Abstract
Apolipoprotein (apo) E was initially described as a lipid transport protein and major ligand for low density lipoprotein (LDL) receptors with a role in cholesterol metabolism and cardiovascular disease. It has since emerged as a major risk factor (causative gene) for Alzheimer’s disease and other neurodegenerative disorders. Detailed understanding of the structural features of the three isoforms (apoE2, apoE3, and apoE4), which differ by only a single amino acid interchange, has elucidated their unique functions. ApoE2 and apoE4 increase the risk for heart disease: apoE2 increases atherogenic lipoprotein levels (it binds poorly to LDL receptors), and apoE4 increases LDL levels (it binds preferentially to triglyceride-rich, very low density lipoproteins, leading to downregulation of LDL receptors). ApoE4 also increases the risk for neurodegenerative diseases, decreases their age of onset, or alters their progression. ApoE4 likely causes neurodegeneration secondary to its abnormal structure, caused by an interaction between its carboxyl- and amino-terminal domains, called domain interaction. When neurons are stressed or injured, they synthesize apoE to redistribute cholesterol for neuronal repair or remodeling. However, because of its altered structure, neuronal apoE4 undergoes neuron-specific proteolysis, generating neurotoxic fragments (12–29 kDa) that escape the secretory pathway and cause mitochondrial dysfunction and cytoskeletal alterations, including tau phosphorylation. ApoE4-associated pathology can be prevented by small-molecule structure correctors that block domain interaction by converting apoE4 to a molecule that resembles apoE3 both structurally and functionally. Structure correctors are a potential therapeutic approach to reduce apoE4 pathology in both cardiovascular and neurological disorders.
Keywords: Alzheimer’s disease, ApoE, Atherosclerosis, Cholesterol, Small-molecule structure correctors
Early characterization of plasma lipoproteins highlighted the importance of their components, referred to as apolipoproteins, in controlling lipoprotein metabolism and cholesterol homeostasis [1]. Apolipoprotein (apo) E was late to be recognized as a critical protein constituent of lipoproteins [2]. First described in the early 1970s as a minor apolipoprotein in very low density lipoproteins (VLDL), apoE was subsequently identified as a major apolipoprotein in cholesterol-rich VLDL (β-VLDL) in cholesterol-fed animals [2–8] and found to be enriched in a subclass of high density lipoproteins (HDL; HDL1, HDLc) [8, 9]. Originally referred to as the arginine-rich apoprotein [2], the protein became known as apoE in 1982 [10].
ApoE has three common alleles encoded by the apoE gene on chromosome 19 [2]. These alleles occur at different frequencies in humans (ε2, 5–10 %; ε3, 65–70 %; and ε4, 15–20 %) and give rise to three homozygous (apoE2/2, apoE3/3, and apoE4/4) and three heterozygous (apoE3/2, apoE4/2, and apoE4/3) phenotypes. The structural basis for the three isoforms occurs through amino acid interchanges (single base changes in the apoE gene) at residues 112 and 158: apoE2 has cysteines at both sites, apoE4 has arginines, and apoE3 has cysteine (Cys)-112 and arginine (Arg)-158 [2, 11, 12]. All other animals, including the great apes, have a single isoform that has arginines at the residues equivalent to 112 and 158 [12, 13].
Plasma apoE is synthesized primarily by liver hepatocytes, which account for ~75 % of the body’s apoE production. In normolipidemic subjects, the plasma concentration of apoE is approximately 4–8 mg/dl. The second most common organ synthesizing apoE is the brain, where it is produced primarily by astrocytes, but also by oligodendrocytes, microglia, and neurons, especially injured or stressed neurons [see refs. 14–16 for more discussion]. In the brain, apoE is synthesized in situ and does not cross the blood brain barrier from the peripheral circulation [17]. Various cells throughout the body, including macrophages, also synthesize apoE [2, 11].
Structure and function of apoE isoforms
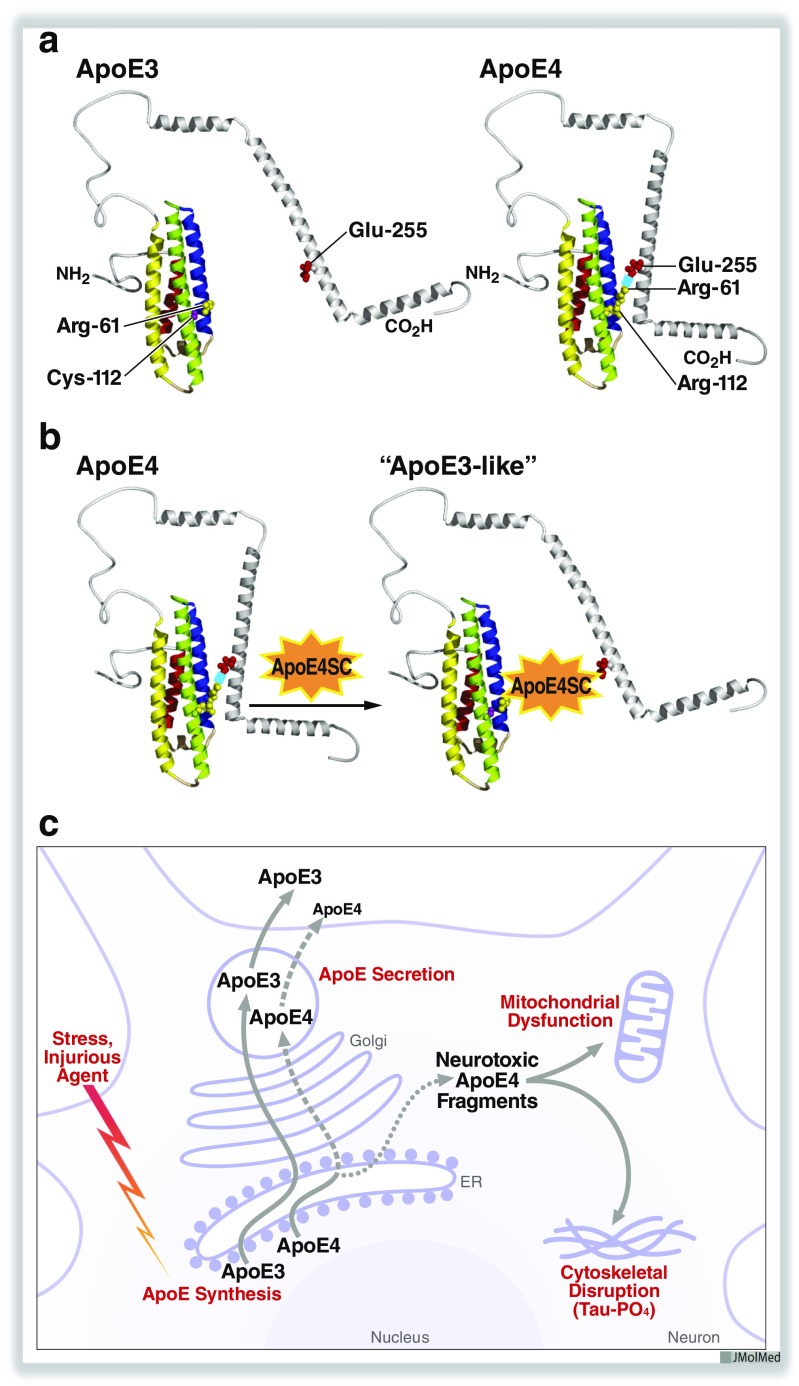
ApoE, a 34-kDa protein of 299 amino acids with a single glycosylation site at threonine-194 [18], has two structural domains separated by a hinge region. The amino-terminal domain (amino acids 1–191) contains the low density lipoprotein (LDL) receptor binding region (amino acids 136–150) [11, 19, 20]. The carboxyl-terminal domain (amino acids ~225–299) contains the lipid binding region (amino acids ~240–260) [11, 13]. The tertiary structure of the amino-terminal domains of apoE4, apoE3, and apoE2, solved by x-ray crystallography, consists of four helices arranged in anti-parallel fashion [13]. The carboxyl-terminal domain has amphipathic α-helices that bind to lipids (Fig. 1a).
Fig. 1.
Structures of apoE3 and apoE4. a ApoE4 displays domain interaction caused by an ionic interaction between Arg-61 and Glu-255. This structural feature of apoE4 (apoE4 > apoE3 > apoE2) alters its function in cardiovascular and neurological disorders. b Small-molecule apoE4 structure correctors block domain interaction and convert apoE4 to an apoE3-like molecule structurally and functionally. c Injury of neurons induces apoE expression. When neurons are stressed or injured, they synthesize apoE to function in lipid redistribution for neuronal repair and remodeling. In this model, apoE4 is recognized as structurally abnormal and undergoes proteolytic cleavage, generating several neurotoxic fragments (12–29 kDa) that escape the secretory pathway, enter the cytosol, and cause mitochondrial dysfunction and tau phosphorylation (Tau-PO4), ultimately causing cell death. ApoE4SC, apoE4 structure corrector. Modified from ref. 38. Copyright 2012 American Chemical Society
ApoE3 and apoE4 bind to the LDL receptor with similar affinity (~20-fold greater than that of apoB100, the other LDL receptor ligand). The key amino acids for receptor binding were identified by site-directed mutagenesis and by the existence of naturally occurring receptor-defective human mutations in type III hyperlipoproteinemic patients [11]. ApoE plays a major role in regulating cholesterol homeostasis by mediating the uptake of VLDL, intermediate density lipoproteins, and chylomicron remnants [2, 11, 12, 21]. ApoE2, however, defectively binds to the LDL receptor (~2 % of normal activity), because it has a cysteine at residue 158 rather than an arginine, as in apoE3 and apoE4. As shown by x-ray crystallography of the amino-terminal domain, Cys-158 prevents normal receptor binding by altering the conformation of the side chains in the critical basic residues in the 136–150 region. In apoE3, Arg-158 forms a salt bridge with aspartic acid–154; however, in apoE4, with Cys-158, this salt bridge is disrupted, and aspartic acid–154 interacts with Arg-150, altering the entire receptor binding region [11, 19, 20, 22].
ApoE4 has an arginine at residue 112 (Arg-112). This produces a property of apoE4 referred to as domain interaction, in which Arg-112 causes the side chain of Arg-61 to extend away from the amino-terminal domain [23, 24], enabling it to interact ionically with glutamic acid (Glu)–255 in the carboxyl-terminal domain (Fig. 1a). Domain interaction is less likely to occur in apoE3 or apoE2, because in these isoforms, with Cys-112, the side chain of Arg-61 is more tucked into the helical domain of the amino terminus [11, 24]. Domain interaction is unlikely to be an all-or-none property. Protein structure is dynamic, and it is likely that there is a gradient in the propensity of apoE to display domain interaction (apoE4 > apoE3 > apoE2).
ApoE and cardiovascular disease
ApoE2
Because it binds defectively to LDL receptors, apoE2 homozygosity can precipitate type III hyperlipoproteinemia [2, 11, 21, 22]. This disorder occurs only when another condition—diabetes, obesity, hypothyroidism, or estrogen deficiency—leads to overproduction of VLDL or fewer LDL receptors, overwhelming the limited ability of apoE2 to mediate the clearance of triglyceride- and cholesterol-rich β-VLDL. Other dominant and recessive mutations in apoE that affect residues in or around the receptor binding region also cause type III hyperlipoproteinemia [2, 11, 22]. The defective receptor binding can precipitate hyperlipidemia in the context of other genetic or environmental factors and increase the risk for atherosclerosis, as the β-VLDL that accumulate in the plasma are highly atherogenic and cause cholesterol accumulation, especially in peripheral arteries.
ApoE4
ApoE4 increases plasma LDL levels and the risk for atherosclerosis [11, 12, 25]. The lipoprotein-binding preference of apoE4 to large (30–80 nm), triglyceride-rich VLDL, is associated with elevated LDL levels. (ApoE3 and apoE2 preferentially bind to small, 9–16-nm spherical HDL particles enriched in phospholipids and surface proteins, primarily apoAI.) The enrichment of VLDL with apoE4 accelerates their clearance from the plasma by receptor-mediated endocytosis in the liver; as a result, LDL receptors are downregulated, and plasma LDL levels increase [26–28]. Hepatic clearance of apoE-enriched lipoproteins involves LDL receptors and LDL receptor-related proteins (members of the LDL receptor family) and heparan sulfate proteoglycans—all of which interact with high affinity with apoE [for review, see refs. 29, 30].
How does the amino acid difference at residue 112 in the amino-terminal domain—arginine in apoE4 and cysteine in apoE3 and apoE2—alter the lipoprotein preference of apoE4 when lipid binding is mediated by residues 240–260 in the carboxyl-terminal domain? The reason is the structural effect of domain interaction. When domain interaction is disrupted with small molecules that prevent the Arg-61–Glu-255 ionic interaction or by site-directed mutagenesis (Arg-61 to threonine), apoE4’s binding preference shifts from VLDL to smaller, phospholipid-rich HDL [23, 24, 31]. In fact, apoE4 domain interaction is associated with decreased phospholipid-binding capacity [32]. Domain interaction may stabilize an extended helical structure involving the amino- and carboxyl-terminal domains of apoE4 [33–35], allowing it to interact with larger VLDL and accommodating interaction with lipoprotein particles with less curvature.
ApoE4 and neurological disease
ApoE4 is the major genetic risk factor (or causative gene) for Alzheimer’s disease (AD) and other neurological disorders, including poor clinical outcomes after traumatic brain injury, or stroke, frontotemporal dementia, Down syndrome, certain patients with Parkinson’s disease, and Lewy body disease [for review, see refs. 12, 36–38]. ApoE4 dramatically affects AD, and 65–80 % of all AD patients carry at least one apoE4 allele. ApoE4 increases the risk of developing AD by 4-fold (one allele) to 14-fold (two alleles) compared with apoE3/3 homozygosity, and it decreases the age of onset by 8 years for each apoE4 allele (onset by mid-1960s with two alleles). Importantly, apoE4 alleles are not rare: ~25 % of people worldwide have at least one apoE4 allele.
Multiple factors acting through various pathways cause cognitive decline and neurodegeneration. Several mechanisms have been proposed, but the so-called amyloid hypothesis has received the most attention [for review, see refs. 39–41]. Rare mutations in the gene for the human amyloid precursor protein and the enzymes (secretases) that generate elevated levels of the amyloid beta (Aβ) peptide are related to the development of early-onset AD [42–44]. These genetic variants explain 1–2 % of AD cases; the majority of AD is sporadic. Mouse models of AD in which these variants are markedly overexpressed display some of the pathological features of AD, including increased Aβ and amyloid plaques, augmented tau phosphorylation, loss of synaptic connections, and impaired learning and memory, but not the significant neurodegeneration seen in AD [37, 43, 45, 46]. Aβ accumulation is considered neurotoxic.
Mechanisms for apoE4’s involvement in the amyloid pathway
There is no consensus concerning the mechanisms by which apoE4 affects the amyloid pathway [39–41]. Several lines of evidence suggest that apoE4 accelerates Aβ deposition to form amyloid plaques [47]. Others suggest that apoE4 is deficient in Aβ clearance [48, 49]. Some studies show that apoE3 binds Aβ to a greater extent than apoE4 and mediates the uptake and degradation of Aβ by receptor-mediated endocytosis in the brain, whereas apoE4 is less efficient in Aβ clearance and is associated with increased Aβ levels [39–41]. Others have shown that Aβ does not interact with apoE at all and suggest that, by an unknown mechanism, apoE4 competes with an Aβ clearance mechanism and thus increases Aβ levels [50].
Lipidation of apoE-containing lipoproteins in the brain has been postulated to modulate amyloid deposition and Aβ clearance by delivering Aβ to microglia and astrocytes for degradation or to the blood brain barrier for transport out of the brain [39, 40, 51, 52]. ApoE mediates lipoprotein interaction with various members of the LDL receptor family involved in these processes [39, 40]. Lipidation of apoE in the brain occurs through the activity of specific ATP-binding cassette transporters (ABC), including ABCA1 [53] and possibly ABCA7 [54], ABCG1 [55, 56], and ABCG4 [56], which are expressed in the brain. ApoE4 levels [57, 58] and the degree of apoE4 lipidation [32] are decreased in the brain compared with apoE3 and apoE2, and this has been correlated with AD risk. Overexpression of ABCA1 in an AD mouse model reduces amyloid deposition and improves Aβ clearance [59]. Alternatively, decreased ABCA1 expression reduced apoE levels and lipidation and increased amyloid deposition [60, 61]. However, the importance of ABCA1 activity in the human brain remains unknown, and existing data are conflicting. Recently, a population-based study demonstrated that a loss-of-function mutation in ABCA1 was associated with low plasma levels of apoE (no data on brain apoE levels) and increased risk of AD [62]. On the other hand, lipidation of apoE- and apoAI-containing lipoproteins in the brain may act independently of Aβ metabolism by protecting the integrity of the blood brain barrier and maintaining normal cerebrovascular function [63]. Whether modulation of brain lipoprotein lipidation represents a viable therapeutic approach remains to be determined.
Regardless of the reason, apoE4 carriers have increased numbers of amyloid plaques in their brains [37, 39, 64]. However, the plaque accumulation commences early in life, before cognitive impairment is evident, and does not entirely correlate with AD; about one-third of individuals with high levels of amyloid plaques are cognitively normal. AD in apoE4 carriers correlates better with the accumulation of phosphorylated tau in the hippocampus, which is critically important in cognition [65]. ApoE4 enhances tau phosphorylation and neurofibrillary tangle formation.
ApoE’s direct effects on neuropathology independent of Aβ
Studies from the Gladstone Institutes showed that apoE4 acts directly and independently of Aβ on a parallel path leading to neuropathology [12, 36–38]. By itself, apoE4 has many detrimental effects on neuronal cells in vitro and in vivo—including mitochondrial dysfunction due to decreased levels and activities of various electron transport enzymes and ATP synthase [66, 67]; increased tau phosphorylation and the formation of intracellular inclusions resembling neurofibrillary tangles [68, 69]; impairment in mitochondrial motility [67], intracellular trafficking of apoE [70], neurite outgrowth [67, 71], and synaptogenesis [72]; accelerated neuropathology involving loss of GABAergic hippocampal interneurons [69]; and impaired learning and memory in apoE4 mouse models [69, 73–75]. These detrimental effects are reversed when apoE4 domain interaction is blocked by site-directed mutagenesis (Arg-61 to threonine) or by small molecules that “correct” the structure of apoE4 [for review, see refs. 36–38].
Generation of apoE4 neurotoxic fragments
ApoE4 is highly susceptible to neuron-specific proteolysis, which generates 12–29-kDa neurotoxic fragments [68, 76, 77] (Fig. 1c). In response to injury or stress, neurons synthesize apoE, presumably to facilitate the rapid transport of cholesterol and other lipids for membrane repair and remodeling. Neuronal apoE synthesis is highly regulated by neuron-specific mRNA splicing [78]. We hypothesize that domain interaction causes apoE4 to be “sensed” by the cell as abnormal, targeting it for proteolytic cleavage in the endoplasmic reticulum or Golgi apparatus. ApoE3 is much less susceptible to this proteolysis, which does not occur in astrocytes or hepatocytes that also produce apoE. The initial cleavage removes the carboxyl-terminal 27–30 amino acids, generating a toxic 29-kDa fragment; subsequent proteolysis results in amino-terminal cleavage, generating 12–20-kDa fragments, some of which are also toxic.
These fragments escape the secretory pathway and enter the cytosol, where they stimulate tau phosphorylation and interact with mitochondria, causing mitochondrial dysfunction and neurodegeneration [36–38] (Fig. 1c). Fragments containing the receptor binding region (residues 136–150) and the lipid binding region (residues 240–260) are the minimal structure of apoE responsible for translocation, mitochondrial localization, and neurotoxicity [79]. The receptor binding region behaves like a protein translocation domain, as in viral proteins (region of a protein enriched in arginines and lysines that facilitates translocation of a protein across a membrane). The hydrophobicity of the lipid binding region mediates the interaction of the fragments with mitochondria and subsequent neurotoxicity. The critical residues in these regions of apoE have been identified by site-directed mutagenesis. In the brain, the apoE fragments are much more abundant in patients with AD than in age-matched, nondemented controls [76, 80]. The fragments also occur in the brains of mice expressing apoE4 in neurons.
Identifying structure correctors to block apoE4 domain interaction
Small-molecule structure correctors that block the ionic interaction of Arg-61 and Glu-255 in apoE4 (Fig. 1b) have been identified with a cellular fluorescence resonance energy transfer (FRET) assay in which the amino-terminal domain of apoE4 is labeled with green fluorescent protein and the carboxyl-terminal domain is labeled with Escherichia coli dihydrofolate reductase. The FRET assay measures the ability of an active structure corrector to prevent the FRET emission signal that occurs in apoE4 when the amino- and carboxyl-terminal domains are in close proximity [67]. Testing apoE4 structure correctors with this assay revealed downstream functional effects in neurons. For example, active structure correctors restore mitochondrial cytochrome c oxidase levels, which are depleted in apoE4-expressing cultured neurons [67]. Structure–activity relationships have been established for such compounds at low nanomolar levels.
Retarding apoE4-associated neuropathology in vivo in mice
Overexpression of apoE4 in transgenic mice or expression of apoE4 in targeted-replacement mice causes central nervous system pathology, including increased tau phosphorylation, loss of interneurons, and impaired learning and memory [69, 73–75]. A prototypical apoE4 structure corrector, PY-101, has been used in the apoE4 transgenic mice as a proof-of-concept that blocking apoE4 domain interaction can reverse the detrimental effects associated with apoE4 [38]. After intraperitoneal or subcutaneous administration of PY-101 (30–50 mg/kg body weight for 10 consecutive days), apoE4 fragment levels in the brain and hippocampus decreased by 20–25 %, and mitochondrial cytochrome c oxidase levels increased by 50–55 %. Preclinical studies are in process, and potent small molecules with drug-like properties have been identified. Thus, apoE4 is a promising drug target for apoE4-associated neuropathology in AD and other disorders. As an additive or ancillary therapy, apoE structure correctors might also help reduce plasma LDL levels and risk for coronary artery disease.
Acknowledgments
This work was supported, in part, by The Wellcome Trust Seeding Drug Discovery Award 101786. We thank Sylvia Richmond and Charlotte Pfeiffer for manuscript preparation, Stephen Ordway and Crystal Herron for editorial assistance, and John C. W. Carroll for graphics.
References
- 1.Fredrickson DS, Levy RI, Lees RS (1967) Fat transport in lipoproteins—an integrated approach to mechanisms and disorders. N Engl J Med 276: 34–44, 94–103, 148–156, 215–225, 273–281 [DOI] [PubMed]
- 2.Mahley RW (1988) Apolipoprotein E: cholesterol transport protein with expanding role in cell biology. Science 240:622–630 [DOI] [PubMed]
- 3.Shore VG, Shore B (1973) Heterogeneity of human plasma very low density lipoproteins. Separation of species differing in protein components. Biochemistry 12:502–507 [DOI] [PubMed]
- 4.Shore VG, Shore B, Hart RG (1974) Changes in apolipoproteins and properties of rabbit very low density lipoproteins on induction of cholesteremia. Biochemistry 13:1579–1585 [DOI] [PubMed]
- 5.Mahley RW, Weisgraber KH, Innerarity T (1974) Canine lipoproteins and atherosclerosis. II. Characterization of the plasma lipoproteins associated with atherogenic and nonatherogenic hyperlipidemia. Circ Res 35:722–733 [DOI] [PubMed]
- 6.Mahley RW, Weisgraber KH, Innerarity T, Brewer HB Jr, Assmann G (1975) Swine lipoproteins and atherosclerosis. Changes in the plasma lipoproteins and apoproteins induced by cholesterol feeding. Biochemistry 14:2817–2823 [DOI] [PubMed]
- 7.Mahley RW, Weisgraber KH, Innerarity T (1976) Atherogenic hyperlipoproteinemia induced by cholesterol feeding in the Patas monkey. Biochemistry 15:2979–2985 [DOI] [PubMed]
- 8.Mahley RW (1985) Atherogenic lipoproteins and coronary artery disease: concepts derived from recent advances in cellular and molecular biology. Circulation 72:943–948 [DOI] [PubMed]
- 9.Mahley RW, Innerarity TL, Rall SC Jr, Weisgraber KH (1984) Plasma lipoproteins: apolipoprotein structure and function. J Lipid Res 25:1277–1294 [PubMed]
- 10.Zannis VI, Breslow JL, Utermann G, Mahley RW, Weisgraber KH, Havel RJ, Goldstein JL, Brown MS, Schonfeld G, Hazzard WR et al (1982) Proposed nomenclature of apoE isoproteins, apoE genotypes, and phenotypes. J Lipid Res 23:911–914 [PubMed]
- 11.Mahley RW, Rall SC Jr (2000) Apolipoprotein E: far more than a lipid transport protein. Annu Rev Genomics Hum Genet 1:507–537 [DOI] [PubMed]
- 12.Mahley RW, Weisgraber KH, Huang Y (2009) Apolipoprotein E: structure determines function, from atherosclerosis to Alzheimer’s disease to AIDS. J Lipid Res 50:S183–S188 [DOI] [PMC free article] [PubMed]
- 13.Weisgraber KH (1994) Apolipoprotein E: structure–function relationships. Adv Protein Chem 45:249–302 [DOI] [PubMed]
- 14.Xu Q, Bernardo A, Walker D, Kanegawa T, Mahley RW, Huang Y (2006) Profile and regulation of apolipoprotein E (apoE) expression in the CNS in mice with targeting of green fluorescent protein gene to the apoE locus. J Neurosci 26:4985–4994 [DOI] [PMC free article] [PubMed]
- 15.Uchihara T, Duyckaerts C, He Y, Kobayashi K, Seilhean D, Amouyel P, Hauw J-J (1995) ApoE immunoreactivity and microglial cells in Alzheimer’s disease brain. Neurosci Lett 195:5–8 [DOI] [PubMed]
- 16.Boyles JK, Pitas RE, Wilson E, Mahley RW, Taylor JM (1985) Apolipoprotein E associated with astrocytic glia of the central nervous system and with nonmyelinating glia of the peripheral nervous system. J Clin Invest 76:1501–1513 [DOI] [PMC free article] [PubMed]
- 17.Linton MF, Gish R, Hubl ST, Bütler E, Esquivel C, Bry WI, Boyles JK, Wardell MR, Young SG (1991) Phenotypes of apolipoprotein B and apolipoprotein E after liver transplantation. J Clin Invest 88:270–281 [DOI] [PMC free article] [PubMed]
- 18.Wernette-Hammond ME, Lauer SJ, Corsini A, Walker D, Taylor JM, Rall SC Jr (1989) Glycosylation of human apolipoprotein E. The carbohydrate attachment site is threonine 194. J Biol Chem 264:9094–9101 [PubMed]
- 19.Wilson C, Mau T, Weisgraber KH, Wardell MR, Mahley RW, Agard DA (1994) Salt bridge relay triggers defective LDL receptor binding by a mutant apolipoprotein. Structure 2:713–718 [DOI] [PubMed]
- 20.Wilson C, Wardell MR, Weisgraber KH, Mahley RW, Agard DA (1991) Three-dimensional structure of the LDL receptor-binding domain of human apolipoprotein E. Science 252:1817–1822 [DOI] [PubMed]
- 21.Mahley RW, Rall SC Jr (2001) Type III hyperlipoproteinemia (dysbetalipoproteinemia): the role of apolipoprotein E in normal and abnormal lipoprotein metabolism. In: Scriver CR, Beaudet AL, Sly WS, Valle D, Childs B, Kinzler KW, Vogelstein B (eds) The Metabolic and Molecular Bases of Inherited Disease. McGraw-Hill, New York, pp 2835–2862
- 22.Mahley RW, Huang Y, Rall SC Jr (1999) Pathogenesis of type III hyperlipoproteinemia (dysbetalipoproteinemia): questions, quandaries, and paradoxes. J Lipid Res 40:1933–1949 [PubMed]
- 23.Dong L-M, Weisgraber KH (1996) Human apolipoprotein E4 domain interaction. Arginine 61 and glutamic acid 255 interact to direct the preference for very low density lipoproteins. J Biol Chem 271:19053–19057 [DOI] [PubMed]
- 24.Dong L-M, Wilson C, Wardell MR, Simmons T, Mahley RW, Weisgraber KH, Agard DA (1994) Human apolipoprotein E. Role of arginine 61 in mediating the lipoprotein preferences of the E3 and E4 isoforms. J Biol Chem 269:22358–22365 [PubMed]
- 25.Bennet AM, Di Angelantonio E, Ye Z, Wensley F, Dahlin A, Ahlbom A, Keavney B, Collins R, Wiman B, de Faire U et al (2007) Association of apolipoprotein E genotypes with lipid levels and coronary risk. JAMA 298:1300–1311 [DOI] [PubMed]
- 26.Gregg RE, Zech LA, Schaefer EJ, Stark D, Wilson D, Brewer HB Jr (1986) Abnormal in vivo metabolism of apolipoprotein E4 in humans. J Clin Invest 78:815–821 [DOI] [PMC free article] [PubMed]
- 27.Weintraub MS, Eisenberg S, Breslow JL (1987) Dietary fat clearance in normal subjects is regulated by genetic variation of apolipoprotein E. J Clin Invest 80:1571–1577 [DOI] [PMC free article] [PubMed]
- 28.Li H, Dhanasekaran P, Alexander ET, Rader DJ, Phillips MC, Lund-Katz S (2013) Molecular mechanisms responsible for the differential effects of apoE3 and apoE4 on plasma lipoprotein–cholesterol levels. Arterioscler Thromb Vasc Biol 33:687–693 [DOI] [PMC free article] [PubMed]
- 29.Mahley RW, Ji Z-S (1999) Remnant lipoprotein metabolism: key pathways involving cell-surface heparan sulfate proteoglycans and apolipoprotein E. J Lipid Res 40:1–16 [PubMed]
- 30.Mahley RW, Huang Y (2007) Atherogenic remnant lipoproteins: role for proteoglycans in trapping, transferring, and internalizing. J Clin Invest 117:94–98 [DOI] [PMC free article] [PubMed]
- 31.Ye S, Huang Y, Müllendorff K, Dong L, Giedt G, Meng EC, Cohen FE, Kuntz ID, Weisgraber KH, Mahley RW (2005) Apolipoprotein (apo) E4 enhances amyloid β peptide production in cultured neuronal cells: apoE structure as a potential therapeutic target. Proc Natl Acad Sci U S A 102:18700–18705 [DOI] [PMC free article] [PubMed]
- 32.Xu Q, Brecht WJ, Weisgraber KH, Mahley RW, Huang Y (2004) Apolipoprotein E4 domain interaction occurs in living neuronal cells as determined by fluorescence resonance energy transfer. J Biol Chem 279:25511–25516 [DOI] [PubMed]
- 33.Hatters DM, Budamagunta MS, Voss JC, Weisgraber KH (2005) Modulation of apolipoprotein E structure by domain interaction. Differences in lipid-bound and lipid-free forms. J Biol Chem 280:34288–34295 [DOI] [PubMed]
- 34.Peters-Libeu CA, Newhouse Y, Hatters DM, Weisgraber KH (2006) Model of biologically active apolipoprotein E bound to dipalmitoylphosphatidylcholine. J Biol Chem 281:1073–1079 [DOI] [PubMed]
- 35.Nguyen D, Dhanasekaran P, Nickel M, Nakatani R, Saito H, Phillips MC, Lund-Katz S (2010) Molecular basis for the differences in lipid and lipoprotein binding properties of human apolipoproteins E3 and E4. Biochemistry 49:10881–10889 [DOI] [PMC free article] [PubMed]
- 36.Mahley RW, Huang Y (2012) Apolipoprotein E sets the stage: response to injury triggers neuropathology. Neuron 76:871–885 [DOI] [PMC free article] [PubMed]
- 37.Huang Y, Mucke L (2012) Alzheimer mechanisms and therapeutic strategies. Cell 148:1204–1222 [DOI] [PMC free article] [PubMed]
- 38.Mahley RW, Huang Y (2012) Small-molecule structure correctors target abnormal protein structure and function: the structure corrector rescue of apolipoprotein E-associated neuropathology. J Med Chem 55:8997–9008 [DOI] [PMC free article] [PubMed]
- 39.Holtzman DM, Herz J, Bu G (2012) Apolipoprotein E and apolipoprotein E receptors: normal biology and roles in Alzheimer disease. Cold Spring Harb Perspect Med 2:a006312 [DOI] [PMC free article] [PubMed]
- 40.Bu G (2009) Apolipoprotein E and its receptors in Alzheimer’s disease: pathways, pathogenesis and therapy. Nat Rev Neurosci 10:333–344 [DOI] [PMC free article] [PubMed]
- 41.Kim J, Basak JM, Holtzman DM (2009) The role of apolipoprotein E in Alzheimer’s disease. Neuron 63:287–303 [DOI] [PMC free article] [PubMed]
- 42.Campion D, Dumanchin C, Hannequin D, Dubois B, Belliard S, Puel M, Thomas-Anterion C, Michon A, Martin C, Charbonnier F et al (1999) Early-onset autosomal dominant Alzheimer disease: prevalence, genetic heterogeneity, and mutation spectrum. Am J Hum Genet 65:664–670 [DOI] [PMC free article] [PubMed]
- 43.Palop JJ, Mucke L (2010) Amyloid-β-induced neuronal dysfunction in Alzheimer’s disease: from synapses toward neural networks. Nat Neurosci 13:812–818 [DOI] [PMC free article] [PubMed]
- 44.Bertram L, Lill CM, Tanzi RE (2010) The genetics of Alzheimer disease: back to the future. Neuron 68:270–281 [DOI] [PubMed]
- 45.Ashe KH, Zahs KR (2010) Probing the biology of Alzheimer’s disease in mice. Neuron 66:631–645 [DOI] [PMC free article] [PubMed]
- 46.Zahs KR, Ashe KH (2010) ‘Too much good news’—are Alzheimer mouse models trying to tell us how to prevent, not cure, Alzheimer’s disease? Trends Neurosci 33:381–389 [DOI] [PubMed]
- 47.Wisniewski T, Castaño EM, Golabek A, Vogel T, Frangione B. Acceleration of Alzheimer’s fibril formation by apolipoprotein E in vitro. Am J Pathol. 1994;145:1030–1035. [PMC free article] [PubMed] [Google Scholar]
- 48.Castellano JM, Kim J, Stewart FR, Jiang H, DeMattos RB, Patterson BW, Fagan AM, Morris JC, Mawuenyega KG, Cruchaga C, et al. Human apoE isoforms differentially regulate brain amyloid-β peptide clearance. Sci Transl Med. 2011;3:89ra57. doi: 10.1126/scitranslmed.3002156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.LaDu MJ, Falduto MT, Manelli AM, Reardon CA, Getz GS, Frail DE. Isoform-specific binding of apolipoprotein E to β-amyloid. J Biol Chem. 1994;269:23403–23406. [PubMed] [Google Scholar]
- 50.Verghese PB, Castellano JM, Garai K, Wang Y, Jiang H, Shah A, Bu G, Frieden C, Holtzman DM. ApoE influences amyloid-β (Aβ) clearance despite minimal apoE/Aβ association in physiological conditions. Proc Natl Acad Sci U S A. 2013;110:E1807–E1816. doi: 10.1073/pnas.1220484110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Koldamova R, Fitz NF, Lefterov I. The role of ATP-binding cassette transporter A1 in Alzheimer’s disease and neurodegeneration. Biochim Biophys Acta. 2010;1801:824–830. doi: 10.1016/j.bbalip.2010.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Stukas S, Robert J, Wellington Cheryl L. High-density lipoproteins and cerebrovascular integrity in Alzheimer’s disease. Cell Metab. 2014;19:574–591. doi: 10.1016/j.cmet.2014.01.003. [DOI] [PubMed] [Google Scholar]
- 53.Vitali C, Wellington CL, Calabresi L. HDL and cholesterol handling in the brain. Cardiovasc Res. 2014;103:405–413. doi: 10.1093/cvr/cvu148. [DOI] [PubMed] [Google Scholar]
- 54.Quazi F, Molday RS. Differential phospholipid substrates and directional transport by ATP-binding cassette proteins ABCA1, ABCA7, and ABCA4 and disease-causing mutants. J Biol Chem. 2013;288:34414–34426. doi: 10.1074/jbc.M113.508812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Burgess BL, Parkinson PF, Racke MM, Hirsch-Reinshagen V, Fan J, Wong C, Stukas S, Theroux L, Chan JY, Donkin J, et al. ABCG1 influences the brain cholesterol biosynthetic pathway but does not affect amyloid precursor protein or apolipoprotein E metabolism in vivo. J Lipid Res. 2008;49:1254–1267. doi: 10.1194/jlr.M700481-JLR200. [DOI] [PubMed] [Google Scholar]
- 56.Wang N, Lan D, Chen W, Matsuura F, Tall AR. ATP-binding cassette transporters G1 and G4 mediate cellular cholesterol efflux to high-density lipoproteins. Proc Natl Acad Sci U S A. 2004;101:9774–9779. doi: 10.1073/pnas.0403506101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Riddell DR, Zhou H, Atchison K, Warwick HK, Atkinson PJ, Jefferson J, Xu L, Aschmies S, Kirksey Y, Hu Y, et al. Impact of apolipoprotein E (apoE) polymorphism on brain apoE levels. J Neurosci. 2008;28:11445–11453. doi: 10.1523/JNEUROSCI.1972-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Fryer JD, DeMattos RB, McCormick LM, O’Dell MA, Spinner ML, Bales KR, Paul SM, Sullivan PM, Parsadanian M, Bu G, et al. The low density lipoprotein receptor regulates the level of central nervous system human and murine apolipoprotein E but does not modify amyloid plaque pathology in PDAPP mice. J Biol Chem. 2005;280:25754–25759. doi: 10.1074/jbc.M502143200. [DOI] [PubMed] [Google Scholar]
- 59.Wahrle SE, Jiang H, Parsadanian M, Kim J, Li A, Knoten A, Jain S, Hirsch-Reinshagen V, Wellington CL, Bales KR, et al. Overexpression of ABCA1 reduces amyloid deposition in the PDAPP mouse model of Alzheimer disease. J Clin Invest. 2008;118:671–682. doi: 10.1172/JCI33622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hirsch-Reinshagen V, Maia LF, Burgess BL, Blain J-F, Naus KE, McIsaac SA, Parkinson PF, Chan JY, Tansley GH, Hayden MR, et al. The absence of ABCA1 decreases soluble apoE levels but does not diminish amyloid deposition in two murine models of Alzheimer disease. J Biol Chem. 2005;280:43243–43256. doi: 10.1074/jbc.M508781200. [DOI] [PubMed] [Google Scholar]
- 61.Koldamova R, Staufenbiel M, Lefterov I. Lack of ABCA1 considerably decreases brain apoE level and increases amyloid deposition in APP23 mice. J Biol Chem. 2005;280:43224–43235. doi: 10.1074/jbc.M504513200. [DOI] [PubMed] [Google Scholar]
- 62.Nordestgaard LT, Tybjærg-Hansen A, Nordestgaard BG, Frikke-Schmidt R. Loss-of-function mutation in ABCA1 and risk of Alzheimer’s disease and cerebrovascular disease. Alzheimers Dement. 2015;11:1430–1438. doi: 10.1016/j.jalz.2015.04.006. [DOI] [PubMed] [Google Scholar]
- 63.Zlokovic BV. Cerebrovascular effects of apolipoprotein E: implications for Alzheimer disease. JAMA Neurol. 2013;70:440–444. doi: 10.1001/jamaneurol.2013.2152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tiraboschi P, Hansen LA, Masliah E, Alford M, Thal LJ, Corey-Bloom J. Impact of APOE genotype on neuropathologic and neurochemical markers of Alzheimer disease. Neurology. 2004;62:1977–1983. doi: 10.1212/01.WNL.0000128091.92139.0F. [DOI] [PubMed] [Google Scholar]
- 65.Giannakopoulos P, Herrmann FR, Bussière T, Bouras C, Kövari E, Perl DP, Morrison JH, Gold G, Hof PR. Tangle and neuron numbers, but not amyloid load, predict cognitive status in Alzheimer’s disease. Neurology. 2003;60:1495–1500. doi: 10.1212/01.WNL.0000063311.58879.01. [DOI] [PubMed] [Google Scholar]
- 66.Chen H-K, Ji Z-S, Dodson SE, Miranda RD, Rosenblum CI, Reynolds IJ, Freedman SB, Weisgraber KH, Huang Y, Mahley RW. Apolipoprotein E4 domain interaction mediates detrimental effects on mitochondria and is a potential therapeutic target for Alzheimer disease. J Biol Chem. 2011;286:5215–5221. doi: 10.1074/jbc.M110.151084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Chen H-K, Liu Z, Meyer-Franke A, Brodbeck J, Miranda RD, McGuire JG, Pleiss MA, Ji Z-S, Balestra ME, Walker DW, et al. Small molecule structure correctors abolish detrimental effects of apolipoprotein E4 in cultured neurons. J Biol Chem. 2012;287:5253–5266. doi: 10.1074/jbc.M111.276162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Huang Y, Liu XQ, Wyss-Coray T, Brecht WJ, Sanan DA, Mahley RW. Apolipoprotein E fragments present in Alzheimer’s disease brains induce neurofibrillary tangle-like intracellular inclusions in neurons. Proc Natl Acad Sci U S A. 2001;98:8838–8843. doi: 10.1073/pnas.151254698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Andrews-Zwilling Y, Bien-Ly N, Xu Q, Li G, Bernardo A, Yoon SY, Zwilling D, Yan TX, Chen L, Huang Y. Apolipoprotein E4 causes age- and Tau-dependent impairment of GABAergic interneurons, leading to learning and memory deficits in mice. J Neurosci. 2010;30:13707–13717. doi: 10.1523/JNEUROSCI.4040-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Brodbeck J, McGuire J, Liu Z, Meyer-Franke A, Balestra ME, Jeong D-e, Pleiss M, McComas C, Hess F, Witter D, et al. Structure-dependent impairment of intracellular apolipoprotein E4 trafficking and its detrimental effects are rescued by small-molecule structure correctors. J Biol Chem. 2011;286:17217–17226. doi: 10.1074/jbc.M110.217380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Nathan BP, Bellosta S, Sanan DA, Weisgraber KH, Mahley RW, Pitas RE. Differential effects of apolipoproteins E3 and E4 on neuronal growth in vitro. Science. 1994;264:850–852. doi: 10.1126/science.8171342. [DOI] [PubMed] [Google Scholar]
- 72.Brodbeck J, Balestra ME, Saunders AM, Roses AD, Mahley RW, Huang Y. Rosiglitazone increases dendritic spine density and rescues spine loss caused by apolipoprotein E4 in primary cortical neurons. Proc Natl Acad Sci U S A. 2008;105:1343–1346. doi: 10.1073/pnas.0709906104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Buttini M, Orth M, Bellosta S, Akeefe H, Pitas RE, Wyss-Coray T, Mucke L, Mahley RW. Expression of human apolipoprotein E3 or E4 in the brains of Apoe–/– mice: isoform-specific effects on neurodegeneration. J Neurosci. 1999;19:4867–4880. doi: 10.1523/JNEUROSCI.19-12-04867.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Raber J, Wong D, Yu G-Q, Buttini M, Mahley RW, Pitas RE, Mucke L. Apolipoprotein E and cognitive performance. Nature. 2000;404:352–354. doi: 10.1038/35006165. [DOI] [PubMed] [Google Scholar]
- 75.Andrews-Zwilling Y, Gillespie AK, Kravitz AV, Nelson AB, Devidze N, Lo I, Yoon SY, Bien-Ly N, Ring K, Zwilling D, et al. Hilar GABAergic interneuron activity controls spatial learning and memory retrieval. PLoS ONE. 2012;7 doi: 10.1371/journal.pone.0040555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Harris FM, Brecht WJ, Xu Q, Tesseur I, Kekonius L, Wyss-Coray T, Fish JD, Masliah E, Hopkins PC, Scearce-Levie K, et al. Carboxyl-terminal-truncated apolipoprotein E4 causes Alzheimer’s disease-like neurodegeneration and behavioral deficits in transgenic mice. Proc Natl Acad Sci U S A. 2003;100:10966–10971. doi: 10.1073/pnas.1434398100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Brecht WJ, Harris FM, Chang S, Tesseur I, Yu G-Q, Xu Q, Fish JD, Wyss-Coray T, Buttini M, Mucke L, et al. Neuron-specific apolipoprotein E4 proteolysis is associated with increased tau phosphorylation in brains of transgenic mice. J Neurosci. 2004;24:2527–2534. doi: 10.1523/JNEUROSCI.4315-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Xu Q, Walker D, Bernardo A, Brodbeck J, Balestra ME, Huang Y. Intron-3 retention/splicing controls neuronal expression of apolipoprotein E in the CNS. J Neurosci. 2008;28:1452–1459. doi: 10.1523/JNEUROSCI.3253-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Chang S, Ma TR, Miranda RD, Balestra ME, Mahley RW, Huang Y. Lipid- and receptor-binding regions of apolipoprotein E4 fragments act in concert to cause mitochondrial dysfunction and neurotoxicity. Proc Natl Acad Sci U S A. 2005;102:18694–18699. doi: 10.1073/pnas.0508254102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Jones PB, Adams KW, Rozkalne A, Spires-Jones TL, Hshieh TT, Hashimoto T, von Armin CAF, Mielke M, Bacskai BJ, Hyman BT. Apolipoprotein E: isoform specific differences in tertiary structure and interaction with amyloid-β in human Alzheimer brain. PLoS ONE. 2011;6 doi: 10.1371/journal.pone.0014586. [DOI] [PMC free article] [PubMed] [Google Scholar]