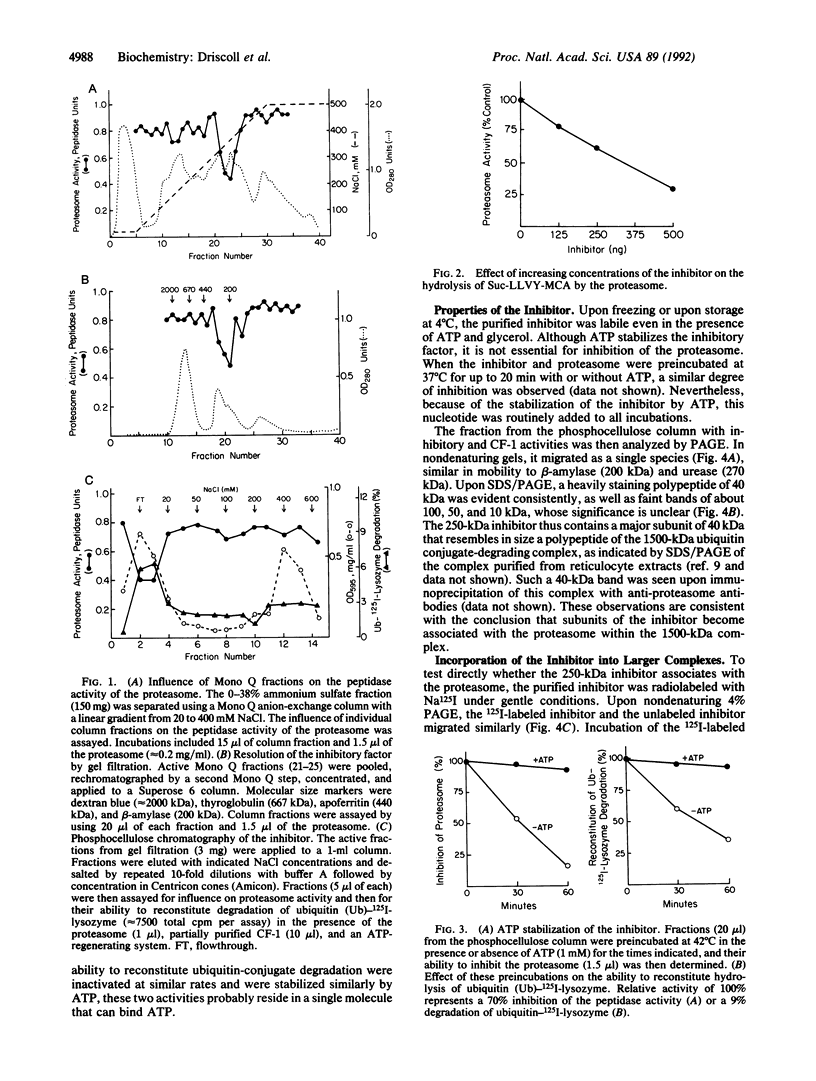
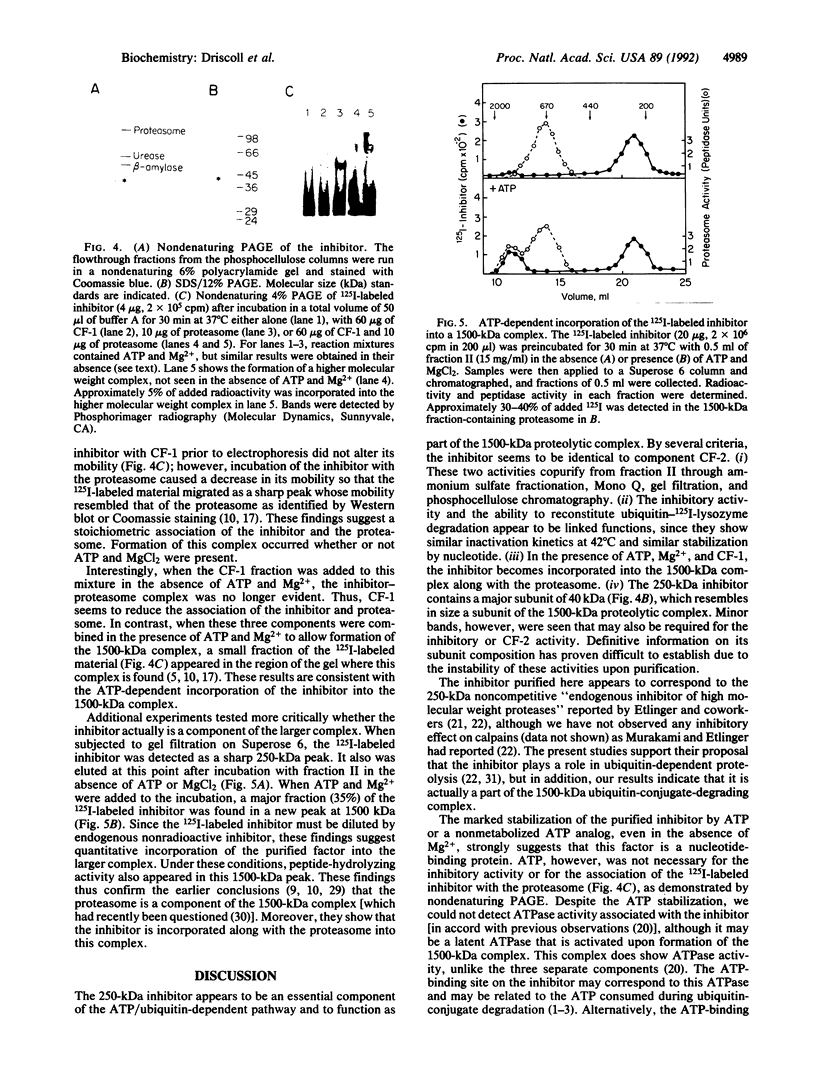
Abstract
Proteins conjugated to ubiquitin are degraded by a 26S (1500-kDa) proteolytic complex that, in reticulocyte extracts, can be formed by the association of three factors: CF-1, CF-2, and CF-3. One of these factors, CF-3, has been shown to be the proteasome, a 650-kDa multicatalytic protease complex. We have purified a 250-kDa inhibitor of the proteasome and shown that it corresponds to CF-2. In the presence or absence of ATP, this factor inhibited hydrolysis by the proteasome of both fluorogenic tetrapeptides and protein substrates. When the inhibitor, proteasome, and CF-1 were incubated together in the presence of ATP and Mg2+, degradation of ubiquitin-125I-lysozyme occurred. Both the inhibitory activity and the ability to reconstitute ubiquitin-125I-lysozyme degradation were very labile at 42 degrees C, but both activities were stabilized by ATP or a nonhydrolyzable ATP analog. SDS/PAGE indicated that the 250-kDa inhibitor fraction contained a major subunit of 40 kDa (plus some minor bands). The 125I-labeled inhibitor and purified proteasome formed a complex. When CF-1, ATP, and Mg2+ were also present, the 125I-labeled inhibitor along with the proteasome formed a complex of 1500 kDa. The inhibitor (CF-2) thus appears to be an ATP-binding component that regulates proteolysis within the 1500-kDa complex.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Armon T., Ganoth D., Hershko A. Assembly of the 26 S complex that degrades proteins ligated to ubiquitin is accompanied by the formation of ATPase activity. J Biol Chem. 1990 Dec 5;265(34):20723–20726. [PubMed] [Google Scholar]
- Arrigo A. P., Tanaka K., Goldberg A. L., Welch W. J. Identity of the 19S 'prosome' particle with the large multifunctional protease complex of mammalian cells (the proteasome). Nature. 1988 Jan 14;331(6152):192–194. doi: 10.1038/331192a0. [DOI] [PubMed] [Google Scholar]
- Ciechanover A., Heller H., Elias S., Haas A. L., Hershko A. ATP-dependent conjugation of reticulocyte proteins with the polypeptide required for protein degradation. Proc Natl Acad Sci U S A. 1980 Mar;77(3):1365–1368. doi: 10.1073/pnas.77.3.1365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciehanover A., Hod Y., Hershko A. A heat-stable polypeptide component of an ATP-dependent proteolytic system from reticulocytes. Biochem Biophys Res Commun. 1978 Apr 28;81(4):1100–1105. doi: 10.1016/0006-291x(78)91249-4. [DOI] [PubMed] [Google Scholar]
- Dahlmann B., Kuehn L., Rutschmann M., Reinauer H. Purification and characterization of a multicatalytic high-molecular-mass proteinase from rat skeletal muscle. Biochem J. 1985 May 15;228(1):161–170. doi: 10.1042/bj2280161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Driscoll J., Goldberg A. L. Skeletal muscle proteasome can degrade proteins in an ATP-dependent process that does not require ubiquitin. Proc Natl Acad Sci U S A. 1989 Feb;86(3):787–791. doi: 10.1073/pnas.86.3.787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Driscoll J., Goldberg A. L. The proteasome (multicatalytic protease) is a component of the 1500-kDa proteolytic complex which degrades ubiquitin-conjugated proteins. J Biol Chem. 1990 Mar 25;265(9):4789–4792. [PubMed] [Google Scholar]
- Etlinger J. D., McMullen H., Rieder R. F., Ibrahim A., Janeczko R. A., Marmorstein S. Mechanisms and control of ATP-dependent proteolysis. Prog Clin Biol Res. 1985;180:47–60. [PubMed] [Google Scholar]
- Eytan E., Ganoth D., Armon T., Hershko A. ATP-dependent incorporation of 20S protease into the 26S complex that degrades proteins conjugated to ubiquitin. Proc Natl Acad Sci U S A. 1989 Oct;86(20):7751–7755. doi: 10.1073/pnas.86.20.7751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fagan J. M., Waxman L., Goldberg A. L. Skeletal muscle and liver contain a soluble ATP + ubiquitin-dependent proteolytic system. Biochem J. 1987 Apr 15;243(2):335–343. doi: 10.1042/bj2430335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falkenburg P. E., Haass C., Kloetzel P. M., Niedel B., Kopp F., Kuehn L., Dahlmann B. Drosophila small cytoplasmic 19S ribonucleoprotein is homologous to the rat multicatalytic proteinase. Nature. 1988 Jan 14;331(6152):190–192. doi: 10.1038/331190a0. [DOI] [PubMed] [Google Scholar]
- Ganoth D., Leshinsky E., Eytan E., Hershko A. A multicomponent system that degrades proteins conjugated to ubiquitin. Resolution of factors and evidence for ATP-dependent complex formation. J Biol Chem. 1988 Sep 5;263(25):12412–12419. [PubMed] [Google Scholar]
- Goldberg A. L. The mechanism and functions of ATP-dependent proteases in bacterial and animal cells. Eur J Biochem. 1992 Jan 15;203(1-2):9–23. doi: 10.1111/j.1432-1033.1992.tb19822.x. [DOI] [PubMed] [Google Scholar]
- Hershko A., Ciechanover A. The ubiquitin pathway for the degradation of intracellular proteins. Prog Nucleic Acid Res Mol Biol. 1986;33:19-56, 301. doi: 10.1016/s0079-6603(08)60019-7. [DOI] [PubMed] [Google Scholar]
- Hershko A., Heller H., Elias S., Ciechanover A. Components of ubiquitin-protein ligase system. Resolution, affinity purification, and role in protein breakdown. J Biol Chem. 1983 Jul 10;258(13):8206–8214. [PubMed] [Google Scholar]
- Hershko A., Heller H. Occurrence of a polyubiquitin structure in ubiquitin-protein conjugates. Biochem Biophys Res Commun. 1985 May 16;128(3):1079–1086. doi: 10.1016/0006-291x(85)91050-2. [DOI] [PubMed] [Google Scholar]
- Hershko A. Ubiquitin-mediated protein degradation. J Biol Chem. 1988 Oct 25;263(30):15237–15240. [PubMed] [Google Scholar]
- Hough R., Pratt G., Rechsteiner M. Purification of two high molecular weight proteases from rabbit reticulocyte lysate. J Biol Chem. 1987 Jun 15;262(17):8303–8313. [PubMed] [Google Scholar]
- Hough R., Pratt G., Rechsteiner M. Ubiquitin-lysozyme conjugates. Identification and characterization of an ATP-dependent protease from rabbit reticulocyte lysates. J Biol Chem. 1986 Feb 15;261(5):2400–2408. [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Matthews W., Driscoll J., Tanaka K., Ichihara A., Goldberg A. L. Involvement of the proteasome in various degradative processes in mammalian cells. Proc Natl Acad Sci U S A. 1989 Apr;86(8):2597–2601. doi: 10.1073/pnas.86.8.2597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGuire M. J., Croall D. E., DeMartino G. N. ATP-stimulated proteolysis in soluble extracts of BHK 21/C13 cells. Evidence for multiple pathways and a role for an enzyme related to the high-molecular-weight protease, macropain. Arch Biochem Biophys. 1988 Apr;262(1):273–285. doi: 10.1016/0003-9861(88)90189-0. [DOI] [PubMed] [Google Scholar]
- McGuire M. J., DeMartino G. N. Purification and characterization of a high molecular weight proteinase (macropain) from human erythrocytes. Biochim Biophys Acta. 1986 Sep 26;873(2):279–289. doi: 10.1016/0167-4838(86)90055-5. [DOI] [PubMed] [Google Scholar]
- Murakami K., Etlinger J. D. Endogenous inhibitor of nonlysosomal high molecular weight protease and calcium-dependent protease. Proc Natl Acad Sci U S A. 1986 Oct;83(20):7588–7592. doi: 10.1073/pnas.83.20.7588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orino E., Tanaka K., Tamura T., Sone S., Ogura T., Ichihara A. ATP-dependent reversible association of proteasomes with multiple protein components to form 26S complexes that degrade ubiquitinated proteins in human HL-60 cells. FEBS Lett. 1991 Jun 24;284(2):206–210. doi: 10.1016/0014-5793(91)80686-w. [DOI] [PubMed] [Google Scholar]
- Seelig A., Kloetzel P. M., Kuehn L., Dahlmann B. Molecular interaction of the proteasome (multicatalytic proteinase). Evidence that the proteasome is not a constituent of the '26 S' multienzyme complex. Biochem J. 1991 Nov 15;280(Pt 1):225–232. doi: 10.1042/bj2800225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Speiser S., Etlinger J. D. ATP stimulates proteolysis in reticulocyte extracts by repressing an endogenous protease inhibitor. Proc Natl Acad Sci U S A. 1983 Jun;80(12):3577–3580. doi: 10.1073/pnas.80.12.3577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka K., Ichihara A. Involvement of proteasomes (multicatalytic proteinase) in ATP-dependent proteolysis in rat reticulocyte extracts. FEBS Lett. 1988 Aug 15;236(1):159–162. doi: 10.1016/0014-5793(88)80306-5. [DOI] [PubMed] [Google Scholar]
- Tanaka K., Ii K., Ichihara A., Waxman L., Goldberg A. L. A high molecular weight protease in the cytosol of rat liver. I. Purification, enzymological properties, and tissue distribution. J Biol Chem. 1986 Nov 15;261(32):15197–15203. [PubMed] [Google Scholar]
- Waxman L., Fagan J. M., Goldberg A. L. Demonstration of two distinct high molecular weight proteases in rabbit reticulocytes, one of which degrades ubiquitin conjugates. J Biol Chem. 1987 Feb 25;262(6):2451–2457. [PubMed] [Google Scholar]
- Wilk S., Orlowski M. Evidence that pituitary cation-sensitive neutral endopeptidase is a multicatalytic protease complex. J Neurochem. 1983 Mar;40(3):842–849. doi: 10.1111/j.1471-4159.1983.tb08056.x. [DOI] [PubMed] [Google Scholar]