Abstract
Background
Inhibition of mechanistic target of rapamycin (mTOR) has emerged as a viable means to lengthen lifespan and healthspan in mice, although it is still unclear whether these benefits will extend to other mammalian species. We previously reported results from a pilot experiment wherein common marmosets (Callithrix jacchus) were treated orally with rapamycin to reduce mTOR signaling in vivo in line with previous reports in mice and humans. Further, long-term treatment did not significantly alter body weight, daily activity, blood lipid concentrations, or glucose metabolism in this cohort.
Methods
In this study, we report on the molecular consequences of rapamycin treatment in marmosets on mechanisms that regulate protein homeostasis (proteostasis) in vivo. There is growing appreciation for the role of proteostasis in longevity and for the role that mTOR plays in regulating this process. Tissue samples of liver and skeletal muscle from marmosets in our pilot cohort were assessed for expression and activity of components of the ubiquitin-proteasome system, macroautophagy, and protein chaperones.
Results
Rapamycin treatment was associated with increased expression of PSMB5, a core subunit of the 20S proteasome, but not PSMB8 which is involved in the formation of the immunoproteasome, in the skeletal muscle and liver. Surprisingly, proteasome activity measured in these tissues was not affected by rapamycin. Rapamycin treatment was associated with an increased expression of mitochondria-targeted protein chaperones in skeletal muscle, but not liver. Finally, autophagy was increased in skeletal muscle and adipose, but not liver, from rapamycin-treated marmosets.
Conclusions
Overall, these data show tissue-specific upregulation of some, but not all, components of the proteostasis network in common marmosets treated with a pharmaceutical inhibitor of mTOR.
Keywords: healthspan, protein chaperone, proteasome, immunoproteasome, autophagy
Due to the rapidly growing population of the aged and elderly, there is increasing need to discover means to address age-related diseases and pathologies. A fundamental goal of aging research is to understand the underlying mechanisms of aging in part to develop therapeutic interventions to improve healthy aging. In pursuit of this goal, mechanistic target of rapamycin (mTOR) has emerged as a targetable cellular signaling pathway to improve both healthspan and lifespan. mTOR has a central and evolutionarily conserved role in the regulation of longevity among eukaryotes including the common laboratory models yeast, nematodes, flies and mice (1–5). Inhibition of mTOR using the macrolide rapamycin was shown to be the first pharmaceutical intervention capable of reproducible extension of lifespan in mice (6,7). Rapamycin extends lifespan of both male and female mice in multiple different genetic backgrounds, extends lifespan whether administered chronically or intermittently, and extends lifespan in mice whether treatments were begun early or late in life (6–11). There is also growing evidence that rapamycin can improve markers of healthspan in both normally aging mice and mouse models of age-related disease (11–15). It is important to then clarify whether rapamycin, or more generally mTOR inhibition, could be used to prevent or delay age-related diseases clinically. Rapamycin and its analogs are currently used as part of chemotherapeutic treatments for organ transplant, prevention of cardiac stent closure, neuro-genetic diseases and against some forms of cancer (16). However, the effects of long-term mTOR inhibition in healthy human subjects is relatively unknown, though several recent studies suggest a potential to improve some aspects of age-related dysfunction (17–19).
We have utilized the common marmoset (Callithrix jacchus) as a pre-clinical translational model to explore the physiological effects of chronic mTOR inhibition in a healthy non-human primate population. These new-world monkeys are of relatively small size and short lifespan compared to other non-human primates commonly used in biomedical research (20–22). Unlike rodents, marmosets have a diverse range of age-related pathologies that are similar to other primates including humans (21,22). These are among the most pertinent reasons as to why marmosets have been suggested as a valuable model organism that could be used to bridge translation of anti-aging interventions from mouse studies to humans. We have previously reported results from a pilot study testing a treatment protocol utilizing an oral administration of rapamycin to marmosets that achieves clinically therapeutic blood concentrations of rapamycin and significant reduction of mTOR signaling (23). We have also shown that long-term (14 months) rapamycin treatment in this pilot study does not significantly alter body weight, daily activity, blood lipid concentrations, or markers of glucose metabolism in the subject marmosets (24).
Here, we report the molecular effects on the regulation of proteostasis in vivo that are associated with long-term rapamycin treatment to marmosets. Longevity and healthspan has been attributed, at least in part, to the regulation of protein homeostasis or proteostasis (25,26). For effective proteostasis, a balance is required between the production and maintenance of properly folded protein complexes and their eventual degradation or recycling. Dysregulation of proteostasis can lead to the accumulation of non-native intracellular protein aggregates or misfolded proteins that can contribute to cellular dysfunction and the process of aging itself (25,27). It is widely accepted that mTOR plays a key role in protein synthesis and autophagy (28). There is also growing appreciation for a role for mTOR in both the regulation of protein folding via molecular chaperones and the ubiquitin-proteasome system (29,30). In this study, we delineate whether long-term rapamycin treatment of marmosets alters the in vivo regulation of a wide range of molecular markers of proteostasis, including the ubiquitin-proteasome system, protein chaperones, and autophagy.
Methods
Animal subjects
The animal subjects and study design have been described previously (23,24). In brief, male and female common marmosets (Callithrix jacchus) between the ages of 7.1 and 9.1 years of age were each treated once daily by oral dosing of 0.40 mg active rapamycin/day delivered as eudragit-encapsulated rapamycin in a yogurt vehicle via syringe. This dosage is roughly equivalent to 1 mg rapamycin/kg body weight. Animals were housed as pairs at the Southwest National Primate Research Center (SNPRC), and all housing and husbandry were approved by SNPRC Institutional Animal Care and Use Committee. At the end of trial, animals were sacrificed, and tissues were collected immediately, flash frozen, and stored at −80°C until further analysis. At sacrifice, multiple tissues including muscle, liver, kidney, heart, brain, adipose, spleen, plasma, etc. were frozen, stored, and shared with collaborators. Our studies here focus on liver and muscle due largely as 1) an attempt to recapitulate previous studies on proteostasis in rapamycin-treated mice (11,31) and 2) the availability of significant quantities of these tissues in house.
Immunoblot
Total protein was extracted from frozen tissue samples by homogenization in RIPA buffer with additional protease and phosphatase inhibitors (Thermo Scientific, Rockford, IL, USA), followed by centrifuged at 14,000 g at 4°C for 15 min and with the resultant supernatant used for all studies. Equal amounts of protein samples were separated by electrophoresis using SDS-PAGE and then transferred to a polyvinylidene fluoride membrane (Millipore, Billerica, MA, USA). Primary antibodies and their sources used in this study: phosphorylated and total ribosomal protein S6, Calnexin, Hsp40, Hsp60, Hsp70, LC3-B, GAPDH (Cell Signaling, Beverly, MA) and PSMB5, PSMB8, Clp protease (ClpP) (Abcam, Cambridge, MA). Alkaline phosphatase-conjugated secondary antibodies (Santa Cruz Biotechnology, Santa Cruz, CA) followed by ECL reagent were used to visualize protein bands which were using ImageJ. When possible, protein samples from all subjects involved in study were analyzed on a single immunoblot for individual protein tested. For LC3B assessment, data were collected over multiple immunoblots; for each immunoblot, both control and rapamycin-treated samples were analyzed concurrently. For all, data were normalized to relative Ponceau S staining of membrane following all antibody visualization.
Proteasome activity
Frozen skeletal muscle samples were prepared with Dounce homogenizer in a buffer containing 50 mM Tris-CL (pH 7.4), 1 mM DTT, 5 mM MgCl2 either with (26S activity) or without (20S activity) 0.25 mM ATP. Supernatant from these preparations was used for assays. Triplicate samples of tissue homogenates were assayed in 96 well clear bottom black plates at a final protein content of 2 µg in 100 µL buffer. For each sample, 1 µL SDS (3%) and 50 µM Suc-LLVY-AMC substrate were added to each well. To assess non-specific proteasome activity, replicate samples were run in the presence of the proteasome inhibitor MG132 at concentration of 10 µM. Fluorescent signal of samples was acquired with SpectraMax microplate reader maintained at 37°C and rate of activity was calculated over the linear portion of this readout.
Statistical analysis
To compare differences between control and rapamycin-treated groups, Student's t-test was utilized with a p≤0.05 deemed statistically significant. Pearson's correlation test was utilized to assess relationship of inhibition of mTOR between tissues in rapamycin-treated animals. All results are presented as means±standard error unless stated otherwise.
Results
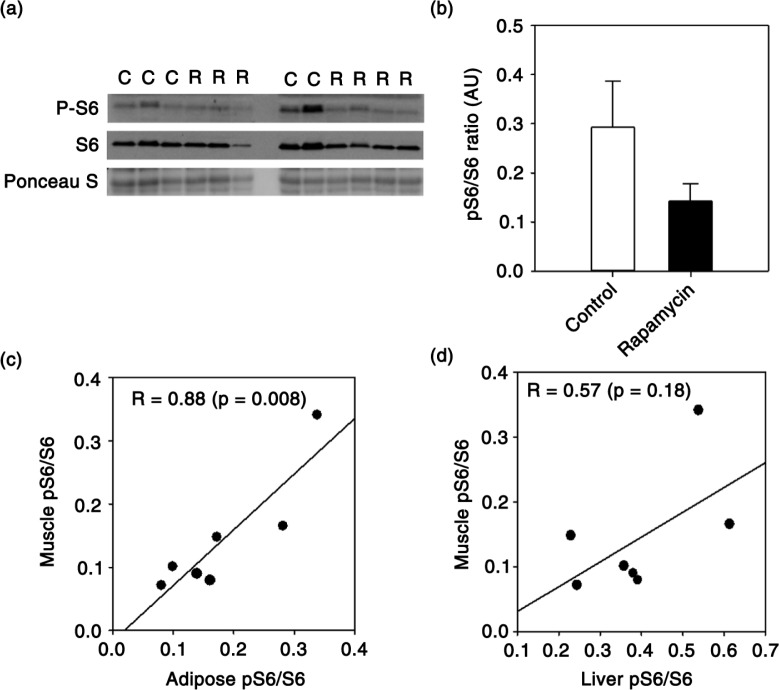
We previously reported that chronic treatment of marmosets with eudragit-encapsulated rapamycin inhibited mTORC1 in peripheral blood mononuclear cells, liver and adipose (23). In tissues collected from those same animals, we show here a similar reduction in skeletal muscle mTORC1 signaling as represented by reduced phosphorylation of ribosomal protein S6 (Fig. 1a, b). Using data collected previously from these same animals (23), we then asked whether individual marmosets treated with rapamycin showed the same relative reduction of mTORC1 across multiple tissues. Even in our small sample set (7 rapamycin-treated marmosets), we found that the degree of mTORC1 inhibition in skeletal muscle induced by rapamycin treatment was positively correlated with that found in adipose tissue (Fig. 1c) and also tended to be correlated with that in liver (Fig. 1d). The degree of mTORC1 inhibition was also significantly correlated between liver and adipose tissue (R = 0.81, p = 0.03). There was no relationship among tissues in degree of mTORC1 inhibition in control animals (data not shown).
Fig. 1.
(a) Immunoblot showing phosphorylated and total ribosomal protein S6 from skeletal muscle of control (lanes indicated with ‘C’) and rapamycin-treated (lanes indicated with ‘R’) marmosets. (b) Quantification of relative levels of phosphorylation of ribosomal protein S6 (S6) in skeletal muscle generated from immunoblot in A. Data are presented as mean values (± SEM) for indicated groups (c–d). Plot comparing relative levels of phosphorylated/total S6 ratio in muscle with that found in adipose and liver collected from the same rapamycin-treated marmosets (n = 7 total rapamycin-treated animals). Circles represent values generated from samples collected from an individual animal and line is regression line. Values in each panel give Pearson's correlation coefficient for indicated relationship and p value. Data from liver and adipose were presented previously in (23,24).
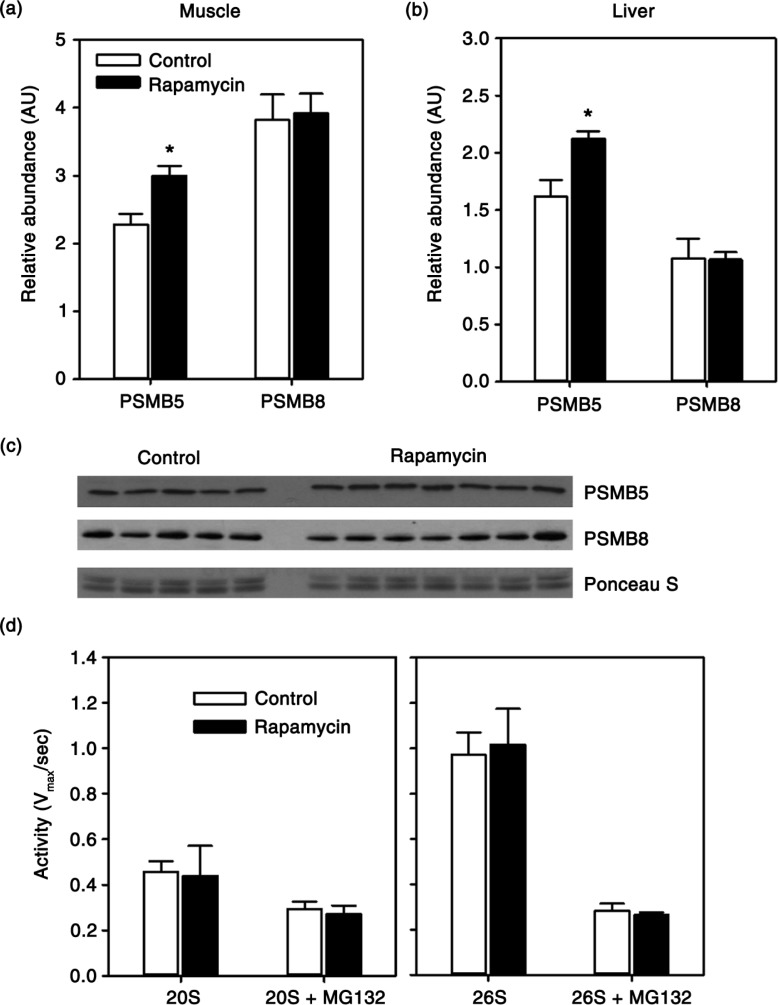
Due to availability of tissue, and basing on previous studies in mice (11,31), we focused the remainder of our study on skeletal muscle and liver. There is growing evidence that mTOR plays a role in the regulation of protein degradation through the ubiquitin-proteasome system (30,32). Here, we found that expression of PSMB5, a catalytic component of the constitutively active 20S proteasome, showed a 31% increase (p = 0.01) in skeletal muscle of rapamycin-treated animals (Fig. 2a, c). We found a similar increase (p = 0.02) in PSMB5 expression in the liver of these animals (Fig. 2b). We also addressed whether rapamycin upregulated the expression of PSMB8, a component of the immunoproteasome that responds to cytokines released in an inflammatory response. In both skeletal muscle and liver, there was no statistically significant change in expression of PSMB8 with rapamycin treatment. Next, we tested whether this increase in constitutive expression of the PSMB5 component of the 20S proteasome was associated with a change in proteasome activity. Surprisingly, we found no effect of rapamycin treatment on 20S-mediated cleavage of a fluorescent peptide substrate in skeletal muscle tissue homogenates (Fig. 2d). Similarly, we found no effect of rapamycin on 26S-mediated peptide cleavage (Fig. 2d).
Fig. 2.
Quantification of relative abundance of proteasome subunits PSMB5 and PSMB8 in skeletal muscle (a) or liver (b) from control and rapamycin-treated marmosets. Data are presented as values for each protein normalized using Ponceau S staining of immunoblot as a loading control. (c) Representative immunoblot for skeletal muscle data presented in (a). (d) Rate of 20S or 26S-mediated cleavage of fluorescent peptide (Suc-LLVY-AMC) with or without proteasome inhibitor MG132 in samples from skeletal muscle of control (n = 5) and rapamycin-treated (n = 7) marmosets. Data are presented as mean values (± SEM) for indicated groups. Asterisks represent p < 0.05 for Student's t-test comparing control to rapamycin values.
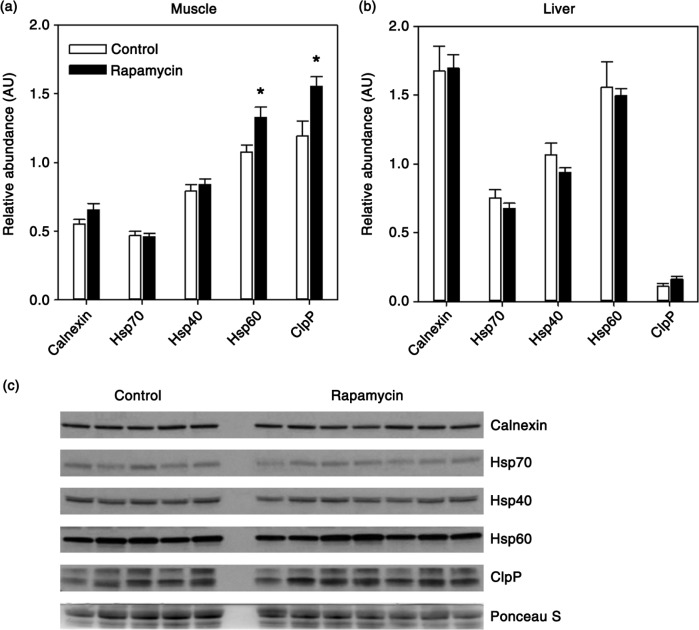
Chaperone-mediated folding of protein structure is an important step in preserving protein homeostasis without degradation of proteins. We assessed the expression of a broad range of these cellular chaperones in both muscle and liver. Calnexin, located in the endoplasmic reticulum, is a calcium-dependent chaperone that aids in correctly folding proteins destined for the secretory pathway (33). Hsp70, along with its co-chaperone Hsp40, plays a role in protein folding, refolding, complex formation, and sequestration of toxic protein aggregates (34). Hsp60 and ClpP are mitochondria-localized chaperones responsible for maintenance of the mitochondrial proteome and are upregulated as a part of the mitochondrial unfolded protein response (35,36). We found no effect of rapamycin on the relative expression of calnexin, Hsp70, of Hsp40 in either muscle or liver (Fig. 3a, c). However, both of the mitochondrial-targeted protein chaperones, Hsp60 (+23%) and ClpP (+30%), were significantly increased in muscle from rapamycin-treated marmosets (Fig. 3a). In contrast, these chaperones were unaffected by rapamycin in the liver (Fig. 3b).
Fig. 3.
Quantification of relative levels of indicated molecular chaperones in skeletal muscle (a) or liver (b) from control (n = 5) and rapamycin-treated (n = 7) marmosets. Data are presented as mean values (± SEM) for indicated groups. Data are presented as values for each protein normalized using Ponceau S staining of immunoblot as a loading control. Asterisks represent p < 0.05 for Student's t-test comparing control to rapamycin values. (c) Representative immunoblot for skeletal muscle data presented in (a).
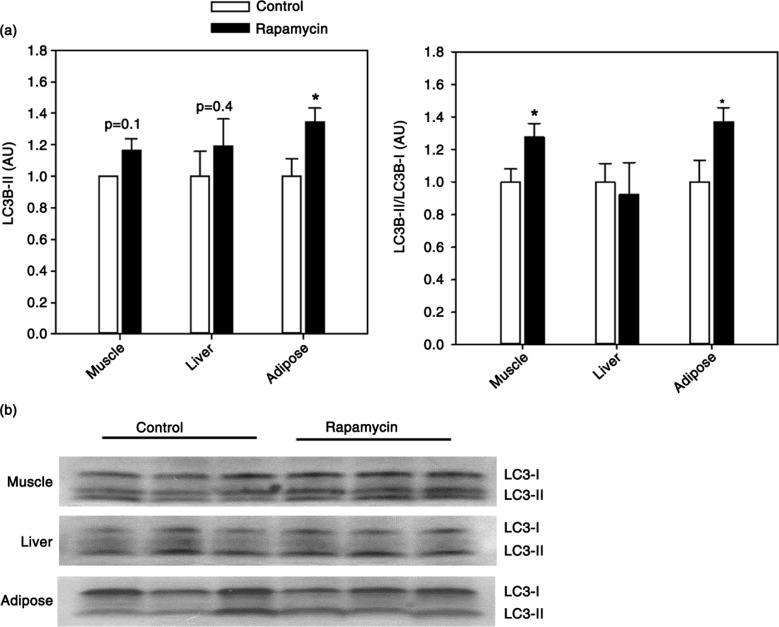
Autophagy, an intracellular process by which cellular macromolecules and organelles are recycled, is one of the key functional outputs of mTOR signaling. Previous studies show upregulation of autophagy in rapamycin-treated yeast, flies, and rodents (11,37,38). LC3B is an ubiquitin-like protein that conjugates to phosphatidylethanolamine phospholipids during the formation of the autophagosome forming the autophagosome membrane-bound isoform LC3B-II. We found no significant alteration in total LC3B-II levels in either the muscle or liver (Fig. 4). An increase in the ratio of LC3B-II to the precursor LC3B-I can be used as one of the hallmarks of the process of autophagy (39). We found rapamycin treatment associated with a significant increase in LC3B-II/LC3BI ratio in muscle, but not liver, from rapamycin-treated animals indicative of increased autophagy in this tissue (Fig. 4). Because of the strong relationship between autophagy and mTOR signaling, we addressed the effect of rapamycin on autophagy in an additional tissue collected from marmosets. As predicted, we found that rapamycin is associated with elevated levels of autophagy (both LC3B-II content and LC3B-II/LC3B-I ratio) in adipose tissue from these animals (Fig. 4).
Fig. 4.
(a) Quantification of LC3B-II levels (left) and LC3B-II/LC3B-I ratio (right) in skeletal muscle, liver, or adipose tissue from control and rapamycin-treated marmosets. Data are presented as mean values (± SEM) for indicated groups and normalized to control-treated samples in each case for clarity of presentation. Asterisks represent p < 0.05 for Student's t-test comparing control to rapamycin values. (b) Representative immunoblots for data presented in (a).
Discussion
Our results show that long-term pharmaceutical inhibition of mTOR signaling in the common marmoset induces mild, though measurable, activation of some of the molecular components responsible for the regulation of proteostasis in vivo. In general, these data hint at potential tissue-specific regulation of these processes. For example, while we show here evidence for upregulation of autophagy consistent with mTOR inhibition in skeletal muscle and adipose tissue, we found no evidence of any change in autophagy in liver. Similarly, we found rapamycin associated with increased expression of mitochondrial chaperones in muscle but not liver. One possible explanation for this tissue-specificity could be differences among tissues in the degree of mTOR inhibition in response to rapamycin. In this and our previous studies, rapamycin reduced S6 phosphorylation to a greater degree in adipose (65%) and muscle (52%) than that measured in liver (34%) of the marmoset. In mice treated with rapamycin orally, a similar disparity of mTOR inhibition among tissues has also been noted (40). At least part of this could be explained by tissue differences in bioavailability and metabolism of rapamycin in the liver (41). It may be important to address whether these tissue differences might be minimized by alternative doses of rapamycin or by use of different mTOR inhibitors.
In tissues from rapamycin-treated marmosets, we observed a general increase in constitutive 20S proteasome expression, but no change in the expression of a component of the immunoproteasome. However, we found no effect on either 20S or 26S proteasome activity. These findings are in contrast to a previous study suggesting chronic treatment of mice with rapamycin increased both proteasome activity and expression of the immunoproteasome (31). On the other hand, others have shown a reduction in immunoproteasome expression and no significant change in 20S proteasome expression in response to rapamycin treatment (42). There are considerable differences among all these studies in terms of species, age of subjects, means of drug administration, etc. that could be partial causes of the equivocal findings in regard to rapamycin and the regulation of proteasome function. However, it will be important to clearly delineate whether the regulation of proteasome expression and activity is a critical component of the pro-longevity effect of mTOR inhibition.
It was curious that we found no effect of rapamycin on proteasome activity despite evidence for increased levels of the 20S proteasome subunit PSMB5. One possible explanation could be that the specific activity of individual 20S units is reduced by rapamycin treatment; that is, there are more 20S subunits assembled, but the individual activity of each is reduced. Osmulski and Gaczynska showed that rapamycin is a potent inhibitor of proteasome activity in vitro (43). Alternatively, this discrepancy could result due to the relatively non-stressful cellular environment that might be expected in the case of healthy marmosets maintained in the lab. It should be noted that the 20S and 26S immunoproteasome are induced under stressful conditions (44). The increased capacity to respond to stress could explain, at least in part, a potential mechanism linking increased longevity and healthspan associated with the regulation of proteostasis (45,46).
Molecular chaperones play a broad role in proteostasis through their functions of protein folding, refolding, and targeting for degradation. Interestingly, we found rapamycin increased only those chaperones thought to be localized to the mitochondria. Hsp60 has been shown to have pro-survival effects in organisms exposed to oxidative stress and protects against cardiac ischemia and reperfusion injury (47–49). However, RNAi-mediated downregulation of Hsp60 and ClpP is associated with reduced C. elegans longevity (50). More generally, the regulation of the mitochondrial proteome is key to maintaining proper mitochondrial function. As mitochondrial dysfunction is associated with numerous age-related pathologies (and likely the aging process itself), it raises an interesting possibility that rapamycin may prolong longevity by mediating mitochondrial proteostasis. mTOR has a known role in mitochondrial biogenesis, but future studies will be needed to clarify exactly how this pathway preserves mitochondrial proteostasis.
Our data suggest that, at least for some tissues, there is a general upregulation of multiple effectors of proteostasis in marmosets treated with rapamycin. The processes that make up the proteostasis network are known to have some degree of cooperativity in the cell. For example, autophagy and proteasomes have been shown to work together in degrading cytoplasmic proteins marked by ubiquitination (51). In mouse brain, rapamycin was shown to upregulate autophagy that, in turn, induces protein chaperone expression including that of Hsp60 (52). In our study, it may be possible that the induction of autophagy, for instance, is responsible for the increased expression of mitochondrial chaperones. This would provide evidence for increased mitochondrial protection through upregulation of autophagy. Induction of autophagy has been attributed as the main effector of the benefits resulting from mTOR inhibition in several animal models of disease. However, further investigation will be required to address what role chaperone or proteasome regulation may play in these benefits.
In this and our previous studies (23,24), we show that rapamycin-treated marmosets demonstrate clinically relevant blood concentrations of rapamycin, significant reduction of mTOR across multiple tissues, little evidence for metabolic dysfunction or altered lipid homeostasis, and evidence for increased cellular proteostasis in vivo. These studies strengthen the case for using the common marmoset as a pre-clinical translation model in which to test interventions thought to delay aging. In the case of rapamycin, concerns regarding the side-effects of this drug likely limit its widespread use among a healthy population. Even with recent studies suggesting positive immune benefits in geriatric patients free of disease (17), long-term effects of this drug on longevity are ostensibly impossible to test in humans. As a relatively short-lived species, the common marmoset represents a useful model in which we can test the hypothesis that mTOR inhibition will extend longevity and improve healthy aging in primates.
Acknowledgements
This material is the result of work supported with resources and the use of facilities at South Texas Veterans Health Care System, San Antonio, Texas. The contents do not represent the views of the U.S. Department of Veterans Affairs or the United States Government.
Authors’ contributions
Conception and design: ML, CR, ST and ABS. Acquisition and analysis of data: ML, YL and ABS. Writing and review of manuscript: ML, ST and ABS.
Conflicts of interest and funding
The authors disclose no potential conflicts of interest. ML was supported throughout this project as an awardee of the Medical Student Training in Aging Research (MSTAR) program supported by the American Federation for Aging Research and the National Institute on Aging. The lab of ABS is supported by a grant from the National Institute of Health (R01 AG050797), a grant from the American Heart Association (15BGIA23220016) and the Geriatric Research, Education and Clinical Center of the South Texas Veterans Health Care System.
References
- 1.Bjedov I, Toivonen JM, Kerr F, Slack C, Jacobson J, Foley A, et al. Mechanisms of life span extension by rapamycin in the fruit fly Drosophila melanogaster. Cell Metab. 2010;11(1):35–46. doi: 10.1016/j.cmet.2009.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Vellai T, Takacs-Vellai K, Zhang Y, Kovacs AL, Orosz L, Muller F. Genetics: influence of TOR kinase on lifespan in C. elegans. Nature. 2003;426(6967):620. doi: 10.1038/426620a. [DOI] [PubMed] [Google Scholar]
- 3.McCormick MA, Delaney JR, Tsuchiya M, Tsuchiyama S, Shemorry A, Sim S, et al. A comprehensive analysis of replicative lifespan in 4,698 single-gene deletion strains uncovers conserved mechanisms of aging. Cell Metab. 2015;22(5):895–906. doi: 10.1016/j.cmet.2015.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lamming DW, Ye L, Katajisto P, Goncalves MD, Saitoh M, Stevens DM, et al. Rapamycin-induced insulin resistance is mediated by mTORC2 loss and uncoupled from longevity. Science. 2012;335(6076):1638–43. doi: 10.1126/science.1215135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wu JJ, Liu J, Chen Edmund B, Wang Jennifer J, Cao L, Narayan N, et al. Increased mammalian lifespan and a segmental and tissue-specific slowing of aging after genetic reduction of mTOR expression. Cell Rep. 2013;4(5):913–20. doi: 10.1016/j.celrep.2013.07.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Harrison DE, Strong R, Sharp ZD, Nelson JF, Astle CM, Flurkey K, et al. Rapamycin fed late in life extends lifespan in genetically heterogeneous mice. Nature. 2009;460(7253):392–5. doi: 10.1038/nature08221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Miller RA, Harrison DE, Astle CM, Baur JA, Boyd AR, de Cabo R, et al. Rapamycin, but not resveratrol or simvastatin, extends life span of genetically heterogeneous mice. J Gerontol A Biol Sci Med Sci. 2011;66A(2):191–201. doi: 10.1093/gerona/glq178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Miller RA, Harrison DE, Astle CM, Fernandez E, Flurkey K, Han M, et al. Rapamycin-mediated lifespan increase in mice is dose and sex dependent and metabolically distinct from dietary restriction. Aging Cell. 2014;13(3):468–77. doi: 10.1111/acel.12194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Neff F, Flores-Dominguez D, Ryan DP, Horsch M, Schroder S, Adler T, et al. Rapamycin extends murine lifespan but has limited effects on aging. J Clin Invest. 2013;123(8):3272–91. doi: 10.1172/JCI67674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fok WC, Chen Y, Bokov A, Zhang Y, Salmon AB, Diaz V, et al. Mice fed rapamycin have an increase in lifespan associated with major changes in the liver transcriptome. PLoS One. 2014;9(1):e83988. doi: 10.1371/journal.pone.0083988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang Y, Bokov A, Gelfond J, Soto V, Ikeno Y, Hubbard G, et al. Rapamycin extends life and health in C57BL/6 Mice. J Gerontol A Biol Sci Med Sci. 2014;69A(2):119–30. doi: 10.1093/gerona/glt056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hasty P, Livi CB, Dodds SG, Jones D, Strong R, Javors M, et al. eRapa restores a normal life span in a FAP mouse model. Cancer Prev Res. 2014;7(1):169–78. doi: 10.1158/1940-6207.CAPR-13-0299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fischer KE, Gelfond JA, Soto VY, Han C, Someya S, Richardson A, et al. Health effects of long-term rapamycin treatment: the impact on mouse health of enteric rapamycin treatment from four months of age throughout life. PLoS One. 2015;10(5):e0126644. doi: 10.1371/journal.pone.0126644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Caccamo A, Majumder S, Richardson A, Strong R, Oddo S. Molecular interplay between Mammalian Target of Rapamycin (mTOR), Amyloid-β, and Tau: effects on cognitive impairments. J Biol Chem. 2010;285(17):13107–20. doi: 10.1074/jbc.M110.100420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Majumder S, Caccamo A, Medina DX, Benavides AD, Javors MA, Kraig E, et al. Lifelong rapamycin administration ameliorates age-dependent cognitive deficits by reducing IL-1β and enhancing NMDA signaling. Aging Cell. 2012;11(2):326–35. doi: 10.1111/j.1474-9726.2011.00791.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ehninger D, Neff F, Xie K. Longevity, aging and rapamycin. Cell Mol Life Sci. 2014;71(22):4325–46. doi: 10.1007/s00018-014-1677-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mannick JB, Del Giudice G, Lattanzi M, Valiante NM, Praestgaard J, Huang B, et al. mTOR inhibition improves immune function in the elderly. Sci Transl Med. 2014;6(268):268ra179. doi: 10.1126/scitranslmed.3009892. [DOI] [PubMed] [Google Scholar]
- 18.Hurez V, Dao V, Liu A, Pandeswara S, Gelfond J, Sun L, et al. Chronic mTOR inhibition in mice with rapamycin alters T, B, myeloid, and innate lymphoid cells and gut flora and prolongs life of immune-deficient mice. Aging Cell. 2015;14(6):945–56. doi: 10.1111/acel.12380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Benjamin D, Colombi M, Moroni C, Hall MN. Rapamycin passes the torch: a new generation of mTOR inhibitors. Nat Rev Drug Discov. 2011;10(11):868–80. doi: 10.1038/nrd3531. [DOI] [PubMed] [Google Scholar]
- 20.Tardif S, Bales K, Williams L, Moeller EL, Abbott D, Schultz-Darken N, et al. Preparing new world monkeys for laboratory research. ILAR J. 2006;47(4):307–15. doi: 10.1093/ilar.47.4.307. [DOI] [PubMed] [Google Scholar]
- 21.Tardif SD, Mansfield KG, Ratnam R, Ross CN, Ziegler TE. The marmoset as a model of aging and age-related diseases. ILAR J. 2011;52(1):54–65. doi: 10.1093/ilar.52.1.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ross CN, Davis K, Dobek G, Tardif SD. Aging phenotypes of common marmosets (Callithrix jacchus) J Aging Res. 2012;2012:567143. doi: 10.1155/2012/567143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tardif S, Ross C, Bergman P, Fernandez E, Javors M, Salmon A, et al. Testing efficacy of administration of the antiaging drug rapamycin in a nonhuman primate, the common marmoset. J Gerontol A Biol Sci Med Sci. 2015;70(5):577–87. doi: 10.1093/gerona/glu101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ross C, Salmon A, Strong R, Fernandez E, Javors M, Richardson A, et al. Metabolic consequences of long-term rapamycin exposure on common marmoset monkeys (Callithrix jacchus) Aging. 2015;7(11):964–73. doi: 10.18632/aging.100843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Labbadia J, Morimoto RI. The biology of proteostasis in aging and disease. Annu Rev Biochem. 2015;84:435–64. doi: 10.1146/annurev-biochem-060614-033955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Koga H, Kaushik S, Cuervo AM. Protein homeostasis and aging: the importance of exquisite quality control. Ageing Res Rev. 2011;10(2):205–15. doi: 10.1016/j.arr.2010.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.López-Otín C, Blasco MA, Partridge L, Serrano M, Kroemer G. The hallmarks of aging. Cell. 2013;153(6):1194–217. doi: 10.1016/j.cell.2013.05.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Diaz-Troya S, Perez-Perez ME, Florencio FJ, Crespo JL. The role of TOR in autophagy regulation from yeast to plants and mammals. Autophagy. 2008;4(7):851–65. doi: 10.4161/auto.6555. [DOI] [PubMed] [Google Scholar]
- 29.Conn CS, Qian SB. mTOR signaling in protein homeostasis: less is more? Cell Cycle. 2011;10(12):1940–7. doi: 10.4161/cc.10.12.15858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhao J, Zhai B, Gygi SP, Goldberg AL. mTOR inhibition activates overall protein degradation by the ubiquitin proteasome system as well as by autophagy. Proc Natl Acad Sci USA. 2015;112(52):15790–7. doi: 10.1073/pnas.1521919112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pickering AM, Lehr M, Miller RA. Lifespan of mice and primates correlates with immunoproteasome expression. J Clin Invest. 2015;125(5):2059–68. doi: 10.1172/JCI80514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang Y, Nicholatos J, Dreier JR, Ricoult SJ, Widenmaier SB, Hotamisligil GS, et al. Coordinated regulation of protein synthesis and degradation by mTORC1. Nature. 2014;513(7518):440–3. doi: 10.1038/nature13492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lamriben L, Graham JB, Adams BM, Hebert DN. N-glycan based ER molecular chaperone and protein quality control system: the calnexin binding cycle. Traffic. 2016;17(4):308–26. doi: 10.1111/tra.12358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Radons J. The human HSP70 family of chaperones: where do we stand? Cell Stress Chaperones. 2016;21(3):379–404. doi: 10.1007/s12192-016-0676-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cheng MY, Hartl FU, Martin J, Pollock RA, Kalousek F, Neupert W, et al. Mitochondrial heat-shock protein hsp60 is essential for assembly of proteins imported into yeast mitochondria. Nature. 1989;337(6208):620–5. doi: 10.1038/337620a0. [DOI] [PubMed] [Google Scholar]
- 36.Cole A, Wang Z, Coyaud E, Voisin V, Gronda M, Jitkova Y, et al. Inhibition of the mitochondrial protease ClpP as a therapeutic strategy for human acute myeloid leukemia. Cancer Cell. 2015;27(6):864–76. doi: 10.1016/j.ccell.2015.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ravikumar B, Vacher C, Berger Z, Davies JE, Luo S, Oroz LG, et al. Inhibition of mTOR induces autophagy and reduces toxicity of polyglutamine expansions in fly and mouse models of Huntington disease. Nat Genet. 2004;36(6):585–95. doi: 10.1038/ng1362. [DOI] [PubMed] [Google Scholar]
- 38.Kamada Y, Funakoshi T, Shintani T, Nagano K, Ohsumi M, Ohsumi Y. Tor-mediated induction of autophagy via an Apg1 protein kinase complex. J Cell Biol. 2000;150(6):1507–13. doi: 10.1083/jcb.150.6.1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Klionsky DJ, Abdalla FC, Abeliovich H, Abraham RT, Acevedo-Arozena A, Adeli K, et al. Guidelines for the use and interpretation of assays for monitoring autophagy. Autophagy. 2012;8(4):445–544. doi: 10.4161/auto.19496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liu Y, Diaz V, Fernandez E, Strong R, Ye L, Baur JA, et al. Rapamycin-induced metabolic defects are reversible in both lean and obese mice. Aging. 2014;6(9):742–54. doi: 10.18632/aging.100688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gallant-Haidner HL, Trepanier DJ, Freitag DG, Yatscoff RW. Pharmacokinetics and metabolism of sirolimus. Ther Drug Monit. 2000;22(1):31–5. doi: 10.1097/00007691-200002000-00006. [DOI] [PubMed] [Google Scholar]
- 42.Zhang HM, Fu J, Hamilton R, Diaz V, Zhang Y. The mammalian target of rapamycin modulates the immunoproteasome system in the heart. J Mol Cell Cardiol. 2015;86:158–67. doi: 10.1016/j.yjmcc.2015.07.027. [DOI] [PubMed] [Google Scholar]
- 43.Osmulski PA, Gaczynska M. Rapamycin allosterically inhibits the proteasome. Mol Pharmacol. 2013;84(1):104–13. doi: 10.1124/mol.112.083873. [DOI] [PubMed] [Google Scholar]
- 44.Pickering AM, Koop AL, Teoh CY, Ermak G, Grune T, Davies KJ. The immunoproteasome, the 20S proteasome and the PA28alphabeta proteasome regulator are oxidative-stress-adaptive proteolytic complexes. Biochem J. 2010;432(3):585–94. doi: 10.1042/BJ20100878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Salmon AB, Leonard S, Masamsetti V, Pierce A, Podlutsky AJ, Podlutskaya N, et al. The long lifespan of two bat species is correlated with resistance to protein oxidation and enhanced protein homeostasis. FASEB J. 2009;23(7):2317–26. doi: 10.1096/fj.08-122523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Perez VI, Buffenstein R, Masamsetti V, Leonard S, Salmon AB, Mele J, et al. Protein stability and resistance to oxidative stress are determinants of longevity in the longest-living rodent, the naked mole-rat. Proc Natl Acad Sci USA. 2009;106(9):3059–64. doi: 10.1073/pnas.0809620106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kaetsu A, Fukushima T, Inoue S, Lim H, Moriyama M. Role of heat shock protein 60 (HSP60) on paraquat intoxication. J Appl Toxicol. 2001;21(5):425–30. doi: 10.1002/jat.774. [DOI] [PubMed] [Google Scholar]
- 48.Wu CW, Biggar KK, Zhang J, Tessier SN, Pifferi F, Perret M, et al. Induction of antioxidant and heat shock protein responses during torpor in the gray mouse lemur, microcebus,urinus. Genomics Proteomics Bioinformatics. 2015;13(2):119–26. doi: 10.1016/j.gpb.2015.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hollander JM, Lin KM, Scott BT, Dillmann WH. Overexpression of PHGPx and HSP60/10 protects against ischemia/reoxygenation injury. Free Radical Biol Med. 2003;35(7):742–51. doi: 10.1016/s0891-5849(03)00400-3. [DOI] [PubMed] [Google Scholar]
- 50.Durieux J, Wolff S, Dillin A. The cell-non-autonomous nature of electron transport chain-mediated longevity. Cell. 2011;144(1):79–91. doi: 10.1016/j.cell.2010.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Qin ZH, Wang Y, Kegel KB, Kazantsev A, Apostol BL, Thompson LM, et al. Autophagy regulates the processing of amino terminal huntingtin fragments. Hum Mol Genet. 2003;12(24):3231–44. doi: 10.1093/hmg/ddg346. [DOI] [PubMed] [Google Scholar]
- 52.Sheng R, Liu XQ, Zhang LS, Gao B, Han R, Wu YQ, et al. Autophagy regulates endoplasmic reticulum stress in ischemic preconditioning. Autophagy. 2012;8(3):310–25. doi: 10.4161/auto.18673. [DOI] [PubMed] [Google Scholar]