., Kotenko S.V., Sideras P., Lehr H.A., Tepe M. IL-28A (IFN-λ2) modulates lung DC function to promote Th1 immune skewing and suppress allergic airway disease. EMBO Mol. Med. 2011;3:348–361. doi: 10.1002/emmm.201100142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Königshoff M., Kramer M., Balsara N., Wilhelm J., Amarie O.V., Jahn A., Rose F., Fink L., Seeger W., Schaefer L. WNT1-inducible signaling protein-1 mediates pulmonary fibrosis in mice and is upregulated in humans with idiopathic pulmonary fibrosis. J. Clin. Invest. 2009;119:772–787. doi: 10.1172/JCI33950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kontaki H., Talianidis I. Lysine methylation regulates E2F1-induced cell death. Mol. Cell. 2010;39:152–160. doi: 10.1016/j.molcel.2010.06.006. [DOI] [PubMed] [Google Scholar]
- Kontaki H., Talianidis I. Cross-talk between post-translational modifications regulate life or death decisions by E2F1. Cell Cycle. 2010;9:3836–3837. doi: 10.4161/cc.9.19.13384. [DOI] [PubMed] [Google Scholar]
- Kouskouti A., Scheer E., Staub A., Tora L., Talianidis I. Gene-specific modulation of TAF10 function by SET9-mediated methylation. Mol. Cell. 2004;14:175–182. doi: 10.1016/s1097-2765(04)00182-0. [DOI] [PubMed] [Google Scholar]
- Lamouille S., Xu J., Derynck R. Molecular mechanisms of epithelial-mesenchymal transition. Nat. Rev. Mol. Cell Biol. 2014;15:178–196. doi: 10.1038/nrm3758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W., Rui H., Wang J., Lin S., He Y., Chen M., Li Q., Ye Z., Zhang S., Chan S.C. Axin is a scaffold protein in TGF-beta signaling that promotes degradation of Smad7 by Arkadia. EMBO J. 2006;25:1646–1658. doi: 10.1038/sj.emboj.7601057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu G., Friggeri A., Yang Y., Milosevic J., Ding Q., Thannickal V.J., Kaminski N., Abraham E. miR-21 mediates fibrogenic activation of pulmonary fibroblasts and lung fibrosis. J. Exp. Med. 2010;207:1589–1597. doi: 10.1084/jem.20100035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massagué J. TGFβ signalling in context. Nat. Rev. Mol. Cell Biol. 2012;13:616–630. doi: 10.1038/nrm3434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore B.B., Hogaboam C.M. Murine models of pulmonary fibrosis. Am. J. Physiol. Lung Cell. Mol. Physiol. 2008;294:L152–L160. doi: 10.1152/ajplung.00313.2007. [DOI] [PubMed] [Google Scholar]
- Moustakas A., Heldin P. TGFβ and matrix-regulated epithelial to mesenchymal transition. Biochim. Biophys. Acta. 2014;1840:2621–2634. doi: 10.1016/j.bbagen.2014.02.004. [DOI] [PubMed] [Google Scholar]
- Munro S., Khaire N., Inche A., Carr S., La Thangue N.B. Lysine methylation regulates the pRb tumour suppressor protein. Oncogene. 2010;29:2357–2367. doi: 10.1038/onc.2009.511. [DOI] [PubMed] [Google Scholar]
- Nikolaou K., Tsagaratou A., Eftychi C., Kollias G., Mosialos G., Talianidis I. Inactivation of the deubiquitinase CYLD in hepatocytes causes apoptosis, inflammation, fibrosis, and cancer. Cancer Cell. 2012;21:738–750. doi: 10.1016/j.ccr.2012.04.026. [DOI] [PubMed] [Google Scholar]
- Nikolaou K.C., Moulos P., Chalepakis G., Hatzis P., Oda H., Reinberg D., Talianidis I. Spontaneous development of hepatocellular carcinoma with cancer stem cell properties in PR-SET7-deficient livers. EMBO J. 2015;34:430–447. doi: 10.15252/embj.201489279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogunjimi A.A., Briant D.J., Pece-Barbara N., Le Roy C., Di Guglielmo G.M., Kavsak P., Rasmussen R.K., Seet B.T., Sicheri F., Wrana J.L. Regulation of Smurf2 ubiquitin ligase activity by anchoring the E2 to the HECT domain. Mol. Cell. 2005;19:297–308. doi: 10.1016/j.molcel.2005.06.028. [DOI] [PubMed] [Google Scholar]
- Pradhan S., Chin H.G., Estève P.O., Jacobsen S.E. SET7/9 mediated methylation of non-histone proteins in mammalian cells. Epigenetics. 2009;4:383–387. doi: 10.4161/epi.4.6.9450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarris M., Nikolaou K., Talianidis I. Context-specific regulation of cancer epigenomes by histone and transcription factor methylation. Oncogene. 2014;33:1207–1217. doi: 10.1038/onc.2013.87. [DOI] [PubMed] [Google Scholar]
- Simonsson M., Heldin C.H., Ericsson J., Grönroos E. The balance between acetylation and deacetylation controls Smad7 stability. J. Biol. Chem. 2005;280:21797–21803. doi: 10.1074/jbc.M503134200. [DOI] [PubMed] [Google Scholar]
- Subramanian K., Jia D., Kapoor-Vazirani P., Powell D.R., Collins R.E., Sharma D., Peng J., Cheng X., Vertino P.M. Regulation of estrogen receptor alpha by the SET7 lysine methyltransferase. Mol. Cell. 2008;30:336–347. doi: 10.1016/j.molcel.2008.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatarakis A., Margaritis T., Martinez-Jimenez C.P., Kouskouti A., Mohan W.S., 2nd, Haroniti A., Kafetzopoulos D., Tora L., Talianidis I. Dominant and redundant functions of TFIID involved in the regulation of hepatic genes. Mol. Cell. 2008;31:531–543. doi: 10.1016/j.molcel.2008.07.013. [DOI] [PubMed] [Google Scholar]
- ten Dijke P., Hill C.S. New insights into TGF-beta-Smad signalling. Trends Biochem. Sci. 2004;29:265–273. doi: 10.1016/j.tibs.2004.03.008. [DOI] [PubMed] [Google Scholar]
- Travis M.A., Sheppard D. TGF-β activation and function in immunity. Annu. Rev. Immunol. 2014;32:51–82. doi: 10.1146/annurev-immunol-032713-120257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valcourt U., Kowanetz M., Niimi H., Heldin C.H., Moustakas A. TGF-beta and the Smad signaling pathway support transcriptomic reprogramming during epithelial-mesenchymal cell transition. Mol. Biol. Cell. 2005;16:1987–2002. doi: 10.1091/mbc.E04-08-0658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakefield L.M., Hill C.S. Beyond TGFβ: roles of other TGFβ superfamily members in cancer. Nat. Rev. Cancer. 2013;13:328–341. doi: 10.1038/nrc3500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wynn T.A. Integrating mechanisms of pulmonary fibrosis. J. Exp. Med. 2011;208:1339–1350. doi: 10.1084/jem.20110551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia H., Diebold D., Nho R., Perlman D., Kleidon J., Kahm J., Avdulov S., Peterson M., Nerva J., Bitterman P., Henke C. Pathological integrin signaling enhances proliferation of primary lung fibroblasts from patients with idiopathic pulmonary fibrosis. J. Exp. Med. 2008;205:1659–1672. doi: 10.1084/jem.20080001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J., Wang A.H., Oses-Prieto J., Makhijani K., Katsuno Y., Pei M., Yan L., Zheng Y.G., Burlingame A., Brückner K., Derynck R. Arginine methylation initiates BMP-induced Smad signaling. Mol Cell. 2013;51:5–19. doi: 10.1016/j.molcel.2013.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan X., Liu Z., Chen Y. Regulation of TGF-beta signaling by Smad7. Acta Biochim. Biophys. Sin. (Shanghai) 2009;41:263–272. doi: 10.1093/abbs/gmp018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J., Huang J., Dasgupta M., Sears N., Miyagi M., Wang B., Chance M.R., Chen X., Du Y., Wang Y. Reversible methylation of promoter-bound STAT3 by histone-modifying enzymes. Proc. Natl. Acad. Sci. USA. 2010;107:21499–21504. doi: 10.1073/pnas.1016147107. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.