Abstract
The present research was designed to explore indigenous probiotic Lactic acid bacteria from traditional fermented foods and beverages of North-western Himalayas for their probiotic potential. It was achieved through a step-by step approach focused on the technological characterization, evaluation of the probiotic traits and adherence ability. Fifty one LAB isolates from traditional fermented foods and beverages were initially screened for their technological properties and among them twenty isolates were selected. These isolates were further characterized and identified using 16S rRNA gene sequencing as Lactobacillus brevis (7 isolates), Lactobacillus casei (5), Lactobacillus paracasei (2), Lactobacillus buchneri (1), Lactobacillus plantarum (1) and Lactobacillus sp. (3). Identified isolates were evaluated by in vitro methods including survival in gastrointestinal tract, antibiotic susceptibility, antimicrobial activity, cell surface characteristics, exopolysacharride production and haemolytic activity. The results of these experiments were used as input data for Principal Component Analysis; thus, to select the most promising probiotic isolates. Three isolates (L. brevis PLA2, L. paracasei PLA8 and L. brevis PLA16) were found to be most technological relevant and promising probiotic candidates in comparison to commercial probiotic strains. L. brevis PLA2 was selected as best isolate with probiotic potential by in vitro adherence to the human intestinal HT-29 cell line.
Electronic supplementary material
The online version of this article (doi:10.1007/s13197-016-2231-y) contains supplementary material, which is available to authorized users.
Keywords: Probiotics, LAB, Lactobacillus, Traditional fermented foods, PCA
Introduction
Human intestinal tract contains total bacterial population of around 1013 and 1014 cfu/g. Complex symbiosis between the beneficial and pathogenic microorganisms is necessary to maintain the optimal gut balance to prevent pathogenesis. Increase in the need of balanced intestinal microbiota and health promoting effects of probiotics; have received attention in recent years. Probiotics have been defined as “live microorganisms which when administered in adequate amounts confer a health benefits on the host” (FAO/WHO, 2001) and usually belong to the genus Lactobacillus or Bifidobacterium. Lactobacilli are a group of Lactic acid bacteria (LAB) and are found in the gastrointestinal tract soon after birth. In healthy humans, lactobacilli are normally present in the oral cavity (103–104 cfu/g), the ileum (103–107 cfu/g), colon (104–108 cfu/g), and are abundant in vagina (107–108 cfu/g) as reported by Bernardeau et al. (2008) and Merk et al. (2005).
LAB from different fermented foods and beverages are highly valued for their probiotic properties. Spontaneously fermented foods may constitute a reservoir for new LAB strains with potential probiotic characteristics (Taked et al. 2011). Traditionally fermented products have been considered as the best carrier for probiotics. A large number of potentially probiotic microorganisms are present in different kinds of food products which are still unknown and uncharacterized (Ramosa et al. 2013, Sonar and Halami, 2014). In north-west (NW) Himalayas, the people prepare and consume a number of traditional foods and beverages. An attempt has been made for the characterization of LAB with probiotic potentials from the traditional fermented foods and beverages of tribal regions of North-western Himalayas (Himachal Pradesh). The present study is focused on exploration and characterization of indigenous fermented foods and beverages from north-west Himalayas for the presence of Lactobacilli and study the probiotic potential of selected isolates.
Materials and methods
The research was divided into different steps, as follows: a) Isolation of Lactobacilli from fermented foods and beverages; b) biochemical, technological and genotypic characterization of the Lactobacillus isolates; c) probiotic characterization of the relevant isolates; d) Principal Component Analysis (PCA) to select the promising probiotic isolates; e) in vitro adherence to the human intestinal HT-29 cell line for the selection of most promising isolates.
Lactobacillus isolation and culture conditions
A total number of 51 LAB isolates were isolated from various traditional fermented foods and beverages (Chhang, Phab, Arak, Sudung, Chilra, Pole, Bhaturoo, Seera and Churpe) which were collected from different tribal regions of Himachal Pradesh shown in Table 1. 10 g/ml of each sample was diluted with 90 ml of a sterile saline solution (0.9 % NaCl) and homogenized for 60 s. The homogenates were serially diluted in saline solution and plated onto MRS (Man Rogosa Sharpe, Hi-Media) medium and incubated at 30 °C for 24–48 h. Isolates were presumptively identified as LAB by culturing on MRS agar and examined for colony and cell appearance, catalase activity, motility, Gram and endospore characters. All LAB isolates were grown routinely in MRS broth, and stored in Elliker agar plates at 4 °C.
Table 1.
Lactobacillus spp. isolated from fermented foods and beverages
| S. No | Fermented products | Characteristics | Strains identified | Accession Numbers |
|---|---|---|---|---|
| 1 | Rice Channg | Alcoholic beverage | L. brevis strain PLA1 | KJ726646 |
| 2 | Jau Channg | Alcoholic beverage | L. brevis strain PLA2 | KJ726647 |
| 3 | Phab | Traditional starter | L. brevis strain PLA3 | KJ726648 |
| 4 | Dheli | Traditional starter | L. brevis strain PLA4 | KJ726649 |
| 5 | Churpe (churu milk) | Dried cheese | L. casei strain PLA5 | KJ726650 |
| 6 | Sudung (Black Grape wine) | Alcoholic beverage | L. casei strain PLA6 | KJ726651 |
| 7 | Kronto | Fermented Must | L. brevis strain PLA7 | KM410930 |
| 8 | Churpe | Dried cheese | L. casei strain PLA8 | KJ726652 |
| 9 | Sudung (Green grape wine) | Alcoholic beverage | L. buchneri strain PLA9 | KJ726653 |
| 10 | Seera | Cereal based fermented product (Wheat) | L. casei strain PLA10 | KJ726654 |
| 11 | Chilra | Pancakes (Barley) | L. paracasei strain PLA11 | KJ726655 |
| 12 | Chilra | Pancakes (Buckwheat) | L. casei strain PLA12 | KJ726656 |
| 13 | Pole | Fermented fried bread | L. paracasei subsp. Tolerans strain PLA13 | KJ726657 |
| 14 | Pole | Fermented fried bread | L. brevis strain PLA14 | KJ726658 |
| 15 | Khameer | Fermented dough | L. kefiri strain PLA15 | KM410931 |
| 16 | Babroo | Fermented fried disk | L. brevis strain PLA16 | KJ726659 |
| 17 | Chulli | Alcoholic beverage | Lactobacillus sp. PLA17 | KJ726660 |
| 18 | Arak | Alcoholic beverage | Lactobacillus sp. PLA18 | KJ726661 |
| 19 | Bhatoroo | Indigenous bread | L. plantarum strain PLA19 | KM657203 |
| 20 | Seera | Cereal based fermented product (wheat) | L. casei strain PLA20 | KM657204 |
Biochemical characterization
Gas production from glucose was checked for lactobacilli sp. in order to determine homo or hetero-fermentative isolates. Arginine hydrolysis and citrate utilization were analysed. Lactobacilli were inoculated into carbohydrate fermentation medium containing different sugars (sucrose, raffinose, trihalose, xylose, maltose, fructose, galactose, ribose, dextrose, mannitol, starch and lactose) for determination of carbohydrate fermentation pattern. Growth of lactobacilli at different pH values (2.5, 3.5, 8.5 and 9.5), NaCl concentration (2, 4 and 6.5 %) and temperature (15, 37 and 45 °C) was measured.
Genotypic identification by 16S rRNA gene sequencing
Twenty lactobacilli isolates were selected for 16S rRNA gene sequence analysis using the universal primers 27F (5′-AGAGTTTGATCCTGGCTCAG-3′) and 1492R (5′-TACGGYTACCTTGTTACGACTT-3′). 16S rRNA gene sequence was analysed with the Bioedit software and the resulted sequence was compared with already sequenced organisms by BLAST (Basic Local Alignment Search Tool) program on the NCBI website http://www.ncbi.nlm.nih.gov to determine the closest known relatives. Sequence was submitted in National Center for Biological Information (NCBI) using BankIt sequence submission tool and accession numbers were obtained. Multiple sequence analysis was done with Clustal Omega. Phylogenetic tree was constructed using MEGA BLAST 6 and similarities among the sequences were compared by the Neighbor-joining method.
Resistance to biological barriers in stimulated GI tract
Tolerance to acidic pH, bile salt and lysozyme
Acid (pH 2, 3 and 7) and bile (0.5, 1 and 2 %) tolerance at 3 h and 12 h, respectively was assayed by the method of Maragkoudakis et al. (2006). Survival of isolates in the presence of different concentration of lysozyme (50,100 and 150 μg/ml) for 3 h was determined. Survival of lactobacilli in various biological barriers were calculated in terms of Log cfu/mL in comparison to the probiotic reference strains i.e. Lactobacillus casei Shirota (P1) and Lactobacillus rhamnosus (P2) which were procured from University of Helsinki, Finland.
Antibiotic susceptibility
Bacterial antibiotic resistance was determined on MRS agar by disk diffusion method (Turchi et al. 2013). The following antimicrobial agents have been tested: inhibitors of cell wall synthesis- Penicillin G (P; 10 Units), Ampicillin (A; 10 μg), Vancomycin (VA; 30 μg), Carbenicillin (CB; 100 μg), Oxacilin (OX; 5 μg), Cefotaxime (CTX; 30 μg); inhibitor of nucleic acid synthesis- Norfloxacin (NX; 10 μg); inhibitor of protein synthesis-Chloramphenicol (C; 30 μg), Erythromycin (E; 15 μg), Clindamycin (CD; 2 μg), Gentamicin (GEN; 10 μg); inhibitor of folate synthesis- Co-trimoxazole (COT; 25 μg), Co-trimazine (CM; 25 μg). Diameters of inhibition zones (mm) were measured after incubation at 30 °C for 24 h.
Antimicrobial activity
Antimicrobial effects of lactobacilli on Gram positive bacteria (Staphylococcus aureus and Bacillus subtilis) and Gram negative bacteria (Yersinia enterocolitica, Pseudomonas aeruginosa, Shigella dysenteriae and Escherichia coli) as test organisms were determined by the bit disk method and well diffusion assay (Schillinger and Lucke, 1989). Diameter of the inhibition zone was measured in millimetres (mm).
Determination of cell surface characteristics
Autoaggregation assay
Autoaggregation of Lactobacillus isolates were determined as described by Reniero et al. (1992) as the autoaggregation percentage. Autoaggregation percentage at 3 h and 24 h was determined using the following equation:
Coaggregation assay
The coaggregation test was performed as a method described by Handley et al. (1987). The coaggregation (%) at 3 h and 24 h was calculated according to the equation as given below:
where x and y represent each of the two strains in the control tubes and (x + y) in the mixture at 3 h and 24 h.
Cell surface hydrophobicity
The bacterial adhesion to hydrocarbons (BATH) with different hydrocarbons n-hexadecane, xylene, octane and toluene was performed according to Rosenberg et al. (1980). Results were reported as a percentage from 3 replicates according to the formula:
Whereas, A0 and A are absorbance before and after mixing with solvents at A600 nm.
Exopolysacharride production
Exopolysacharride production was evaluated according to Mora et al. (2002).
Haemolytic activity
For haemolytic activity tests, the isolates were subcultured in MRS and then streaked on Columbia agar plates, containing 5 % of sheep blood (Maragkoudakis et al. 2009). Isolates that producing green-hued zones around the colonies or did not produce any effect on the blood plates were considered non haemolytic. The lactobacilli isolates showing blood lysis zones around the colonies were classified as haemolytic (β-haemolysis).
Bile salt hydrolase activity
Bile salt deconjugation ability of lactobacilli were determined by the qualitative direct plate assay (Ha et al. 2006). Overnight grown cultures were spotted on MRS agar plates containing 0.37 g/L CaCl2 and 0.5 % sodium salt of taurodeoxycholic acid (TDCA). Plates were incubated at 30 °C for 72 h. Presence of halos around colonies or a white opaque colony indicated positive bile salt hydrolase activity.
Data processing and statistical analysis
Statistical analysis was carried out with SPSS Inc. software (version 19.0). One way analysis of variance (ANOVA) was used to study significant difference between means, with significance level at p < 0.01/0.05. 2-sided Tukey’s HSD test was used to perform multiple comparisons between means. All data presented are mean values of two determinations and three replicates.
Principal component analysis
Statistical differences among the isolates were pointed out through the Principal Component Analysis (PCA) done by the method given by Perricone et al. (2014). PCA makes it possible to distinguish between various Lactobacilli isolates and to identify the most potential probiotic isolate (Ghosh and Chattopadhyay, 2011). The relationship among the isolates was determined by Hierarchical Cluster Analysis (HCA) and PCA using XLSTAT™ software. The cases introduced in the analysis were the 40 identified lactobacilli isolates along with the two standard probiotic strains while the discriminating variables were acid and bile tolerance, hydrophobicity, auto and co-aggregation, antimicrobial and antibiotic assay. Results of the qualitative and quantitative probiotic characterization were converted into three coded values (0, 1 and 2) and used as input data for PCA. PCA was done by using varimax rotation.
In vitro adherence to the human intestinal cell lines
Growth and maintenance of HT-29 cells
The human adenocarcinoma cell line HT-29 (mucus secreting) was cultured by method given by Messaoudi et al. (2012).
Adhesion capacity to HT-29 cell line
Bacterial adhesion capacity to the HT-29 cell line was investigated for the 3 selected lactobacilli as per the method described by Jacobsen et al. (1999). The adhesion score was measured by enumerating adhered bacteria per 20 different microscopic fields as described by Duary et al. (2011).
Results
Isolation and identification
Fifty one presumptive LAB isolates from different fermented foods and beverages from various tribal regions of Himachal Pradesh were screened. Among the 20 putative LAB isolates showed positive catalase activity and 10 were positive for endospore staining. Thus, Gram negative, endospore and catalase positive isolates were discarded and remaining 30 isolates were further characterized. In order to reduce the number of isolates for further examinations, the other biochemical and technological identification of the isolates was done.
Biochemical and genotypic characterization
Seventeen Lactobacillus isolates were homo-fermentative as the end product of fermentation was only lactic acid and rest of the 13 isolates were heterofermentative as they produced gas (carbon dioxide) from glucose fermentation. All isolates grew at 37 °C but showed variable growth at 15 °C and 45 °C as shown in Table 2. Considerable good growth was observed at 2 % and 4 % NaCl concentration but Lactobacillus isolates grew variably at 6.5 % NaCl concentration.
Table 2.
Physiological characteristics of Lactobacillus spp. from traditional fermented foods and beverages
| Isolates | pH | Temperature (°C) | NaCl (%) | Homo/hetero fermentation | Lactic acid | Acetic acid | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 2.5 | 3.5 | 8.5 | 9.5 | 15 | 37 | 45 | 2 | 4 | 6.5 | ||||
| L1 | + | ++ | +++ | +++ | +++ | +++ | ++ | +++ | +++ | − | Hetero | ++ | ++ |
| L5 | − | − | ++ | + | +++ | +++ | +++ | +++ | +++ | + | Hetero | + | ++ |
| L6 | − | − | +++ | − | +++ | +++ | +++ | +++ | +++ | − | Hetero | − | + |
| L9 | − | − | ++ | − | ++ | +++ | ++ | +++ | +++ | + | Hetero | + | + |
| L11 | + | + | +++ | ++ | +++ | +++ | ++ | +++ | +++ | + | Homo | ++ | + |
| L14 | − | − | +++ | − | − | +++ | ++ | +++ | +++ | ++ | Homo | − | ++ |
| L16 | + | + | +++ | ++ | +++ | +++ | ++ | +++ | +++ | + | Hetero | ++ | ++ |
| L19 | + | + | +++ | +++ | − | +++ | ++ | +++ | +++ | − | Hetero | + | + |
| L21 | − | − | +++ | − | +++ | +++ | ++ | +++ | +++ | − | Homo | + | + |
| L22 | + | + | +++ | ++ | +++ | +++ | ++ | +++ | +++ | + | Homo | − | ++ |
| L23 | − | − | +++ | +++ | +++ | +++ | ++ | +++ | +++ | + | Homo | + | ++ |
| L24 | + | + | +++ | − | +++ | +++ | +++ | +++ | +++ | + | Homo | + | +++ |
| L26 | _ | _ | ++ | − | +++ | +++ | − | +++ | +++ | − | Homo | − | − |
| L28 | − | − | +++ | ++ | +++ | +++ | ++ | +++ | +++ | + | Hetero | ++ | +++ |
| L29 | − | + | +++ | + | +++ | +++ | − | +++ | +++ | − | Hetero | + | +++ |
| L31 | + | + | +++ | +++ | ++ | +++ | ++ | +++ | +++ | − | Hetero | − | +++ |
| L34 | − | + | ++ | − | +++ | +++ | ++ | +++ | +++ | + | Hetero | ++ | +++ |
| L36 | − | − | ++ | ++ | − | +++ | − | +++ | +++ | − | Hetero | − | ++ |
| L38 | − | − | ++ | ++ | − | +++ | − | +++ | +++ | − | Homo | − | ++ |
| L40 | − | − | ++ | ++ | + | +++ | − | +++ | +++ | − | Homo | + | + |
+ Mild growth; ++ moderate growth; +++ good growth; − no growth
The low acidic pH (2.5 and 3.5) of medium exerted a strong effect on lactobacilli microflora, as the growth of 10 isolates was completely inhibited at low pH. On the basis of technological characterization 20 Lactobacillus isolates were selected and the carbohydrate fermentation of various sugars (lactose, sucrose, dextrose, mannitol, galactose, raffinose, maltose, starch and arabinose) was observed. All the sugars were variably utilized by 20 Lactobacillus isolates (data not shown).
Twenty most technologically relevant isolates (1, 5, 6, 9, 11, 14, 16, 19, 21, 22, 23, 24, 26, 28, 29, 31, 34, 36, 38 and 40) were finally selected and sequenced for 16S rDNA identification and were analysed for their probiotic traits. These 20 isolates were identified as Lactobacillus brevis (seven isolates), Lactobacillus casei (five isolates), Lactobacillus paracasei (two isolates), Lactobacillus buchneri, Lactobacillus kefiri, Lactobacillus plantarum and Lactobacillus sp. (3 isolates).
Phylogenetic analysis
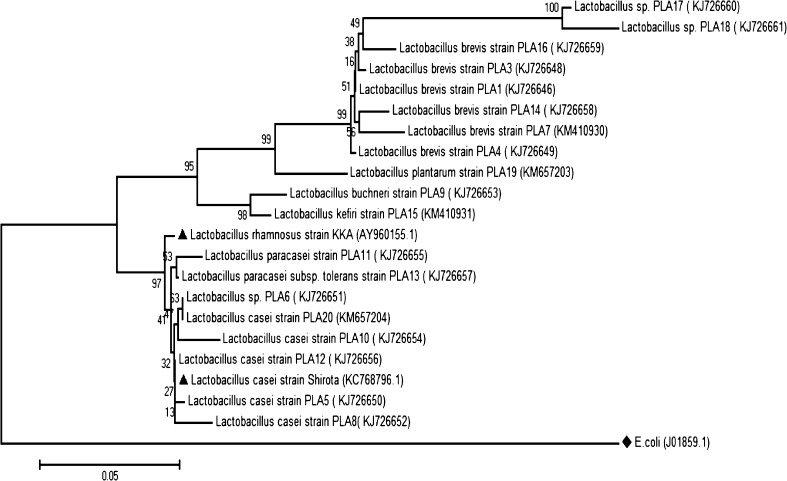
Phylogenetic tree of the 20 Lactobacillus isolates were constructed based on 16S rDNA sequences (Fig. 1). The identification of the predominant microflora in the traditional fermented foods with probiotic potential by 16S rDNA revealed that Lactobacillus brevis and Lactobacillus casei were the main fermenting organisms. In addition Lactobacillus buchneri and Lactobacillus paracasei were the other lactic acid bacterial species associated with food fermentations having probiotic characteristics.
Fig 1.
Phylogenetic trees of the 20 Lactobacillus isolates were constructed based on 16S rDNA sequences
Taxonomic identification of Lactobacillus isolates was performed by comparing the resulting sequences of each isolate using online tool BLAST (http://blast.ncbi.nlm.nih.gov/Blast.cgi) database. The isolates were identified based on the highest hit scores (>99 % sequence identity) and the sequences of all lactobacilli were deposited in the National Center for Biotechnology Information (NCBI) nucleotide sequence database. Partial sequences were aligned using multiple sequence alignment software ClustalW2.
Resistance to biological barriers in stimulated GI tract
Tolerance to acidic pH
The results given in Table 3 showed that most isolates were quite tolerant to pH 3.0. However, the critical limit to survive the exposure to acidic conditions was pH 2.0. The incubation in PBS buffer at pH 2 resulted in significant decreases in the survival rate of the selected probiotic isolates. Among all the isolates, L16, L22, L29, L31, L23, L28 and L9 showed the highest viability (Log cfu/ml) to the acidic pH, whereas L1 and L6 were the most acid-sensitive isolates as compared to the commercial probiotic strains L. rhamnosus GG and L. casei Shirota (P1 and P2).
Table 3.
Tolerance of biological barriers in stimulated GI tract and cell surface characteristics of isolated Lactobacillus spp.
| pH (log CFU/ml) | Bile concentration (log CFU/ml) | Autoaggregation* (%) | Coaggregation* (%) | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Isolates | pH 7.0 | pH 3.0 | pH 2.0 | 0.5 % | 1 % | 2 % | 3 h | 24 h | 3 h | 24 h |
| L1 | 8.47 ± 0.17b | 7.32 ± 0.12a | 7.04 ± 0.07a | 8.35 ± 0.09b | 8.30 ± 0.05b | 7.82 ± 0.21a | 13.35 ± 0.04klmn | 59.35 ± 0.39 | 5.84 ± 0.04 | 50.19 ± 0.09 |
| L5 | 8.72 ± 0.05c | 8.43 ± 0.06b | 8.28 ± 0.05a | 8.64 ± 0.08b | 8.60 ± 0.09b | 8.16 ± 0.16a | 10.75 ± 0.04hijkl | 47.42 ± 0.03 | 16.45 ± 0.03 | 42.76 ± 0.04 |
| L6 | 7.98 ± 0.10a | 7.86 ± 0.09a | 7.81 ± 0.12a | 8.07 ± 0.05c | 8.03 ± 0.03c | 8.01 ± 0.08c | 7.65 ± 0.04abdefghi | 41.39 ± 0.05 | 13.35 ± 0.03 | 45.57 ± 0.03 |
| L9 | 8.93 ± 0.05b | 8.70 ± 0.08b | 8.45 ± 0.13a | 8.84 ± 0.06b | 8.80 ± 0.02b | 8.37 ± 0.10a | 40.61 ± 0.02p | 72.94 ± 0.04 | 15.44 ± 0.04 | 61.68 ± 0.02 |
| L11 | 7.72 ± 0.06c | 7.57 ± 0.05b | 7.01 ± 0.03a | 7.66 ± 0.05b | 7.61 ± 0.05b | 7.26 ± 0.16a | 9.15 ± 0.02fghijk | 50.86 ± 0.02d | 18.23 ± 0.02 | 42.21 ± 0.18 |
| L14 | 7.93 ± 0.05c | 7.22 ± 0.10b | 6.94 ± 0.05a | 7.82 ± 0.05bc | 7.67 ± 0.03ab | 7.57 ± 0.09a | 12.19 ± 0.02ijklm | 65.07 ± 0.06f | 19.84 ± 0.02 | 61.13 ± 0.03 |
| L16 | 8.54 ± 0.04c | 8.25 ± 0.05b | 8.07 ± 0.05a | 8.46 ± 0.06bc | 8.40 ± 0.06b | 8.01 ± 0.03a | 16.10 ± 0.02mno | 68.22 ± 0.03 | 23.43 ± 0.2 | 72.11 ± 0.01 |
| L19 | 6.10 ± 0.07b | 5.76 ± 0.19a | 5.19 ± 0.14a | 6.05 ± 0.07b | 5.91 ± 0.02b | 5.32 ± 0.11a | 8.23 ± 0.03efghi | 54.09 ± 0.08 | 15.43 ± 0.02 | 56.21 ± 0.02 |
| L21 | 8.40 ± 0.04b | 8.18 ± 0.06a | 8.06 ± 0.04a | 8.34 ± 0.05b | 8.32 ± 0.06b | 7.76 ± 0.14a | 8.67 ± 0.02efghi | 72.05 ± 0.03 | 22.04 ± 0.02 | 68.62 ± 0.02 |
| L22 | 8.60 ± 0.04a | 8.52 ± 0.04a | 8.49 ± 0.07a | 8.79 ± 0.06b | 8.75 ± 0.07b | 8.71 ± 0.06b | 18.74 ± 0.02o | 71.05 ± 0.02g | 16.23 ± 0.02 | 66.21 ± 0.02 |
| L23 | 8.17 ± 0.05b | 8.06 ± 0.03b | 7.93 ± 0.05a | 8.07 ± 0.05b | 8.03 ± 0.04b | 7.48 ± 0.19a | 11.94 ± 0.02ijklm | 51.83 ± 0.02 | 14.16 ± 0.02 | 45.21 ± 0.02 |
| L24 | 9.20 ± 0.36b | 8.68 ± 0.19ab | 8.35 ± 0.31a | 9.11 ± 0.36ab | 9.08 ± 0.03ab | 8.58 ± 0.09a | 13.18 ± 0.02jklmn | 52.72 ± 0.03 | 16.14 ± 0.02 | 67.21 ± 0.02 |
| L26 | 7.89 ± 0.05b | 7.72 ± 0.05a | 7.57 ± 0.06a | 7.80 ± 0.01b | 7.76 ± 0.02b | 7.22 ± 0.10a | 9.53 ± 0.03ghijk | 51.23 ± 0.02e | 3.10 ± 0.01 | 11.02 ± 0.02 |
| L28 | 9.52 ± 0.05c | 9.21 ± 0.05b | 9.07 ± 0.03a | 9.44 ± 0.03b | 9.40 ± 0.03b | 8.79 ± 0.21a | 10.23 ± 0.02hijk | 48.04 ± 0.02 | 19.34 ± 0.03 | 39.21 ± 002 |
| L29 | 8.72 ± 0.04c | 8.58 ± 0.06b | 8.33 ± 0.06a | 8.66 ± 0.05b | 8.63 ± 0.05b | 8.34 ± 0.15a | 9.32 ± 0.23fghijk | 51.04 ± 0.02de | 28.53 ± 0.02 | 61.21 ± 0.02 |
| L31 | 7.82 ± 0.05b | 7.66 ± 0.05b | 7.25 ± 0.14a | 7.76 ± 0.08b | 7.70 ± 0.10b | 7.04 ± 0.11a | 11.53 ± 0.02ijklm | 71.11 ± 0.02g | 29.55 ± 0.02 | 67.21 ± 0.02 |
| L34 | 8.08 ± 0.05c | 7.89 ± 0.11b | 7.62 ± 0.05a | 7.99 ± 0.01b | 7.93 ± 0.02b | 7.58 ± 0.12a | 11.76 ± 0.03ijklm | 57.23 ± 0.02 | 18.32 ± 0.02 | 49.71 ± 0.01 |
| L36 | 8.07 ± 0.05c | 7.75 ± 0.12b | 6.81 ± 0.16a | 7.96 ± 0.10b | 7.92 ± 0.06b | 7.56 ± 0.12a | 11.84 ± 0.01ijklm | 51.35 ± 0.04e | 13.73 ± 0.02 | 68.12 ± 0.02 |
| L38 | 9.53 ± 0.07c | 8.71 ± 0.03b | 8.05 ± 0.04a | 9.46 ± 0.09b | 9.40 ± 0.10b | 8.36 ± 0.14a | 12.08 ± 0.03ijklm | 53.35 ± 0.03 | 18.06 ± 0.03 | 51.22 ± 0.02 |
| L40 | 7.87 ± 0.05b | 7.49 ± 0.18a | 7.21 ± 0.10a | 7.78 ± 0.07b | 7.73 ± 0.06b | 7.34 ± 0.11a | 17.05 ± 0.03no | 71.07 ± 0.03g | 13.47 ± 0.02 | 65.61 ± 0.01 |
| P1 | 8.09 ± 0.02c | 7.85 ± 0.04b | 6.84 ± 0.03a | 7.91 ± 0.11b | 7.84 ± 0.04b | 7.49 ± 0.12a | 15.07 ± 0.06lmno | 65.02 ± 0.03f | 12.02 ± 0.03 | 61.03 ± 0.03 |
| P2 | 9.21 ± 0.36c | 8.64 ± 0.08b | 7.89 ± 0.02a | 9.07 ± 0.36b | 9.32 ± 0.03b | 8.16 ± 0.01a | 18.02 ± 0.02o | 69.02 ± 0.03 | 21.02 ± 0.03 | 68.03 ± 0.03 |
Values represented as mean ± SD; for each row, different subscripts lowercase letters indicate significantly different at p < 0.05, as measured by 2-sided Tukey’s HSD between bile %, 0.5, 1, 2, control and between pH 2, pH 3 and pH 7
P1 and P2 (L. casei Shirota and L. rhamnosus) are probiotic reference strains
*Values represented as mean ± SD; for 3 h column, different subscripts lowercase letters indicate significantly different at p < 0.00001; for column 24 h, similar superscript letters are not significantly differ while remaining unsuperscript are significantly differ at p < 0.00001, as measured by 2-sided Tukey’s HSD between different isolates
Resistance to bile salt
Most of the isolates retained their viability with negligible reduction in viable counts after exposure to bile (< 1 log cycle) as shown in Table 3. However, no significant difference was observed in the loss of viability of the 6 Lactobacillus isolates (L6, L11, L22, L29, L23 and L36) at 0.5 and 1 % of bile concentration in comparison to the log CFU of control whereas negligible i.e. 0.5–1 log reduction in the viability was observed at 2 % bile concentration. Out of 20 tested isolates, 4 Lactobacillus isolates (L38, L19, L26 and L31) demonstrated approximately 3–4 log reduction in viable cell counts after their exposure to bile salts at 1 and 2 % bile concentrations as compared to the control (without bile salt). Amongst all the isolates, L23 and L22 possessed the highest viability at all the bile concentrations in comparison to the reference probiotic strains (P1 and P2) and thus these were considered to be the most bile tolerant isolates as no significant (p < 0.05) differences in the viability was found.
Lysozyme resistance
Viability (Data not shown) of lactobacilli in the presence of different concentration of lysozyme was checked. No such difference in the survival percentage was observed at 50 and 100 μg/ml of lysozyme concentration as compared to the reference strains P1 and P2 but there was a decline in viability (Log cfu/mL) at 150 μg/ml. Isolates L40 and L26 were found to be most lysozyme sensitive isolates.
Antibiotic susceptibility
Variations in the susceptibility of lactobacilli to different antibiotics were observed. All of the Lactobacillus isolates displayed resistant to vancomycin. Most of the isolates were found to be resistant to Co-trimoxazole and Co-trimazine. Susceptibility against 14 antibiotics was checked and variability in the susceptibility of Lactobacillus with the antibiotics was found as shown in Table 4.
Table 4.
Antibiotic susceptibility of Lactobacillus spp. from traditional foods and beverages
| Isolates | P | CD | COT | E | V | S | A | CB | CTX | C | CM | GEN | NX | OX |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| L1 | S | S | S | S | R | S | S | S | S | R | R | S | R | S |
| L5 | R | R | R | R | R | R | R | S | R | R | R | S | S | R |
| L6 | R | R | R | R | R | S | R | R | R | R | R | R | R | R |
| L9 | R | R | R | R | R | R | S | S | S | R | R | S | R | R |
| L11 | S | S | S | S | R | S | S | S | S | R | R | S | R | R |
| L14 | R | S | R | S | R | S | S | S | S | S | R | S | R | S |
| L16 | S | S | S | S | R | S | S | S | S | R | R | S | S | S |
| L19 | R | R | R | R | R | R | S | S | S | S | R | S | S | R |
| L21 | R | R | R | R | R | R | S | S | R | S | S | R | R | S |
| L22 | R | R | R | S | R | S | S | S | S | S | R | R | R | S |
| L23 | R | S | R | S | R | S | S | S | R | S | R | R | R | S |
| L24 | S | S | S | S | R | S | S | S | S | S | R | R | R | S |
| L26 | R | R | R | R | R | R | R | R | R | R | R | R | R | R |
| L28 | R | S | R | R | R | R | S | S | S | S | S | S | R | S |
| L29 | S | R | R | R | R | S | S | S | S | R | R | R | S | S |
| L31 | R | R | R | R | R | R | S | R | S | S | R | S | S | S |
| L34 | R | R | R | R | R | R | S | S | S | S | R | R | R | S |
| L36 | S | S | S | S | R | R | S | S | S | S | S | S | S | S |
| L38 | R | S | S | S | R | S | S | S | R | S | S | S | S | S |
| L40 | S | R | R | R | R | R | S | S | R | R | R | S | S | S |
| P1 | S | R | R | R | R | R | S | S | S | R | R | S | S | S |
| P2 | S | R | R | R | R | S | S | S | S | R | R | S | S | S |
(R-Resistant; S-Sensitive) [P-Penicillin G (10 Units); CD-Clindamycin (2 μg); COT-Co-trimoxazole (25 μg); E-Erythromycin (15 μg); VA-Vancomycin (30 μg); A-Ampicillin (10 μg); CB-Carbenicillin (100 μg); CTX-Cefotaxime (30 μg); C Chloramphenicol (30 μg); CM-Co-trimazine (25 μg); GEN-Gentamicin (10 μg); NX-Norfloxacin (10 μg) and OX-Oxacilin (5 μg)]
Antimicrobial activity
The antimicrobial activities of Lactobacillus isolates were screened using bit disk method against all the pathogens (Data not shown). All isolates showed good zone of inhibition in bit disc assay against the various pathogens were further supported by agar well diffusion assay method (Table 5).
Table 5.
Inhibition of pathogens (mm) by Lactobacillus spp. using well diffusion method
| Isolates | S. aureus | B. subtilis | P. aeruginosa | S. dysenteriae | E. coli |
|---|---|---|---|---|---|
| L1 | 18.0 ± 0.2 | 20.5 ± 0.1 | 9.1 ± 0.15 | 17.2 ± 0.1 | 10.0 ± 0.1 |
| L5 | 16.1 ± 0.1 | 18.3 ± 0.2 | - | 7.2 ± 0.25 | 9.02 ± 0.2 |
| L6 | 13.2 ± 0.2 | 15.1 ± 0.1 | 11.0 ± 0.5 | 8.3 ± 0.15 | 8.03 ± 0.05 |
| L9 | 9.2 ± 0.15 | 15.5 ± 0.15 | 12.5 ± 0.01 | 13.2 ± 0.15 | 12.2 ± 0.1 |
| L11 | 16.3 ± 0.15 | 17.2 ± 0.2 | 9.2 ± 0.05 | 10.1 ± 0.5 | 14.0 ± 0.05 |
| L14 | 17.2 ± 0.15 | 13.0 ± 0.3 | 8.4 ± 0.05 | 12.4 ± 0.1 | 8.5 ± 0.2 |
| L16 | 13.1 ± 0.15 | 16.1 ± 0.1 | 10.3 ± 0.1 | 13.1 ± 0.1 | 10.1 ± 0.1 |
| L19 | 17.5 ± 0.15 | 15.5 ± 0.05 | - | 12.4 ± 0.25 | 12.2 ± 0.15 |
| L21 | 15.2 ± 0.25 | 16.3 ± 0.1 | 10.2 ± 0.05 | 11.0 ± 0.2 | 9.2 ± 0.15 |
| L22 | 14.1 ± 0.2 | 15.1 ± 0.25 | 11.1 ± 0.1 | 13.0 ± 0.15 | 9.5 ± 0.5 |
| L23 | 13.4 ± 0.15 | 18.5 ± 0.1 | 9.5 ± 0.2 | 14.0 ± 0.15 | 9.3 ± 0.01 |
| L24 | 16.5 ± 0.1 | 15.0 ± 0.21 | 10.1 ± 0.1 | 11.0 ± 0.1 | - |
| L26 | 13.5 ± 0.05 | 16.1 ± 0.1 | - | 9.2 ± 0.1 | 14.5 ± 0.25 |
| L28 | 11.3 ± 0.1 | 15.2 ± 0.05 | - | 7.2 ± 0.1 | 8.0 ± 0.05 |
| L29 | 15.0 ± 0.05 | 16.5 ± 0.1 | - | 8.1 ± 0.15 | 9.3 ± 0.1 |
| L31 | - | 19.4 ± 0.25 | 10.3 ± 0.1 | 11.4 ± 0.5 | 16.5 ± 0.1 |
| L34 | - | 24.3 ± 0.1 | - | - | 12.3 ± 0.15 |
| L36 | 8.5 ± 0.15 | 11.2 ± 0.15 | 9.2 ± 0.25 | 12.3 ± 0.5 | 9.3 ± 0.05 |
| L38 | 11.1 ± 0.15 | 14.5 ± 0.1 | - | - | 11.4 ± 0.1 |
| L40 | 14.0 ± 0.15 | 20.2 ± 0.2 | 13.7 ± 0.2 | 16.4 ± 0.1 | 12.5 ± 0.15 |
| P1 | - | 11.1 ± 0.15 | - | 13.0 ± 0.15 | 10.5 ± 0.1 |
| P2 | 13.5 ± 0.1 | 12.0 ± 0.15 | 9.6 ± 0.1 | 9.7 ± 0.1 | - |
The results are expressed as the mean of triplicate samples from three independent experiments ±SD
All 20 isolates indicated the antimicrobial activity against E. coli (with an inhibition zone of 8.0–16.5 mm in diameter), P. aeruginosa (8.4–13.7 mm), S. aureus (8.5–18.0 mm), B. subtilis (9.4–24.3 mm), S. dysenteriae (7.2–16.4 mm) and Y. enterocolitica (9.5 mm). However, nine isolates; L3, L4, L17, L18, L20, L32, L33, L37 and L39 did not indicate the inhibitory activity towards S. aureus, B. subtilis and P. aeruginosa. P1 and P2, the reference strains recognized as probiotic, showed inhibitory properties against B. subtilis (11.1 mm), S. dysenteriae (13.0 mm), E. coli (10.5 mm) and S. aureus (13.5 mm), B. subtilis (12.0 mm), S. dysenteriae (9.7 mm) and P. aeruginosa (9.6 mm), respectively. None of the isolates except L28 showed any inhibitory activity against Y. enterocolitica.
Determination of cell surface characteristics
Autoaggregation and co-aggregation assay
In general, the probiotic strains showed higher autoaggregation abilities than pathogenic strains. All isolates tested showed higher percentages of aggregation after 24 h of incubation in comparison to 0 h and 3 h.
A high range of variation was observed in autoaggregation capacity among the tested isolates at 3 h ranging from 7.65 % (for L6) to 40.61 % (L9); while after 24 h, the isolates showed a very high significant variability (p < 0.00001) ranging from L6 (41.39 %) to L9 (72.94 %) as depicted in Table 3. The probiotic control strains (P1 and P2) showed moderate adhesion capacity ranging from 15.07 to 18.02 for 3 h and comparatively higher at 24 h i.e. 65.02 to 69.02 %, respectively. L9 after 3 h and L21, L16, L22, L31 and L40 after 24 h showed significantly (p < 0.00001) higher autoaggregation than the P1 and P2. The isolates with the best autoaggregation abilities after 24 h of incubation at 30 °C, were L9, L21, L22, L31 and L40 (>70.0 %) while the least autoaggregative isolate was L5 and L6 (<10 %) under the same conditions.
A broad range of variation in co-aggregation with L. monocytogenes, depending on the isolate and the incubation time was noticed. All tested isolates exhibited some coaggregation properties, 5 isolates (L16, L21, L28, L29 and L31) exhibited highest coaggregative interactions with the L. monocytogenes ranging from 23.43 to 29.55 % at 3 h of incubation and the degree of coaggregation gradually enhanced by time. Twelve isolates (L1, L9, L14, L16, L19, L21, L22, L24, L29, L31, L36 and L40) exhibited highest coaggregation activity i.e. > 60 % at 24 h. L16 was recorded to possess proportionally high percentage of co-aggregation i.e. 72.11 % having high potential capability to adhere to epithelial cells and mucosal surfaces. The probiotic control strains (P1 and P2) showed moderate adhesion capacity ranging from 12.02 to 21.02 for 3 h and it increased to 61.03 to 68.03 %, respectively in 24 h. Most of the Lactobacillus isolates exhibited significantly (p < 0.00001) higher coaggregation over the P1 and P2 (Table 3).
Bacterial adhesion to hydrocarbons (BATH)
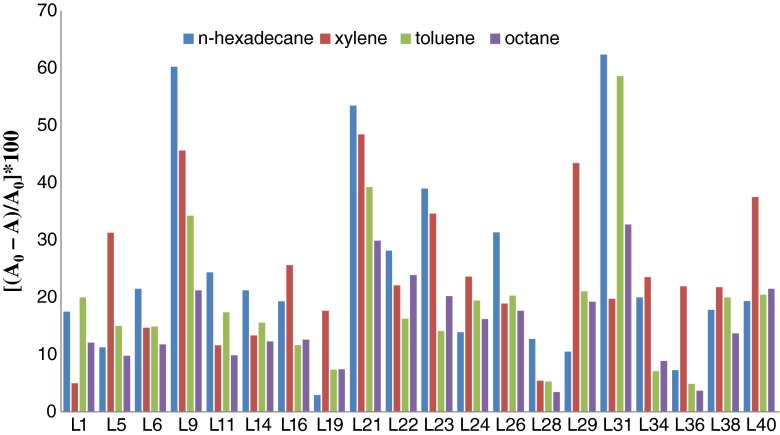
According to the Statistical analysis (p < 0.05), the isolates L9, L21, L31 and L40 showed greater affinity towards the solvents tested in the present study. Some Lactobacillus isolates showed high degree of relative hydrophobicity (>45 %), as judged by bacterial adherence to hydrocarbons, indicating their hydrophobic nature. Among all the Lactobacillus isolates tested, L31 showed maximum affinity towards n-hexadecane.
With different hydrocarbons, maximum adhesion was with hexadecane (62.33 %), followed by toluene (58.65 %), xylene (48.41 %) and octane (32.72 %) as shown in Fig. 2. In comparison to the standard strains (P1 and P2) L9, L21 and L31 had significantly maximum hydrophobicity for n-hexadecane, followed by L9, L21, L29 and L40 for xylene, L31 for toluene and L21 and L31 for octane. The isolates L9 (60.23 %), L21 (53.45 %) and L31 (62.35 %) showed the highest hydrophobicity index (HI) in n-hexadecane in comparison to P1 (49 %) and P2 (52.10 %).
Fig 2.
Hydrophobic index (HI %) of the Lactobacillus spp. (mean values, n = 4) to hexadecane, xylene, octane and toluene
Except four isolates, all the Lactobacillus isolates produce EPS on skimmed milk-ruthedium red plates. Presence of ropy white coloured mucous producing lactobacilli are the EPS producers. None of the examined isolates exhibited α and β haemolytic activity. All of the lactobacilli (20 isolates) were ɤ-haemolytic (i.e. no haemolysis). All the tested Lactobacillus isolates were negative for BSH activity as none of them showed precipitation (granulation) zones on agar plates.
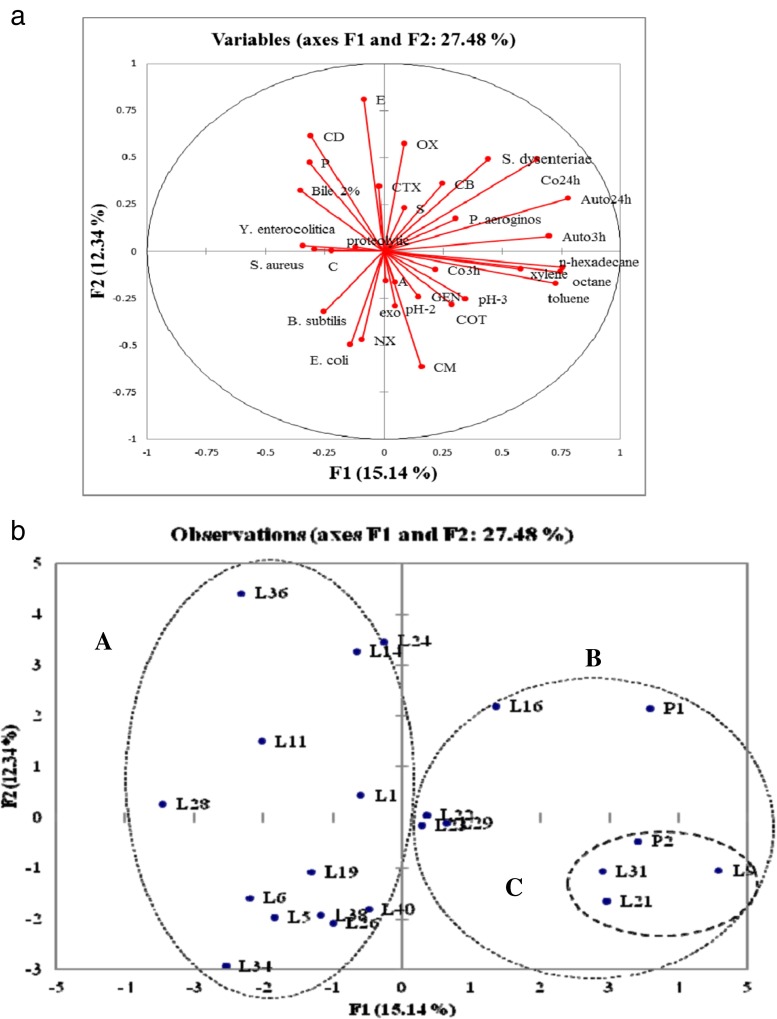
Principal component analysis (PCA)
The final step of the probiotic characterization was the selection of the most promising probiotic isolates through the Principal Component Analysis. Analysis of PCA revealed that first four principal components (PCs) explaining 27.5 % of total variation, while PC1 and PC2 accounting for 15.1 % and 12.3 % respectively in Figs. 3a and b. Figure 3a represents the homogeneous distribution of variables on the plane of principal components. Three main groups (A, B and C) could be pointed out based on the position of the variables in the factorial space of PCA (Fig. 3b), C being the best one for the probiotic traits. Group C (L9, L21 and L31) were quite promising as they possessed the probiotic traits at their highest levels showing same properties as a probiotic strain P2 and possessed the probiotic attributes superior than P1(Fig. 3b). Moreover, the isolates L16, L22, L23 and L29 were also interesting, as they showed the superior trend over P1 (probiotic reference strain). Thus the isolates L9 (L. brevis PLA2), L21 (L. paracasei PLA8) and L31 (L. brevis PLA16) were selected as representative of these three promising groups. This gives an clear idea that isolate (L31, L21 and L9) has more similar or efficient probiotic activity as that of reference probiotic strains i.e. P1 and P2.
Fig 3.
a Variable projection of the PC1 and PC2 analysed by PCA b Projection of the 22 isolates in the space of PC1 and PC2
In-vitro adherence to the human intestinal cell lines
The three finally selected isolates (L. brevis PLA2, L. paracasei PLA8 and L. brevis PLA16) on the basis of probiotic characterization and PCA were further examined for their ability to adhere to HT-29 cells. All the tested isolates showed good percentage of adherence (15.26–24.36 %) as compared to the standard strain (L. casei Shirota) after 4 h suggesting that all these isolates demonstrate higher adherence properties than L. casei Shirota having 7.81 % percentage adherence.
All the 3 isolates adhered to HT-29 cell lines at different adhesion capacity. On comparative evaluation with L. casei Shirota (7.81 ± 0.25), L. brevis PLA2 (24.36 ± 0.5), L. paracasei PLA8 (21.21 ± 0.25) and L. brevis PLA16 (15.26 ± 0.5) were the most adhesive isolates based on their respective percent adhesion. The observations with regard to per cent adhesion obtained using HT-29 can be further corroborated by the adhesion score i.e. 499.1 ± 19, 475.2 ± 28 and 361.6 ± 22 in L. brevis PLA2, L. paracasei PLA8 and L. brevis PLA16, respectively as compared to the standard probiotic strain L. casei Shirota (162.1 ± 16).
Discussion
Potential positive impact of probiotic lactic acid bacteria isolated from fermented foods on human health has been pointed out by various scientific findings (Morelli, 2007). Lactobacillus spp. are found to be more tolerant to acid environment than the other genera of lactic acid bacteria (cocci). Hence, this property makes Lactobacillus spp. abundant in the final phases of many food fermentations (Devirgiliis et al. 2009). This might be the main reason that most of the Lactobacillus isolates from traditional fermented foods of North-western Himalayas were of Lactobacillus spp.
Resistance to gastric conditions is one of the prerequisite in vitro tests for the evaluation of the probiotic potential of Lactobacillus strains. The in vitro survival test revealed that Lactobacillus isolates (L1, L19, L21, L26 and L40) were most resistant to pH 2 after 3 h of exposure. These results were similar to those of the previous studies, where Lactobacillus strains were viable even after being exposed to pH values of 2.5–4.0, but showed reduced viability at lower pH values (Wang et al. 2010). The variation in acid tolerance of lactic acid bacteria has been linked to the difference in induction of H+-ATPase activity resulting in the removal of protons (H+), alkanization of the external environment, changes in the composition of the cell envelope (Cotter and Hill, 2003).
Bile plays an important role in specific and nonspecific defense mechanisms of the gut. Human bile concentration varies from 0.5 to 2 % depending on the type of food being consumed. In the present study, isolates (L22 and L23) exhibited best bile tolerance at 2.0 % bile concentration which are in agreement with previous studies (Tulumoglu et al. 2013; Wang et al. 2010). The adaptation to bile salts is related to changes in carbohydrate fermentation, glycosidase activity, exopolysaccharide production, the composition of membrane proteins and fatty acids and increased adhesion to human mucus (Quezada et al. 2013).
Sensitivity of probiotics to commonly prescribed antibiotics at the low concentrations is desirable. However, resistance of the probiotic strains to some antibiotics could be used for both preventive and therapeutic purposes in controlling intestinal infections. Lactobacillus isolates were found to be resistant to vancomycin (intrinsic, chromosomally encoded and non-transmissible) which was due to the presence of D-Ala-D-lactate in their peptidoglycan instead of the normal dipeptide D-Ala-D-Ala, which is the target of the antibiotic (Coppola et al. 2005). The antibiotic susceptibility profiles of Lactobacillus isolates have been documented by many researchers (Zoumpopoulou et al. 2008; Ammor et al. 2007). However, our study indicated that there are lesser chances of occurrence of such transferable resistance gene in the Lactobacillus isolates from fermented foods.
Use of Lactobacillus as biopreservatives has been observed in several studies, owing to their antagonistic activities toward various foodborne pathogens (Vizoso-Pinto et al. 2006). The antimicrobial activity of lactic acid bacteria is due to production of acids, diminished pH levels, competition for substrates, the production of bacteriocins and bacteriocin-like substances. The Lactobacillus isolates in this study showed antagonistic potential toward almost all of the pathogenic bacteria; however, they did not significantly inhibit the growth of Y. enterocolitica. Simsek et al. (2006) and Iyer et al. (2013) conducted a similar study and reported the antibacterial activity of lactobacilli isolated from fermented products against various Gram positive and negative pathogens. Lactobacillus isolates (L1, L11, L14 and L19) having antimicrobial activity may be effectively used in various food products to prevent pathogenic bacterial contamination that may occur during the manufacturing processes and transportation.
Lactobacilli are able to interfere with pathogens by different mechanisms: the competitive dispensation of pathogens from the cell surface, co-aggregation with certain pathogenic bacteria, adherence to epithelial cells and biofilm formation based on autoaggregation and surface hydrophobicity (Dunne et al., 2001). Analyses of surface properties, showed that L21, L22, L31 and L40 exhibited the highest auto-aggregation and co-aggregation values (>65 %). It was observed that the correlations between autoaggregation and coaggregation of lactobacilli isolates were significant. The proteins present in the culture supernatant and lipoproteins and polysaccharides located on the cell surfaces of Lactobacillus are involved in cell aggregation (Aslim et al. 2007). The large differences in the cell surface hydrophobicity of Lactobacillus isolates in present study could be due to variation in the level of expression of cell surface proteins among strains of a species as well as due to environmental conditions which could affect the expression of surface proteins (Ramiah et al. 2007).
Lactobacillus strains often produce polymeric substances such as exopolysaccharides (EPS) which enhance the colonization of probiotic bacteria in gastrointestinal tract and helps in modifying the rheological properties and texture of products (Kanmani et al. 2013). None of the isolates from fermented foods and beverages in the present investigation displayed bile salt hydrolase activity which was similar to the findings reported by Botes et al. (2008) and Quezada et al. (2013). Most often, bile salt hydrolase activity has been detected in strains indigenous to the gastrointestinal tract rich in conjugated and unconjugated bile acids (Martoni et al. 2008).
Studies of bacterial adhesion in-vivo is generally difficult, in vitro adhesive models can be used for this assessment (Tuomola and Salminen, 1998). The colonic cell line HT-29 displays characteristics of enterocyte differentiation and has been used for in vitro adhesion assays (Quezada et al. 2013). L. brevis PLA2 and L. paracasei PLA8 showed best ability to adhere to intestinal cells. The adhesion mechanisms are not fully understood; however bacterial cell-surface associated proteins with mucous and intestinal cell binding properties have been identified and characterized in various adhesive probiotic strains (Sanchez et al. 2008).
Conclusion
The present study demonstrated the capacity of two Lactobacillus isolates as a promising candidate for probiotic applications. The present work provides a preliminary selection of these two potential isolates i.e. L. brevis PLA2 and L. paracasei PLA8 which could be used as probiotic, due to their high tolerance to various conditions at GIT environment in comparison to the commercial probiotic strain. In addition, they were able to adhere to human colonic cell line (HT-29). This also indicates that the selected promising probiotic Lactobacillus isolates can be further commercialized for potential human health benefits. Therefore, these isolates could lead to the development of new traditional probiotic food products which may indeed convey a favourable impact in rural economy, especially the tribal and less developed regions.
Electronic Supplementary Materials
(DOC 47 kb)
Acknowledgments
The authors would like to acknowledge the University Grants Commission, New Delhi for providing financial assistance. Authors also acknowledge Prof. S. S Kanwar, Department of Biotechnology, HPU, Summerhill, Shimla, India for providing all necessary facilities for cell line adherence analysis.
Footnotes
Research highlights
• Lactobacillus spp. from fermented foods and beverage of North-western Himalayas has been characterized for its probiotic attributes.
• Principal component analysis (PCA) was used to select the most promising isolate.
• Isolates PLA2, 8 and 16 were the most promising probiotic candidates.
• L. brevis PLA2 and L. paracasei PLA8 showed best in-vitro cell adhesion property with HT-29 cell lines.
References
- Ammor MS, Belen Florez A, Mayo B. Antibiotic resistance in non-enterococcal lactic acid bacteria and bifidobacteria. Food Microbiol. 2007;24:559–570. doi: 10.1016/j.fm.2006.11.001. [DOI] [PubMed] [Google Scholar]
- Aslim B, Onal D, Beyatli Y. Factors influencing autoaggregation and aggregation of Lactobacillus delbrueckii subsp. bulgaricus isolated from handmade yogurt. J Food Prot. 2007;70:223–227. doi: 10.4315/0362-028x-70.1.223. [DOI] [PubMed] [Google Scholar]
- Bernardeau M, Vernoux JP, Henri-Dubernet S, Gueguen M. Safety assessment of dairy microorganisms: the Lactobacillus genus. Int J Food Microbiol. 2008;126:278–285. doi: 10.1016/j.ijfoodmicro.2007.08.015. [DOI] [PubMed] [Google Scholar]
- Botes M, Loos B, van Reenen CA, Dicks LMT. Adhesion of the probiotic strains Enterococcus mundtii ST4SA and Lactobacillus plantarum 423 to Caco-2 cells under conditions simulating the intestinal tract, and in the presence of antibiotics and anti-inflammatory medicaments. Arch Microbiol. 2008;190:573–584. doi: 10.1007/s00203-008-0408-0. [DOI] [PubMed] [Google Scholar]
- Coppola R, Succi M, Tremonte P, Reale A, et al. Antibiotic susceptibility of L. rhamnosus strains isolated from Parmigiano Reggiano cheese. Lait. 2005;85:193–204. doi: 10.1051/lait:2005007. [DOI] [PubMed] [Google Scholar]
- Cotter PD, Hill C. Surviving the acid test: responses of gram positive bacteria to low pH. Microbiol Mol Biol Rev. 2003;67:429–453. doi: 10.1128/MMBR.67.3.429-453.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devirgiliis C, Coppola D, Barile S, Colonna B, Perozzi G. Characterization of the Tn916 conjugative transposon in a food-borne strain of Lactobacillus paracasei. Appl Environ Microbiol. 2009;75:3866–3871. doi: 10.1128/AEM.00589-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duary RK, Rajput YS, Batish VK, Grover S. Assessing the adhesion of putative indigenous probiotic lactobacilli to human colonic epithelial cells. Indian J Med Res. 2011;134:664–671. doi: 10.4103/0971-5916.90992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunne C, O‟ Mahony L, Murphy L, Thornton G, Morrissey D et al (2001) In vitro selection criteria for probiotic bacteria of human origin: correlation with in vivo findings. Am J Clin Nutr 73:386–392. [DOI] [PubMed]
- FAO⁄WHO . Evaluation of health and nutritional properties of powder milk and live lactic acid bacteria. FAO: Food and Agriculture Organization of the United Nations and World Health Organization expert consultation report. Rome; 2001. [Google Scholar]
- Ghosh D, Chattopadhyay P. Application of principal component analysis (PCA) as a sensory assessment tool for fermented food products. J Food Sci Technol. 2011;49:328–334. doi: 10.1007/s13197-011-0280-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ha CG, Cho JK, Lee CH, Chai YG, Ha YA, Shin SH. Cholesterol lowering effect of Lactobacillus plantarum isolated from human faeces. J Microbiol Biotechnol. 2006;16:1201–1209. [Google Scholar]
- Iyer BK, Singhal RK, Ananthanarayan L. Characterization and in vitro probiotic evaluation of lactic acid bacteria isolated from idli batter. J Food Sci Technol. 2013;50:1114–1121. doi: 10.1007/s13197-011-0445-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Handley PS, Harty DWS, Wyatt JE, Brown CR, Doran JP, Gibbs ACC. A comparison of the adhesion, co-aggregation and cell-surface hydrophobicity properties of fibrillar and fimbriate strains of Streptococcus salivarius. J Gen Microbiol. 1987;133:3207–3217. doi: 10.1099/00221287-133-11-3207. [DOI] [PubMed] [Google Scholar]
- Jacobsen CN, Rosenfeldt Nielsen V, Hayford AE, Moller PL, Michaelsen KF, et al. Screening of probiotic activities of forty-seven strains of Lactobacillus spp. by in vitro techniques and evaluation of the colonization ability of five selected strains in humans. Appl Environ Microbiol. 1999;11:4949–4956. doi: 10.1128/aem.65.11.4949-4956.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanmani P, Suganya K, Yuvaraj N, Pattukumar V, Paari KA, Arul V. Synthesis and functional characterization of antibiofilm exopolysaccharide produced by Enterococcus faecium MC13 isolated from the gut of fish. Appl Biochem Biotechnol. 2013;169:1001–1015. doi: 10.1007/s12010-012-0074-1. [DOI] [PubMed] [Google Scholar]
- Maragkoudakis PA, Zoumpopoulou G, Miaris C, Kalantzopoulos G, Pot B, Tsakalidou E. Probiotic potential of Lactobacillus strains isolated from dairy products. Int Dairy J. 2006;16:189–199. doi: 10.1016/j.idairyj.2005.02.009. [DOI] [Google Scholar]
- Maragkoudakis PA, Mountzouris KC, Psyrras D, Cremonese S, Fischer J, Cantor MD, Tsakalidou E (2009) Functional properties of novel protective lactic acid bacteria and application in raw chicken meat against Listeria monocytogenes and Salmonella enteritidis. Int J Food Microbiol 130:219–26 [DOI] [PubMed]
- Martoni CBJ, Urbanska AM, Prakash S. Microencapsulated bile salt hydrolase producing Lactobacillus reuteri for oral targeted delivery in the gastrointestinal tract. Appl Microbiol Biotechnol. 2008;81:225–233. doi: 10.1007/s00253-008-1642-8. [DOI] [PubMed] [Google Scholar]
- Merk K, Borelli C, Korting HC. Lactobacilli bacteria host interactions with special regard to the urogenital tract. Int J Med Microbiol. 2005;295:9–18. doi: 10.1016/j.ijmm.2004.11.006. [DOI] [PubMed] [Google Scholar]
- Messaoudi S, Madib A, Prévost H, Feuilloley M, Manaic M, et al. In vitro evaluation of the probiotic potential of Lactobacillus salivarius SMXD51. Anaerobe. 2012;18:584–585. doi: 10.1016/j.anaerobe.2012.10.004. [DOI] [PubMed] [Google Scholar]
- Mora D, Fortina MG, Parini C, Ricci G, Gatti M, et al. Genetic diversity and technological properties of Streptococcus thermophilus strains isolated from dairy products. J Appl Microbiol. 2002;93:278–287. doi: 10.1046/j.1365-2672.2002.01696.x. [DOI] [PubMed] [Google Scholar]
- Morelli L. In vitro assessment of probiotic bacteria: from survival to functionality. Int Dairy J. 2007;17:1278–1283. doi: 10.1016/j.idairyj.2007.01.015. [DOI] [Google Scholar]
- Perricone M, Bevilacqua A, Corbo MR, Sinigaglia M. Technological characterization and probiotic traits of yeasts isolated from Altamura sourdough to select promising microorganisms as functional starter cultures for cereal-based products. Food Microbiol. 2014;38:26–35. doi: 10.1016/j.fm.2013.08.006. [DOI] [PubMed] [Google Scholar]
- Quezada SM, Chenoll E, Vieites JM, et al. Isolation, identification and characterisation of three novel probiotic strains (Lactobacillus paracasei CNCM I-4034, Bifidobacterium breve CNCM I-4035 and Lactobacillus rhamnosus CNCM I-4036) from the faeces of exclusively breast-fed infants. Bri J Nutr. 2013;109:51–62. doi: 10.1017/S0007114512005211. [DOI] [PubMed] [Google Scholar]
- Ramiah K, van Reenena CA, Dicks LM. Expression of the mucus adhesion genes Mub and MapA, adhesion-like factor EF-Tu and bacteriocin gene plaA of Lactobacillus plantarum 423, monitored with real-time PCR. Int J Food Microbiol. 2007;116:405–409. doi: 10.1016/j.ijfoodmicro.2007.02.011. [DOI] [PubMed] [Google Scholar]
- Ramosa CL, Thorsen L, Schwana RF, Jespersen L. Strain-specific probiotics properties of Lactobacillus fermentum, Lactobacillus plantarum and Lactobacillus brevis isolates from Brazilian food products. Food Microbiol. 2013;36:22–29. doi: 10.1016/j.fm.2013.03.010. [DOI] [PubMed] [Google Scholar]
- Reniero R, Cocconcelli P, Bottazzi V, Morelli L. High frequency of conjugation in Lactobacillus mediated by an aggregation-promoting factor. J Gen Microbiol. 1992;138:763–768. doi: 10.1099/00221287-138-4-763. [DOI] [Google Scholar]
- Rosenberg M, Gutnick D, Rosenberg E. Adherence of bacteria to hydrocarbons: a simple method for measuring cell hydrophobicity. FEMS Microbiol Lett. 1980;9:29–33. doi: 10.1111/j.1574-6968.1980.tb05599.x. [DOI] [Google Scholar]
- Sanchez B, Bressollier P, Urdaci MC. Exported proteins in probiotic bacteria: adhesion to intestinal surfaces, host immunomodulation and molecular crosstalking with the host. FEMS Immunol Med Microbiol. 2008;54:1–17. doi: 10.1111/j.1574-695X.2008.00454.x. [DOI] [PubMed] [Google Scholar]
- Schillinger U, Lucke FK. Antibacterial activity of Lactobacillus sake isolated from meat. Appl Environ Microbiol. 1989;55:1901–1906. doi: 10.1128/aem.55.8.1901-1906.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simsek O, Hilmicon A, Tulumoglu S. Isolating lactic starter cultures with antimicrobial activity for sourdough processes. Food Control. 2006;17:263–270. doi: 10.1016/j.foodcont.2004.10.011. [DOI] [Google Scholar]
- Sonar NR, Halam PM. Phenotypic identification and technological attributes of native lactic acid bacteria present in fermented bamboo shoot products from North-East India. J Food Sci Technol. 2014;51:4143–4148. doi: 10.1007/s13197-014-1456-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taked S, Yamasaki K, Takeshita M, Kikuchi Y, Tsend-Ayush C, et al. The investigation of probiotic potential of lactic acid bacteria isolated from traditional Mongolian dairy products. Anim Sci J. 2011;82:571–579. doi: 10.1111/j.1740-0929.2011.00874.x. [DOI] [PubMed] [Google Scholar]
- Tulumoglu S, Yuksekdag ZN, Beyatli Y, Simsek O, Cinar B, Yaşar E. Probiotic properties of lactobacilli species isolated from children's feces. Anaerobe. 2013;24:36–42. doi: 10.1016/j.anaerobe.2013.09.006. [DOI] [PubMed] [Google Scholar]
- Tuomola EM, Salminen SJ. Adhesion of some probiotic and dairy Lactobacillus strains to Caco-2 cell cultures. Int J Food Microbiol. 1998;41:45–51. doi: 10.1016/S0168-1605(98)00033-6. [DOI] [PubMed] [Google Scholar]
- Turchi B, Mancini S, Fratini F, Pedonese F, Nuvoloni R, et al. Preliminary evaluation of probiotic potential of Lactobacillus plantarum strains isolated from Italian food products. W J Microbiol Biotechnol. 2013;29:1913–1922. doi: 10.1007/s11274-013-1356-7. [DOI] [PubMed] [Google Scholar]
- Vizoso-Pinto MG, Franz CM, Schillinger U, Holzapfel WH. Lactobacillus spp. with in vitro probiotic properties from human feces and traditional fermented products. Int J Food Microbiol. 2006;109:205–214. doi: 10.1016/j.ijfoodmicro.2006.01.029. [DOI] [PubMed] [Google Scholar]
- Wang CY, Lin PR, Ng CC, Shyu YT. Probiotic properties of Lactobacillus strains isolated from the feces of breast-fed infants and Taiwanese pickled cabbage. Anaerobe. 2010;16:578–585. doi: 10.1016/j.anaerobe.2010.10.003. [DOI] [PubMed] [Google Scholar]
- Zoumpopoulou G, Foligne B, Christodoulou K, Grangette C, Pot B, Tsakalidou E. Lactobacillus fermentum ACA-DC 179 displays probiotic potential in vitro and protects against trinitrobenzene sulfonic acid (TNBS)-induced colitis and salmonella infection in murine models. Int J Food Microbiol. 2008;121:18–26. doi: 10.1016/j.ijfoodmicro.2007.10.013. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOC 47 kb)