Abstract
Aptamers are single-stranded DNA or RNA oligonucleotides that can bind with exquisitely high affinity and specificity to target molecules and are thus often referred to as ‘nucleic acid’ antibodies. Oligonucleotide aptamers are derived through a process of directed chemical evolution called SELEX (Systematic Evolution of Ligands by Exponential enrichment). This chemical equivalent of Darwinian evolution was first described in 1990 by Tuerk & Gold and Ellington & Szostak and has since yielded aptamers for a wide-range of applications, including biosensor technologies, in vitro diagnostics, biomarker discovery, and therapeutics. Since the inception of the original SELEX method, numerous modifications to the protocol have been described to fit the choice of target, specific conditions or applications. Technologies such as high-throughput sequencing methods and microfluidics have also been adapted for SELEX. In this chapter, we outline key steps in the SELEX process for enabling the rapid identification of RNA aptamers for in vivo applications. Specifically, we provide a detailed protocol for the selection of chemically-optimized RNA aptamers using the original in vitro SELEX methodology. In addition, methods for performing next-generation sequencing of the RNAs from each round of selection, based on Illumina sequencing technology, are discussed.
Keywords: In vitro SELEX, RNA aptamers, Mutant T7 RNA polymerase, Modified nucleotides, 2′-Fluoro pyrimidines, Recombinant proteins, Next-generation sequencing, Illumina sequencing
1. Introduction
Aptamers are highly-structured, single-stranded nucleic acid ligands whose binding properties are comparable to those of antibody/antigen interactions (thus are often referred to as nucleic acid antibodies) [1,2]. Fig. 1 shows the predicted tertiary structure of a chemically stabilized RNA aptamer (A9g) to prostate specific membrane antigen (PSMA) [3,4]. Aptamers are generated using an iterative selection process that partitions oligonucleotides on the basis of their binding or functional/catalytic activities. This in vitro process termed Systematic Evolution of Ligands by Exponential enrichment or SELEX was first described in 1990 [5,6]. The SELEX process relies on established technologies such as 1) DNA synthetic methods that enable the generation of large populations of random oligonucleotides, 2) polymerase chain reaction (PCR) to facilitate the robust amplification of low abundance oligonucleotide sequences and 3) affinity purification methods that permit capturing of target-specific sequences. In a typical SELEX experiment, the first step is to synthesize a random sequence DNA library of ~20–100 nucleotides in length containing flanking constant sequences required for PCR amplification. The DNA library typically contains between 1012 to 1015 different sequences. In the case of DNA SELEX [7–10], the single-stranded DNA (ssDNA) library for each round of selection is prepared by the strand separation of double-strand (ds) PCR products. In the case of RNA SELEX [5,6], the single stranded RNA library for each round of selection is prepared by in vitro transcription of dsDNA templates using T7 RNA polymerases. This chapter will focus on detailing the SELEX and next-generation sequencing (NGS) methods for enabling the rapid development of chemically-optimized RNA aptamers for therapeutic applications. The SELEX library (Sel3; TriLink BioTechnologies Inc.) has been optimized for maximal RNA yields during in vitro transcription.
Fig. 1.
Predicted tertiary structure of PSMA RNA aptamer A9g. The predicted tertiary structure (3D) of A9g was obtained as described in Rockey et al. (2011). The constructed PDB files were then processed using the Swiss PDBViewer to generate the 3D structure. This aptamer is chemically modified with 2′ fluoro chemistry for clinical applications. A9g is a non-competitive inhibitor of PSMA enzymatic activity. It inhibits PSMA-mediated migration and invasion of prostate cancer cells in culture and dissemination in a mouse model of metastatic bone disease.
The development of aptamers as therapeutics has primarily involved their tendency to inhibit their targets upon binding [1,11–14], although aptamers have also been developed to activate cell-surface receptors [15]. Several properties render aptamers attractive candidates for therapeutic applications. For example, like antibodies, aptamers can be designed to have high affinity and specificity for their target. However, aptamers are relatively small in size (8–15 kDa) and therefore generally display better tissue penetration and superior target-to-noise ratios compared to their protein counterparts. In addition, chemically-optimized aptamers are non-immunogenic. Aptamer development has been greatly facilitated by recent progress in standard automated solid-phase synthesis, which enables the synthesis of kilogram (kg) quantities of aptamers at relatively low cost (less than $200/ g for unmodified aptamers). Finally, activity of aptamers can be easily reversed by an antidote (e.g., an oligonucleotide of complementary sequence), a feature that holds great utility for drug design and development [11]. Given the above properties, aptamers are considered as potent and versatile next-generation ligands for the development of state-of-the-art diagnostic tools and therapeutic agents. Currently, one aptamer-based drug, pegaptanib (Macugen) is already approved for treating macular degeneration, while several others are being evaluated clinically [1,14,16].
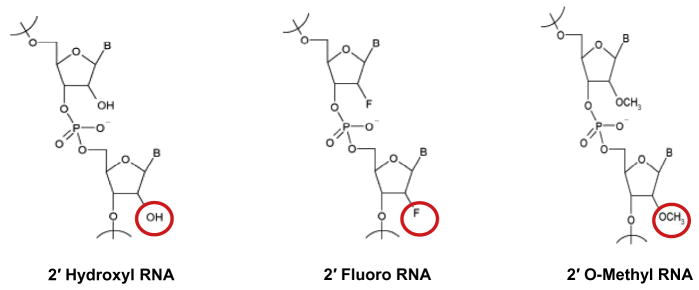
Therapeutic aptamers are typically modified to contain fluoro or O-methyl groups at the 2′-position of their sugar moieties (Fig. 2), rendering them resistant to degradation by serum nucleases and reducing the inherent immunogenicity of unmodified RNA [1]. The modified nucleotide triphosphates (NTPs) are typically incorporated during the selection process to reduce the possibility of post-selection modifications impairing aptamer function [17–19]. However, for modified NTPs to be useful in SELEX, they must not only serve as good substrates for polymerases, but the modified oligonucleotides into which they are incorporated must also be viable templates for PCR amplification. Efficient incorporation of 2′-fluoro pyrimidine NTPs during the selection process has been made possible by the development of mutant polymerases that readily incorporate these modified nucleotides into nascent DNA [20–23]. Such modified nucleotides are also well-tolerated by reverse transcriptases (RT) [1]. Therapeutic RNA aptamers containing 2′ fluoro-modified NTPs are often modified post-selection by introduction of 2′-O-methyl purines and pyrimidines in order to further enhance stability and improve safety [24]. This is an empirical process that can result in reduced [17–19] or, in rare cases, improved function [17–19] of the original aptamer(s).
Fig. 2.
Commonly used 2′-ribose modified nucleotides in aptamer selection experiments. The mutated T7 (Y639F) RNA polymerase is able to easily incorporate the 2′OH and 2′F modifications during selection, while the 2′Ome modification is normally incorporated synthetically after selection has occurred. (Modified nucleotides and aptamer libraries can be purchased from TriLink BioTechnologies, Inc. – http://www.trilinkbiotech.com/.)
A recent breakthrough in SELEX has been the application of NGS technologies (e.g. Illumina, 454), which, together with bioinformatics analysis, expedite the identification of ‘winner’ sequences and allow researchers to track aptamer evolution [25–30]. SELEX technologies are quite diverse, allowing for generation of aptamers against a variety of targets ranging from small molecules and peptides to proteins and cells (cell-SELEX) [1,10–12,27,31–34]. In a particularly notable application, in vivo SELEX has been used to identify aptamers targeting entire tissues [35,36]. The use of SELEX against such diverse targets has been described elsewhere [37–40]. In this chapter, we provide detailed methods for the rapid development of chemically-optimized RNA aptamers against recombinant, purified protein targets.
2. Materials
2.1. Reagents for synthesis of the DNA aptamer library
1.0 M Tris–HCl pH 8.0 (Sigma SLBF9645), pH 8.0.
50 mM MgCl2.
100 μM Sel3 Template Oligo (5′-TCGGGCGAGTCGTCTGN20 CCGCATCCTCCTCCC-3′; TriLink Biotechnologies, Inc.) (see note #1).
100 μM Sel3 5′-Primer (5′-TAATACGACTCACTATAGGGAG GAGGATGCGG-3′; TriLink BioTechnologies Inc.).
10 mM dNTP Mix. (Invitrogen #4893)
10× buffer Taq DNA polymerase buffer (Denville Scientific #CB3702-7).
Taq DNA polymerase 5 Units/μL (Denville Scientific #CB4050-1).
Microcentrifuge tubes DNase/RNase free (USA Scientific # 1615-5500).
QIAquick Gel Extraction Kit (Qiagen #28706).
Isotemp Heating blocks (Fisher Scientific).
Molecular biology grade agarose (RPI Corp. # Cas 9012-36-6).
TAE buffer (Fisher Scientific #BP1335-4).
6× gel loading dye (New England Biolabs Inc. # B7021S).
100 base pair DNA ladder (New England Biolabs Inc. #N3231).
Minigel System; 7 × 10 cm gel size (Fisher Scientific #FB-SB-710).
PowerPac Basic Power Supply (Bio-Rad #164-5050).
Ethidium Bromide (Sigma #E1510).
Molecular Imager Gel Doc XR System (BioRad #107-8170).
2.2. Reagents for RNA aptamer transcription and purification
5× T7 RNAP Buffer: 20% w/v PEG-8000 (Sigma P5413), 200 mM Tris–HCl pH 8.0 (Life Technologies 15568-025), 60 mM MgCl2 (Sigma M8266), 5 mM spermidine HCl (Sigma 233994), 25 mM DTT (Sigma 646563).
10× rNTP mix (w/2′Fs): 30 mM 2′F-C/U with 10 mM 2′OH-G/A (2′F-CTP: TriLink N-1008-013008; 2′F-UTP: TriLink N1010) (2′OH-ATP Roche 14470220; 2′OH-GTP Roche 14611221).
Inorganic pyrophosphatase (IPPase) (Thermo Fisher Scientific cat #EF0221).
T7 (Y639F) Polymerase [20].
DNAse1 10 Units/μL (Roche #04716728001).
Chloroform (Fisher Scientific # CAS 67-66-3).
Ammonium Persulfate; APS (Sigma # A3678).
TEMED (Sigma Aldrich #T9281).
10% acrylamide gel: 115 g Urea (rpi U20200-1000), 62.5 mL 40% acrylamide:bis 29:1 (Bio-Rad 161-0146), 12.5 mL 10× TBE (rpi, T32024-4000) and bring to 250 mL with dH2O. Filter and store at 4 °C.
20 × 20 cm Vertical Electrophoresis System (Fisher Biotech FB-VE20-1).
Power supply (Bio-rad model 3000Xi).
Spacers and combs (see note #2).
Acetone (Sigma # 320110).
75% Ethanol (Sigma #459844).
1% Alconox (Sigma #242985).
2× Formamide gel loading buffer. 0.01 g Xylene Cyanol (Sigma Aldrich # X4126), 0.01 g Bromophenol Blue (Sigma Aldrich #B5525), 500 μL 10x TBE (rpi, T32024-4000), 10 mL formamide (Amresco #0464-500).
Fluor-coated TLC plate (Ambion #10110).
0.5 M EDTA (Ambion #AM9260G).
0. mM EDTA 10 mM TE pH 7.5 (Affymetrix #75793).
0.2 μm cellulose acetate Centrex MF filter (Whatman #10467013).
0.45 μm Pore SFCA Membrane (Corning #431220).
10 kDa MWCO regenerated cellulose centrifugal filter (Millipore UFC801024).
15 mL tubes (Celltreat #229410).
Nanodrop 1000 (Thermo Scientific).
Handheld 254 nm UV lamp.
2.3. Reagents for RNA library selection
0.1 M DTT (Sigma #D9779).
10× Binding Buffer: 100 mL 1 M HEPES (Sigma #H3375), 150 mL 5 M NaCl (Sigma #S9888), 10 mL 1 M CaCl2 (Sigma #C3881), 240 mL MQ H2O.
Linear acrylamide 5 mg/mL (Ambion AM9520).
10 M ammonium acetate: 770 g ammonium acetate (Amresco 01023) in 1 L distilled H2O. Solution should be filter-sterilized.
0.45 μm Nitrocellulose-mixed Esters Syringe Filters (Fisher #DDE04025SM).
0.2 μm cellulose acetate Centrex MF filter (Whatman #10467013).
Phenol:Chloroform:Isoamyl Alcohol (25:24:1, v/v) (Fisher #15593-031).
Chloroform (Fisher Scientific # CAS 67-66-3).
Ethanol 200 proof (Sigma #459844).
2.4. Reagents for reverse transcription and SELEX PCR
10 mM dNTP Mix. (Invitrogen #4893).
SuperScript® III Reverse Transcriptase (RT) (Thermofisher #18080-044).
100 μM Sel3 3′-primer (5′-TCGGGCGAGTCGTCTG-3′; TriLink BioTechnologies, Inc.).
Taq DNA polymerase 5 Units/μL (Denville Scientific #CB4050-1).
Molecular biology grade agarose (RPI Corp. # Cas 9012-36-6).
TAE buffer (Fisher Scientific #BP1335-4).
6× gel loading dye (New England Biolabs Inc. # B7021S).
100 base pair DNA ladder (New England Biolabs Inc. #N3231).
Minigel System; 7 × 10 cm gel size (Fisher Scientific #FB-SB-710).
PowerPac Basic Power Supply (Bio-Rad #164-5050).
Ethidium Bromide (Sigma #E1510).
Molecular Imager Gel Doc XR System (BioRad #107-8170).
2.5. Reagents for Illumina deep sequencing
Qiagen gel extraction kit (Qiagen #28704).
10 mM dNTP Mix. (Invitrogen #4893).
SuperScript® III Reverse Transcriptase (RT) (Thermofisher #18080-044).
Taq DNA polymerase 5 Units/μL (Denville Scientific #CB4050-1).
Molecular biology grade agarose (RPI Corp. # Cas 9012-36-6).
TAE buffer (Fisher Scientific #BP1335-4).
6× gel loading dye (New England Biolabs Inc. # B7021S).
100 base pair DNA ladder (New England Biolabs Inc. #N3231).
Minigel System; 7 × 10 cm gel size (Fisher Scientific #FB-SB-710).
PowerPac Basic Power Supply (Bio-Rad #164–5050).
Ethidium Bromide (Sigma #E1510).
Molecular Imager Gel Doc XR System (BioRad #107-8170).
NGS 5′ (5′-AATGATACGGCGACCACCGAGATCTACACTCTTTCC CTACACGACGCTCTTCCGATCT-8 nt Barcode-GGGAGGAC GATGCGG-3′);
NGS 3′ (5′-CAAGCAGAAGACGGCATACGAGCTCTTCC GATCTT CGGGCGAGTCGTCTG-3′).
3. Methods
3.1. Synthesis of the initial (round 0) double stranded DNA aptamer library
3.1.1. Annealing and elongation
Combine 9 μL of 100 mM Tris-HCl pH 8.0,15 μL of 50mM MgCl2, 20 μL (2 nmol) of 100 mM Sel3 5′ primer, 10 μL (1 nmol) of 100 mM Sel3 template oligo, and bring the volume up to 90 μL with MQ H2O (36 μL) to make master mix 1 (MM1).
Aliquot MM1 into five PCR tubes with 18 μL/tube, then place into the thermal cycler and run the following protocol: (1) 95 °C for 5 min; (2) 25 °C for 20 min.
In a separate microcentrifuge tube add 50 μL of 10× PCR buffer, 10 μL of 10 mM dNTP mix, 4 μL of Choice Taq and bring the volume up to 412 with MQ H2O (348 μL) to make master mix 2 (MM2).
Heat MM2 to 95 °C for 5 min and then allow to cool to ambient temperature for 20 min.
Add 82 μL of MM2 to each tube containing MM1, then place into the thermal cycler and run the following protocol: (1) 72 °C for 30 min; (2) 25 °C for 10 min; and (3) hold at 4 °C. To make the Sel3 DS DNA template oligo (Sel3 DS).
3.1.2. Purification of the double-stranded DNA aptamer library
To purify the Sel3 DS use the QIAquick Gel Extraction Kit (Qiagen #28704) as described below.
-
Use 4 Qiagen Miniprep (Qiagen #27115) columns to purify the DNA duplexes:
Add 5 vol of Buffer PB (component of QIAquick Gel extraction kit) to the Sel3 DS reaction and mix.
Add 750 μL of the mixture to each column and spin in microcentrifuge at maximum speed for 1 min.
Re-run flow-through in the spin column to collect any DNA that may have passed through and spin again for 1 min.
Discard flow-through.
Add 500 μL Buffer PE (component of QIAquick Gel extraction kit) to each column and spin for 1 min and discard flow-through.
Spin again for 1 min to dry filter.
Transfer columns to fresh micro centrifuge tubes and add 60 μL MQ H2O to each.
Incubate at room temp for 1 min and then spin for 1 min.
Pool all the samples together.
To determine presence of the purified Sel3 DS samples run 5–10 μL of purified sample on a 3% agarose gel. The size of the DNA duplex for an N20 library should be 70–80 bp.
If the proper band size is seen on the gel, determine concentration of the purified Sel3 DS by UV spectrometry.
3.2. Generation of the round 0 RNA aptamer library
3.2.1. In vitro transcription
For the round 0 transcription of RNA aptamers, combine 50 μL of 5X T7 RNAP Buffer, 25 μL of 10× rNTP mix (w/2′-Fs), 2 μL of IPPI, 2 μL of the T7 (Y639F) Polymerase (see note #3), 125 pmol of the purified Sel3 DS, and bring to volume up to 250 μL with MQ H2O. For subsequent rounds use 62.5 pmol of the Sel3 DS and scale other reagents down to 125 μL.
Incubate transcription at 37 °C for 4 h to overnight.
3.2.2. Purification of round 0 RNA aptamer library
To the 250 μL transcription reaction add 1 μL DNase I and incubate at 37 °C for 10 min.
After the 10 min, add 250 μL of chloroform to the mixture, vortex, and then spin at maximum speed for 10 min.
Carefully pipette top aqueous phase into a fresh microfuge tube.
Add one volume of 2× formamide RNA loading dye and heat the RNA-dye solution to 65 °C for 10 min prior to loading onto the acrylamide gel.
3.2.3. RNA gel purification (see note #4)
To generate the solidifying gel-solution add 75 μL of 10% APS and 25 μL of TEMED to 25 mL of the 10% acrylamide gel.
Mix the gel solution by inverting and quickly add to the assemble glass plates by using a 25 mL pipette, add the wells quickly, and let the gel solidify for about 20 min.
Place the glass plates into the gel apparatus (Fig. 3) and fill it with 0.5× TBE. Then preheat polymerized gel at 24W for 30 min.
After the 30 min, wash out the wells to remove urea and add the RNA-Dye solution to each well.
Let the gel run at 24W for 30–45 min or until the dye band is close to the bottom of the gel.
Carefully separate the plates and place the gel on a piece of plastic wrap.
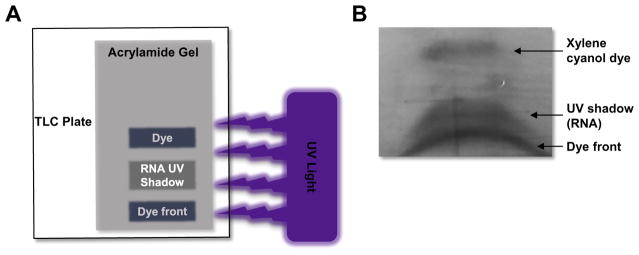
Then place the gel/plastic wrap on a fluor-coated TLC plate (Ambion #10110) and use the handheld UV lamp to detect the RNA aptamer library by UV shadowing (Fig. 4). Quickly excise the UV shadow band using a clean razor blade.
To elute RNA, place the excised gel fragment into a 15 mL conical tube containing 8 mL of 0.1 mM EDTA 10 mM TE pH 7.5. Gently shake/rotate at 37 °C for at least 1 h. Perform the elution twice with an additional 8 mL of 0.1 mM EDTA 10 mM TE pH 7.5.
To remove any residual gel fragments from the 16 mL of eluted RNA use either a 0.2 μm cellulose acetate Centrex MF filter or a 0.45 cellulose acetate syringe filter.
Transfer the 16 mL of eluted RNA to 10 kDa MWCO cellulose centrifugal filter and centrifuge at ~4000× g for 15 min. Discard the flow through (see note #5).
Wash the concentrated eluted RNA by adding 4 mL 0.1 mM EDTA 10 mM TE pH 7.5 to the 10 kDa MWCO filter and centrifuge at ~4000× g for 10–15 min. Repeat the wash step twice.
Centrifuge at ~4000× g for 30 min to concentrate the RNA down to ~100–200 μL.
Determine the concentration of purified Sel3 RNA by UV spectrometry.
-
Fold the RNA library
Dilute the purified RNA Sel3 library to concentrations of 1–10 μM in Binding buffer and proceed to fold by incubation at (1) 95 °C for 5–10 min; (2) 65 °C for 10–15 min; and (3) 37 °C for 20 min.
Folded RNA can be stored at −20 °C. Thaw at 37 °C for 20 min prior to use.
Fig. 3.
Gel running apparatus (Fisher Biotech FB-VE20-1, left) and high-voltage power supply (Bio-Rad model 3000Xi, right).
Fig. 4.
Visual representation of UV shadowing. (A) Schematic of set-up for UV-shadowing of RNA. The denaturing urea-PAGE gel (shown in grey) is positions on top of the TLC plate (shown in white). UV light (purple) is shined on top of the gel. A shadow will appear in the place where the RNA aptamer is on the gel. The RNA aptamer runs between the top dye (xylene cyanol) and the dye front (bromophenol blue). (B) UV shadow of an RNA aptamer visualized using the TLC plate method. The RNA is run on a denaturing Urea-PAGE gel until the dye front reaches the bottom of the gel. The RNA band (shadow) can be easily excised using a clean razor blade and the RNA eluted from the gel in 1X TE buffer.
3.3. First round of selection
-
Negative selection step (preclear) (see note #6)
Dilute control proteins used in the negative selection step (e.g. serum, IgG, Albumin) in 1× binding buffer for a final concentration of 10–1000 pmol (volumes may vary based the ratio of protein:RNA that is used for the negative selection) (see note # 7).
Mix the diluted control proteins and ~2000 pmol of the folded, purified RNA from above and incubate at 37 °C for 10–30 min.
To the protein/aptamer solution above, add a nitrocellulose disk for 10–30 min. The nitrocellulose binds the control protein and any aptamers that are bound to the control protein.
Discard the nitrocellulose disk and add the residual solution to a syringe and use it to force the solution through a 0.45 μm Nitrocellulose-mixed esters syringe filter, collecting the flow-through. This flow-through is referred to as the precleared solution, and should be devoid of aptamers that bind either the pre-cleared proteins or nitrocellulose.
-
Selection steps
Add 100–1000 pmol of your selected protein target to the precleared solution and incubated at 37 °C for 10–30 min.
Add a nitrocellulose disk (2 × 2 cm) to the solution and incubate at 37 °C for 10–30 min. The nitrocellulose disk will retain the aptamer protein complexes.
Transfer the nitrocellulose disk to a clean conical.
Wash the nitrocellulose disk by adding 10 mL of 1× binding buffer and incubating at 37 °C for 10–30 min. Repeat the wash step 2 more times.
To dry the nitrocellulose disk, place it in a 0.2 μm cellulose acetate Centrex MF filter and centrifuge at ~4000× g for 1 min.
-
Extraction of selected aptamers
Placed dried nitrocellulose disk in a microcentrifuge tube, add 300 μL of Phenol:Chloroform:Isoamyl Alcohol, and vortex for 1 min.
Add 300 μL double distilled H2O (ddH2O), vortex for 1 min, and centrifuge at maximum speed for 10 min.
Collect the aqueous (top) layer and place into a new microcentrifuge tube. Add 300 μL of chloroform, vortex for 1 min, and centrifuge at maximum speed for 10 min.
Collect the aqueous layer and place in a new microcentrifuge tube. Add 5 μL linear acrylamide, 1/10 vol 10 M ammonium acetate and 2.5 vol 100% ethanol. Mix well by vortexing.
Incubate the solution at −20 °C overnight.
Centrifuge the solution at maximum speed for 15 min in a refrigerated centrifuge (4 °C). Afterward, there should be a translucent pellet at the bottom of the tube.
Remove the supernatant and wash the pellet with 1 mL 95% ethanol.
Centrifuge the solution at 4 °C for 5 min at full speed.
Discard the supernatant, air-dry the pellet, and dissolve in 25 μL ddH2O. This solution contains your recovered aptamer RNA.
3.4. Reverse transcription and PCR (see note #8)
To make the reverse transcription reaction, combine 10 μL 5× FS buffer, 1 μL 0.1 M DTT, 1 μL 100 μM Sel3 3′-primer, 31 μL PCR-grade H2O, and 5 μL recovered aptamer RNA (from Section 3.3) in a PCR tube. Using a thermal cycler, incubate this “RT MM” solution at 65 °C for 5 min and then, 22 °C for 5 min. Hold at 25 °C.
Add 1 μL 10 mM dNTP mix and 1 μL Superscript III reverse transcriptase to the RT MM solution and incubate at 55 °C for 60 min. Incubate at 72 °C for 15 min to denature the enzyme then hold at 4 °C.
In a fresh microcentrifuge tube, combine 50 μL 10× Taq polymerase buffer, 20 μL 10 mM dNTP mix, 5 μL 100 μM Sel3 5′-primer, 5 μL 100 μM Sel3 3′-primer, 25 units of Taq polymerase, 25 μL of the RT reaction (from step 2) and bring to 500 μL with PCR-grade H2O.
Divide the PCR mix into five PCR tubes (100 μL each) and run the protocol: (1) 95 °C for 2 min; (2) 22–25 cycles of 95 °C for 30 s, 55 °C for 30 s and 72 °C for 5 s; (3) 72 °C for 5 min and (4) hold at 4 °C.
-
Proceed to Section 3.1.2 and repeat for several rounds.
3.5. High-throughput sequencing
The following protocol is based on Illumina Sequencing technology, although other Next Generation Sequencing (NGS) methods can also be used. DNA or RNA from any given round of selection can be used (see note #10).
For the selected RNA aptamer material, first perform an RT reaction as described in Section 3.4.1–3.4.2, but substitute the Sel3 3′-primer with the NGS-3′ primer.
Using the reverse-transcribed RNA aptamer material above or dsDNA from a given selection round, follow the Selex PCR from Section 3.4.3, using the NGS primers. A control with no DNA aptamer material should also be run with each sample. If rounds of selection are going to be multiplexed for NGS, the forward or 5′-NGS primers should contain unique barcodes.
Run the resulting PCR reactions on a 1.5% agarose gel containing ethidium bromide.
-
Extract the barcoded DNA using the Qiagen gel extraction kit
Using a UV lamp and a clean, sharp scalpel, quickly identify and excise the desired DNA fragments from the agarose gel. Cut close to the band to minimize excision of unwanted DNA fragments.
Determine the mass of the gel slice by weighing the tube and subtracting the weight of an equivalent empty tube. Add 3 vol Buffer QG, assuming a mass-volume equivalence of 1mg = 1 μL.
Incubate the tube at 50 °C for roughly 10 min until the gel slice has completely dissolved. Vortex the tube every 2 min during the incubation.
Add an equal volume of isopropanol to the dissolved sample tube and invert.
Transfer the contents of the tube to a QIAquick spin column placed in the 2 mL collection tube provided and centrifuge for 60 s at maximum speed in a benchtop centrifuge. If the volume of the dissolved fragment exceeds 800 μL, use more than one column.
Discard flow-through and replace QIAquick column in the collection tube. Add 0.5 mL of Buffer QG to QIAquick column, centrifuge for 1 min, and discard flow-through.
To wash the column-bound DNA, add 0.75 mL of Buffer PE, centrifuge 1 min, and discard flow-through. Re-centrifuge the QIAquick column for an additional 1 min at ≥10,000× g (~13,000 rpm) to remove residual ethanol.
Place QIAquick column into a clean 1.5 mL microcentrifuge tube and add 30–50 μL of PCR grade ddH2O to the center of the QIAquick column and centrifuge the column for 1 min at maximum speed.
Perform quality/size analysis prior to Next Generation Sequencing (NGS) (see note #11 and 12). To obtain an accurate concentration of the DNA sample submitted for sequencing use a commercially available Qubit Assay and follow manufacturer’s recommendations. (www.invitrogen.com/qubit). Sample quality can also be determined using an Agilent bioanalyzer (Agilent Model 2100 Bioanalyzer).
For downstream data analysis, please refer to “Analyzing HT-SELEX data with the Galaxy Project tools – A web based bioinformatics platform for biomedical research” [41] and http://www.medicine.uiowa.edu/humangenetics/genomics/.
4. Notes
A library with a variable region of 20 nucleotides can contain a maximum of ~1012 (420 = 1.1 × 1012) distinct sequences, and the population of the library produced under the conditions described is ~1014. Hence, these conditions allow for complete coverage of the sequence complexity of the library to ~100-fold multiplicity. However, libraries in which sequence coverage is not complete (i.e., with 30–60 nt variable regions) have also yielded aptamers with high affinities for their targets.
Combs and spacers can be constructed from plastic file folders (Staples #486060), allowing for ultra-thin gels with larger wells.
The optimal amount of Y639F T7 RNA polymerase used for in vitro transcription is determined experimentally, and is dependent the activity of a given enzyme preparation. The Y639F T7 RNA polymerase enzyme can also be acquired from Epicentre (#TH950K).
RNA purification can also be performed using commercially available RNA purification columns [42]. However, column purification is not recommended for aptamer selection in this protocol as it favors retention of aberrant transcripts (i.e., smaller transcripts produced by early transcription termination).
The 10 kDa MWCO cellulose centrifugal filters hold only 5 mL of buffer. The 16 mL of eluted RNA can be applied to the same filter in sequential steps until the total volume is reduce to ~100–200 μL.
The negative (preclear) is unique to a given selection. For example, if the target protein is tagged, an appropriate negative selection step will be to preclear against the tag. This will ensure that the resulting aptamers are specific for the target and not the tag. Common control proteins used in the in vitro selection process include serum, human IgG and BSA.
Typically, the ratio of protein:RNA is varied to favor the selection of high affinity binders. In the case of the negative selection a high protein:RNA ratio will ensure the elimination of sequences that bind with high affinity to the negative control proteins.
RT/PCR amplification step: One step kits can also be used at this step in the protocol (e.g., SuperScript™ III One-Step RT-PCR System with Platinum ®Taq High Fidelity-ThermoFisher #12574-018). This enables the RT and PCR steps to be completed in the same reaction, reducing potential contamination during the PCR amplification step and expediting the SELEX process.
The progress of the selection can be monitored during the selection process by performing a Complexity Assay [26]. Furthermore, the % sequence enrichment of the RNAs in a given round can be determined from the NGS data [41].
Several different next generation sequencing platforms are available. The Illumina technology uses clonal amplification and sequencing by synthesis (SBS) chemistry to enable rapid, accurate sequencing. This process simultaneously identifies DNA bases while incorporating them into a nucleic acid chain. Each base emits a unique fluorescent signal as it is added to the growing strand, which is used to determine the order of the DNA sequence. Importantly, the Illumina technology sequences large stretches of DNA (covering the length of the aptamer libraries) and offers high throughput and flexibility to scale studies and sequence multiple samples simultaneously [43,44].
Samples for NGS can be submitted to the University of Iowa Genomics Core Facility (http://www.medicine.uiowa.edu/humangenetics/genomics/). The facility is also set up to perform quality analysis with the Agilent Bioanalyzer (http://www.medicine.uiowa.edu/humangenetics/genomics/qc/).
Due to the high sequence complexity of the initial library, the quality/size analysis of the samples will result in a broader peak than in subsequent rounds of selection.
Disclosures
Dr. Anton McCaffrey is the Sr. Director of Research Biology at TriLink BioTechnologies Inc.
Acknowledgments
This work was supported by the National Institutes of Health (R01CA138503 and R21DE019953 to PHG), Mary Kay Foundation (9033-12 and 001-09 to PHG), Elsa U. Pardee Foundation (E2766 to PHG), and the Roy J. Carver Charitable Trust (RJCCT 01-224 to PHG). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Keefe AD, Pai S, Ellington A. Aptamers as therapeutics. Nat Rev Drug Discovery. 2010;9(7):537–550. doi: 10.1038/nrd3141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Diafa S, Hollenstein M. Generation of aptamers with an expanded chemical repertoire. Molecules. 2015;20(9):16643–16671. doi: 10.3390/molecules200916643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dassie JP, et al. Targeted inhibition of prostate cancer metastases with an RNA aptamer to prostate-specific membrane antigen. Mol Ther. 2014;22(11):1910–1922. doi: 10.1038/mt.2014.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rockey WM, et al. Rational truncation of an RNA aptamer to prostate-specific membrane antigen using computational structural modeling. Nucleic Acid Ther. 2011;21(5):299–314. doi: 10.1089/nat.2011.0313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ellington AD, Szostak JW. In vitro selection of RNA molecules that bind specific ligands. Nature. 1990;346(6287):818–822. doi: 10.1038/346818a0. [DOI] [PubMed] [Google Scholar]
- 6.Tuerk C, Gold L. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science. 1990;249(4968):505–510. doi: 10.1126/science.2200121. [DOI] [PubMed] [Google Scholar]
- 7.Tasset DM, Kubik MF, Steiner W. Oligonucleotide inhibitors of human thrombin that bind distinct epitopes. J Mol Biol. 1997;272(5):688–698. doi: 10.1006/jmbi.1997.1275. [DOI] [PubMed] [Google Scholar]
- 8.Bruno JG, Kiel JL. In vitro selection of DNA aptamers to anthrax spores with electrochemiluminescence detection. Biosens Bioelectron. 1999;14(5):457–464. doi: 10.1016/s0956-5663(99)00028-7. [DOI] [PubMed] [Google Scholar]
- 9.Jenison RD, et al. Oligonucleotide inhibitors of P-selectin-dependent neutrophil-platelet adhesion. Antisense Nucleic Acid Drug Dev. 1998;8(4):265–279. doi: 10.1089/oli.1.1998.8.265. [DOI] [PubMed] [Google Scholar]
- 10.Blank M, et al. Systematic evolution of a DNA aptamer binding to rat brain tumor microvessels. Selective targeting of endothelial regulatory protein pigpen. J Biol Chem. 2001;276(19):16464–16468. doi: 10.1074/jbc.M100347200. [DOI] [PubMed] [Google Scholar]
- 11.Rusconi CP, et al. RNA aptamers as reversible antagonists of coagulation factor IXa. Nature. 2002;419(6902):90–94. doi: 10.1038/nature00963. [DOI] [PubMed] [Google Scholar]
- 12.Giangrande PH, et al. Distinct roles of E2F proteins in vascular smooth muscle cell proliferation and intimal hyperplasia. Proc Natl Acad Sci USA. 2007;104(32):12988–12993. doi: 10.1073/pnas.0704754104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mi J, et al. RNA aptamer-targeted inhibition of NF-kappa B suppresses non-small cell lung cancer resistance to doxorubicin. Mol Ther. 2008;16(1):66–73. doi: 10.1038/sj.mt.6300320. [DOI] [PubMed] [Google Scholar]
- 14.Lee JW, Kim HJ, Heo K. Therapeutic aptamers: developmental potential as anticancer drugs. BMB Rep. 2015;48(4):234–237. doi: 10.5483/BMBRep.2015.48.4.277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McNamara JO, et al. Multivalent 4-1BB binding aptamers costimulate CD8+ T cells and inhibit tumor growth in mice. J Clin Invest. 2008;118(1):376–386. doi: 10.1172/JCI33365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sundaram P, et al. Therapeutic RNA aptamers in clinical trials. Eur J Pharm Sci. 2013;48(1–2):259–271. doi: 10.1016/j.ejps.2012.10.014. [DOI] [PubMed] [Google Scholar]
- 17.Ruckman J, et al. 2′-Fluoropyrimidine RNA-based aptamers to the 165-amino acid form of vascular endothelial growth factor (VEGF165). Inhibition of receptor binding and VEGF-induced vascular permeability through interactions requiring the exon 7-encoded domain. J Biol Chem. 1998;273(32):20556–20567. doi: 10.1074/jbc.273.32.20556. [DOI] [PubMed] [Google Scholar]
- 18.Floege J, et al. Novel approach to specific growth factor inhibition in vivo: antagonism of platelet-derived growth factor in glomerulonephritis by aptamers. Am J Pathol. 1999;154(1):169–179. doi: 10.1016/S0002-9440(10)65263-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Adler A, et al. Post-SELEX chemical optimization of a trypanosome-specific RNA aptamer. Comb Chem High Throughput Screening. 2008;11(1):16–23. doi: 10.2174/138620708783398331. [DOI] [PubMed] [Google Scholar]
- 20.Sousa R, Padilla R. A mutant T7 RNA polymerase as a DNA polymerase. EMBO J. 1995;14(18):4609–4621. doi: 10.1002/j.1460-2075.1995.tb00140.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Siegmund V, et al. Screening mutant libraries of T7 RNA polymerase for candidates with increased acceptance of 2′-modified nucleotides. Chem Commun (Camb) 2012;48(79):9870–9872. doi: 10.1039/c2cc35028a. [DOI] [PubMed] [Google Scholar]
- 22.Chelliserrykattil J, Ellington AD. Evolution of a T7 RNA polymerase variant that transcribes 2′-O-methyl RNA. Nat Biotechnol. 2004;22(9):1155–1160. doi: 10.1038/nbt1001. [DOI] [PubMed] [Google Scholar]
- 23.Padilla R, Sousa R. A Y639F/H784A T7 RNA polymerase double mutant displays superior properties for synthesizing RNAs with non-canonical NTPs. Nucleic Acids Res. 2002;30(24):e138. doi: 10.1093/nar/gnf138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Burmeister PE, et al. 2′-Deoxy purine, 2′-O-methyl pyrimidine (dRmY) aptamers as candidate therapeutics. Oligonucleotides. 2006;16(4):337–351. doi: 10.1089/oli.2006.16.337. [DOI] [PubMed] [Google Scholar]
- 25.Thiel KW, et al. Delivery of chemo-sensitizing siRNAs to HER2+-breast cancer cells using RNA aptamers. Nucleic Acids Res. 2012;40(13):6319–6337. doi: 10.1093/nar/gks294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Thiel WH, et al. Rapid identification of cell-specific, internalizing RNA aptamers with bioinformatics analyses of a cell-based aptamer selection. PLoS One. 2012;7(9):e43836. doi: 10.1371/journal.pone.0043836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Huang YZ, et al. RNA aptamer-based functional ligands of the neurotrophin receptor, TrkB. Mol Pharmacol. 2012;82(4):623–635. doi: 10.1124/mol.112.078220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Thiel WH, et al. Nucleotide bias observed with a short SELEX RNA aptamer library. Nucleic Acid Ther. 2011;21(4):253–263. doi: 10.1089/nat.2011.0288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cho M, et al. Quantitative selection of DNA aptamers through microfluidic selection and high-throughput sequencing. Proc Natl Acad Sci USA. 2010;107(35):15373–15378. doi: 10.1073/pnas.1009331107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hoinka J, et al. Large scale analysis of the mutational landscape in HT-SELEX improves aptamer discovery. Nucleic Acids Res. 2015;43(12):5699–5707. doi: 10.1093/nar/gkv308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Swensen JS, et al. Continuous, real-time monitoring of cocaine in undiluted blood serum via a microfluidic, electrochemical aptamer-based sensor. J Am Chem Soc. 2009;131(12):4262–4266. doi: 10.1021/ja806531z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jenison RD, et al. High-resolution molecular discrimination by RNA. Science. 1994;263(5152):1425–1429. doi: 10.1126/science.7510417. [DOI] [PubMed] [Google Scholar]
- 33.McKeague M, DeRosa MC. Challenges and opportunities for small molecule aptamer development. J Nucleic Acids. 2012;2012 doi: 10.1155/2012/748913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lorger M, et al. Targeting the variable surface of African trypanosomes with variant surface glycoprotein-specific, serum-stable RNA aptamers. Eukaryot Cell. 2003;2(1):84–94. doi: 10.1128/EC.2.1.84-94.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cheng C, et al. In vivo SELEX for identification of brain-penetrating aptamers. Mol Ther Nucleic Acids. 2013;2:e67. doi: 10.1038/mtna.2012.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mi J, et al. In vivo selection of tumor-targeting RNA motifs. Nat Chem Biol. 2010;6(1):22–24. doi: 10.1038/nchembio.277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stoltenburg R, Reinemann C, Strehlitz B. SELEX – a (r)evolutionary method to generate high-affinity nucleic acid ligands. Biomol Eng. 2007;24(4):381–403. doi: 10.1016/j.bioeng.2007.06.001. [DOI] [PubMed] [Google Scholar]
- 38.Szeto K, et al. High-throughput binding characterization of RNA aptamer selections using a microplate-based multiplex microcolumn device. Anal Bioanal Chem. 2014;406(11):2727–2732. doi: 10.1007/s00216-014-7661-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Blind M, Blank M. Aptamer selection technology and recent advances. Mol Ther Nucleic Acids. 2015;4(1):e223. doi: 10.1038/mtna.2014.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ohuchi S. Cell-SELEX technology. BioRes Open Access. 2012;1(6):265–272. doi: 10.1089/biores.2012.0253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Thiel WH, Giangrande PH. Analyzing HT-SELEX data with the galaxy project tools – a web based bioinformatics platform for biomedical research. Methods. 2015 doi: 10.1016/j.ymeth.2015.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhou J, Rossi JJ. Aptamer-targeted RNAi for HIV-1 therapy. Methods Mol Biol. 2011;721:355–371. doi: 10.1007/978-1-61779-037-9_22. [DOI] [PubMed] [Google Scholar]
- 43.Liu L, et al. Comparison of next-generation sequencing systems. BioMed Res Int. 2012;2012 doi: 10.1155/2012/251364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Metzker ML. Sequencing technologies—the next generation. Nat Rev Genet. 2010;11(1):31–46. doi: 10.1038/nrg2626. [DOI] [PubMed] [Google Scholar]