Abstract
Herpesviral mRNAs are produced and translated by cellular machinery, rendering them susceptible to the network of regulatory events that impact translation. In response, these viruses have evolved to infiltrate and hijack translational control pathways as well as to integrate specialized host translation strategies into their own repertoire. They are robust systems to dissect mechanisms of mammalian translational regulation and continue to offer insight into cis-acting mRNA features that impact assembly and activity of the translation apparatus. Here, I discuss recent advances revealing the extent to which the three herpesvirus subfamilies regulate both host and viral translation, thereby dramatically impacting the landscape of protein synthesis in infected cells.
Keywords: herpesvirus, translation, eIF4F, protein kinase R, unfolded protein response, uORF
INTRODUCTION
Herpesviruses are ancient and remarkably widespread viruses that are extremely well adapted to the human population, as evidenced by their near-ubiquitous seroprevalence. Based on their distinct biological properties and genomic sequences, these nuclear replicating, large dsDNA viruses are divided into three subfamilies: the alpha-, beta-, and gammaherpesviruses. Because their genes are transcribed and processed by the cellular machinery, herpesviral mRNAs structurally resemble those of their host. These conserved structural features, including the 5′ 7-methylguanosine cap and 3′ poly(A) tail, direct the loading of translation factors to enable protein synthesis. Thus, herpesviral gene expression is susceptible to the extensive network of regulatory pathways that control cellular translation, many of which are designed to restrict translation upon viral infection. To counteract this shutdown, herpesviruses manipulate translation factors and signaling cascades, as well as enact other broadly acting mechanisms to control mRNA abundance and access to polysomes. In this manner, each of the three subfamilies of herpesviruses dramatically impacts the translational landscape of the cell during infection.
Here, I focus on recent advances regarding the strategies herpesviruses have evolved to interface with, manipulate, and co-opt cellular translation machinery. I begin by summarizing how the translation machinery is efficiently recruited to viral mRNAs, particularly when the infected cell attempts translational shutdown to prevent viral amplification. I then discuss noncanonical mechanisms herpesviruses employ to expand and regulate their coding capacity, many of which have emerging parallels in eukaryotic gene expression control. As is often the case in virology, dissecting these virus-host interactions continues to advance our understanding of how cellular gene expression is regulated during homeostasis and how specific deregulatory events impact both infectious and noninfectious disease.
OVERVIEW OF TRANSLATION INITIATION
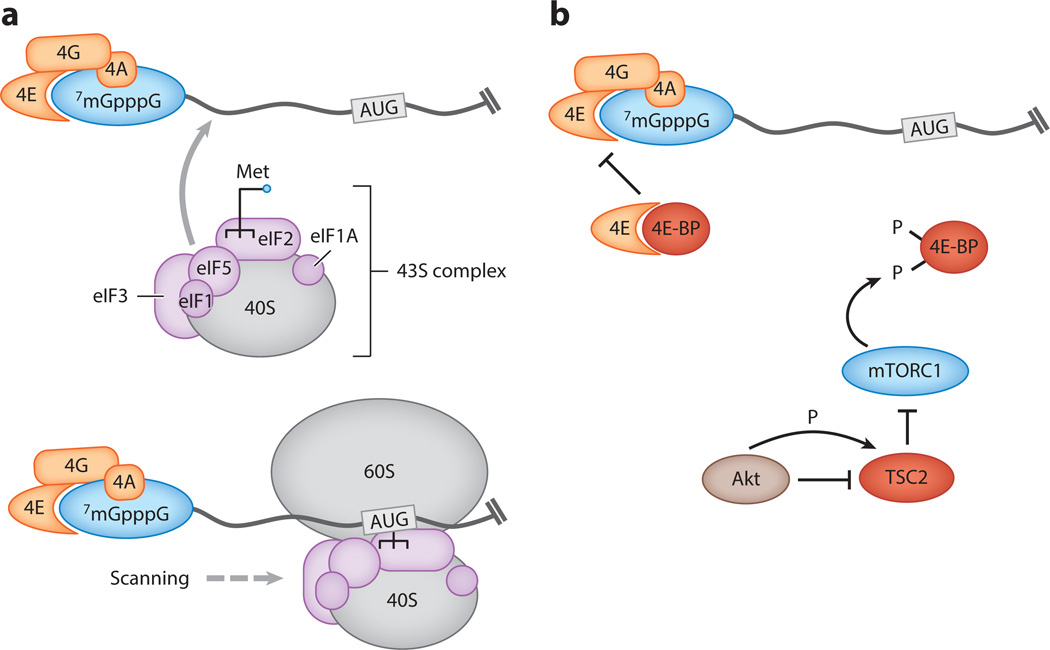
Initiation of protein synthesis from cellular and most herpesviral mRNAs involves the ordered assembly of translation factors on the mRNA cap, an event that serves as a pivotal control point for translational regulation (Figure 1a). The beginning of this assembly process involves a group of three proteins—the eukaryotic translation initiation factor 4E (eIF4E) cap-binding protein, the eIF4G scaffold protein, and the eIF4A RNA helicase—which together constitute the eIF4F complex (1). Assembly of eIF4F on the cap leads to binding of the 40S ribosomal subunit in association with additional initiation factors (eIF1, eIF1A, eIF3, eIF5, and eIF2 · GTP) that make up the 43S preinitiation complex, which is loaded with the charged initiator-methionine tRNA. The 40S subunit then scans through the 5′ untranslated region (UTR) of the mRNA in search of an appropriately positioned start codon. Following start codon recognition is recruitment of the 60S ribosome, release of initiation factors, and subsequent translation of the open reading frame (ORF) by the 80S ribosome. In addition to cap binding, the eIF4G component of eIF4F also coordinates interactions between the mRNA ends through an association with the poly(A)-binding protein (PABP), forming a looped structure that likely helps ensure recruitment of translation machinery to transcripts with intact termini.
Figure 1.
Regulation of translation initiation complex assembly. (a) Initiation begins with loading of the eIF4F complex, comprising the eIF4E cap-binding protein, eIF4G scaffold, and eIF4A helicase, onto the cap. This leads to binding of the 40S ribosomal subunit in association with the initiation factors (eIF1, eIF1A, eIF3, eIF5, and eIF2 · GTP) that make up the 43S preinitiation complex, which is loaded with the charged initiator-methionine tRNA (Met). The 40S subunit then scans through the 5′ untranslated region of the mRNA until it reaches the start codon, whereupon the 60S ribosome joins and translation of the open reading frame begins. (b) Assembly of the eIF4F complex blocked by the interaction of eIF4E with unphosphorylated eIF4E-binding proteins (4E-BPs). This inhibitory interaction is prevented by the phosphorylation of 4E-BP by the mammalian target of rapamycin kinase complex 1 (mTORC1). In turn, mTORC1 is regulated by its inhibitor TSC2, which itself is inhibited upon phosphorylation by the kinase Akt.
Translation is an intensely energy- and resource-consuming process; thus, under conditions of cell stress, including nutrient deprivation or infection, cells attempt to block eIF4F assembly to restrict translational output (1). Formation of the eIF4F complex is negatively regulated by the interaction of its eIF4E cap-binding component with unphosphorylated eIF4E-binding proteins (4E-BPs). In unstressed cells, this inhibitory interaction is antagonized by the phosphorylation of 4E-BP by the mammalian target of rapamycin kinase complex 1 (mTORC1), as phosphorylated 4E-BP1 cannot bind eIF4E (Figure 1b) (2). As described below, herpesviruses infiltrate the intricate network of positive and negative regulatory factors that control the efficiency of translation initiation, thereby facilitating expression of viral proteins under conditions of infection-induced cell stress.
VIRAL STIMULATION OF TRANSLATION COMPLEX ASSEMBLY
The importance of eIF4F assembly and active mTORC1 for viral translation is underscored by the reiterative targeting of their pathway components during herpesvirus infection (Table 1). Herpes simplex virus 1 (HSV-1) activates eIF4F assembly through at least two distinct mechanisms. HSV-1 encodes ICP6—an eIF4F stimulatory protein that binds free eIF4E, serving as a molecular chaperone to drive eIF4E incorporation into eIF4F and thereby facilitate translation (3). HSV-1 also encodes the US3 kinase, which inactivates a negative regulator of the 4E-BP kinase mTORC1, enabling constitutive mTORC1 activity. The viral US3 kinase mimics the cellular signaling kinase Akt and phosphorylates the mTORC1 inhibitor TSC2 at the same sites that Akt does, leading to TSC2 inactivation (4). This liberates mTORC1 and enables it to hyperphosphorylate 4E-BP1, thereby enhancing eIF4F assembly on viral mRNAs. Although HSV-1 infection leads to the transient activation of Akt as well (5), it is the activation of US3 that enables constitutive mTORC1 activation throughout the course of infection (4).
Table 1.
Herpesviral proteins that stimulate eIF4F assembly
| Virus | Protein | Activity |
|---|---|---|
| HSV | ICP6 | Chaperone; promotes eIF4E assembly into eIF4F |
| US3 | AKT kinase mimic; phosphorylates TSC2 | |
| HCMV | pUL38 | Binds/inhibits TSC2 |
| KSHV | vGPCR | Phosphorylates TSC2 |
| EBV | LMP2A | Activates AKT, mTORC1 |
Abbreviations: EBV, Epstein-Barr virus; HCMV, human cytomegalovirus; HSV, herpes simplex virus; KSHV, Kaposi’s sarcoma–associated herpesvirus.
mTORC1 activation, often via TSC2 inhibition, is a mechanism common to multiple other herpesviruses during infection. Examples include TSC2 inactivation upon interaction with the human cytomegalovirus (HCMV) pUL38 protein (6), phosphorylation of TSC2 by the Kaposi’s sarcoma–associated herpesvirus (KSHV) viral G protein–coupled receptor (vGPCR) protein (7, 8), and activation of Akt and mTORC by Epstein-Barr virus (EBV)-encoded latent membrane protein 2A (LMP2A) (9). Collectively, these observations suggest a central role for the mTORC kinase network in modulating viral translation.
Interestingly, there is not an absolute requirement for eIF4F during HCMV infection, as it is increasingly dispensable for the translation of viral mRNAs as infection progresses. This has been determined using Torin, a specific inhibitor of mTORC1 that results in the accumulation of unphosphorylated 4E-BP, thereby disrupting the assembly of eIF4F (10). Addition of Torin at the beginning of infection results in a robust decrease in the synthesis of viral late proteins (although reports differ on its effects during early protein accumulation) (11, 12). However, whenmTORC1 activity is blocked by Torin at later times after HCMV infection has been “established” (>48 hpi), the inhibition of viral gene expression is much less pronounced (11). These observations suggest that viral mRNA translation eventually becomes resistant to the 4E-BP1-mediated inhibition of eIF4F assembly. This hypothesis is bolstered by a recent report that treatment with Torin, or selective inhibition of the eIF4A helicase component of eIF4F using hippuristanol, is detrimental to viral gene expression when the inhibitor is added early but not when it is added late in HCMV infection (13).
It has been shown that eIF4F is preferentially required for the translation of host mRNAs that contain long, structured 5′ UTRs, as well as pyrimidine-rich elements termed TOP or PRTE elements (see sidebar, Plasticity in the Translation Complex) (14, 15). Such motifs are lacking in the 5′ UTRs of mapped HCMV mRNAs, in agreement with the observation that their translation may not be entirely eIF4F dependent. Furthermore, it should be noted that the inhibitory effect of Torin (added at the start of infection) is rescued by depletion of 4E-BP1 during murine cytomegalovirus (MCMV) infection but not during HCMV infection, indicating that the HCMV requirement for mTORC1 activity cannot be solely attributed to its ability to hyperphosphorylate 4E-BP1 (11, 12).
PLASTICITY IN THE TRANSLATION COMPLEX.
An emerging theme is that at least a subset of the canonical translation initiation factors are not broadly required for translation, but instead are needed by only select types of mRNA (14–16). This has now been clearly established for the eIF4A RNA helicase component of eIF4F, whose targets have been mapped genome wide. Inhibiting eIF4A selectively impacts transcripts with long, structured 5′ UTRs (among other features)—for example, those frequently found in growth regulatory factors and oncogenes (94, 95). Whether, for the remaining transcripts, eIF4F functions in the absence of eIF4A catalytic activity or engages alternative isoforms of eIF4F components remains unknown. In this regard, viral mRNAs that become resistant to eIF4F inhibition in a temporal manner during infection may provide a robust model to dissect eIF4F plasticity (11–13). Such discrimination between particular mRNA transcripts is not limited to the initiation machinery; it can also be observed for individual ribosomal protein subunits during RNA virus infection (122). Thus, the composition of the translational apparatus is likely to vary and may well be an important source of translational control.
How might cytomegalovirus late mRNAs be translated in the presence of the hypophosphorylated inhibitory form of 4E-BP1 or in the absence of the eIF4A helicase? One hypothesis is that late in infection viral mRNAs may recruit alternate translation factors that are eIF4E independent and thus resistant to 4E-BPs—for example, the cellular cap-binding complex, composed of CBP80 and CBP20, that is loaded onto mRNA caps in the nucleus (13). Alternatively, viral mRNAs may preferentially recruit low residual levels of active eIF4F. In this regard, HCMV infection increases production of the eIF4F components eIF4E, eIF4G, and eIF4A, as well as cytoplasmic PABP (PABPC), after 48 hpi (17, 18). In addition, HCMV may also protect viral mRNAs via its pUL69 protein, which binds viral mRNA caps through the eIF4A1 helicase and PABPC, leading to exclusion of 4E-BP1 from the cap-binding complex (19).
During KSHV infection, additional translational stimulation is achieved through the ORF45 protein, a robust activator of ribosomal S6 kinase (RSK) (20, 21). Using a targeted approach, it was shown that ORF45-mediated RSK activation leads to the phosphorylation of eIF4B (22), an accessory factor that likely assists with ribosome recruitment to mRNA and that also stimulates activity of the eIF4A helicase (23). The ORF45-RSK signaling axis phosphorylates eIF4B in KSHV-infected cells, which stimulates its interaction with the mRNA cap and increases the association of total mRNA with polysomes (22). In contrast, inhibiting this cascade correlates with reduced viral gene expression (22, 24). Although RSK has many targets, the observation that eIF4B overexpression during infection increases viral gene expression suggests that eIF4B activation through ORF45 contributes to KSHV mRNA translation.
MANAGEMENT OF CELLULAR STRESS RESPONSES
Protein Kinase R
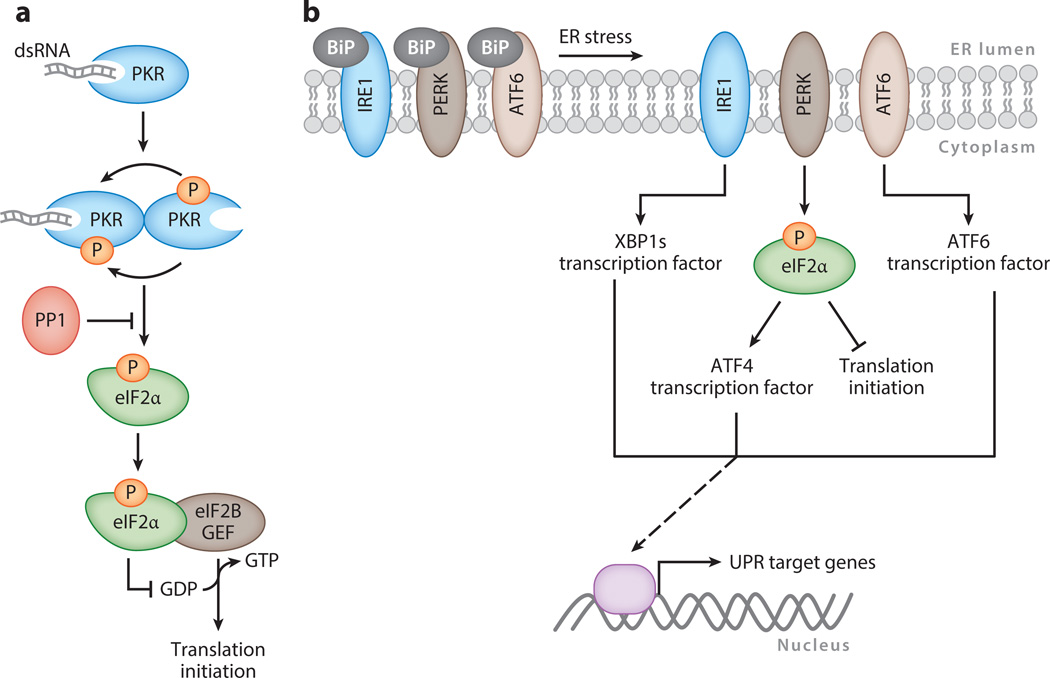
Viral infection can be viewed as an unrelenting form of cell stress, and a key host response to stress is to restrict translation by phosphorylating the eIF2α component of the eIF2-tRNAiMet-GTP ternary complex at serine 51. Phosphorylated eIF2α binds the guanine nucleotide exchange factor eIF2B, thereby inhibiting the GDP-to-GTP exchange required for translation initiation. Several stress-responsive cellular kinases can target eIF2α, although viral infection is most frequently linked to stimulation of the dsRNA-activated protein kinase [protein kinase R (PKR)] (Figure 2a). DNA viruses often produce dsRNA as a by-product of overlapping transcription from both strands of the genome, which triggers PKR activation (25, 26). Binding of dsRNA to PKR facilitates its dimerization and autophosphorylation, which significantly increases its kinase activity and induces translational arrest via eIF2α phosphorylation (27). Although not discussed extensively here, shutdown of protein synthesis during infection is often also augmented through the coincident induction of 2′,5′-oligoadenylate synthetase (OAS), which activates RNaseL to degrade mRNA and rRNA (28).
Figure 2.
Cellular stress response pathways that control translation. (a) Protein kinase R (PKR) is activated upon binding dsRNA and undergoes dimerization and autophosphorylation, which dramatically increases its kinase activity. It then phosphorylates eIF2α. Phosphorylated eIF2αbinds the guanine nucleotide exchange factor (GEF) eIF2B in a manner that blocks the GDP-to-GTP exchange required for translation initiation. The phosphatase PP1 inhibits eIF2αphosphorylation. (b) The unfolded protein response (UPR) is activated in response to endoplasmic reticulum (ER) stress. The ER-resident sensors IRE1, PERK, and ATF6 are held in an inactive state by the BiP chaperone. They are released upon ER stress, leading to translation inhibition and induction of transcription factors that induce UPR target genes.
HSV-1 encodes three mechanistically and kinetically distinct inhibitors of PKR: the RNase virion host shutoff (vhs), ICP34.5, and US11 (Table 2). Early in infection, PKR activation is blocked by the vhs RNase, which is brought in with the viral tegument. The importance of blocking PKR activity was demonstrated by the impaired replication of vhs-null viruses in cells lacking the PKR inhibitor mitogen-activated protein kinase kinase (MEK), which have increased PKR activity (29). Transient expression of wild-type, but not catalytically inactive vhs is sufficient to reduce PKR phosphorylation (29), further demonstrating the potent role of vhs in downregulating PKR activity. Although it remains unknown how vhs blocks PKR activity, its action could be linked to the widespread degradation by vhs of viral mRNAs with the capacity to form dsRNA, or to the degradation of cellular mRNAs whose protein products potentiate PKR activation.
Table 2.
Herpesviral proteins that counteract or usurp components of the PKR and UPR pathwaysa
| Pathway | Virus | Protein | Activity |
|---|---|---|---|
| PKR | HSV-1 | vhs | Degrades mRNA; proposed targets include viral mRNAs that could form dsRNA and/or cellular factors involved in PKR response |
| HSV-1/PRV | ICP34.5/IE180 | Directs the PP1 phosphatase to dephosphorylate the PKR target eIF2α | |
| HSV-1 | US11 | Binds PKR and prevents its phosphorylation | |
| HCMV | TRS1/IRS1 | Bind PKR and induce its nuclear relocalization; prevent cytoplasmic PKR phosphorylation (proposed) |
|
| MCMV | M142/M143 | ||
| EBV | EBER1/EBER2 | Short noncoding RNAs that bind PKR and prevent its activation | |
| UPR | HCMV | UL38 | Activates ATF4, which induces genes involved in UPR recovery and translation; suppresses IRE1-mediated JNK phosphorylation to limit apoptosis |
| MCMV | M50 | Binds IRE1 and induces its degradation | |
| EBV | LMP1 | Activates PERK, leading to ATF4 induction (LMP1 promoter is ATF4 responsive) |
|
| HSV-1 | gB | Inhibits PERK |
See text for additional examples that have not yet been ascribed to particular viral proteins.
Abbreviations: EBV, Epstein-Barr virus; HCMV, human cytomegalovirus; HSV-1, herpes simplex virus 1; MCMV; murine cytomegalovirus; PKR, protein kinase R; PRV, pseudorabies virus; UPR, unfolded protein response.
At the transition between early and late gene expression, vhs is inactivated and the HSV-1 ICP34.5 protein instead blocks PKR. Rather than inhibiting the initial PKR activation, ICP34.5 promotes dephosphorylation of the PKR target eIF2α via the cellular protein phosphatase PP1 (30). PP1 has multiple cellular targets, but ICP34.5 binds both PP1 and eIF2α and serves as a bridge to specifically direct the phosphatase to eIF2α via their respective binding motifs (31). The PP1-linked inhibition of eIF2α phosphorylation is also a strategy employed by the IE180 protein during infection with the alphaherpesvirus pseudorabies virus (32). Finally, at late stages of HSV-1 infection, the US11 protein binds PKR in an RNA-dependent manner and prevents its phosphorylation. US11 also inhibits the RNaseL activator OAS (33).
During HCMV or MCMV infection, PKR is counteracted by the virally encoded dsRNA-binding proteins TRS1 and IRS1 or M142 and M143, respectively (34, 35). HCMV IRS1 and TRS1, expressed from inverted repeats, exhibit significant sequence similarity, and the expression of only one of the two encoded proteins is necessary to counteract PKR during HCMV infection (36). IRS1 and TRS1 are identical over much of their N-terminal regions, which contain their RNA-binding domain (37), but use their more divergent C termini to interact with PKR and induce its nuclear relocalization (38). Although nuclear sequestration of PKR is one probable mechanism by which these proteins block its activation, the observation that the remaining cytoplasmic pool of PKR is largely unphosphorylated suggests that additional inhibitory mechanisms are also in place (38). Both HCMV proteins can also inhibit OAS activation, although its RNaseL target still remains inactive in a TRS1/IRS1 double mutant, suggesting TRS1 and IRS1 are not the sole proteins involved in counteracting RNaseL (36, 39). Indeed, HCMV ORF94 has been shown to dampen OAS expression as well and thus might compensate for this phenotype in cells lacking TRS1 or IRS1 (40).
The best-characterized PKR inhibitors during gammaherpesvirus infection are a set of RNA polymerase III–transcribed noncoding RNAs expressed during both latent and lytic EBV infection, termed EBER1 and EBER2. The EBERs form ~160-nt stem-loop structures that bind PKR with similar affinity as its dsRNA activators, yet they prevent subsequent PKR activation in cell-free systems (41–43). However, the contribution of EBERs toward PKR inhibition during EBV infection remains unclear, as during latency they cannot counteract IFN-α-induced PKR activation (44). Other PKR inhibitory proteins are also encoded by EBV (EB) and KSHV (vIRF2), both of which have been shown to bind PKR in vitro and inhibit its kinase activation (45, 46).
The Unfolded Protein Response
Another cell stress response commonly activated during infection that impacts protein production is the unfolded protein response (UPR) (Figure 2b). The UPR is governed by three endoplasmic reticulum (ER) lumen–resident sensors: PKR-like ER kinase (PERK), activating transcription factor 6 (ATF6), and inositol-requiring enzyme 1 (IRE1) (47). These sensors are held in an inactive state by the BiP/GRP78 chaperone until their release during ER stress. Upon release, they coordinate to inhibit accumulation of improperly folded proteins in the ER lumen—a common occurrence during infection when protein production outpaces ER capacity. Similar to PKR, activated PERK phosphorylates eIF2α to restrict new translation and, together with IRE1, upregulates or activates the transcription factors ATF4, ATF6, and XBPs. Collectively, these factors induce production of ER chaperones as well as enzymes that reduce oxidative stress and mediate ER-associated protein degradation (47).
During infection, when the ER is inundated with an influx of viral proteins and ER stress is insurmountable, the UPR will drive cells into apoptosis. To permit cell survival, viruses must counteract the apoptotic or translational inhibitory functions of the UPR, yet strive to benefit from activities that augment ER function (Table 2). These activities have been most extensively studied for cytomegaloviruses. Components of all three UPR sensor pathways are activated during infection with HCMV and MCMV, yet in a manner that leads to atypical downstream consequences that favor the viral life cycle (48, 49). For example, only a subset of the genes normally induced by the UPR-responsive transcription factors ATF6 and XBP1 are upregulated; in general, these include genes encoding proteins such as chaperones that could assist with viral protein production. Furthermore, PERK activation during HCMV infection causes only limited eIF2α phosphorylation and no global translational attenuation, whereas the HCMV pUL38 protein activates the PERK-responsive ATF4 transcription factor and induces host genes important for UPR recovery and translation (48, 50). HCMV pUL38 further promotes cell survival through the suppression of IRE1-mediated phosphorylation of the stress-activated c-Jun N-terminal kinase (JNK), which is linked to ER stress–induced apoptosis (50). The IRE1 pathway is also inhibited during MCMV infection through binding of the viral M50 protein to IRE1 in a manner that induces IRE1 degradation (51). Finally, HCMV both transcriptionally and translationally induces the ER chaperone BiP/GRP78, which enables proper virion assembly and egress (52, 53).
EBV provides another example of viral UPR manipulation. During latent EBV infection, the oncogenicLMP1protein activates PERK (as well asATF6and IRE1), leading to subsequent eIF2α phosphorylation and induction of the transcription factor ATF4 (54). Although this presumably attenuates translation, the viral LMP1 promoter itself is ATF4 responsive, and thus LMP1 is selectively upregulated. Interestingly, during lytic reactivation of EBV, the UPR-responsive transcription factor XBP1 transcriptionally induces the R and Z immediate early promoters of EBV and is required for constitutive lytic gene expression (55). Thus, EBV hijacks several of the transcriptional regulatory activities induced by the UPR to drive viral gene synthesis.
Additional documented examples of herpesviruses interfacing with the UPR include inhibition of PERK by HSV-1 gB (56) and activation of PERK and IRE1 by varicella zoster virus (VZV) (57).
REPROGRAMMING OF THE TRANSLATIONAL REPERTOIRE DURING INFECTION
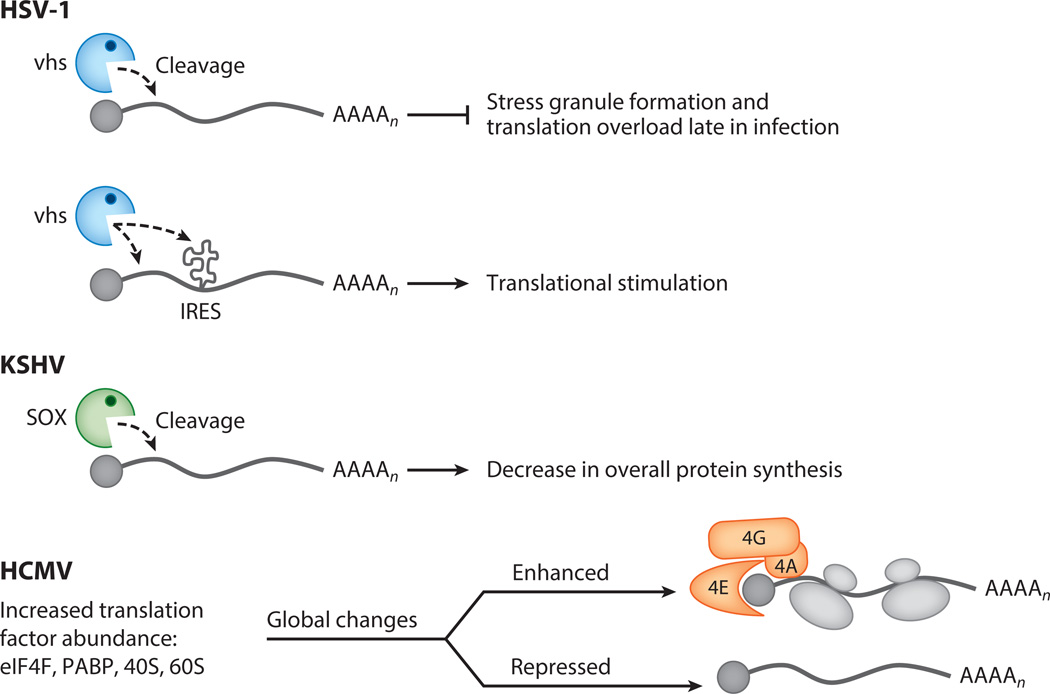
Beyond altering the cellular response to stress, herpesviruses possess a number of additional strategies to dramatically reshape the translational landscape. Lytic infection with alpha- and gammaherpesviruses leads to widespread decreases in cytoplasmic mRNA abundance due to expression of virally encoded mRNA-specific endonucleases—the alphaherpesvirus nuclease vhs and the gammaherpesvirus nuclease SOX (Figure 3). Although nonhomologous, these nucleases share several key features, including broad targeting of both viral and cellular mRNAs for endonucleolytic cleavage and ensuing degradation of the intermediates by the cellular Xrn1 exonuclease (58). This global shutoff of cellular gene expression has generally been presumed to facilitate viral access to translation machinery and dampen immune sensing. However, the observation that most viral transcripts do not escape cleavage during infection with the alphaherpesvirus HSV-1 or the gammaherpesvirus murine herpesvirus 68 (MHV68) has made it difficult to predict the role of widespread mRNA degradation in viral translation. Interestingly, for both alpha- and gammaherpesviruses, the effects are indeed profound but distinct.
Figure 3.
Herpesvirus infection alters the global translational landscape of an infected cell. During herpes simplex virus 1 (HSV-1) and Kaposi’s sarcoma–associated herpesvirus (KSHV) infection the pool of mRNAs available for translation is significantly reduced due to mRNA degradation by the viral endonucleases vhs and SOX, respectively. In addition, vhs has the ability to stimulate translation of select mRNAs, such as those containing internal ribosome entry site (IRES) elements, in a manner independent of its mRNA degradation function. During human cytomegalovirus (HCMV) infection, translation is broadly impacted through increased abundance of multiple translation factors. This leads to both large-scale enhancement and repression of mRNAs associated with polysomes.
In the case of HSV-1 vhs, a clear link has been established between mRNA degradation and liberation of translational machinery. Infection with vhs-null HSV-1 causes the induction of stress granules that accumulate stalled translation initiation events, and causes a specific defect in the accumulation of viral late gene products independent of eIF2α phosphorylation (59, 60). This late gene–specific defect is not linked to the nature or structure of the 5′ leader sequence; instead, late viral mRNAs are excluded due to overloading of the translational machinery in the absence of vhs-imposed control of mRNA accumulation early in infection (61). This effect is cell type dependent, suggesting variations in translational capacity and viral requirements to regulate mRNA abundance between cell lineages. Interestingly, HSV-1 vhs also participates in translational regulation in ways independent of its mRNA-degrading activity. Although the underlying mechanisms remain unknown, vhs boosts the translation of certain types of internal ribosome entry site (IRES)-driven genes expressed in a bicistronic context, as well as viral mRNAs with select 5′ leader sequences, independent of mRNA levels (62).
Degradation of host and viral mRNAs is similarly widespread during infection with the gammaherpesvirusMHV68 through the activity of its SOX ortholog. However, infection of cells with an MHV68 mutant with impaired SOX activity does not dampen late gene translation (63). Instead, increased viral mRNA levels in the absence of SOX-induced degradation lead to a corresponding increase in protein levels for most genes tested. Surprisingly, this increase is ultimately detrimental to the viral life cycle, as it skews viral protein composition in progeny virions as well as alters cell surface binding and immediate early gene expression during the subsequent round of replication. The predominant outcome of gammaherpesvirus infection in vivo is latency, yet failure to regulate protein abundance through mRNA degradation during the lytic cycle appears to shift this balance in favor of lytic cycle entry, at least in cultured cells (63, 64). The SOX mutant virus also displays replication defects in multiple cell types and in vivo, suggesting that the ability to fine-tune viral protein abundance via mRNA degradation is key to gammaherpesvirus replication (63).
One potential factor contributing to the distinct phenotypes of vhs- versus SOX-mutant viruses is the kinetics of viral replication. HSV-1 replicates significantly faster than MHV-68 and may thus impose a more immediate burden on the cellular translational apparatus. Alternatively, the preferred cell types for these viruses may possess distinct translational capacities, causing the viruses to evolve divergent needs for translational control through mRNA degradation.
Unlike infection with members of the other herpesvirus subfamilies, infection with the beta-herpesviruses does not impose a generalized reduction in protein synthesis but instead stimulates translation by increasing translation factor abundance and eIF4F assembly. Mohr and colleagues (65) recently revealed the surprising extent to which these phenotypes impact the global translational landscape in an infected cell (Figure 3). One notable finding was that HCMV infection enhances polysome recruitment of mRNAs involved in ribosome biogenesis and correspondingly increases 40S and 60S ribosome subunit concentrations. Remarkably, polysome profiling coupled to microarray analysis showed that this does not lead to a generalized increase in cellular mRNA translation, but instead results in large and selective alterations to the cellular translational repertoire (65). Nearly equal numbers of cellular mRNAs in the analysis were translationally repressed as were translationally enhanced, and each group was enriched for products with specific functions, many linked to processes important for viral replication or antiviral responses. Furthermore, this HCMV-impacted gene set significantly overlapped with genes regulated by pathophysiological states such as cancer, a disease with similar increases in translation factor abundance. Many of the HCMV-imposed changes to the mRNA polysome profile could be linked to the activity of the viral mTORC activator pUL38 (65), although HCMV possesses additional translational control strategies as well.
How is HCMV infection–induced translational selectivity achieved? One possibility is the influence of HCMV infection on the activity or expression of specific sets of translation factors. For example, HCMV-induced increases in eIF4E availability and eIF4F assembly correlate well with increases in the polysome association of mRNAs whose 5′ leader sequences are shorter than average and/or contain TOP elements (65, 66). In addition, host-imposed antiviral responses may affect the polysome association of other mRNAs. A surprising example is cellular eIF6, whose translational enhancement was shown to restrict HCMV gene expression and replication, in line with observations indicating its overexpression inhibits 60S joining (65, 67).
VIRAL TRIUMPH OVER THE CELLULAR INTRON ADVANTAGE
Herpesviral mRNAs closely resemble host cell mRNAs, with the important exception that relatively few viral mRNAs contain spliced introns. Splicing plays central roles in mRNA nuclear quality control and export, as well as in the deposition of the exon junction complex (EJC), several components of which are linked to translational stimulation (68). These gene expression challenges associated with unspliced transcripts are at least partially resolved by a protein conserved across the herpesvirus subfamilies (named ICP27 in HSV-1, pUL69 in HCMV, EB/SM in EBV, and ORF57/MTA in KSHV). Here, I focus on the translational stimulatory activity of these orthologs, which is generally distinct from their functions in the nucleus (69–72). Interestingly, these orthologs do not stimulate translation via a broad-acting mechanism but are restricted to a subset of intronless mRNA targets. This processing distinction may offer an opportunity for the viruses to selectively enhance translation of transcripts common in herpesviruses but rare in the host.
ICP27, EB, pUL69, and ORF57 cosediment with polysomes in infected cells and associate with translation factors, in agreement with their role in facilitating translation of viral mRNAs (19, 69, 71–73). In the case of KSHV, ORF57-stimulated translation requires the interaction of ORF57 with the cellular protein PYM (69). In uninfected cells, PYM helps recruit the 40S ribosome to spliced mRNAs via its interactions with the EJC components Y14 and Magoh (74, 75). However, when bound by ORF57, PYM is recruited to intronless mRNAs in the absence of other EJC components, where it is proposed to act as a bridge between the 40S ribosome and other mRNA-bound translation factors such as PABPC, eIF4A, and eIF4G (69). As described above, during HCMV infection, translation is stimulated by pUL69 through its interactions with eIF4A1 and PABPC, which lead to eviction of the translational inhibitor 4E-BP1 from the cap-binding complex (19). BothHSV-1 ICP27 and EBV EB also enhance translation of unspliced viral mRNAs (70, 72, 76), although whether this occurs through mechanisms similar to those used by HCMV or KSHV remains unknown.
The assumption in each of the cases mentioned above is that translational enhancement requires a direct association of a viral protein with its target mRNA. For example, binding-induced enhancement has been demonstrated for ICP27 using theMS2 tethering system (71). That said, an unresolved question is how specific mRNAs are targeted, as not all viral transcripts undergo translational enhancement in the presence of these proteins. It is also interesting that the translational stimulatory effects of EBV EB, HSV-1 ICP27, and HCMV pUL69 appear most pronounced for genes expressed with late kinetics (19, 70, 72). This may have evolved because many late genes encode viral structural components that must be produced at high levels for robust particle assembly. However, in a yeast three-hybrid screen of an HSV-1 genomic library, ICP27 was shown to associate with a wide array of viral mRNAs from all kinetic classes, with no conserved sequence motif apparent on bound transcripts (77).
It is unclear whether KSHV ORF57 displays a more robust translation phenotype on late mRNAs, although the set of genes whose mRNA abundance is most ORF57 dependent does not cluster in a particular kinetic class (78). Furthermore, like ICP27, the ORF57 mRNA binding profile is not restricted to late transcripts. An initial UV cross-linking and immunoprecipitation (CLIP) analysis established an association between KSHV ORF57 and at least 11 KSHV mRNAs (79). This number has been expanded by a recent high-throughput HITS-CLIP analysis, which showed >200 clusters of ORF57-bound sequences across the KSHV genome, as well as sequence clusters within >700 cellular genes (80). ORF57 binding was not limited to intronless mRNAs, and a subset of cellular binding sites were enriched at the boundaries of the 5′-most exon-intron junction. For a number of these interactions, ORF57 binding impacts nuclear processing and target stability (80), which may similarly be the case for ICP27 target binding, as both orthologs have established functions in nuclear mRNA processing. Whether ORF57 or ICP27 remains stably associated with most of these targets after their export into the cytoplasm is unknown. Thus, the extent of correlation between their translational enhancement phenotype and mRNA target binding is an issue for future study.
REGULATION OF SELF-SYNTHESIS AND RECODING
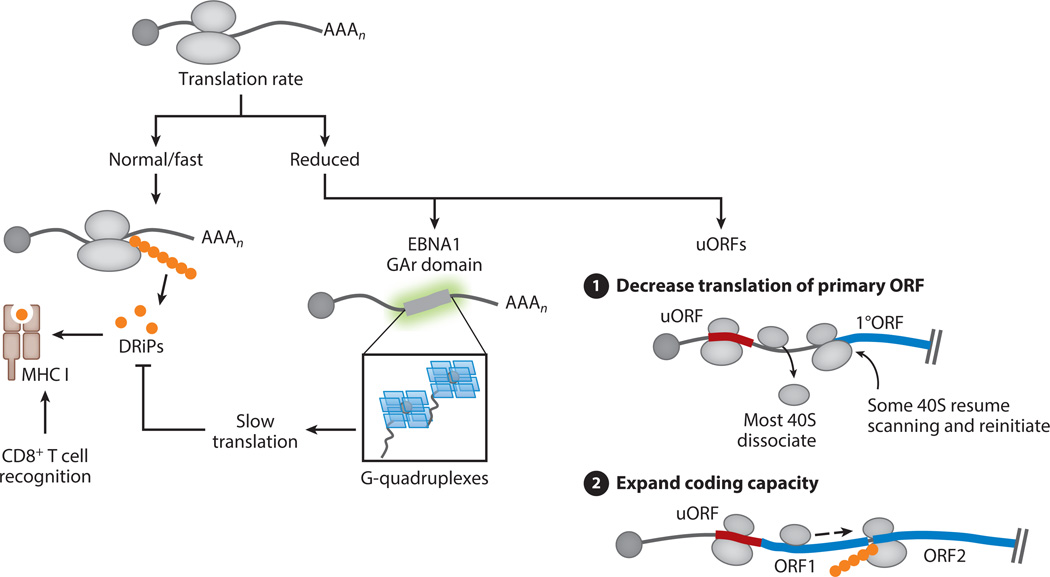
All herpesviruses undergo cycles of both latency and lytic replication. A key feature of viral latency is long-term persistence while avoiding the sentinel of circulating immune cells. This presents a challenge for the latent virus, as viral genome maintenance often requires the presence of at least one viral protein, whose peptide presentation on major histocompatibility complex (MHC) I at the cell surface would be a source of epitopes for CD8+ T cell recognition. Although the stability of many viral proteins might preclude production of epitopes on a time and abundance scale necessary for robust presentation and detection, evidence suggests that the majority of antigenic MHC I peptides may be derived from rapidly degraded defective ribosomal products (DRiPs) generated during translation (81–83). The production of DRiPs has been linked to high protein translation rate (84); therefore, one strategy to avoid DRiP-mediated antigen presentation would be to maintain tight translational control over latently expressed proteins (Figure 4). A fascinating example is illustrated by the regulated self-synthesis of the EBV-encoded latency factor EBNA1.
Figure 4.
Controlling the rate of translation as a means to impact antigen processing and regulate viral gene expression. Many antigenic MHC I peptides are generated from rapidly degraded defective ribosomal products (DRiPs) made during translation. Viral proteins may avoid DRiP presentation by reducing their translation rate. The EBNA1 latency protein of Epstein-Barr virus contains a glycine-alanine repeat (GAr) domain that folds into G-quadruplex structures, which decrease EBNA1 translation and correlate with reduced antigen presentation. Viral protein translational efficiency can also be controlled by the presence of short upstream open reading frames (uORFs). (❶) uORFs decrease the translation rate of the primary ORF (1°ORF) by engaging scanning ribosomes, enabling only a subset to reinitiate at the downstream ORF. (❷) uORFs have also been shown to increase viral coding capacity by enabling ribosomes to bypass the 5′ primary ORF and reinitiate at a downstream internal ORF, rendering mRNAs functionally polycistronic. Rapidly degraded peptides produced from uORFs may also be a source of DRiPs.
EBNA1 is essential for viral genome maintenance and is expressed in all EBV-associated malignancies. An unusual feature of EBNA1 is its overrepresentation of purines, particularly within its internal glycine-alanine repeat (GAr) domain. The GAr domain potently restricts EBNA1 translation, as well as translation of reporter mRNAs to which it is appended in any reading frame, suggesting a cis-acting mRNA-based mechanism (85–87). Ribosome footprinting experiments demonstrated that GAr-encoding mRNAs do not display a reduced accumulation of ribosomes at the initiator AUG, but instead, translation prematurely terminates within the 5′ region of the mRNA (88). These and other findings pointed to a role for RNA structure in mediating the effects of GAr-encoding domains, and these sequences were recently shown to adopt unusual G-quadruplex structures (89).
G-quadruplexes are four-stranded, stacked guanine tetrads formed by the coplanar arrangement of four G bases stabilized by Hoogsteen hydrogen bonding. The role of G-quadruplexes in mediating GAr-encoding domain function was confirmed by the demonstration that destabilizing G-quadruplexes with targeted antisense oligonucleotides relieved translational repression (89). In contrast, stabilizing G-quadruplexes using the small molecule pyridostatin further inhibited EBNA1 translation both in vivo and in vitro, presumably by steric hindrance of the ribosome (89). In addition, many studies have linked the presence of the GAr-encoding domain to restricted antigen presentation of EBNA1 (85, 90), and recent findings link this phenotype to the formation of the GAr domain G-quadruplex, showing that this structure impacts EBNA1-specific CD8+ T cell epitope production (89) as well as priming of antigen-specific T cells in vivo (Figure 4) (91).
In addition to EBNA1, other gammaherpesvirus mRNAs that encode latency proteins with purine-rich sequence stretches cause similar phenotypes (92, 93), and computational analyses predict a number of these stretches may similarly possess G-quadruplex structures (89). Thus, studies on G-quadruplexes and how they modulate viral and cellular mRNA expression may lead to the development of small molecule G-quadruplex inhibitors to enhance immune-based killing of latently infected cells. In this regard, recent studies have defined the subset of mRNAs whose translation requires the eIF4A helicase; among their hallmark features are long, structured 5′ UTRs and computationally predicted G-quadruplex structures (94, 95). These structures were also found to cluster in many cellular oncogenes and transcriptional activators, perhaps explaining the ability of eIF4A expression to contribute to oncogenesis in a T cell acute lymphoblastic leukemia model (94, 95). Given that EBV- and KSHV-induced oncogenesis is primarily driven by cellular changes during latency, analyzing the contribution of eIF4A toward viral latency antigen expression and viral persistence should be of significant interest.
In addition to translational attenuation, the repeat region of transcripts encoding EBNA1 and the major KSHV latency protein LANA1 have recently been shown to cause remarkably efficient protein recoding through programmed ribosome frameshifting and alternate initiation (96, 97). LANA contains an internal repeat region nearly identical to that of EBNA1 at the nucleotide level, but positioned in a different reading frame, generating alternate reading frame (ARF) isoforms with serine/arginine repeats similar to those found in neurodegenerative disorders (98). EBNA1 also undergoes recoding to produce an ARF with a repeat region similar to the major LANA isoform (96). Both EBVARF and KSHVARF can be detected during infection by using reporter assays, and some of these isoforms display distinct subcellular localization (96, 97, 99). Whether these isoforms exhibit altered functionality or contribute to viral oncogenesis remains an important future question. Regardless, recoding provides an interesting example of a viral strategy to expand coding capacity without increasing genome size. These viral latency factors may also provide valuable models to study the recoding that may similarly occur in repeat-containing human genes associated with hereditary disorders.
Additional translational regulation during infection can occur through the use of IRES elements. These complex RNA structures recruit subsets of translation initiation factors and the 40S ribosome in a cap-independent manner. The subset of translation factors assembled on an IRES varies depending on the RNA sequence and structure. However, given the cap-independent nature of IRES-driven translation, nearly all characterized IRES elements bypass the need for the complete set of eIF4F complex components. A notable exception to this observation has recently been documented for one of the latency-associated proteins of KSHV, termed vFLIP. The vFLIP protein, which is involved in inhibiting FAS-induced apoptosis through the activation of NF-κB (100, 101), is translated as a downstream gene from a polycistronic mRNA by an IRES element located within the coding region of the upstream gene (102–104). The vFLIP IRES was shown to recruit the complete eIF4F complex, and that translation required eIF4A, as well as a functional eIF4E cap-binding protein and its ability to interact with full-length eIF4G (105). Based on data from a combination of RNA binding assays and translation inhibitors, the authors proposed that eIF4F is recruited to the vFLIP IRES via the direct interaction between the IRES, the 40S ribosome subunit, and eIF3, and that this interaction tethers ribosomes to the mRNA (105). These observations expand the known functional requirements for IRES elements and support additional activities for the cap-binding protein in translational stimulation (106).
EXPANSION AND REGULATION OF VIRAL CODING CAPACITY THROUGH UPSTREAM AND SMALL OPEN READING FRAMES
The combined application of ribosome profiling, high-throughput mass spectrometry, and genome and mRNA sequencing during infection has yielded dramatically expanded and detailed maps of HCMV, KSHV, and EBV gene expression (99, 107, 108). In particular, ribosome profiling has enabled quantification of ongoing translation at single-nucleotide resolution through the sequencing of ribosome-protected mRNA footprints (109). This technique was first applied in virology to HCMV, revealing a remarkable 751 translated ORFs within its ~240-kb genome—more than double previous estimates of its coding capacity (108). Similarly, ribosome profiling during KSHV infection identified 50 new translated genome segments, expanding the coding repertoire of the virus by 45% (107).
How were all these ORFs missed in previous annotations, and how might viruses, with their genome size constraints, encode such an abundance of distinct proteins? The simple explanations involve size and context: Classical and in silico annotations generally recognize ORFs that initiate with the canonical AUG start codon and are a minimum of 100 amino acids long. Yet, in most cases, novel translation products are much shorter, do not necessarily initiate with an AUG, and derive from regions of the genome considered noncoding; they may be embedded in 5′ leader regions (UTRs) of previously annotated mRNAs (termed upstream open reading frames, or uORFs) or within noncoding RNAs (107, 108). Thus, viral genomes are extraordinarily translationally dense, having expanded their peptide repertoire through the incorporation of a multitude of short ORFs.
The clear challenge now is to determine whether and how these short translation products contribute to the viral life cycle, especially given that they tend to be extremely labile and thus have low steady-state abundance. Several recent reports have ascribed functional relevance to at least a subset of these “peptide” ORFs. For example, one short ORF embedded in a KSHV RNA (previously annotated as noncoding) is antisense to the major lytic transactivator RTA (110). This 45 amino acid ORF, termed viral small peptide 1 (vSP-1), binds a region of RTA involved in mediating its ubiquitin-linked proteasomal degradation, thereby stabilizing the RTA protein (110). An additional example is a uORF in the 5′ leader sequence of the HCMV gp48 gene, whose encoded peptide attenuates gp48 translation by inhibiting termination at its own stop codon, thereby preventing leaky scanning by the 40S ribosomal subunit to the gp48AUG (111).
In the majority of cases, however, it may be the act of translating these short ORFs, rather than their encoded proteins, that is of functional relevance. This is most clearly illustrated by the uORFs. These are generally >20 codons long and are present in many cellular mRNAs, where they are proposed to function as translational “speed bumps” to decrease protein production from the major coding regions of genes. After translation of a uORF, ribosomes have the potential to reengage, usually with reduced efficiency, at a downstream AUG (112, 113). Reinitiation is presumably enabled because uORFs are sufficiently short that not all of the initiation factors have dissociated by the time ribosomes have reached the stop codon. Thus, after termination and release of the 60S ribosomal subunit, the 40S subunit can continue to scan, reacquire the eIF2-tRNAiMet-GTP ternary complex, and reinitiate at a downstream start codon. uORFs are extremely abundant and translationally engaged during HCMV and KSHV infection, implicating them as a major source of translational regulation for many viral genes (107, 108). Evidence indicates that this regulation can occur in at least two ways: (a) by temporally controlling viral translation and (b) by enabling ribosomes to access internal “full-length” ORFs on polycistronic mRNAs (Figure 4).
Temporal control is suggested by the primarily late engagement of uORFs during HCMV and KSHV infection (107, 108). For HCMV, this is at least partially due to the increased inclusion of uORF-containing 5′ leader sequences on several viral mRNAs during the course of infection (108). However, uORF-linked temporal control of translation may occur even in cases in which uORF inclusion is constitutive. This idea is based on evidence from the well-characterized GCN4 mRNA in Saccharomyces cerevisiae, whose translation is regulated by a series of uORFs that are differentially engaged during times of nutrient deprivation, depending on the level of eIF2α phosphorylation (114, 115). When eIF2α is unphosphorylated, the ternary complex is abundant and ribosomes that resume scanning after translation of the 5′ uORF have sufficient time to charge with eIF2 and reinitiate, either at a subsequent uORF (thus further decreasing translation of the primary ORF) or at the primary ORF. However, when eIF2α is phosphorylated (common during the course of infection), it sequesters the guanine nucleotide exchange factor eIF2B, thus limiting the pool of ternary complexes and increasing the distance the 40S subunit may need to scan before acquisition of the ternary complex (1, 116). Therefore, translation of the primary ORF could be impacted, either positively or negatively, during the course of infection depending on multiple factors: the distance between the primary ORF and the uORF, the number of uORFs present in the 5′ leader sequence, and the cellular eIF2α phosphorylation status. Currently, there is only a very limited understanding of the mechanisms and consequences of this type of fine-tuning of viral protein levels during infection.
A second function of uORFs may be to expand viral coding capacity by enabling translation of multiple functional proteins from a single mRNA. In this regard, the ability of uORFs to render mRNAs functionally polycistronic has been documented for the KSHV ORF35–37 locus. Translation of both ORF35 (which encodes a protein of unknown function) and the downstream ORF36 (which encodes the viral protein kinase) occurs from the same mRNA, whereas ORF37 is expressed from a distinct monocistronic transcript (117, 118). The polycistronic mRNA contains two uORFs within its 5′ mRNA leader sequence, the second of which (uORF2) overlaps with the ORF35 start codon. Although ORF35AUG is flanked by a strong Kozak consensus sequence, translational engagement of the overlapping uORF2 allows ribosomes to frequently bypass ORF35AUG and reinitiate at the downstream ORF36 gene (117). Translation of the 5′-most uORF1 further reduces ribosome engagement at ORF35AUG, resulting in fairly balanced initiation rates for both ORF35 and ORF36 from the same mRNA by leaky scanning and termination-reinitiation mechanisms, respectively (118).
While the ORF35/36 case is one of the few documented examples of a uORF enabling polycistronic translation, it is possible that a similar mechanism drives expression of multiple proteins from other uORF-containing viral mRNAs or at least a subset of the ~4,000 human mRNAs containing uORFs that overlap a primary ORF (8, 23–27). In this manner, uORFs have clear potential to expand the coding capacity of viral and cellular transcriptomes, as has been documented for C/EBPα and C/EBPβ protein isoforms and the innate mitochondrial antiviral signaling (MAVS) immune regulator (119, 120). An intriguing hypothesis is that the translation machinery may increasingly engage uORFs during pathogenic stress, thereby producing “hidden” isoforms of proteins involved in antiviral or stress responses. This possibility may be resolved by future experiments to monitor how uORF usage across the mammalian and viral transcriptomes is impacted during infection and other types of stress.
Regardless of whether viral peptides encoded by uORFs and short ORFs display individual functions, the mere act of their production likely has antigenic consequences. Similar to DRiPs, small peptide ORFs tend to be rapidly degraded and thus may serve as an important source of MHC I cargo. Indeed, it has recently been shown that human T cells from HCMV-positive, but not HCMV-negative, donors exhibit robust immune responses to several short ORFs embedded within the HCMV beta 2.7 “noncoding” RNA (121). Therefore, these peptides are produced during HCMV infection in humans and expand the range of antigenic epitopes displayed by infected cells.
CONCLUDING REMARKS
Herpesviruses have evolved a remarkable diversity of mechanisms to infiltrate the cellular translational control network. Yet, fundamental questions remain regarding how these changes are driven and what their ultimate consequences may be for the infected cell. In each of the translational enhancement strategies described above, the issue of selectivity is still poorly understood. Aside from a reduced frequency of splicing events, herpesviral mRNAs do not contain obvious features that distinguish them from host transcripts. Even in the case of unspliced mRNAs, ribonucleoprotein complexes that might differentiate spliced cellular transcripts from unspliced viral transcripts would be largely stripped off during an initial pioneering round of translation. Therefore, how do viruses control the translational environment to favor their infection? The answer to this question likely involves a complex series of events whose composite outcome creates a translational landscape that benefits the virus. The relatively slow replication time of many herpesviruses and their persistence during latency would indicate that rates and abundances of cellular mRNA translation are fine-tuned rather than severely blunted to ensure cell survival for the duration of infection. Furthermore, an ongoing challenge is to determine whether and how herpesviral mRNAs (of any subfamily) can be structurally or chemically distinguished from the bulk cellular mRNA pool to facilitate their robust translation late in infection.
One emerging area of research that may shed light on this issue is the role of alternative ribosome subunits in translation of particular subsets of mRNA. The fact that cap-dependent translation of some RNA virus proteins requires specialized ribosome subunits highlights how cellular translation factors can selectively impact translation of specific mRNAs (122). Deciphering how this functional plasticity is used by the cell as part of its antiviral defense arsenal, as well as how viruses have evolved to reorganize the translation factor repertoire to favor their own replication, will be an exciting challenge for the future.
SUMMARY POINTS.
Assembly of the eIF4F complex is a key point of translational regulation during herpesvirus infection, as evidenced by the diversity of mechanisms these viruses have evolved to ensure eIF4F activity.
Infection with each of the three herpesvirus subfamilies causes large-scale alterations in the pool of mRNAs undergoing translation. This occurs through manipulation of signaling pathway components including those involved in stress responses, increasing the abundance of translation initiation factors and ribosome subunits, and modulating host and viral mRNA abundance through virally encoded mRNA endonucleases.
Most viral mRNAs are intronless but are efficiently translated due to the translational stimulatory activity of viral proteins with specificity for unspliced transcripts.
Many beta- and gammaherpesvirus latency protein–encoding mRNAs possess structural features such as G-quadruplexes that reduce the speed of translation, which correlates with a decrease in the abundance of viral peptides available for antigenic presentation by MHC I.
Ribosome profiling experiments, coupled with transcriptomics and mass spectrometry, have revealed a striking abundance of previously undocumented, but translated, ORFs in KSHV and HCMV. The majority of these are <100 amino acids long, and they may impact the translational regulation of longer ORFs.
FUTURE ISSUES.
How do the structure and sequence composition of viral mRNA 5′ UTRs impact their requirement for specific translation factors, including eIF4F, and why do some viral mRNAs become resistant to eIF4F depletion late in infection?
Do herpesviral mRNAs recruit or require specialized ribosomal proteins for translation, as has been observed for some RNA viruses?
What features of viral intronless mRNAs enable their specific translational enhancement by proteins such as KSHV ORF57, EBV EB, HSV-1 ICP27, and HCMV pUL69?
How do the myriad of uORFs mapped across herpesvirus genomes affect translational regulation of the downstream ORF, and is the regulation temporally controlled and related to cellular stress responses?
What functions, if any, can be assigned to the translated short ORFs in KSHV and HCMV—particularly those embedded within larger coding regions or in RNAs previously annotated as noncoding?
To what extent do the widespread changes in the cellular translational landscape induced during herpesvirus infection impact viral biology and pathogenesis?
Acknowledgments
I apologize to investigators whose research on this topic could not be cited owing to space constraints. I thank Dr. James Leung for helpful discussions, careful reading of the manuscript, and expert editorial assistance. I am funded by a W.M. Keck Foundation Distinguished Young Investigator Award, a Burroughs Wellcome Foundation Investigator in the Pathogenesis of Infectious Disease Award, and National Institutes of Health grants CA160556 and CA136367.
Glossary
- Polysome
a cluster of multiple ribosomes bound to an mRNA
- Eukaryotic translation initiation factor 4F (eIF4F) complex
comprises the eIF4A RNA helicase, the eIF4E cap-binding protein, and the eIF4G scaffolding protein
- eIF4E-binding proteins (4E-BPs)
translational repressors that bind in their unphosphorylated state to eIF4E and prevent its assembly into eIF4F
- Mammalian target of rapamycin kinase complex 1 (mTORC1)
promotes eIF4F assembly by hyperphosphorylating 4E-BPs to prevent their inhibitory interaction with eIF4E
- Torin
a potent inhibitor of mTORC
- Hippuristanol
a small molecule inhibitor of the eIF4A RNA helicase
- Eukaryotic translation initiation factor 2α(eIF2α)
the regulatory subunit of eIF2, which mediates binding of tRNAMet to the ribosome; its phosphorylation during stress blocks eIF2B activity
- Protein kinase R (PKR)
activated by dsRNA, leading to eIF2α phosphorylation and translation inhibition
- Unfolded protein response (UPR)
activated by ER stress; reduces translation, upregulates chaperones, and can cause apoptosis
- Exon junction complex (EJC)
deposited on mRNA during splicing and impacts translation and mRNA surveillance
- Defective ribosomal product (DRiP)
rapidly degraded newly synthesized protein that provides peptide ligands for MHC I
- G-quadruplex
stable RNA structure formed by non-Watson-Crick interactions between stacked guanine tetrads
- Upstream open reading frame (uORF)
short ORF present upstream of the major coding region of an mRNA that can regulate translation
- Peptide ORF
a very short ORF whose translation product may be functional or rapidly degraded
Footnotes
DISCLOSURE STATEMENT
The author is not aware of any affiliations, memberships, funding, or financial holdings that might be perceived as affecting the objectivity of this review.
LITERATURE CITED
- 1.Jackson RJ, Hellen CU, Pestova TV. The mechanism of eukaryotic translation initiation and principles of its regulation. Nat. Rev. Mol. Cell Biol. 2010;11:113–127. doi: 10.1038/nrm2838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ma XM, Blenis J. Molecular mechanisms of mTOR-mediated translational control. Nat. Rev. Mol. Cell Biol. 2009;10:307–318. doi: 10.1038/nrm2672. [DOI] [PubMed] [Google Scholar]
- 3.Walsh D, Mohr I. Assembly of an active translation initiation factor complex by a viral protein. Genes Dev. 2006;20:461–472. doi: 10.1101/gad.1375006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Chuluunbaatar U, Roller R, Feldman ME, Brown S, Shokat KM, Mohr I. Constitutive mTORC1 activation by a herpesvirus Akt surrogate stimulates mRNA translation and viral replication. Genes Dev. 2010;24:2627–2639. doi: 10.1101/gad.1978310. A viral kinase mimics the substrate specificity of Akt to activate mTORC1.
- 5.Benetti L, Roizman B. Protein kinase B/Akt is present in activated form throughout the entire replicative cycle of ΔUS3 mutant virus but only at early times after infection with wild-type herpes simplex virus 1. J. Virol. 2006;80:3341–3348. doi: 10.1128/JVI.80.7.3341-3348.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Moorman NJ, Cristea IM, Terhune SS, Rout MP, Chait BT, Shenk T. Human cytomegalovirus protein UL38 inhibits host cell stress responses by antagonizing the tuberous sclerosis protein complex. Cell Host Microbe. 2008;3:253–262. doi: 10.1016/j.chom.2008.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Arias C, Walsh D, Harbell J, Wilson AC, Mohr I. Activation of host translational control pathways by a viral developmental switch. PLOS Pathog. 2009;5:e1000334. doi: 10.1371/journal.ppat.1000334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sodhi A, Chaisuparat R, Hu J, Ramsdell AK, Manning BD, et al. The TSC2/mTOR pathway drives endothelial cell transformation induced by the Kaposi’s sarcoma-associated herpesvirus G protein-coupled receptor. Cancer Cell. 2006;10:133–143. doi: 10.1016/j.ccr.2006.05.026. [DOI] [PubMed] [Google Scholar]
- 9.Moody CA, Scott RS, Amirghahari N, Nathan CO, Young LS, et al. Modulation of the cell growth regulator mTOR by Epstein-Barr virus-encoded LMP2A. J. Virol. 2005;79:5499–5506. doi: 10.1128/JVI.79.9.5499-5506.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Thoreen CC, Kang SA, Chang JW, Liu Q, Zhang J, et al. An ATP-competitive mammalian target of rapamycin inhibitor reveals rapamycin-resistant functions of mTORC1. J. Biol. Chem. 2009;284:8023–8032. doi: 10.1074/jbc.M900301200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Clippinger AJ, Maguire TG, Alwine JC. The changing role of mTOR kinase in the maintenance of protein synthesis during human cytomegalovirus infection. J. Virol. 2011;85:3930–3939. doi: 10.1128/JVI.01913-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Moorman NJ, Shenk T. Rapamycin-resistant mTORC1 kinase activity is required for herpesvirus replication. J. Virol. 2010;84:5260–5269. doi: 10.1128/JVI.02733-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Lenarcic EM, Ziehr B, De Leon G, Mitchell D, Moorman NJ. Differential role for host translation factors in host and viral protein synthesis during human cytomegalovirus infection. J. Virol. 2014;88:1473–1483. doi: 10.1128/JVI.02321-13. Late in infection, HCMV RNAs but not cellular mRNAs become resistant to eIF4F disruption.
- 14.Hsieh AC, Liu Y, Edlind MP, Ingolia NT, Janes MR, et al. The translational landscape of mTOR signalling steers cancer initiation and metastasis. Nature. 2012;485:55–61. doi: 10.1038/nature10912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Thoreen CC, Chantranupong L, Keys HR, Wang T, Gray NS, Sabatini DM. A unifying model for mTORC1-mediated regulation of mRNA translation. Nature. 2012;485:109–113. doi: 10.1038/nature11083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee AS, Kranzusch PJ, Cate JH. eIF3 targets cell-proliferation messenger RNAs for translational activation or repression. Nature. 2015;522:111–114. doi: 10.1038/nature14267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Perez C, McKinney C, Chulunbaatar U, Mohr I. Translational control of the abundance of cytoplasmic poly(A) binding protein in human cytomegalovirus-infected cells. J. Virol. 2011;85:156–164. doi: 10.1128/JVI.01778-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Walsh D, Perez C, Notary J, Mohr I. Regulation of the translation initiation factor eIF4F by multiple mechanisms in human cytomegalovirus-infected cells. J. Virol. 2005;79:8057–8064. doi: 10.1128/JVI.79.13.8057-8064.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Aoyagi M, Gaspar M, Shenk TE. Human cytomegalovirus UL69 protein facilitates translation by associating with the mRNA cap-binding complex and excluding 4EBP1. PNAS. 2010;107:2640–2645. doi: 10.1073/pnas.0914856107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kuang E, Tang Q, Maul GG, Zhu F. Activation of p90 ribosomal S6 kinase by ORF45 of Kaposi’s sarcoma-associated herpesvirus and its role in viral lytic replication. J. Virol. 2008;82:1838–1850. doi: 10.1128/JVI.02119-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kuang E, Wu F, Zhu F. Mechanism of sustained activation of ribosomal S6 kinase (RSK) and ERK by Kaposi sarcoma-associated herpesvirus ORF45: Multiprotein complexes retain active phosphorylated ERK and RSK and protect them from dephosphorylation. J. Biol. Chem. 2009;284:13958–13968. doi: 10.1074/jbc.M900025200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kuang E, Fu B, Liang Q, Myoung J, Zhu F. Phosphorylation of eukaryotic translation initiation factor 4B (EIF4B) by open reading frame 45/p90 ribosomal S6 kinase (ORF45/RSK) signaling axis facilitates protein translation during Kaposi sarcoma-associated herpesvirus (KSHV) lytic replication. J. Biol. Chem. 2011;286:41171–41182. doi: 10.1074/jbc.M111.280982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Parsyan A, Svitkin Y, Shahbazian D, Gkogkas C, Lasko P, et al. mRNA helicases: the tacticians of translational control. Nat. Rev. Mol. Cell Biol. 2011;12:235–245. doi: 10.1038/nrm3083. [DOI] [PubMed] [Google Scholar]
- 24.Fu B, Kuang E, Li W, Avey D, Li X, et al. Activation of p90 ribosomal S6 kinases (RSKs) by ORF45 of Kaposi sarcoma-associated herpesvirus is critical for optimal production of infectious viruses. J. Virol. 2014;89:195–207. doi: 10.1128/JVI.01937-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jacquemont B, Roizman B. RNA synthesis in cells infected with herpes simplex virus. X. Properties of viral symmetric transcripts and of double-stranded RNA prepared from them. J. Virol. 1975;15:707–713. doi: 10.1128/jvi.15.4.707-713.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Langland JO, Jacobs BL. The role of the PKR-inhibitory genes, E3L and K3L, in determining vaccinia virus host range. Virology. 2002;299:133–141. doi: 10.1006/viro.2002.1479. [DOI] [PubMed] [Google Scholar]
- 27.Taylor DR, Lee SB, Romano PR, Marshak DR, Hinnebusch AG, et al. Autophosphorylation sites participate in the activation of the double-stranded-RNA-activated protein kinase PKR. Mol. Cell. Biol. 1996;16:6295–6302. doi: 10.1128/mcb.16.11.6295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Silverman RH. Viral encounters with 2′,5′-oligoadenylate synthetase and RNase L during the interferon antiviral response. J. Virol. 2007;81:12720–12729. doi: 10.1128/JVI.01471-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sciortino MT, Parisi T, Siracusano G, Mastino A, Taddeo B, Roizman B. The virion host shutoff RNase plays a key role in blocking the activation of protein kinase R in cells infected with herpes simplex virus 1. J. Virol. 2013;87:3271–3276. doi: 10.1128/JVI.03049-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.He B, Gross M, Roizman B. The γ134.5 protein of herpes simplex virus 1 complexes with protein phosphatase 1α to dephosphorylate the α subunit of the eukaryotic translation initiation factor 2 and preclude the shutoff of protein synthesis by double-stranded RNA-activated protein kinase. PNAS. 1997;94:843–848. doi: 10.1073/pnas.94.3.843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li Y, Zhang C, Chen X, Yu J, Wang Y, et al. ICP34.5 protein of herpes simplex virus facilitates the initiation of protein translation by bridging eukaryotic initiation factor 2α (eIF2α) and protein phosphatase 1. J. Biol. Chem. 2011;286:24785–24792. doi: 10.1074/jbc.M111.232439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Van Opdenbosch N, Van den Broeke C, De Regge N, Tabares E, Favoreel HW. The IE180 protein of pseudorabies virus suppresses phosphorylation of translation initiation factor eIF2α. J. Virol. 2012;86:7235–7240. doi: 10.1128/JVI.06929-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sanchez R, Mohr I. Inhibition of cellular 2′–5′ oligoadenylate synthetase by the herpes simplex virus type 1 Us11 protein. J. Virol. 2007;81:3455–3464. doi: 10.1128/JVI.02520-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Budt M, Niederstadt L, Valchanova RS, Jonjic S, Brune W. Specific inhibition of the PKR-mediated antiviral response by the murine cytomegalovirus proteins m142 and m143. J. Virol. 2009;83:1260–1270. doi: 10.1128/JVI.01558-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Child SJ, Geballe AP. Binding and relocalization of protein kinase R by murine cytomegalovirus. J. Virol. 2009;83:1790–1799. doi: 10.1128/JVI.01484-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Marshall EE, Bierle CJ, Brune W, Geballe AP. Essential role for either TRS1 or IRS1 in human cytomegalovirus replication. J. Virol. 2009;83:4112–4120. doi: 10.1128/JVI.02489-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hakki M, Geballe AP. Double-stranded RNA binding by human cytomegalovirus pTRS1. J. Virol. 2005;79:7311–7318. doi: 10.1128/JVI.79.12.7311-7318.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hakki M, Marshall EE, De Niro KL, Geballe AP. Binding and nuclear relocalization of protein kinase R by human cytomegalovirus TRS1. J. Virol. 2006;80:11817–11826. doi: 10.1128/JVI.00957-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Child SJ, Hakki M, De Niro KL, Geballe AP. Evasion of cellular antiviral responses by human cytomegalovirus TRS1 and IRS1. J. Virol. 2004;78:197–205. doi: 10.1128/JVI.78.1.197-205.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tan JC, Avdic S, Cao JZ, Mocarski ES, White KL, et al. Inhibition of 2′,5′-oligoadenylate synthetase expression and function by the human cytomegalovirus ORF94 gene product. J. Virol. 2011;85:5696–5700. doi: 10.1128/JVI.02463-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Clarke PA, Schwemmle M, Schickinger J, Hilse K, Clemens MJ. Binding of Epstein-Barr virus small RNAEBER-1 to the double-stranded RNA-activated protein kinase DAI. Nucleic Acids Res. 1991;19:243–248. doi: 10.1093/nar/19.2.243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.McKenna SA, Kim I, Liu CW, Puglisi JD. Uncoupling of RNA binding and PKR kinase activation by viral inhibitor RNAs. J. Mol. Biol. 2006;358:1270–1285. doi: 10.1016/j.jmb.2006.03.003. [DOI] [PubMed] [Google Scholar]
- 43.Sharp TV, Schwemmle M, Jeffrey I, Laing K, Mellor H, et al. Comparative analysis of the regulation of the interferon-inducible protein kinase PKR by Epstein-Barr virus RNAs EBER-1 and EBER-2 and adenovirus VAI RNA. Nucleic Acids Res. 1993;21:4483–4490. doi: 10.1093/nar/21.19.4483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ruf IK, Lackey KA, Warudkar S, Sample JT. Protection from interferon-induced apoptosis by Epstein-Barr virus small RNAs is not mediated by inhibition of PKR. J. Virol. 2005;79:14562–14569. doi: 10.1128/JVI.79.23.14562-14569.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Burysek L, Pitha PM. Latently expressed human herpesvirus 8-encoded interferon regulatory factor 2 inhibits double-stranded RNA-activated protein kinase. J. Virol. 2001;75:2345–2352. doi: 10.1128/JVI.75.5.2345-2352.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Poppers J, Mulvey M, Perez C, Khoo D, Mohr I. Identification of a lytic-cycle Epstein-Barr virus gene product that can regulate PKR activation. J. Virol. 2003;77:228–236. doi: 10.1128/JVI.77.1.228-236.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Brewer JW. Regulatory crosstalk within the mammalian unfolded protein response. Cell. Mol. Life Sci. 2014;71:1067–1079. doi: 10.1007/s00018-013-1490-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Isler JA, Skalet AH, Alwine JC. Human cytomegalovirus infection activates and regulates the unfolded protein response. J. Virol. 2005;79:6890–6899. doi: 10.1128/JVI.79.11.6890-6899.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Qian Z, Xuan B, Chapa TJ, Gualberto N, Yu D. Murine cytomegalovirus targets transcription factor ATF4 to exploit the unfolded-protein response. J. Virol. 2012;86:6712–6723. doi: 10.1128/JVI.00200-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Xuan B, Qian Z, Torigoi E, Yu D. Human cytomegalovirus protein pUL38 induces ATF4 expression, inhibits persistent JNK phosphorylation, and suppresses endoplasmic reticulum stress-induced cell death. J. Virol. 2009;83:3463–3474. doi: 10.1128/JVI.02307-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Stahl S, Burkhart JM, Hinte F, Tirosh B, Mohr H, et al. Cytomegalovirus downregulates IRE1 to repress the unfolded protein response. PLOS Pathog. 2013;9:e1003544. doi: 10.1371/journal.ppat.1003544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Buchkovich NJ, Maguire TG, Yu Y, Paton AW, Paton JC, Alwine JC. Human cytomegalovirus specifically controls the levels of the endoplasmic reticulum chaperone BiP/GRP78, which is required for virion assembly. J. Virol. 2008;82:31–39. doi: 10.1128/JVI.01881-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Buchkovich NJ, Yu Y, Pierciey FJ, Jr, Alwine JC. Human cytomegalovirus induces the endoplasmic reticulum chaperone BiP through increased transcription and activation of translation by using the BiP internal ribosome entry site. J. Virol. 2010;84:11479–11486. doi: 10.1128/JVI.01330-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lee DY, Sugden B. The LMP1 oncogene of EBV activates PERK and the unfolded protein response to drive its own synthesis. Blood. 2008;111:2280–2289. doi: 10.1182/blood-2007-07-100032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bhende PM, Dickerson SJ, Sun X, Feng WH, Kenney SC. X-box-binding protein 1 activates lytic Epstein-Barr virus gene expression in combination with protein kinase D. J. Virol. 2007;81:7363–7370. doi: 10.1128/JVI.00154-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mulvey M, Arias C, Mohr I. Maintenance of endoplasmic reticulum (ER) homeostasis in herpes simplex virus type 1-infected cells through the association of a viral glycoprotein with PERK, a cellular ER stress sensor. J. Virol. 2007;81:3377–3390. doi: 10.1128/JVI.02191-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Carpenter JE, Jackson W, Benetti L, Grose C. Autophagosome formation during varicella-zoster virus infection following endoplasmic reticulum stress and the unfolded protein response. J. Virol. 2011;85:9414–9424. doi: 10.1128/JVI.00281-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gaglia MM, Glaunsinger BA. Viruses and the cellular RNA decay machinery. WIRES RNA. 2010;1:47–59. doi: 10.1002/wrna.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Dauber B, Pelletier J, Smiley JR. The herpes simplex virus 1 vhs protein enhances translation of viral true late mRNAs and virus production in a cell type-dependent manner. J. Virol. 2011;85:5363–5373. doi: 10.1128/JVI.00115-11. mRNA degradation by the vhs endonuclease liberates translation machinery for late gene expression.
- 60.Esclatine A, Taddeo B, Roizman B. Herpes simplex virus 1 induces cytoplasmic accumulation of TIA-1/TIAR and both synthesis and cytoplasmic accumulation of tristetraprolin, two cellular proteins that bind and destabilize AU-rich RNAs. J. Virol. 2004;78:8582–8592. doi: 10.1128/JVI.78.16.8582-8592.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Dauber B, Saffran HA, Smiley JR. The herpes simplex virus 1 virion host shutoff protein enhances translation of viral late mRNAs by preventing mRNA overload. J. Virol. 2014;88:9624–9632. doi: 10.1128/JVI.01350-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Saffran HA, Read GS, Smiley JR. Evidence for translational regulation by the herpes simplex virus virion host shutoff protein. J. Virol. 2010;84:6041–6049. doi: 10.1128/JVI.01819-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Abernathy E, Clyde K, Yeasmin R, Krug LT, Burlingame A, et al. Gammaherpesviral gene expression and virion composition are broadly controlled by accelerated mRNA degradation. PLOS Pathog. 2014;10:e1003882. doi: 10.1371/journal.ppat.1003882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Richner JM, Clyde K, Pezda AC, Cheng BY, Wang T, et al. Global mRNA degradation during lytic gammaherpesvirus infection contributes to establishment of viral latency. PLOS Pathog. 2011;7:e1002150. doi: 10.1371/journal.ppat.1002150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. McKinney C, Zavadil J, Bianco C, Shiflett L, Brown S, Mohr I. Global reprogramming of the cellular translational landscape facilitates cytomegalovirus replication. Cell Rep. 2014;6:9–17. doi: 10.1016/j.celrep.2013.11.045. HCMV impacts the subset of host mRNAs recruited to or excluded from polysomes.
- 66.Park EH, Zhang F, Warringer J, Sunnerhagen P, Hinnebusch AG. Depletion of eIF4G from yeast cells narrows the range of translational efficiencies genome-wide. BMC Genomics. 2011;12:68. doi: 10.1186/1471-2164-12-68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ceci M, Gaviraghi C, Gorrini C, Sala LA, Offenhauser N, et al. Release of eIF6 (p27BBP) from the 60S subunit allows 80S ribosome assembly. Nature. 2003;426:579–584. doi: 10.1038/nature02160. [DOI] [PubMed] [Google Scholar]
- 68.Nott A, Le Hir H, Moore MJ. Splicing enhances translation in mammalian cells: an additional function of the exon junction complex. Genes Dev. 2004;18:210–222. doi: 10.1101/gad.1163204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Boyne JR, Jackson BR, Taylor A, Macnab SA, Whitehouse A. Kaposi’s sarcoma-associated herpesvirus ORF57 protein interacts with PYM to enhance translation of viral intronless mRNAs. EMBO J. 2010;29:1851–1864. doi: 10.1038/emboj.2010.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Fontaine-Rodriguez EC, Knipe DM. Herpes simplex virus ICP27 increases translation of a subset of viral late mRNAs. J. Virol. 2008;82:3538–3545. doi: 10.1128/JVI.02395-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Larralde O, Smith RW, Wilkie GS, Malik P, Gray NK, Clements JB. Direct stimulation of translation by the multifunctional herpesvirus ICP27 protein. J. Virol. 2006;80:1588–1591. doi: 10.1128/JVI.80.3.1588-1591.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ricci EP, Mure F, Gruffat H, Decimo D, Medina-Palazon C, et al. Translation of intronless RNAs is strongly stimulated by the Epstein-Barr virus mRNA export factor EB2. Nucleic Acids Res. 2009;37:4932–4943. doi: 10.1093/nar/gkp497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Fontaine-Rodriguez EC, Taylor TJ, Olesky M, Knipe DM. Proteomics of herpes simplex virus infected cell protein 27: association with translation initiation factors. Virology. 2004;330:487–492. doi: 10.1016/j.virol.2004.10.002. [DOI] [PubMed] [Google Scholar]
- 74.Bono F, Ebert J, Unterholzner L, Guttler T, Izaurralde E, Conti E. Molecular insights into the interaction of PYM with the Mago-Y14 core of the exon junction complex. EMBO Rep. 2004;5:304–310. doi: 10.1038/sj.embor.7400091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Diem MD, Chan CC, Younis I, Dreyfuss G. PYM binds the cytoplasmic exon-junction complex and ribosomes to enhance translation of spliced mRNAs. Nat. Struct. Mol. Biol. 2007;14:1173–1179. doi: 10.1038/nsmb1321. [DOI] [PubMed] [Google Scholar]
- 76.Ellison KS, Maranchuk RA, Mottet KL, Smiley JR. Control of VP16 translation by the herpes simplex virus type 1 immediate-early protein ICP27. J. Virol. 2005;79:4120–4131. doi: 10.1128/JVI.79.7.4120-4131.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Sokolowski M, Scott JE, Heaney RP, Patel AH, Clements JB. Identification of herpes simplex virus RNAs that interact specifically with regulatory protein ICP27 in vivo. J. Biol. Chem. 2003;278:33540–33549. doi: 10.1074/jbc.M302063200. [DOI] [PubMed] [Google Scholar]
- 78.Verma D, Li DJ, Krueger B, Renne R, Swaminathan S. Identification of the physiological gene targets of the essential lytic replicative KSHV ORF57 protein. J. Virol. 2015;89:1688–1702. doi: 10.1128/JVI.02663-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Kang JG, Pripuzova N, Majerciak V, Kruhlak M, Le SY, Zheng ZM. Kaposi’s sarcoma-associated herpesvirus ORF57 promotes escape of viral and human interleukin-6 from microRNA-mediated suppression. J. Virol. 2011;85:2620–2630. doi: 10.1128/JVI.02144-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Sei E, Wang T, Hunter OV, Xie Y, Conrad NK. HITS-CLIP analysis uncovers a link between the Kaposi’s sarcoma associated herpesvirus ORF57 protein and host pre-mRNA metabolism. PLOS Pathog. 2015;11:e1004652. doi: 10.1371/journal.ppat.1004652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Bourdetsky D, Schmelzer CE, Admon A. The nature and extent of contributions by defective ribosome products to the HLA peptidome. PNAS. 2014;111:E1591–E1599. doi: 10.1073/pnas.1321902111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Qian SB, Reits E, Neefjes J, Deslich JM, Bennink JR, Yewdell JW. Tight linkage between translation and MHC class I peptide ligand generation implies specialized antigen processing for defective ribosomal products. J. Immunol. 2006;177:227–233. doi: 10.4049/jimmunol.177.1.227. [DOI] [PubMed] [Google Scholar]
- 83.Yewdell JW. Serendipity strikes twice: the discovery and rediscovery of defective ribosomal products (DRiPS) Cell. Mol. Biol. 2005;51:635–641. [PubMed] [Google Scholar]
- 84.Tellam J, Fogg MH, Rist M, Connolly G, Tscharke D, et al. Influence of translation efficiency of homologous viral proteins on the endogenous presentation of CD8+ T cell epitopes. J. Exp. Med. 2007;204:525–532. doi: 10.1084/jem.20062508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Apcher S, Daskalogianni C, Manoury B, Fahraeus R. Epstein Barr virus-encoded EBNA1 interference with MHC class I antigen presentation reveals a close correlation between mRNA translation initiation and antigen presentation. PLOS Pathog. 2010;6:e1001151. doi: 10.1371/journal.ppat.1001151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Blake N. Immune evasion by gammaherpesvirus genome maintenance proteins. J. Gen. Virol. 2010;91:829–846. doi: 10.1099/vir.0.018242-0. [DOI] [PubMed] [Google Scholar]
- 87.Tellam JT, Lekieffre L, Zhong J, Lynn DJ, Khanna R. Messenger RNA sequence rather than protein sequence determines the level of self-synthesis and antigen presentation of the EBV-encoded antigen, EBNA1. PLOS Pathog. 2012;8:e1003112. doi: 10.1371/journal.ppat.1003112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Cardinaud S, Starck SR, Chandra P, Shastri N. The synthesis of truncated polypeptides for immune surveillance and viral evasion. PLOS ONE. 2010;5:e8692. doi: 10.1371/journal.pone.0008692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Murat P, Zhong J, Lekieffre L, Cowieson NP, Clancy JL, et al. G-quadruplexes regulate Epstein-Barr virus-encoded nuclear antigen 1 mRNA translation. Nat. Chem. Biol. 2014;10:358–364. doi: 10.1038/nchembio.1479. G-quadruplexes in the EBNA1 mRNA slow its translation, thereby potentially reducing antigen presentation.
- 90.Ressing ME, Horst D, Griffin BD, Tellam J, Zuo J, et al. Epstein-Barr virus evasion of CD8+ and CD4+ T cell immunity via concerted actions of multiple gene products. Semin. Cancer Biol. 2008;18:397–408. doi: 10.1016/j.semcancer.2008.10.008. [DOI] [PubMed] [Google Scholar]
- 91.Tellam JT, Zhong J, Lekieffre L, Bhat P, Martinez M, et al. mRNA structural constraints on EBNA1 synthesis impact on in vivo antigen presentation and early priming of CD8+ T cells. PLOS Pathog. 2014;10:e1004423. doi: 10.1371/journal.ppat.1004423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Kwun HJ, da Silva SR, Qin H, Ferris RL, Tan R, et al. The central repeat domain 1 of Kaposi’s sarcoma-associated herpesvirus (KSHV) latency associated-nuclear antigen 1 (LANA1) prevents cis MHC class I peptide presentation. Virology. 2011;412:357–365. doi: 10.1016/j.virol.2011.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Kwun HJ, da Silva SR, Shah IM, Blake N, Moore PS, Chang Y. Kaposi’s sarcoma-associated herpesvirus latency-associated nuclear antigen 1 mimics Epstein-Barr virus EBNA1 immune evasion through central repeat domain effects on protein processing. J. Virol. 2007;81:8225–8235. doi: 10.1128/JVI.00411-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Rubio CA, Weisburd B, Holderfield M, Arias C, Fang E, et al. Transcriptome-wide characterization of the eIF4A signature highlights plasticity in translation regulation. Genome Biol. 2014;15:476. doi: 10.1186/s13059-014-0476-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Wolfe AL, Singh K, Zhong Y, Drewe P, Rajasekhar VK, et al. RNA G-quadruplexes cause eIF4A-dependent oncogene translation in cancer. Nature. 2014;513:65–70. doi: 10.1038/nature13485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Kwun HJ, Toptan T, Ramos da Silva S, Atkins JF, Moore PS, Chang Y. Human DNA tumor viruses generate alternative reading frame proteins through repeat sequence recoding. PNAS. 2014;111:E4342–E4349. doi: 10.1073/pnas.1416122111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Toptan T, Fonseca L, Kwun HJ, Chang Y, Moore PS. Complex alternative cytoplasmic protein isoforms of the Kaposi’s sarcoma-associated herpesvirus latency-associated nuclear antigen 1 generated through noncanonical translation initiation. J. Virol. 2013;87:2744–2755. doi: 10.1128/JVI.03061-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Zaldumbide A, Ossevoort M, Wiertz EJ, Hoeben RC. In cis inhibition of antigen processing by the latency-associated nuclear antigen I of Kaposi sarcoma herpes virus. Mol. Immunol. 2007;44:1352–1360. doi: 10.1016/j.molimm.2006.05.012. [DOI] [PubMed] [Google Scholar]
- 99.Dresang LR, Teuton JR, Feng H, Jacobs JM, Camp DG, II, et al. Coupled transcriptome and proteome analysis of human lymphotropic tumor viruses: insights on the detection and discovery of viral genes. BMC Genomics. 2011;12:625. doi: 10.1186/1471-2164-12-625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Chaudhary PM, Jasmin A, Eby MT, Hood L. Modulation of the NF-κB pathway by virally encoded death effector domains-containing proteins. Oncogene. 1999;18:5738–5746. doi: 10.1038/sj.onc.1202976. [DOI] [PubMed] [Google Scholar]
- 101.Thome M, Schneider P, Hofmann K, Fickenscher H, Meinl E, et al. Viral FLICE-inhibitory proteins (FLIPs) prevent apoptosis induced by death receptors. Nature. 1997;386:517–521. doi: 10.1038/386517a0. [DOI] [PubMed] [Google Scholar]
- 102.Bieleski L, Talbot SJ. Kaposi’s sarcoma-associated herpesvirus vCyclin open reading frame contains an internal ribosome entry site. J. Virol. 2001;75:1864–1869. doi: 10.1128/JVI.75.4.1864-1869.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Grundhoff A, Ganem D. Mechanisms governing expression of the v-FLIP gene of Kaposi’s sarcoma-associated herpesvirus. J. Virol. 2001;75:1857–1863. doi: 10.1128/JVI.75.4.1857-1863.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Low W, Harries M, Ye H, Du MQ, Boshoff C, Collins M. Internal ribosome entry site regulates translation of Kaposi’s sarcoma-associated herpesvirus FLICE inhibitory protein. J. Virol. 2001;75:2938–2945. doi: 10.1128/JVI.75.6.2938-2945.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Othman Z, Sulaiman MK, Willcocks MM, Ulryck N, Blackbourn DJ, et al. Functional analysis of Kaposi’s sarcoma-associated herpesvirus vFLIP expression reveals a new mode of IRES-mediated translation. RNA. 2014;20:1803–1814. doi: 10.1261/rna.045328.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Feoktistova K, Tuvshintogs E, Do A, Fraser CS. Human eIF4E promotes mRNA restructuring by stimulating eIF4A helicase activity. PNAS. 2013;110:13339–13344. doi: 10.1073/pnas.1303781110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Arias C, Weisburd B, Stern-Ginossar N, Mercier A, Madrid AS, et al. KSHV 2.0: a comprehensive annotation of the Kaposi’s sarcoma-associated herpesvirus genome using next-generation sequencing reveals novel genomic and functional features. PLOS Pathog. 2014;10:e1003847. doi: 10.1371/journal.ppat.1003847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Stern-Ginossar N, Weisburd B, Michalski A, Le VT, Hein MY, et al. Decoding human cytomegalovirus. Science. 2012;338:1088–1093. doi: 10.1126/science.1227919. Discovery of hundreds of new ORFs and protein expression control through alternative transcript start sites.
- 109.Ingolia NT, Ghaemmaghami S, Newman JR, Weissman JS. Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science. 2009;324:218–223. doi: 10.1126/science.1168978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Jaber T, Yuan Y. A virally encoded small peptide regulates RTA stability and facilitates Kaposi’s sarcoma-associated herpesvirus lytic replication. J. Virol. 2013;87:3461–3470. doi: 10.1128/JVI.02746-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Cao J, Geballe AP. Translational inhibition by a human cytomegalovirus upstream open reading frame despite inefficient utilization of its AUG codon. J. Virol. 1995;69:1030–1036. doi: 10.1128/jvi.69.2.1030-1036.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Kozak M. Effects of intercistronic length on the efficiency of reinitiation by eucaryotic ribosomes. Mol. Cell. Biol. 1987;7:3438–3445. doi: 10.1128/mcb.7.10.3438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Kozak M. Constraints on reinitiation of translation in mammals. Nucleic Acids Res. 2001;29:5226–5232. doi: 10.1093/nar/29.24.5226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Gaba A, Wang Z, Krishnamoorthy T, Hinnebusch AG, Sachs MS. Physical evidence for distinct mechanisms of translational control by upstream open reading frames. EMBO J. 2001;20:6453–6463. doi: 10.1093/emboj/20.22.6453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Hinnebusch AG. Gene-specific translational control of the yeast GCN4 gene by phosphorylation of eukaryotic initiation factor 2. Mol. Microbiol. 1993;10:215–223. doi: 10.1111/j.1365-2958.1993.tb01947.x. [DOI] [PubMed] [Google Scholar]
- 116.Asano K, Krishnamoorthy T, Phan L, Pavitt GD, Hinnebusch AG. Conserved bipartite motifs in yeast eIF5 and eIF2Bε, GTPase-activating and GDP-GTP exchange factors in translation initiation, mediate binding to their common substrate eIF2. EMBO J. 1999;18:1673–1688. doi: 10.1093/emboj/18.6.1673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Kronstad LM, Brulois KF, Jung JU, Glaunsinger BA. Dual short upstream open reading frames control translation of a herpesviral polycistronic mRNA. PLOS Pathog. 2013;9:e1003156. doi: 10.1371/journal.ppat.1003156. uORFs enable polycistronic gene expression in KSHV.
- 118.Kronstad LM, Brulois KF, Jung JU, Glaunsinger BA. Reinitiation after translation of two upstream open reading frames (ORF) governs expression of the ORF35–37 Kaposi’s sarcoma-associated herpesvirus polycistronic mRNA. J. Virol. 2014;88:6512–6518. doi: 10.1128/JVI.00202-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Brubaker SW, Gauthier AE, Mills EW, Ingolia NT, Kagan JC. A bicistronic MAVS transcript highlights a class of truncated variants in antiviral immunity. Cell. 2014;156:800–811. doi: 10.1016/j.cell.2014.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Calkhoven CF, Muller C, Leutz A. Translational control of C/EBPα and C/EBPβ isoform expression. Genes Dev. 2000;14:1920–1932. [PMC free article] [PubMed] [Google Scholar]
- 121.Ingolia NT, Brar GA, Stern-Ginossar N, Harris MS, Talhouarne GJ, et al. Ribosome profiling reveals pervasive translation outside of annotated protein-coding genes. Cell Rep. 2014;8:1365–1379. doi: 10.1016/j.celrep.2014.07.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Lee AS, Burdeinick-Kerr R, Whelan SP. A ribosome-specialized translation initiation pathway is required for cap-dependent translation of vesicular stomatitis virus mRNAs. PNAS. 2013;110:324–329. doi: 10.1073/pnas.1216454109. The ribosomal protein RPL40 directs transcript-specific translation of vesicular stomatitis virus and select host mRNAs.