Abstract
RNA interference (RNAi) is an extremely useful tool for inhibiting gene expression. It can be triggered by transfected synthetic small interfering RNA (siRNA) or by expressed small hairpin RNA (shRNA). The cellular machinery processes the latter into siRNA in vivo. shRNA is preferred or required in genetic screens and specific RNAi approaches in gene therapy settings. Despite its many successes, the field of shRNAs faces many challenges. Insufficient knockdowns and off-target effects become obstacles for shRNA usage in many applications. Numerous failures are triggered by pitfalls in shRNA design that is often associated with impoverished biogenesis. Here, based on current understanding of the miRNA maturation pathway, we discuss the principles of different shRNA design (pre-miRNA-like, pri-miRNA-like and Ago-shRNA) with an emphasis on the RNA structure. We also provide detailed instructions for an optimal design of pre-miRNA-like shRNA.
1. Introduction
RNA interference (RNAi) is a phenomenon in which double-stranded RNA (dsRNA) inhibits gene expression by degrading mRNA. Since its discovery in 1998 [1], RNAi has revolutionized the way researchers study molecular genetics. The ability of RNAi to target any gene makes it an intriguing platform for developing of next generation therapeutics. In the pursuit to ameliorate the cytotoxic effects resulting from the use of dsRNA, it was discovered that smaller synthetic sequences of 21 to 22 nucleotides, known as small interfering RNA (siRNA), were sufficient to trigger RNAi [2]. However, the main drawback of synthetic siRNAs is their short lifespan, which weakens their ability to regulate gene expression. In order to overcome this limitation two strategies arise: 1) introduce chemical modifications on the backbone of the oligonucleotides to prolong their half-life and 2) express short hairpin-shaped RNA transcripts (shRNA) that can be processed into siRNAs in vivo [3–7].
In parallel to the discovery of RNAi, microRNAs (miRNAs) were identified as an abundant class of small non-coding RNA molecules (~22nt) that are highly conserved among species and can negatively regulate expression of >60% of human genes post-transcriptionally [8]. miRNA are transcribed from the genome into primary transcripts (pri-miRNA) and processed into precursor (pre-miRNA) and mature miRNA by two RNase-III family nucleases, Drosha and Dicer, respectively (reviewed in [9]). Based on the degree of complementarity between the miRNA and its target mRNA, miRNAs can either induce the direct cleavage of the mRNA or initiate a much longer process characterized by translational repression and posterior repression through mRNA decay [10–12].
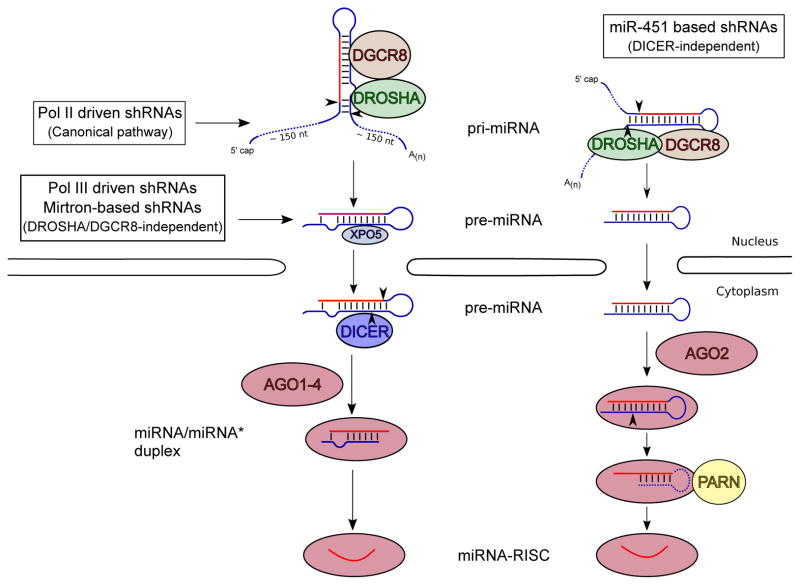
The first line of evidence implicating that miRNA biogenesis and RNAi maturation rely on the same cellular machinery came from the discovery that Dicer was responsible for processing shRNAs into siRNAs [13]. This finding initiated the use of pre-miRNA secondary structures as a scaffold for siRNA production [7]. This first generation of shRNAs (pre-miRNA-like shRNA) was characterized by the use of RNA polymerase III promoters (Pol III). This enabled the precise synthesis of transcripts that folded into pre-miRNA-like stem-loop structures [5]. Mimicking the structures of pri-miRNA opened the door to designing the second generation of shRNAs (pri-miRNA-like shRNA) driven by RNA polymerase II promoters (Pol II) [14, 15] (Fig. 1).
Fig 1. shRNA processing pathways.
The scheme illustrates how pre-miRNA-like shRNA, pri-miRNA-like shRNA and Ago-shRNA designs are sequentially cleaved into their mature form according to their entry points into the miRNA-processing pathway. The figure shows processing triggered by Drosha-DGCR8 complex in the cell nucleus, and after their export to the cytoplasm by Exportin-5, Dicer cleavage and loading into Argonaute proteins to form shRNA-RISC complexes.
These advances illustrated the use of shRNA as a promising tool in gene therapy. Currently, several shRNA drug formulations are under phase I and II clinical trials for treatment of solid tumors and hepatitis C (clinicaltrials.gov 2016). Another widespread application of shRNAs is loss-of-function genetic screening [16–18]. In this regard, efforts of The RNAi Consortium (TRC) have created libraries covering more than 15,000 genes both in human and mice that aim to achieve efficient knockdowns for each gene. Careful screening of these libraries provides a useful tool to identify key molecular targets for the treatment of complex diseases either through gene therapy or conventional drugs.
Nonetheless, the shRNA field still faces many challenges. Unsatisfactory knockdown efficacy and off-target effects rise continue to hamper its applications, often due to flaws in design. Recent advances in the understanding of miRNA biogenesis are often neglected in many shRNA designs. In this article, we summarize and provide guidelines for the optimal design of miRNA-based shRNAs (Table 1), with a focus on RNA structure.
Table 1.
Characteristics shRNA designs
| Design | Structure | Promoter | Depends on Drosha | Depends on Dicer | Advantages / Disadvantages |
|---|---|---|---|---|---|
| Pre-miRNA-like “1st generation” | Pre-miRNA | Pol III | ✗ | ✓ | + Well-characterized system −Competition with endogenous miRNA pathway −Lack promoter-directed specificity |
| Pri-miRNA-like “2nd generation” | Pri-miRNA | Pol II / III | ✓ | ✓ | + Potential promoter-driven cell-type specificity −Complicated processing |
| − Mirtron-based | + Mirtron | Pol II | ✗ | ✓ | + Expressed within host mRNA transcripts − Alternative splicing / specific regulation? |
| − Cluster mimic | + Cluster | Pol II | ✓ | ✓ | + Multiplexed expression of various shRNAs − Interdependencies within individual shRNAs |
| Ago-shRNA | miR-451 | Pol II / III | ✓ | ✗ | + Guide strand is loaded into AGO2 + Restrict off-targets effects of passenger strand −Variable 3′-5′ exonucleolytic activity (23–26 nt mature) |
2. Promoter selection
The starting point for shRNA design is selection of an expression cassette. Like other gene products, shRNAs can be transcribed from any promoter in commercially available expression vectors. However, due to its non-coding nature, certain considerations need to be taken into account to allow transcripts to fold into correct structures, for the recognition and processing by the miRNA biogenesis machinery.
2.1. Pol III promoters
“First generation” shRNA mimics the structure of pre-miRNA, which is a hairpin with 2nt overhangs at the 3′ end. The sequence of shRNA transcripts needs to be well defined to fit such a structure. Pol III promoters initiate transcription at a precise position (23 nt away from the TATA box) and end it within a track of thymidines (T). This unique feature makes it perfect to express exact sequences of pre-miRNA-like shRNAs. Pol II promoters, on the other hand, are coupled with post-transcription processing (capping and polyA addition) and therefore incapable of such task.
The first nucleotide of shRNA should align perfectly with the transcription start site. A common mistake is leaving a gap between these two due to the insertion of cloning sites. This gap creates extra nucleotides on the 5′ end of the intended hairpin structure, jeopardizing its recognition by Dicer and hence lowering shRNA efficacy. Of note, Pol III promoters generally exhibit a strong preference for starting transcription with a purine. To accommodate this, the first nucleotide of Pol III-driven shRNAs should be either G or A [19]. Failure to do so will force transcription to start at a different position nearby, producing shRNAs with suboptimal structures.
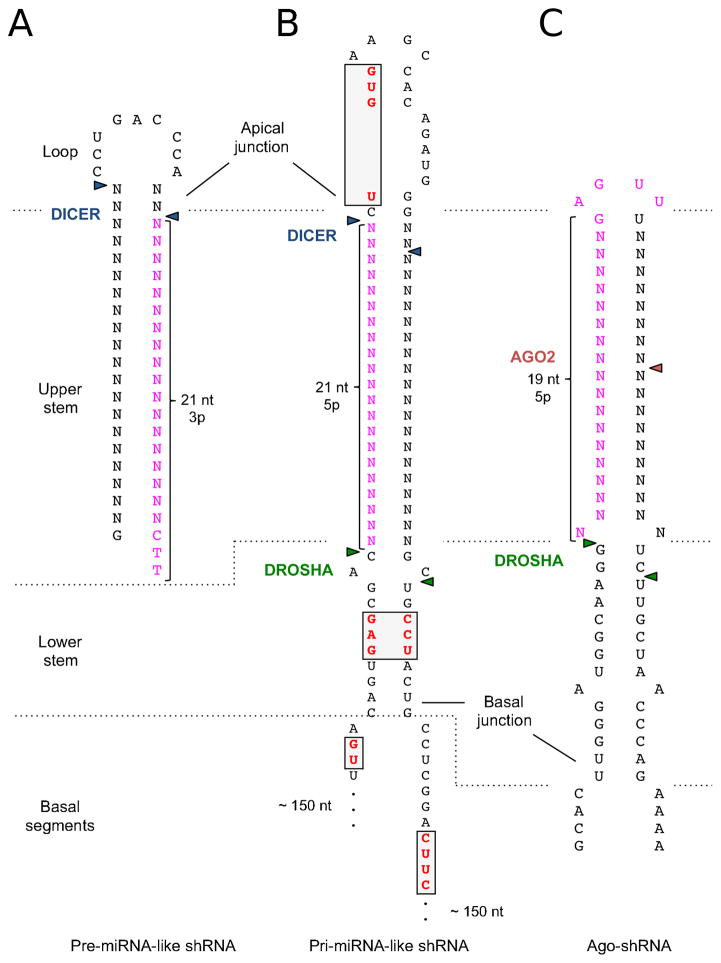
A similar consideration applies to transcriptional termination. A track of five or more Ts in the DNA template is sufficient, leaving most Pol III transcripts with two uridines (U) at the 3′ end [20, 21]. It is important to note that these two Us will automatically be added at the end of any Pol III transcript. To achieve the correct pre-miRNA-like structure, the last nucleotide of the shRNA sequence before the termination signal (≥5 T) should be the end of the hairpin, leaving those two Us from the termination sequence as a 2nt 3′ overhang (Fig. 2A).
Fig 2. Structures involved in the different shRNA designs.
Secondary structures of different shRNA designs. Triangles indicate cleavage sites of the RNases involved in the maturation process. Highlighted in pink is the putative location of shRNA guide sequence, and in red within the grey box are the described motifs. A) Pre-miRNA-like shRNA (“first generation”), hairpin containing miR-22 hairpin B) Pri-miRNA-like shRNA (“second generation”), based on the pri-mir-30a (note incomplete flanking 5′ and 3′ regions) C) Ago-shRNA based on pri-mir-451. Also see Table 1.
The most widely used pol III promoter is U6 [3, 22], followed by H1 [3] or 7SK [23]. While H1 can initiate transcription with pyrimidine or a purine, fidelity of the starting position is less accurate [19]. On the contrary, the U6 promoter is known to be robust and possess a more precise start. Thus, U6-driven shRNAs are more potent, compared to those transcribed by Pol II, in a manner that is dependent on cellular context [24].
The main disadvantage of Pol III promoters hindering their application as therapeutic agents is the toxicity caused by their interference with the endogenous miRNA processing pathways [25]. This concern can be minimized by the use of weaker Pol II promoters at the expense of a possible loss in the therapeutic efficacy.
2.2. Pol II promoters
One of the main features of Pol II promoters is that they allow for extensive customization of gene expression. shRNA can be transcribed from ubiquitous (e.g. CMV or SV40), tissue/cell-type specific (e.g. ALB, INS or Pdx-1), or inducible promoters (e.g. Tet-on, Tet-off). However, poly-adenylylation coupled with Pol II transcription abolishes its ability to express RNA with clear-cut ends. Therefore, it is not surprising that pre-miRNA-like shRNAs driven by Pol II are not recognized by Dicer, and therefore nonfunctional [26]. Instead, the shRNA sequence needs to be embedded into the structure of a primary miRNA (pri-miRNA) [7], in order to be efficiently recognized and excised from the transcript by Drosha [15].
Another advantage of using Pol II and pri-miRNA structures is the ability to express multiple shRNAs simultaneously in one transcript [27]. In fact, more than 40% of human miRNAs are grouped in clusters [28]. However, processing of miRNAs within a cluster is complex as the processivity of each miRNA/shRNA depends on and is influenced by that of others in the cluster [29].
In addition, Pol II promoters can be used to express miRNAs/shRNAs processed through non-canonical pathways that are either Drosha- or Dicer-independent. The first class, mirtrons [30, 31] and mirtron-based shRNAs [32, 33] are precisely located within introns, where the splicing machinery generates pre-miRNA, eluding the need for Drosha processing. In the second class, Ago-shRNAs, which are based on miR-451 [34–36], Dicer cleavage is substituted by Ago2 endonucleolytic activity for maturation (Fig. 1).
Despite several benefits, processing of Pol II-driven shRNAs is not well characterized, and the requirements for its complex maturation are still a matter of study.
3. Processing/maturation
After being transcribed, shRNAs must be processed into siRNAs to exert their inhibitory function. Such process relies on the miRNA biogenesis pathway. Therefore, it is vital to understand how pri/pre-miRNAs are recognized and processed by the miRNA biogenesis machinery.
3.1. Microprocessor cleavage
The first step of miRNA biogenesis takes place in the nucleus [37]. Pri-miRNA transcripts are cleaved co-transcriptionally by the Microprocessor - a complex consisting of one molecule of Drosha and two of its cofactor DGCR8 [38–40]. A typical pri-miRNA contains three components: 1) A loop of variable size 2) A stem of three helical turns (33~35 bp) and 3) single-stranded regions flanking the hairpin. The stem is further divided into two components: upper stem - two helix turns (~22 bp) next to the loop; and lower stem - one helix turn (~11 bp) connected to the single-stranded region (Fig. 2B). Microprocessor recognizes this unique structure and cuts the primary transcript at both sides of the stem between the upper and lower stems. Specifically, Drosha senses the basal junction between single-stranded flanking region and double-stranded lower stem structure, establishing the cleavage site 11 nt away [40–43]. At the same time, the DGCR8 dimer in the complex interacts with the apical junction between the upper stem and the loop to ensure the cleavage fidelity [42, 44, 45]. The released product is pre-miRNA - a hairpin structure with a two-nucleotide 3′ overhang (Fig. 2A).
Features defining a pri-miRNA encompass more than just structure. Sequence motifs like UG and CNNC at the 5′ and the 3′ ends of the basal stem respectively can promote pri-miRNA cleavage by Microprocessor [46, 47]. A bulged GHG motif in the stem and UGUG motif in the loop also contribute to the processing efficiency [46, 48]. About 80% of human miRNAs contain at least one of these sequence motifs, highlighting their role in pri-miRNA-Microprocessor interaction. However, little is known about the factors binding to those sequence motif and the molecular mechanisms determining how they modulate Microprocessor activity.
A recent study revealed that the abundance of miRNA is not always coupled to the level of its primary transcripts (pri-miRNA). Instead, the susceptibility of pri-miRNA to the Microprocessor plays a more decisive role [49]. Therefore, the efficiency of microprocessor cleavage is crucial in determining shRNA abundance and function. However, limited understanding of the underlying mechanism makes it challenging to design an optimal pri-miRNA-like shRNA de novo. Rather, researchers have developed shRNA based on endogenous pri-miRNA structures. Currently, pri-hsa-miR-30a and subsequent optimized backbones have been widely used as templates for shRNA production and candidate library screening [14, 50, 51]. Other pri-miRNA structures like pri-hsa-miR-155 [52] have also been used with many successes. Nonetheless, there is no systematic study to compare the processing efficacy among human pri-miRNAs. It is questionable whether pri-miR-30a or pri-miR-155 is the best starting scaffold. In addition, different cell lines present different processing efficiencies of individual pri-miRNA [49]. This phenomenon might reinforce the idea that complementary motif recognition could result in the modulation of maturation process in a tissue-specific manner, adding an extra layer of complexity for the design of “universal” shRNA platform.
3.2. Dicer cleavage
In the cytoplasm, Dicer cuts hairpin-shaped pre-miRNA-like shRNA near the loop to produce an siRNA duplex [2, 53]. Dicer uses its PAZ domain to dock both the 5′ and 3′ ends of pre-miRNA. The 5′ monophosphate and 2nt 3′ overhang of pre-miRNA are particularly important for this association by providing binding specificity [54]. Functioning as a molecular ruler, human Dicer positions the two catalytic centers of its RNase III domain in ~65Å from the ends of pre-miRNA. Therefore, Dicer cleaves all substrates at a fixed distance (65Å) from the open ends regardless of stem length. How many base pairs fit in such distance depends on the tertiary structure of RNA substrate. For a hairpin stem without mismatches or bulges, this is ~21nt counting from the 5′ end [54–57].
Of note, Dicer is a dsRNA nuclease. A stem shorter than 21 nt will place its catalytic center outside of the duplex region and therefore abolish cleavage. Mismatches or bulges at the cleavage site reduce the processing efficacy and should be avoided. In addition, a short loop (<5 nt) creates tension on nearby stem region, jeopardizing the duplex structure and reducing its accessibility to Dicer cleavage [58]. Taken everything into consideration, pre-miRNA-like shRNA should have well defined ends (2 nt 3′ overhang), a perfectly paired stem of at least 21 nt in length and a loop of ≥6 nt (Fig 2A).
3.3. Cleavage fidelity
Drosha and Dicer cleavages define sequence and function of the siRNA processed from the parental shRNA. It is crucial that these cleavages are precise to ensure the desired siRNA sequences. Achieving high fidelity from both enzymes is important to shRNA design.
As discussed above, there are two models describing how Microprocessor determines its cleavage site along the stem: 1) 11 nt from the basal junction or 2) 22 nt from the apical junction. While the former is clearly the dominant mechanism, the latter also contributes to certain degree in a case-by-case manner [42, 44]. To achieve precise Drosha cleavage, pri-miRNA-like shRNA needs be designed in a way where both models lead to the same cutting position. To this end, shRNA should be embedded in a 22 nt long upper stem together with an 11 nt long lower stem, making the total stem length ~33 nt.
Dicer is also known to produce lots of products with heterogeneous lengths, ranging from 22 to 24 nt [59]. The two main drivers of this heterogeneity are: 1) the presence of asymmetrical structural motifs in the stem and 2) the loop position in the precursor hairpin. The presence of asymmetrical bulges and loops within the two helical turns that span the Dicer anchoring site and the RNase III cleavage site can interfere with the structural ruler [59]. Similar to Drosha, the distance between the terminal loop and the cleavage point servers as a secondary ruler to fine tune the cleavage site [21]. Based on this study, introducing a 2 nt separation from the terminal loop or an apical internal bulge can dramatically increase the fidelity of shRNA processing.
4. RISC formation
Small RNA duplexes need to form a ribonucleoprotein complex called the RNA Induced Silencing Complexes (RISC) to exert their function. In mammals, RISC contains one of four Argonaute proteins at its center (Ago1–4). These have redundant functions as negative regulators of gene expression, but only Ago2 has slicer activity and is able to cleave a complementary target [60–62].
4.1. Ago loading
Ago loading is a process in which double stranded small RNA duplexes are incorporated into RISC where one of the strands will be selected [63]. This is a stepwise process (reviewed in ref [64]). First, double stranded small RNAs associate with an Ago protein as a duplex, forming a non-functional complex named pre-RISC (Fig. 1). The pre-RISC is then transformed into a biologically active mature RISC by conversion of the associated duplex to single-stranded RNA. The strand retained in the Ago protein is designated the guide strand, while the discarded strand is referred to as the passenger strand. Slicer competent Ago (Ago2) accomplishes this step by cleaving the passenger RNA, the products of which are sequentially degraded and removed by endonuclease C3PO (Component 3 Promoter of RISC) [65–67]. Alternatively, a less robust unwinding process (slicer-independent) is responsible for activating of RISC containing non-cleaving Argonautes (hsAgo1/3/4) or cleaving Ago2 associated with slicing-incompetent duplex RNAs as occurs with the majority of endogenous miRNAs [68].
To achieve efficient RISC formation, shRNA can be designed as a perfect stem structure to take advantage of the Ago2 cleavage dependent unwinding [69]. Otherwise, mismatch between positions 2–8 and 12–15 [70, 71] or overall low thermodynamic stability of the shRNA stem [72, 73] will promote efficient slicer-independent unwinding. In fact, it has been shown that presence of a central mismatch between positions 8–10 increases shRNA function [74].
4.2. Strand selection
The decision of which strand is selected as the guide strand is determined by the thermodynamic stability at the end of the duplex [75, 76]. The strand with relatively unstable 5′ end will be incorporated into RISC as the guide. It is critical for shRNA design to ensure loading of the desired strand. Loading of the wrong strand or mixed loading of both strands leads to low yields of inhibitory activity and unwanted off-target effects.
In designing Pol III driven pre-miRNA-like shRNA, it is of a good practice to avoid placing the guide strand in the 5 prime arm (5p) of shRNA. The 5p small RNA generated from Pol III driven shRNA carries a triphosphate group at the 5′ end, which may interfere with its incorporation into Ago2/RISC. Additionally, 5p transcripts starting with G or A are required for efficient Pol III transcription making them structurally unfavorable for Ago2/RISC loading. Instead, the guide sequence should be placed in the 3p arm and begin with U. This is the most preferred nucleotide for Ago2 association, and it also lowers the thermodynamic stability of the 5′ end, further enhancing its preferential loading onto RISC [77, 78].
4.3. Ago-shRNA
Some miRNAs have evolved to bypass certain steps of the canonical maturation pathway. An interesting example of this is hsa-mir-451 that eludes Dicer activity and achieves maturation by cleavage from Argonaute2 (AGO2) [79]. Ago2 directly associates with pre-miR-451, using the 5′ arm as guide and cutting the 3′ arm in the middle. The remaining part of the 3′ arm and loop are removed by an exonuclease PARN, leaving the 5′ arm inside Ago2 to form a mature RISC [80](Fig. 1).
Studies have indicated that this non-canonical maturation pathway requires features different from canonical shRNAs. Two crucial aspects that are defined by the nature of its structure are: 1) the avoidance of Dicer activity that is achieved by a short stem (19 bp) and a short loop (4 nt) and 2) the requirement of the slicer activity of AGO2 that is ensured by extensive base pairing the middle of the stem. Other hallmarks that improve the maturation process on this platform include un-pairing on position 35 and lower G/C content in the distal stem [81].
This platform is of a special interest to shRNA design as it mechanistically ensures the exclusive loading of the 5′ arm strand into Ago2. This not only eliminates undesired loading of the wrong strand, but also avoids loading of the guide strand into slicing-incompetent Argonautes (Ago1, 3 and 4), a potential source of off-target effects [81–85].
5. Selection of target sequence
The last step in shRNA design is selecting the target sequence. It appears to be straightforward, as RNAi is famous for its ability to target “any” sequence. However, many issues still need to be considered to ensure potency and specificity of designed shRNAs.
5.1. Base-pairing between guide strand and target
The model of repression relies on two conditions: 1) the degree of complementarity between guide strand and target, and 2) the identity of Ago involved. Effective gene knockdown can be achieved by invoking the robust cleavage repression mechanism, which requires Ago2 and near-perfect complementarity between guide strand and target. Base-pairing between positions 9 and 11 of the shRNA and its counterpart sequence in the mRNA are particularly important, as any disruption of duplex structure in this region will abolish the slicing activity [60]. While extensive complementarity in both the 5′ and the middle regions of guide strand are crucial for Ago2 mediated cleavage, base pairing at the 3′ end is not required. In fact, mismatches at position 18, 19, 20, 21 promote dislocation and thereby turnover rate of Ago2, enhancing RISC performance towards highly abundant targets [86].
Interestingly, the ternary structure of Ago-guide-target revealed that the first nucleotide of the guide strand does not contribute to the recognition and pairing with target mRNAs [87, 88]. This renders the strategy to place U at the first position of the guide regardless of target sequence, which increases its association with Ago2, thereby enhancing repression efficiency [88, 89].
Mismatches between shRNA-mRNAs and/or the involvement of slicing-incompetent Argonautes (Ago1/3/4) induce target repression through the cleavage-independent pathway, which is characterized by a combination of translational repression and mRNA decay [12]. Although it is the dominant mechanism by which miRNA fine-tunes target mRNAs in animals, cleavage-independent repression (also called miRNA-like repression) is generally weak and therefore not suitable for gene knockdown. In fact, miRNA-like repression is a major source of off-target effects and should be minimized.
5.2. Localization of the target site
In animals, effective miRNA target sites are often found in the 3′ untranslated region (3′UTR). This is because during translation, progressing ribosomes interfere with the weak binding between RISC and its target sequence in the coding region of mRNA [90]. On the contrary, siRNA-induced RISC cleaves target mRNA in a transient manner, which is less affected by translation machinery. Hence, shRNAs can be designed to have a perfect complementarity to target any part of the mRNA sequence.
Another factor that must be considered is the specificity of the target sequence. As many mRNAs from the same gene family share similar sequences, extra care is required in selecting target sequence to allow gene-specific knockdown. Otherwise, targeting a consensus sequence is a good strategy to repress expression from the entire gene family. In addition, alternative splicing and polyA site selection generate mRNA isoforms. To achieve complete knockdown of gene expression, shRNA should target sequences shared among all isoforms.
5.3. Local structure of the target site
Since the RISC complex is unable to unfold structured RNA, target sequences need to be located in a structurally opened area for the RISC to initiate hybridization [91]. However, once the initial interaction has taken place through the 5′-end region, complementary base pairs at the 3′-end stabilize the association, allowing the RISC complex to disrupt nearby secondary structures to further elongate the hybridization [91–94].
Accessibility of target sites can be measured experimentally or estimated by software based on minimum free energy of secondary structure which is often correlated with the AU-richness of surrounding sequences. Of note, RNA binding proteins (e.g. HuR and DND1) associated with nearby sequences can alter the accessibility and mask the target site from RISC binding [95, 96].
6. Off-target effects
One of the major challenges that RNAi technology faces is off-target effects [97]. Factors contributing to these include: 1) miRNA-like repression; 2) incorporating passenger strand into Ago during loading; 3) competition for limiting resources of the endogenous miRNA pathway; and 4) immune responses. Many efforts in addressing these issues came from chemical modifications on the siRNA backbone. However, for expressed shRNA where chemical modification is not available, solutions to these problems rely solely on design.
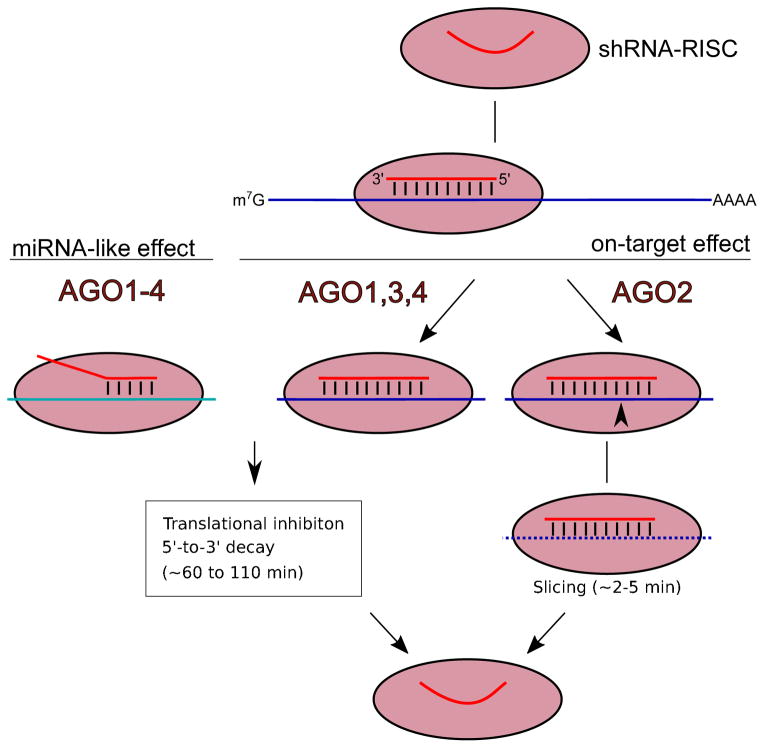
Although the guide strand is usually designed to specifically target one mRNA sequence through perfect complementarity, it will also recognize additional mRNA transcripts through partial base pairing and repress gene expression via the miRNA-like pathway [98, 99] (Fig. 3). As miRNA-like repression only requires the base pairing of as few as 6 nt in position 2–7 (seed region) of guide strand, it is nearly impossible to eliminate the miRNA-like off-targeting completely. One strategy is to take advantage of mechanistic differences between on-targets and off-targets shRNA repression. Inhibition mechanism of the latter, but not the former, relies on a stable association between RISC and target mRNA. Designing the guide sequences with overall weaker binding energy to the target sequence can dramatically reduce the off-target while maintaining a good on-target effect [100]. Another strategy is to combine delivery of the shRNA with RNA decoys that could deplete its excess [101]. In addition, bioinformatics algorithms can predict the off-target effects on a genome-wide scale and therefore eliminate off-target mediated false signals in genetic screens through post run analysis [102, 103].
Fig 3. shRNA on-target and off-target recognition of mRNA.
Once loaded into the RISC complex, shRNA can trigger its inhibitory activity on mRNAs. High complementarity base-pairing induces on-target mRNA cleavage via slicing activity of AGO2,. Translational inhibition can be induced for perfectly paired shRNA-target complexes if AGO1, 3, or 4 are recruited to RISC instead of AGO2. Less extensive base-pairing between the shRNA seed sequence and cellular mRNAs triggers off-target repression, known as the “miRNA-like effect.” This also includes translational inhibition and mRNA decay which can be mediated by any of the Argonaute family members (AGO1–4).
Another source for shRNAs off-target effects is derived from the competition with endogenous miRNAs for limiting factors involved in the processing pathway. To date, principal limiting factors are Argonautes and exportin-5 (XPO5), the latter of which is responsible for nucelocytoplasmatic export of pre-miRNA [104]. This competition with endogenous miRNAs results in the de-repression of their cellular targets, leading to its misregulation. Use of weaker promoters combined with highly potent shRNAs may alleviate this toxic effect. Furthermore, certain innate immune responses can be generated by the recognition of specific motifs both in dsRNA and ssRNA sequences. Some of the motifs that trigger production of inflammatory cytokines are the poly(U)- or the GU-rich sequences [105], which should be avoided in design.
7. Instructions for shRNA design and cloning
Each type of shRNAs discussed here has its pros and cons (Table 1). Nonetheless, Pol III driven pre-miRNA-like shRNA is more prevalent and better proven than others. Details of its processing are well characterized. Hence, we opt to provide here the instructions on how to design and clone pre-miRNA-like shRNA that is driven by the U6 promoter.
7.1 Design of shRNA sequence (Fig. 4A)
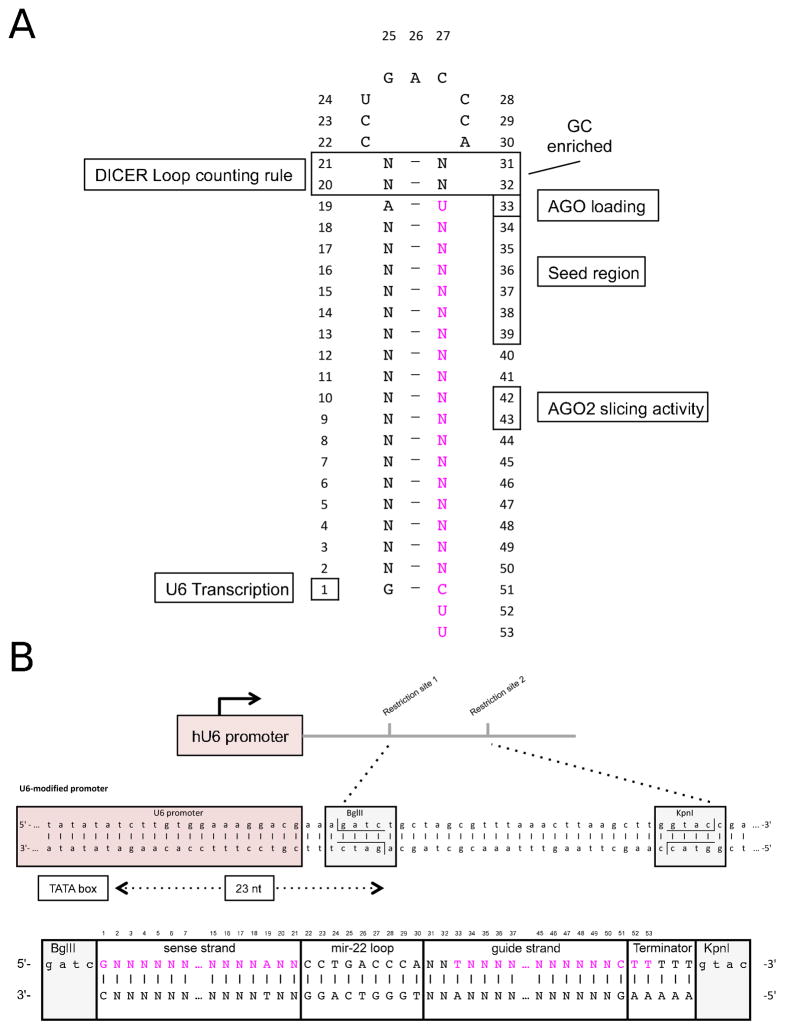
Fig 4. Cloning design for optimal processing of pre-miRNA-like shRNA.
A) shRNA hairpin structure containing a detailed memorandum of the reported sequence and structural features involved in the correct processing by Dicer and loading into AGO proteins. B) Expression though U6 promoter (Pol III) requires a precise start and termination of the transcription of the shRNA hairpin. To achieve this precision on the starting of the transcription, the first nucleotide of the shRNA should be located 23 nt from the TATA box. In this scheme we show the precise location of a BglII restriction site introduced to facilitate this precise cloning. Below, we present an oligonucleotide design containing the passenger sequence, the loop of mir-22, the guide sequence of the shRNA, a poly(T) terminator, and flanked by the required BglII and KpnI overhangs.
Select the sequence in your target gene according to the suggestions in Section 5.2.
Design the 3p arm of shRNA as the guide strand (antisense to target), leaving the 5p arm as passenger strand. The shRNA sequence (from 5′ to 3′) will be in the order of passenger strand, loop, then guide strand.
Start the shRNA sequence with G to accommodate the requirement of U6 promoter (position 1). This G will be the first nucleotide of passenger strand.
Use U as the first nucleotide of the guide strand (position 33) to favor its incorporation into the Argonaute protein during RISC formation and strand selection.
Ensure positions 2 to 18 are perfectly complementary to the target site sequence.
Use C as the 19th nucleotide of the guide strand (position 51), as it will base pair with the first nucleotide of the passenger strand (G).
Design the rest of passenger strand sequence (position 2 to 19) to have perfect complementary to the guide strand (position 34 to 50). Together, they form a duplex that is 19 bp in length.
Design a loop sequence that is free of internal structure. Here we depict the use of the hsa-miR-22 loop, which has been well characterized and tested [21].
Use a 2 bp spacer with an arbitrary sequence between the first nucleotide of guide strand (U) and loop sequence (position 20 to 21), and between the last nucleotide of the passenger strand (A) and loop. This provides a hairpin stem length of 21 bp (position 31 to 32). GC or CG are preferred as strong base pairs to help define a clear boundary between the stem and loop.
Once the guide and the passenger sequences are designed as shown (Fig. 4A), use secondary structure prediction algorithms to make sure that there is no misfolding of the hairpin structure [106, 107]. Some useful bioinformatic tools for the design of shRNAs sequences are available online (Supplementary Table 1).
7.2 Cloning shRNA into expression vector
Currently, Addgene offers a wide variety of vectors suitable for cloning of shRNA designs under different expression cassettes and lentiviral or retroviral vectors (https://www.addgene.org/mammalianrnai/). Of note, not all shRNA expression vectors contain suitable restriction sites to place the first nucleotide of shRNA sequence immediately adjacent to the transcription-starting site. We recommend using the PIII(U6) vector, in which the last two nucleotides of U6 promoter are engineered to contain an Bgl II site (Fig 4B) [108].
-
Digest the PIII(U6) vector with Bgl II and Kpn I restriction enzymes..
Run digestion products on an agarose gel and purify the linearized vector.
-
Synthesize DNA oligo duplex of shRNA with overhang sequences of Bgl II and Kpn I. This can be achieve by following step (Fig. 4B):
Make the sense strand by adding 5 T’s to the end of the shRNA sequence (Passenger-Loop-Guide-5T).
Design the antisense strand by making the reverse complement of the sense strand.
Add GATC to the 5′ end of the sense strand.
Add GTAC to the 3′ end of the sense strand.
Select the target shRNA sequence for your gene of interest based on the previously described guidelines. Take care to ensure that the sense strand should start with G.
Phosphorylate a mix of 1 μL of each sense and antisense oligonucleotides (100 μM) with 1 μL of the T4 Polynucleotide Kinase (PNK) in total volume of 100 μL and incubate at 37°C for 1 hour.
Inactivate the PNK and denature DNA by incubating at 95°C for 5 minutes in a beaker of boiling water. Allow cooling to room temperature for proper annealing.
Ligate 50 ng of the digested vector with the annealed inserts with a 3-fold molar ratio. Ligation can be performed with Quick T4 DNA Ligase at 25°C for 10 minutes. Do not heat inactivate the ligase as it dramatically reduces the transformation efficacy.
Transform 5 μL of the ligation mixture into Escherichia coli competent cells (DH5α or Stellar) according to manufacturer instructions.
Select colonies for screening by preferred method.
8. Concluding remarks
Evidently, there is an intimate relationship between shRNA design and the current comprehension of miRNA pathways. Here, we have presented a set of design guidelines based on the current knowledge of the field. Despite significant advancements in our understanding of the underlying mechanisms over the past decade, many aspects of the pathway remain to be elucidated. Due to these limitations, we have proposed using the Pol III driven pre-miRNA-like shRNA as the optimal silencing tool. Compared to the Pol II-driven pri-miRNA-like shRNAs, pre-miRNA-like shRNAs require less maturation steps and are relatively well defined.
Nonetheless, advantages of pri-miRNA-like shRNA should not be overlooked. The rather complicated processing also provides opportunities to regulate and manipulate the function of shRNA. There is still a lot of work to be done in order to make it a “ready-to-use” platform. To this end, some of the aspects that should be further studied are how the specificity and fidelity of processing are achieved and how these steps are regulated by other cellular factors. Moreover, less is known about mechanisms that affect the half-life of shRNAs once assembled to the RISC complexes.
Although RNAi technologies have advanced, they face the advent of a competing technology known as CRISPR that bears its own technological niches. On one hand, CRISPR-Cas9 provides an extremely powerful tool to study gene function and opens the door to the therapeutic use of gene editing. On the other hand, its caveats such as off-target DNA breaks or chromosome rearrangements have permanent effects which are much more severe than those resulting from off-targeting by RNAi. Of note, RNAi is a mature technology that is already showing its efficacy under clinical trials. It can be still foreseen as the tool of choice for gene therapy approaches requiring the fine-tuning of gene expression. It has been pointed out the possible synergies of both technologies in the genome-wide arrayed screening [18]. Embracing both technologies is the clear choice for a better tomorrow.
Supplementary Material
Supplementary Table 1: Online resources for the shRNA design
Highlights.
Overview of the pros and cons of RNA Pol II/III promoters in transcribing shRNA
Review of the structural and sequence features involved in miRNA/shRNA maturation
Overview on the target sequence selection and off-target effect avoidance
Future challenges and potentialities of miRNA-based shRNAs
Acknowledgments
We thank all the members of the group and specially Angela Dinardo for their helpful discussion and critically reading this manuscript. Authors apologize for any uncited references that were left out due to space limitations. This work was supported by the Intramural Research Program of the National Cancer Institute, National Institutes of Health.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC. Nature. 1998;391:806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- 2.Elbashir SM, Harborth J, Lendeckel W, Yalcin A, Weber K, Tuschl T. Nature. 2001;411:494–498. doi: 10.1038/35078107. [DOI] [PubMed] [Google Scholar]
- 3.Brummelkamp TR, Bernards R, Agami R. Science. 2002;296:550–553. doi: 10.1126/science.1068999. [DOI] [PubMed] [Google Scholar]
- 4.McManus MT, Petersen CP, Haines BB, Chen J, Sharp PA. Rna. 2002;8:842–850. doi: 10.1017/s1355838202024032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Paddison PJ, Caudy AA, Bernstein E, Hannon GJ, Conklin DS. Genes & development. 2002;16:948–958. doi: 10.1101/gad.981002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.McCaffrey AP, Meuse L, Pham TT, Conklin DS, Hannon GJ, Kay MA. Nature. 2002;418:38–39. doi: 10.1038/418038a. [DOI] [PubMed] [Google Scholar]
- 7.Zeng Y, Wagner EJ, Cullen BR. Molecular cell. 2002;9:1327–1333. doi: 10.1016/s1097-2765(02)00541-5. [DOI] [PubMed] [Google Scholar]
- 8.Friedman RC, Farh KK, Burge CB, Bartel DP. Genome research. 2009;19:92–105. doi: 10.1101/gr.082701.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ha M, Kim VN. Nature reviews Molecular cell biology. 2014;15:509–524. doi: 10.1038/nrm3838. [DOI] [PubMed] [Google Scholar]
- 10.Bethune J, Artus-Revel CG, Filipowicz W. EMBO reports. 2012;13:716–723. doi: 10.1038/embor.2012.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Eichhorn SW, Guo H, McGeary SE, Rodriguez-Mias RA, Shin C, Baek D, Hsu SH, Ghoshal K, Villen J, Bartel DP. Molecular cell. 2014;56:104–115. doi: 10.1016/j.molcel.2014.08.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gu S, Kay MA. Silence. 2010;1:11. doi: 10.1186/1758-907X-1-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Siolas D, Lerner C, Burchard J, Ge W, Linsley PS, Paddison PJ, Hannon GJ, Cleary MA. Nature biotechnology. 2005;23:227–231. doi: 10.1038/nbt1052. [DOI] [PubMed] [Google Scholar]
- 14.Stegmeier F, Hu G, Rickles RJ, Hannon GJ, Elledge SJ. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:13212–13217. doi: 10.1073/pnas.0506306102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Silva JM, Li MZ, Chang K, Ge W, Golding MC, Rickles RJ, Siolas D, Hu G, Paddison PJ, Schlabach MR, Sheth N, Bradshaw J, Burchard J, Kulkarni A, Cavet G, Sachidanandam R, McCombie WR, Cleary MA, Elledge SJ, Hannon GJ. Nature genetics. 2005;37:1281–1288. doi: 10.1038/ng1650. [DOI] [PubMed] [Google Scholar]
- 16.Paddison PJ, Silva JM, Conklin DS, Schlabach M, Li M, Aruleba S, Balija V, O’Shaughnessy A, Gnoj L, Scobie K, Chang K, Westbrook T, Cleary M, Sachidanandam R, McCombie WR, Elledge SJ, Hannon GJ. Nature. 2004;428:427–431. doi: 10.1038/nature02370. [DOI] [PubMed] [Google Scholar]
- 17.Pan Q, van der Laan LJ, Janssen HL, Peppelenbosch MP. Trends in biotechnology. 2012;30:206–215. doi: 10.1016/j.tibtech.2012.01.002. [DOI] [PubMed] [Google Scholar]
- 18.Mohr SE, Smith JA, Shamu CE, Neumuller RA, Perrimon N. Nature reviews Molecular cell biology. 2014;15:591–600. doi: 10.1038/nrm3860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ma H, Wu Y, Dang Y, Choi JG, Zhang J, Wu H. Mol Ther Nucleic Acids. 2014;3:e161. doi: 10.1038/mtna.2014.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Braglia P, Percudani R, Dieci G. The Journal of biological chemistry. 2005;280:19551–19562. doi: 10.1074/jbc.M412238200. [DOI] [PubMed] [Google Scholar]
- 21.Gu S, Jin L, Zhang Y, Huang Y, Zhang F, Valdmanis PN, Kay MA. Cell. 2012;151:900–911. doi: 10.1016/j.cell.2012.09.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sui G, Soohoo C, Affar el B, Gay F, Shi Y, Forrester WC, Shi Y. Proceedings of the National Academy of Sciences of the United States of America. 2002;99:5515–5520. doi: 10.1073/pnas.082117599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Czauderna F, Santel A, Hinz M, Fechtner M, Durieux B, Fisch G, Leenders F, Arnold W, Giese K, Klippel A, Kaufmann J. Nucleic acids research. 2003;31:e127. doi: 10.1093/nar/gng127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lebbink RJ, Lowe M, Chan T, Khine H, Wang X, McManus MT. PloS one. 2011;6:e26213. doi: 10.1371/journal.pone.0026213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Grimm D, Streetz KL, Jopling CL, Storm TA, Pandey K, Davis CR, Marion P, Salazar F, Kay MA. Nature. 2006;441:537–541. doi: 10.1038/nature04791. [DOI] [PubMed] [Google Scholar]
- 26.Maczuga P, Koornneef A, Borel F, Petry H, van Deventer S, Ritsema T, Konstantinova P. BMC biotechnology. 2012;12:42. doi: 10.1186/1472-6750-12-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Choi JG, Bharaj P, Abraham S, Ma H, Yi G, Ye C, Dang Y, Manjunath N, Wu H, Shankar P. Molecular therapy : the journal of the American Society of Gene Therapy. 2015;23:310–320. doi: 10.1038/mt.2014.205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Altuvia Y, Landgraf P, Lithwick G, Elefant N, Pfeffer S, Aravin A, Brownstein MJ, Tuschl T, Margalit H. Nucleic acids research. 2005;33:2697–2706. doi: 10.1093/nar/gki567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Truscott M, Islam AB, Frolov MV. Rna. 2016;22:129–138. doi: 10.1261/rna.053264.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ruby JG, Jan CH, Bartel DP. Nature. 2007;448:83–86. doi: 10.1038/nature05983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ladewig E, Okamura K, Flynt AS, Westholm JO, Lai EC. Genome research. 2012;22:1634–1645. doi: 10.1101/gr.133553.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Seow Y, Sibley CR, Wood MJ. Rna. 2012;18:1328–1337. doi: 10.1261/rna.030601.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sibley CR, Seow Y, Curtis H, Weinberg MS, Wood MJ. Nucleic acids research. 2012;40:9863–9875. doi: 10.1093/nar/gks712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cheloufi S, Dos Santos CO, Chong MM, Hannon GJ. Nature. 2010;465:584–589. doi: 10.1038/nature09092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cifuentes D, Xue H, Taylor DW, Patnode H, Mishima Y, Cheloufi S, Ma E, Mane S, Hannon GJ, Lawson ND, Wolfe SA, Giraldez AJ. Science. 2010;328:1694–1698. doi: 10.1126/science.1190809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu YP, Schopman NC, Berkhout B. Nucleic acids research. 2013;41:3723–3733. doi: 10.1093/nar/gkt036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lee Y, Ahn C, Han J, Choi H, Kim J, Yim J, Lee J, Provost P, Radmark O, Kim S, Kim VN. Nature. 2003;425:415–419. doi: 10.1038/nature01957. [DOI] [PubMed] [Google Scholar]
- 38.Denli AM, Tops BB, Plasterk RH, Ketting RF, Hannon GJ. Nature. 2004;432:231–235. doi: 10.1038/nature03049. [DOI] [PubMed] [Google Scholar]
- 39.Gregory RI, Yan KP, Amuthan G, Chendrimada T, Doratotaj B, Cooch N, Shiekhattar R. Nature. 2004;432:235–240. doi: 10.1038/nature03120. [DOI] [PubMed] [Google Scholar]
- 40.Kwon SC, Nguyen TA, Choi YG, Jo MH, Hohng S, Kim VN, Woo JS. Cell. 2016;164:81–90. doi: 10.1016/j.cell.2015.12.019. [DOI] [PubMed] [Google Scholar]
- 41.Han J, Lee Y, Yeom KH, Kim YK, Jin H, Kim VN. Genes & development. 2004;18:3016–3027. doi: 10.1101/gad.1262504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ma H, Wu Y, Choi JG, Wu H. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:20687–20692. doi: 10.1073/pnas.1311639110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Han J, Lee Y, Yeom KH, Nam JW, Heo I, Rhee JK, Sohn SY, Cho Y, Zhang BT, Kim VN. Cell. 2006;125:887–901. doi: 10.1016/j.cell.2006.03.043. [DOI] [PubMed] [Google Scholar]
- 44.Nguyen TA, Jo MH, Choi YG, Park J, Kwon SC, Hohng S, Kim VN, Woo JS. Cell. 2015;161:1374–1387. doi: 10.1016/j.cell.2015.05.010. [DOI] [PubMed] [Google Scholar]
- 45.Zeng Y, Yi R, Cullen BR. The EMBO journal. 2005;24:138–148. doi: 10.1038/sj.emboj.7600491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Auyeung VC, Ulitsky I, McGeary SE, Bartel DP. Cell. 2013;152:844–858. doi: 10.1016/j.cell.2013.01.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fang W, Bartel DP. Molecular cell. 2015;60:131–145. doi: 10.1016/j.molcel.2015.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jensen SM, Schmitz A, Pedersen FS, Kjems J, Bramsen JB. PloS one. 2012;7:e43095. doi: 10.1371/journal.pone.0043095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Conrad T, Marsico A, Gehre M, Orom UA. Cell reports. 2014;9:542–554. doi: 10.1016/j.celrep.2014.09.007. [DOI] [PubMed] [Google Scholar]
- 50.Fellmann C, Hoffmann T, Sridhar V, Hopfgartner B, Muhar M, Roth M, Lai DY, Barbosa IA, Kwon JS, Guan Y, Sinha N, Zuber J. Cell reports. 2013;5:1704–1713. doi: 10.1016/j.celrep.2013.11.020. [DOI] [PubMed] [Google Scholar]
- 51.Knott SR, Maceli AR, Erard N, Chang K, Marran K, Zhou X, Gordon A, El Demerdash O, Wagenblast E, Kim S, Fellmann C, Hannon GJ. Molecular cell. 2014;56:796–807. doi: 10.1016/j.molcel.2014.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Shan ZX, Lin QX, Yang M, Deng CY, Kuang SJ, Zhou ZL, Xiao DZ, Liu XY, Lin SG, Yu XY. Molecular and cellular biochemistry. 2009;323:81–89. doi: 10.1007/s11010-008-9966-3. [DOI] [PubMed] [Google Scholar]
- 53.Bernstein E, Caudy AA, Hammond SM, Hannon GJ. Nature. 2001;409:363–366. doi: 10.1038/35053110. [DOI] [PubMed] [Google Scholar]
- 54.Park JE, Heo I, Tian Y, Simanshu DK, Chang H, Jee D, Patel DJ, Kim VN. Nature. 2011;475:201–205. doi: 10.1038/nature10198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Macrae IJ, Zhou K, Li F, Repic A, Brooks AN, Cande WZ, Adams PD, Doudna JA. Science. 2006;311:195–198. doi: 10.1126/science.1121638. [DOI] [PubMed] [Google Scholar]
- 56.MacRae IJ, Zhou K, Doudna JA. Nature Structural & Molecular Biology. 2007;14:934–940. doi: 10.1038/nsmb1293. [DOI] [PubMed] [Google Scholar]
- 57.Gan J, Shaw G, Tropea JE, Waugh DS, Court DL, Ji X. Mol Microbiol. 2008;67:143–154. doi: 10.1111/j.1365-2958.2007.06032.x. [DOI] [PubMed] [Google Scholar]
- 58.Schopman NC, Liu YP, Konstantinova P, ter Brake O, Berkhout B. Antiviral research. 2010;86:204–211. doi: 10.1016/j.antiviral.2010.02.320. [DOI] [PubMed] [Google Scholar]
- 59.Starega-Roslan J, Krol J, Koscianska E, Kozlowski P, Szlachcic WJ, Sobczak K, Krzyzosiak WJ. Nucleic acids research. 2011;39:257–268. doi: 10.1093/nar/gkq727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Liu J, Carmell MA, Rivas FV, Marsden CG, Thomson JM, Song JJ, Hammond SM, Joshua-Tor L, Hannon GJ. Science. 2004;305:1437–1441. doi: 10.1126/science.1102513. [DOI] [PubMed] [Google Scholar]
- 61.Meister G, Landthaler M, Patkaniowska A, Dorsett Y, Teng G, Tuschl T. Molecular cell. 2004;15:185–197. doi: 10.1016/j.molcel.2004.07.007. [DOI] [PubMed] [Google Scholar]
- 62.Su H, Trombly MI, Chen J, Wang X. Genes & development. 2009;23:304–317. doi: 10.1101/gad.1749809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Czech B, Hannon GJ. Nat Rev Genet. 2011;12:19–31. doi: 10.1038/nrg2916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kawamata T, Tomari Y. Trends Biochem Sci. 2010;35:368–376. doi: 10.1016/j.tibs.2010.03.009. [DOI] [PubMed] [Google Scholar]
- 65.Tian Y, Simanshu DK, Ascano M, Diaz-Avalos R, Park AY, Juranek SA, Rice WJ, Yin Q, Robinson CV, Tuschl T, Patel DJ. Nature structural & molecular biology. 2011;18:658–664. doi: 10.1038/nsmb.2069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ye X, Huang N, Liu Y, Paroo Z, Huerta C, Li P, Chen S, Liu Q, Zhang H. Nature structural & molecular biology. 2011;18:650–657. doi: 10.1038/nsmb.2032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Liu Y, Ye X, Jiang F, Liang C, Chen D, Peng J, Kinch LN, Grishin NV, Liu Q. Science. 2009;325:750–753. doi: 10.1126/science.1176325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Tomari Y, Du T, Zamore PD. Cell. 2007;130:299–308. doi: 10.1016/j.cell.2007.05.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Matranga C, Tomari Y, Shin C, Bartel DP, Zamore PD. Cell. 2005;123:607–620. doi: 10.1016/j.cell.2005.08.044. [DOI] [PubMed] [Google Scholar]
- 70.Kawamata T, Seitz H, Tomari Y. Nature structural & molecular biology. 2009;16:953–960. doi: 10.1038/nsmb.1630. [DOI] [PubMed] [Google Scholar]
- 71.Yoda M, Kawamata T, Paroo Z, Ye X, Iwasaki S, Liu Q, Tomari Y. Nature structural & molecular biology. 2010;17:17–23. doi: 10.1038/nsmb.1733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Gu S, Jin L, Zhang F, Huang Y, Grimm D, Rossi JJ, Kay MA. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:9208–9213. doi: 10.1073/pnas.1018023108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Gu S, Jin L, Huang Y, Zhang F, Kay MA. Current biology : CB. 2012;22:1536–1542. doi: 10.1016/j.cub.2012.06.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wu H, Ma H, Ye C, Ramirez D, Chen S, Montoya J, Shankar P, Wang XA, Manjunath N. PloS one. 2011;6:e28580. doi: 10.1371/journal.pone.0028580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Khvorova A, Reynolds A, Jayasena SD. Cell. 2003;115:209–216. doi: 10.1016/s0092-8674(03)00801-8. [DOI] [PubMed] [Google Scholar]
- 76.Schwarz DS, Hutvagner G, Du T, Xu Z, Aronin N, Zamore PD. Cell. 2003;115:199–208. doi: 10.1016/s0092-8674(03)00759-1. [DOI] [PubMed] [Google Scholar]
- 77.Frank F, Sonenberg N, Nagar B. Nature. 2010;465:818–822. doi: 10.1038/nature09039. [DOI] [PubMed] [Google Scholar]
- 78.Seitz H, Tushir JS, Zamore PD. Silence. 2011;2:4. doi: 10.1186/1758-907X-2-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Yang JS, Maurin T, Robine N, Rasmussen KD, Jeffrey KL, Chandwani R, Papapetrou EP, Sadelain M, O’Carroll D, Lai EC. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:15163–15168. doi: 10.1073/pnas.1006432107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Yoda M, Cifuentes D, Izumi N, Sakaguchi Y, Suzuki T, Giraldez AJ, Tomari Y. Cell reports. 2013;5:715–726. doi: 10.1016/j.celrep.2013.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Yang JS, Maurin T, Lai EC. Rna. 2012;18:945–957. doi: 10.1261/rna.032938.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Herrera-Carrillo E, Harwig A, Liu YP, Berkhout B. Rna. 2014;20:1410–1418. doi: 10.1261/rna.043950.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Ma H, Zhang J, Wu H. Mol Ther Nucleic Acids. 2014;3:e176. doi: 10.1038/mtna.2014.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Shang R, Zhang F, Xu B, Xi H, Zhang X, Wang W, Wu L. Nature communications. 2015;6:8430. doi: 10.1038/ncomms9430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Liu YP, Karg M, Harwig A, Herrera-Carrillo E, Jongejan A, van Kampen A, Berkhout B. RNA biology. 2015;12:92–100. doi: 10.1080/15476286.2015.1017204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.De N, Young L, Lau PW, Meisner NC, Morrissey DV, MacRae IJ. Molecular cell. 2013;50:344–355. doi: 10.1016/j.molcel.2013.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Lewis BP, Burge CB, Bartel DP. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 88.Schirle NT, Sheu-Gruttadauria J, MacRae IJ. Science. 2014;346:608–613. doi: 10.1126/science.1258040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Frank F, Fabian MR, Stepinski J, Jemielity J, Darzynkiewicz E, Sonenberg N, Nagar B. EMBO reports. 2011;12:415–420. doi: 10.1038/embor.2011.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Gu S, Jin L, Zhang F, Sarnow P, Kay MA. Nature structural & molecular biology. 2009;16:144–150. doi: 10.1038/nsmb.1552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Ameres SL, Martinez J, Schroeder R. Cell. 2007;130:101–112. doi: 10.1016/j.cell.2007.04.037. [DOI] [PubMed] [Google Scholar]
- 92.Long D, Lee R, Williams P, Chan CY, Ambros V, Ding Y. Nature structural & molecular biology. 2007;14:287–294. doi: 10.1038/nsmb1226. [DOI] [PubMed] [Google Scholar]
- 93.Kertesz M, Iovino N, Unnerstall U, Gaul U, Segal E. Nature genetics. 2007;39:1278–1284. doi: 10.1038/ng2135. [DOI] [PubMed] [Google Scholar]
- 94.Heale BS, Soifer HS, Bowers C, Rossi JJ. Nucleic Acids Res. 2005;33:e30. doi: 10.1093/nar/gni026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Kedde M, Strasser MJ, Boldajipour B, Oude Vrielink JA, Slanchev K, le Sage C, Nagel R, Voorhoeve PM, van Duijse J, Orom UA, Lund AH, Perrakis A, Raz E, Agami R. Cell. 2007;131:1273–1286. doi: 10.1016/j.cell.2007.11.034. [DOI] [PubMed] [Google Scholar]
- 96.Bhattacharyya SN, Habermacher R, Martine U, Closs EI, Filipowicz W. Cell. 2006;125:1111–1124. doi: 10.1016/j.cell.2006.04.031. [DOI] [PubMed] [Google Scholar]
- 97.Jackson AL, Linsley PS. Nature reviews Drug discovery. 2010;9:57–67. doi: 10.1038/nrd3010. [DOI] [PubMed] [Google Scholar]
- 98.Lin X, Ruan X, Anderson MG, McDowell JA, Kroeger PE, Fesik SW, Shen Y. Nucleic acids research. 2005;33:4527–4535. doi: 10.1093/nar/gki762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Burchard J, Jackson AL, Malkov V, Needham RH, Tan Y, Bartz SR, Dai H, Sachs AB, Linsley PS. Rna. 2009;15:308–315. doi: 10.1261/rna.1326809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Gu S, Zhang Y, Jin L, Huang Y, Zhang F, Bassik MC, Kampmann M, Kay MA. Nucleic acids research. 2014;42:12169–12176. doi: 10.1093/nar/gku854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Mockenhaupt S, Grosse S, Rupp D, Bartenschlager R, Grimm D. Proceedings of the National Academy of Sciences of the United States of America. 2015;112:E4007–4016. doi: 10.1073/pnas.1510476112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Sigoillot FD, Lyman S, Huckins JF, Adamson B, Chung E, Quattrochi B, King RW. Nature methods. 2012;9:363–366. doi: 10.1038/nmeth.1898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Yilmazel B, Hu Y, Sigoillot F, Smith JA, Shamu CE, Perrimon N, Mohr SE. BMC bioinformatics. 2014;15:192. doi: 10.1186/1471-2105-15-192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Grimm D, Wang L, Lee JS, Schurmann N, Gu S, Borner K, Storm TA, Kay MA. The Journal of clinical investigation. 2010;120:3106–3119. doi: 10.1172/JCI43565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Judge AD, Sood V, Shaw JR, Fang D, McClintock K, MacLachlan I. Nature biotechnology. 2005;23:457–462. doi: 10.1038/nbt1081. [DOI] [PubMed] [Google Scholar]
- 106.Gruber AR, Lorenz R, Bernhart SH, Neubock R, Hofacker IL. Nucleic acids research. 2008;36:W70–74. doi: 10.1093/nar/gkn188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Lorenz R, Bernhart SH, Honer Zu Siederdissen C, Tafer H, Flamm C, Stadler PF, Hofacker IL. Algorithms for molecular biology : AMB. 2011;6:26. doi: 10.1186/1748-7188-6-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Amarzguioui M, Rossi JJ, Kim D. FEBS Lett. 2005;579:5974–5981. doi: 10.1016/j.febslet.2005.08.070. [DOI] [PubMed] [Google Scholar]
- 109.Dai Z, Sheridan JM, Gearing LJ, Moore DL, Su S, Wormald S, Wilcox S, O’Connor L, Dickins RA, Blewitt ME, Ritchie ME. F1000 Research. 2014;3:95. doi: 10.12688/f1000research.4204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Janssen S, Giegerich R. Bioinformatics. 2015;31:423–425. doi: 10.1093/bioinformatics/btu649. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Table 1: Online resources for the shRNA design