Abstract
Purpose
Fusarium species are among prevalent airborne fungi and causative agents of human respiratory atopic disorders. We previously identified a 36.5-kDa F. proliferatum component recognized by IgE antibodies in 9 (53%) of the 17 F. proliferatum-sensitized atopic serum samples. The purpose of this study is to characterize the 36.5-kDa allergen of F. proliferatum.
Methods
Characterization of allergens and determination of IgE cross-reactivity were performed by cDNA cloning/expression and immunoblot inhibition studies.
Results
Based on the finding that the 36.5-kDa IgE-binding component reacted with the mouse monoclonal antibody FUM20 against fungal vacuolar serine protease allergens, the cDNA of F. proliferatum vacuolar serine protease (Fus p 9.0101) was subsequently cloned. Nine serum samples from respiratory atopic patients with IgE binding to the vacuolar serine protease allergen of Penicillium chrysogenum (Pen ch 18) also showed IgE-immunoblot reactivity to rFus p 9.0101. The purified rFus p 9.0101 can inhibit IgE and FUM20 binding to the 36.5-kDa component of F. proliferatum. Thus, a novel and important Fus p 9.0101 was identified. The rPen ch 18 can inhibit IgE binding to Fus p 9.0101. It indicates that IgE cross-reactivity between Fus p 9.0101 and Pen ch 18 also exists. Furthermore, neither rFus p 9.0101 K88A nor rPen ch 18 K89A mutants inhibited IgE binding to rFus p 9.0101. Lys88 was considered a critical core amino acid in IgE binding to r Fus p 9.0101 and a residue responsible for IgE cross-reactivity between Fus p 9.0101 and Pen ch 18 allergens.
Conclusions
Results obtained from this study indicate that vacuolar serine protease may be a major allergen of F. proliferatum and an important IgE cross-reactive pan-fungal allergen, and provide important bases for clinical diagnosis of fungal allergy.
Keywords: F. proliferatum, allergen, vacuolar serine protease, IgE cross-reactivity
INTRODUCTION
Airborne fungi which are ubiquitous in our environment have been identified as important causative agents of human respiratory atopic disorders.1,2 Characterization of major allergens of environmental fungi is beneficial in the diagnosis and treatment of clinical fungal allergy.2,3,4
We have identified previously serine proteases as major allergens of prevalent airborne Penicillium (Pen ch 13, Pen ch 18),5,6,7 Aspergillus (Asp f 13, Asp f 18),5,8,9 and Cladosporium (Cla c 9)10 species. Recently, in our characterization of Fusarium allergens, a 36.5-kDa component of F. proliferatum which showed a frequency of IgE binding of 53% (9/17) was identified.11 In addition, the 36.5-kDa component also showed a relatively higher intensity of IgE-immunoblot reactivity than other IgE-binding proteins of F. proliferatum.11 Since it reacts with a MoAb FUM2012 against fungal vacuolar serine protease allergens (data not shown), we putatively considered that the IgE-reacting 36.5-kDa component from F. proliferatum was possibly a vacuolar serine protease. It is important to further characterize the major 36.5-kDa IgE-binding component of F. proliferatum.
Clinically, respiratory atopic patients usually show IgE sensitization to more than 1 fungal species.13,14 The patient may be sensitized individually by different fungal species. Their multiple IgE sensitivity may also be due to IgE cross-reactivity between the homologous allergens from different fungal species. It is crucial to characterize IgE cross-reactivity among allergens from different fungal species. To further characterize important IgE-binding determinants of major fungal allergens, we found that the Lys89 and Phe91 played a significant role in IgE binding to Pen ch 18.15
In this study, to characterize the 36.5-kDa IgE-binding component of F. proliferatum, we cloned the cDNA of the vacuolar serine protease of F. proliferatum, identified its IgE cross-reactivity to the Pen ch 18 allergen. In addition, the Lys88 of the Fusarium vacuolar serine protease was evaluated uthether it plays a critical role in contributing to IgE cross-reactivity between this 36.5-kDa allergen of F. proliferatum (Fus p 9.0101) and the corresponding allergen of P. chrysogenum (Pen ch 18).
MATERIALS AND METHODS
Serum samples
The de-linked residual serum samples used in the present study were obtained from the Biobank at the Taipei Veterans General Hospital. All these serum samples were obtained originally from respiratory atopic patients (allergic asthma and/or atopic rhinitis) who attended the allergy clinics of Taipei Veterans General Hospital and were stored in aliquots at -80℃. This study without written consent has been approved by the Institutional Review Board of Taipei Veterans General Hospital.
Crude extracts of F. proliferatum
The F. proliferatum strain BCRC 30972 was used in this study. It was isolated from the air of Taiwan and provided by the Food Industry Research and Development Institute, Hsinchu, Taiwan. The crude extracts of F. proliferatum were prepared essentially as described previously.11 Briefly, F. proliferatum was cultured in a CYA medium without agitation at 26℃ for 5 days. The CYA medium contains yeast carbon base (Difco Laboratories, Detroit, MI, USA; 11.7 g/L), glucose (Mallinckrodt Baker, Inc., Phillipsburg, NJ, USA; 10 g/L) and casein enzymatic hydrolysate (Sigma Chemical Co., St. Louis, MO, USA; 10 g/L). The protein content of crude extracts was determined with a dye-binding method according to the manufacturer's instructions (Bio-Rad, Richmond, CA, USA).
cDNA cloning
The cDNA encoding the F. proliferatum vacuolar serine protease was isolated with an AffinityScript Multiple Temperature cDNA Synthesis kit (Stratagene, La Jolla, CA, USA) and polymerase chain reactions (PCR) as previously described.6,7,10,11 Primers used in the cloning experiments are listed in Table. The degenerate primers VSP-F-1 and VSP-R-1 were used in the first set of polymerase chain reaction (PCR). They encode conserved amino acid sequences (KNAPWG and MASPHVAG) near the N- and C-termini of fungal serine proteases. The PCR product (first-strand cDNA) was purified electrophoretically on agarose gel, subcloned into the pGem-Teasy vector (Promega, Madison, WI, USA), and then transformed into E. coli Top10F' competent cells. The plasmid DNA was purified, and the nucleotide sequence of the cDNA insert was determined with an automatic sequencer (Applied Biosystems, Foster City, CA, USA).
Table. Primers used in cDNA cloning, expression and site-directed mutagenesis of the vacuolar serine protease of Fusarium proliferatum.
| Name | Nucleotide sequence in 5' to 3'-end orientation |
|---|---|
| VSP-F-1 | 5'-25AA(G/A)A(A/G)(C/T)GC(T/C)CC(T/C/A)TGGGG41-3' |
| VSP-R-1 | 5'-761CC(A/G)GC(A/G)AT(A/G)TG(A/G/T)GG(A/G)(A/G/T)A(A/C/G)GCCAT739-3' |
| FuVSP-GSP1 | 5'-186GGCAGGCCCTCGAAGTCGACGT164-3' (For 5' race) |
| FuVSP-GSP2 | 5'-661ATCTTCGCTCCCGGTCTGAACATT684-3' (For 3' race) |
| AP | 5'-GGCCACGCGTCGACTAGTACT-(dT)16-3' |
| AUAP | 5'-GGCCACGCGTCGACTAGTAC-3' |
| AAP | 5'-GGCCACGCGTCGACTAGTACGGGIIGGGIIGGIIG-3' |
| FuVSP-5' Sma I | 5'-TCCCCCGGGG10GACGGCGAGACCGAGCGACAGGC32-3' |
| FuVSP-3' Hind III | 5'-CCCAAGCTTTTA969GTAGCTACCAGCCTCAACAATCTTCTT943-3' |
| FVSP K88A-f | 5'-256GGTACTGTCGCCGGTGCGAAGTACGGTGTTGC287-3' |
| FVSP K88A-r | 5'-287GCAACACCGTACTTCGCACCGGCGACAGTACC256-3' |
The 5'-and 3'-end RACE (rapid amplification of cDNA ends) reactions were used to obtain the full-length cDNA of the vacuolar serine protease. The primers used were the anchor primers (AP and AAP) together with the gene-specific primers (FuVSP-GSP1 and FuVSP-GSP2) that were synthesized according to the internal sequences of the Fusarium vacuolar serine protease obtained in the previous PCR reaction. FuVSP-GSP2 (for 3'-end RACE) and FuVSP-GSP1 (for 5'-end RACE) cover nucleotides 661 to 684 and 164 to 186, respectively, of the truncated Fusarium vacuolar serine protease cDNA. Primer AP used in the 3'-end RACE reaction contains the sequence of primer AUAP plus a stretch of oligo-(dT). The primer AAP used in the 5'-RACE reaction contains the sequence of primer AUAP and a stretch of oligo-(dG). The first-strand cDNA isolated was used as a template in the 3'-end RACE reaction. An oligo-(dC) was added to the end of the first-strand cDNA with terminal deoxynucleotidyl transferase (Promega) before using as a template in the 5'-RACE reaction. Products of the RACE reactions were purified individually, subcloned, transformed, and sequenced as described above.
Preparation of recombinant F. proliferatum vacuolar serine protease protein
Fungal vacuolar serine proteases were hypothesized to be synthesized as a larger precursor that undergoes both N- and C-terminal cleavage upon maturation.7,16,17,18,19 The F. proliferatum vacuolar serine protease was expressed as 6x His-tagged protein according to the manufacturer's instructions (Qiagen Inc., Valencia, CA, USA). The cDNA of the Fusarium vacuolar serine protease was amplified through PCR. The forward primer (FuVSP-5' Sma I, Table) used in the reaction contains the SmaI restriction site in addition to the cDNA sequence (nucleotides 10-32) encoding the putative N-terminus of the mature Fusarium vacuolar serine protease. We hypothesize that this vacuolar serine protease precursor is cleaved at or near the Tyr (Y) residue encoded by nucleotides 967-969 of the isolated clone, analogous to the proposed C-terminus of the vacuolar serine protease (pepC) of A. nigeri.19 Thus, the reverse primer FuVSP-3' Hind III (Table) contains a Hind III restriction site, a newly added stop codon (TAA) and the sequence encoding the hypothetical C-terminus (KKIVEAGSY, nucleotides 943-969) of the Fusarium vacuolar serine protease. The PCR product was restricted, ligated into the pQE-80 vector, and then transformed into E. coli M15 for protein expression. The recombinant protein of putative mature vacuolar serine protease was affinity-purified with a Ni-NTA resin column (Qiagen) under the denaturing condition according to the manufacturer's instructions. In addition, the purified recombinant 6x His-tagged vacuolar serine protease allergen of P. chrysogenum (Pen ch 18) and the Pen ch 18 K89A mutant were also prepared as previously described.15 Immuno-reactivity of the recombinant proteins to IgE antibodies and MoAb FUM20 was analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE)-immunoblotting.
SDS-PAGE and immunoblotting
Proteins in the crude Fusarium fungal extracts or the purified recombinant proteins were separated by SDS-PAGE8,10,11 and then transferred electrophoretically onto polyvinylidene difluoride (PVDF) membranes (0.45 µm, Millipore, Bedford, MS, USA). Protein components reacting to human IgE antibodies or MoAb FUM20 against fungal vacuolar serine protease allergen12 were determined as previously described.8,10,11,12 The membranes were blocked with 1% skimmed milk and incubated with serum samples for 16 hours at 4℃ or with MoAb FUM20 for 1 hour at room temperature. The membranes were washed, incubated with alkaline phosphatase-conjugated monoclonal anti-human IgE antibodies (Pharmingen, San Diego, CA, USA) or with horseradish peroxidase labeled goat anti-mouse IgG (H+L) (Jackson ImmunoResearch Laboratories, Inc., West Grove, PA, USA), and then developed with enzyme substrates as previously described.8,10,11,12 Serum from a house dust mite-sensitized atopic individual and a MoAb WH920 against house dust mite allergen Der p 7 were used as controls.
Site-directed mutagenesis
The Fusarium vacuolar serine protease mutant carrying single alanine substitute at Lys88 was prepared as previously descried.15 A plasmid encoding the vacuolar serine protease obtained as described above was used as a template in PCR. The primers used in the mutagenesis experiment (FVSP K88A-f and FVSP K88A-r) are shown in Table. The newly synthesized DNA carrying the point mutation was purified and inserted into the pQE80 expression vector (Qiagen) and transformed into E. coli JM109 for the expression of the Fusarium vacuolar serine ptotease K88A mutant. The mutant construct was confirmed by DNA sequencing and affinity-purified with Ni-NTA resin columns (Qiagen) according to the manufacturer's instructions.
Immunoblot inhibition
For immunoblot inhibition studies, IgE-containing serum samples were first reacted with purified recombinant Fusarium vacuolar serine protease, its K88A mutant, rPen ch 18, or rPen ch 18 K89A before incubation with PVDF blots containing F. proliferatum extracts or purified recombinant Fusarium vacuolar serine protease at 4℃ for 16 hours. As controls, the blots were incubated with similar serum samples that had been pre-incubated with equivalent amounts of bovine serum albumin (BSA, Pierce, Rockford, IL, USA). The blots were then washed and incubated with alkaline phosphatase-conjugated monoclonal anti-human IgE antibodies (Pharmingen) and developed with enzyme substrates as previously described.8,10,11
RESULTS
Characterization of an IgE-binding 36.5-kDa F. proliferatum component
In our previous study, a 36.5-kDa F. proliferatum component was found to bind IgE antibodies in 9 (53%) of 17 F. proliferatum-sensitized serum samples from respiratory atopic patients.11 In addition, this 36.5-kDa IgE-binding component also showed immunoblot reactivity to MoAb FUM20 against the vacuolar serine protease fungal allergens of Penicillium, Aspergillus, Rhodotorula, and Cladosporium species12 (data not shown). Results obtained suggest that the 36.5-kDa allergen may be a vacuolar serine protease of F. proliferatum. Thus, in this study the cDNA encoding the vacuolar serine protease of F. proliferatum was cloned.
The full-length cDNA encoding the F. proliferatum vacuolar serine protease was obtained through RT-PCR coupled with the 5'- and 3'-end RACE reactions. The isolated cDNA clone has 1,394 bp (GenBank accession no. KJ462778). The nucleotide and the deduced amino acid sequences (383 residues) of the open reading frame are presented in Fig. 1. A potential polyadenylation signal (AATACA) for mRNAs of higher eukaryotes was found at 13-18 bases upstream from the poly-A tail.
Fig. 1. (A) The nucleotide and deduced amino acid sequences of the vacuolar serine protease protein of F. proliferatum (Fus p 9.0101, GenBank accession no. KJ462778). Numbers to the right indicate the positions of the nucleotides and the deduced amino acid residues of the sequences. The amino acid triad (D45, H77 and S243) which is characteristic of serine proteases is depicted in bold type and shaded. Three potential N-glycosylation sites are in bold letters and underlined. The vertical arrow marks the proposed amino terminus of the mature vacuolar serine protease. The stop codon TAA is denoted with an asterisk. Nucleotides in gray correspond to those synthesized and used as primers for PCR in the cDNA cloning of the vacuolar serine protease of F. proliferatum as shown in Table. The sequences corresponding to primers FuVSP-5'Sma I and FuVSP-3'Hind III used in the preparation of recombinant mature Fusarium vacuolar serine protease protein and its K88A mutant (FVSP K88A-f and FVSP K88A-r) are boxed. (B) Composite alignment of the deduced amino acid sequences of the proposed mature Fus p 9.0101, Pen ch 18, Pen o 18 and the Pen ch 13 fungal serine protease allergens. Dashes denote spaces introduced to optimize the alignment. Identical amino acid residues are denoted with asterisks.
Sequence alignment suggests that this cDNA from F. proliferatum encodes a vacuolar serine protease. Assuming that the mature protein starts from Asp1, a protein of 40,478 Da can be translated from this cDNA. There are 4 cysteines (Cys81, Cys184, Cys215, and Cys308), 3 putative N-glycosylation sites (107NGS109, 147NMS149, and 185NYS187), and 3 active-site residues for serine proteases (Asp45, His77, and Ser243) on this protein (Fig. 1). The encoded protein has 67%-72% sequence identity with that of vacuolar serine proteases from Penicillium, Aspergillus, and Cladosporium species (Pen ch 18; accession no. AF263454; Pen o 18, accession no. AF243425; Pen c 18, accession no. AF245168; Asp f 18, accession no. Y13338; and Cla c 9, accession no. EF407520) plus 54% and 57% sequence identity with those from Rhodotorula mucilaginosa (Rho m 2, accession no. AY547285)21 and Saccharomyces cerevisiae (protease B, accession no. M18097),17 respectively. All these mature fungal serine proteases contain 3 cysteine residues. The most likely candidate for the free cysteine of the Saccharomyces protease B is Cys 81, located near the active-site His77.17 The remaining 2 may be responsible for a di-sulfide bond. Analogous to protease B, the 2 Cys (Cys184, Cys215) may form a disulfide bond.
Antigenicity of the recombinant vacuolar serine protease of F. proliferatum
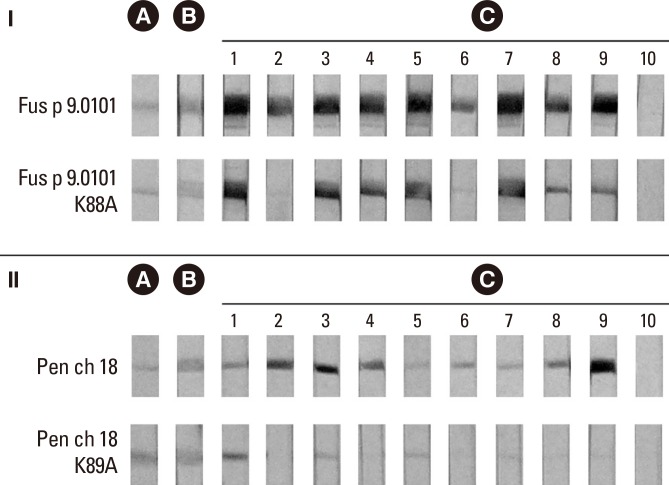
Our results from immunoblotting demonstrated that the recombinant vacuolar serine protease from F. proliferatum (rFus p 9.0101) and the recombinant Pen ch 18 both reacted with MoAb FUM20 (panel B of Fig. 2). In addition, 9 atopic serum samples with IgE binding to Pen ch 18 allergen (Fig. 2, section II, strip nos. 1-9 of panel C) also demonstrated IgE immunoblot reactivity to rFus p 9.0101 (Fig. 2, section I, strip nos. 1-9 of panel C). For control experiments, MoAb WH9 against Der p 7 allergen20 and serum from a house dust mite-atopic subject showed negative immunoblot reactivity to rFus p 9.0101 and rPen ch 18 (data not shown and Fig. 2, strip no. 10 of panel C, respectively). This Fusarium vacuolar serine protease allergen has been designated as Fus p 9.0101 by the IUIS Allergen Nomenclature Sub-committee.
Fig. 2. Antigenicity of the wild-type and mutant fungal vacuolar serine protease allergens. (A) Coomassie blue-stained protein profiles of the recombinant wild-type Fus p 9.0101 and Pen ch 18 allergens as well as the Fus p 9.0101K88A and Pen ch 18 K89A mutants on PVDF membranes. (B) Immunoblot reactivities to MoAb FUM20. (C) Immunoblot reactivities to IgE antibodies in 9 sera from asthmatic patients (strip nos. 1-9) and 1 serum from 1 house dust mite-sensitized asthmatic patient (strip no. 10).
Inhibition of IgE and FUM20-binding to the 36.5-kDa component of F. proliferatum by rFus p 9.0101
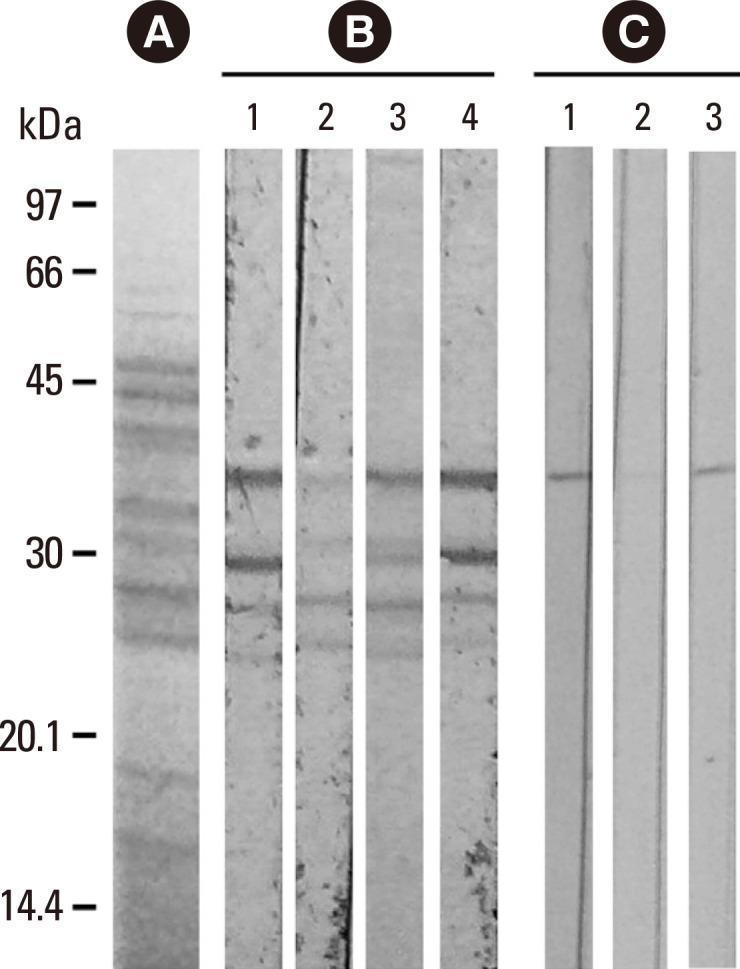
In this study, the intensity of IgE-immunoblot reactivity to the 36.5-kDa and 30-kDa components of F. proliferatum by using serum no. 1 of panel C in Fig. 2 (Fig. 3, strip 1 of panel B) was decreased dose-dependently after pre-absorption of the same serum sample by 20 µg and 5 µg of purified rFus p 9.0101 (Fig. 3, strips 2 and 3 of panel B). For the control experiment, a decrease in the intensity of IgE-immunoblot was not detectable following pre-absorption of the same serum sample by 20 µg of bovine serum albumin (BSA) (Fig. 3, strip 4 of panel B). In addition, the same 36.5-kDa component of F. proliferatum showed positive immunoblot reactivity to MoAb FUM20 (Fig. 3, strip 1 of panel C). MoAb WH9 as a control antibody showed negative immunoblot reactivity (data not shown). FUM20 binding to the 36.5-kDa F. proliferatum component was inhibited by 20 µg of rFus p 9.0101 (Fig. 3, strip 2 of panel C), but not by the same amount of BSA (Fig. 3, strip 3 of panel C). Results obtained indicate that the 36.5-kDa allergen (Fus p 9.0101) is a vacuolar serine protease of F. proliferatum. The 30-kDa component may be a degradation product of the 36.5-kDa vacuolar serine protease of F. proliferatum.
Fig. 3. Immunoblot inhibition of IgE- (panel B) and MoAb FUM20- (panel C) binding to nFus p 9.0101 in crude F. proliferatum extracts with purified rFus p 9.0101 and BSA as inhibitors. (A) Coomassie blue-stained protein profile of F. proliferatum extracts and protein molecular weight markers. (B) IgE binding to the 36.5-kDa component using serum no. 1 from Fig. 2C (lane 1); this binding activity was inhibited dose-dependently by 20 µg (lane 2) and 5 µg (lane 3) of rFus p 9.0101, but not 20 µg of BSA (lane 4). (C) MoAb FUM20 binding to the 36.5-kDa component of F. proliferatum extracts (lane 1); this binding activity was inhibited by 20 µg (lane 2) of rFus p 9.0101, but not 20 µg of BSA (lane 3).
IgE determinant of Fus p 9.0101 and Pen ch 18
Results of Fig. 2 showed that in addition to Fus p 9.0101 and Pen ch 18, MoAb FUM20 reacted with the Fus p 9.0101 K88A mutant (section I, panel B) and the Pen ch 18 K89A mutant (section II, panel B). For control, MoAb WH9 showed negative immunoblot reactivity (data not shown).
In this study, the intensities of bands on the immunoblots were quantified with AlphaEaseFCTM software (version 4.0.0, Alpha Innotech Corpration, San Leandro, CA, USA). IgE binding to the rFus p 9.0101 K88A mutant was decreased by <20% inhibition for serum nos. 1 and 3, by 25%-30% for serum nos. 4, 5 and 7, by 50%-65% for serum nos. 8 and 9, and by about 80% for serum nos. 2 and 6 as compared to those against Fus p 9.0101 (Fig. 2, section I, panel C).
Immunoblot reactivity to IgE antibodies in 7 of the 9 atopic serum samples (strip nos. 2, 3, 4, 6, 7, 8, and 9 of Fig. 2, section II, panel C) tested demonstrated significantly decreased IgE binding (by >70% inhibition) to the rPen ch 18 K89A mutant compared to those to rPen ch 18. For the control experiment, serum sample from a house dust mite-sensitized atopic individual (Fig. 2, sections I and II, panel C, serum no. 10) showed negative immunoblot reacticity to both wild type allergens and their site-directed mutants. Results obtained indicate that the K88 and the K89 play a significant role in IgE binding to Fus p 9.0101 and Pen ch 18 allergens, respectively.
IgE cross-reactivity
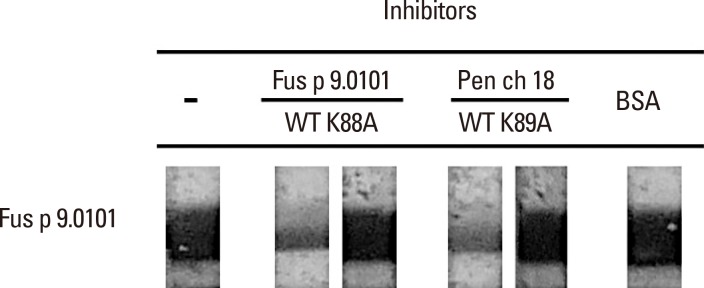
Results in Fig. 4 showed that serum no. 9 from Fig. 2, panel C has IgE-binding activity to rFus p 9.0101. For this serum sample, purified rFus p 9.0101 and rPen ch 18 decreased IgE binding to rFus p 9.0101 when the serum sample was pre-absorbed with 10 µg of the wild type rFus p 9.0101 or rPen ch 18 as indicated (Fig. 4). Pre-absorption of the same serum sample with 10 µg of the purified Fus p 9.0101 K88A mutant, the Pen ch 18 K89A mutant, or BSA did not inhibit its IgE binding to rFus p 9.0101 as indicated.
Fig. 4. Inhibition of IgE-immunoblot reactivity to Fus p 9.0101. IgE-immunoblot experiment was carried out with serum no. 9 from Fig. 2C (lane 9). Serum was pre-absorbed with 10 µg of the wild-type Fus p 9.0101, the wild-type Pen ch 18, the Fus p 9.0101K88A mutant, the Pen ch 18 K89A mutant, or BSA as indicated.
DISCUSSION
In this study, the 36.5-kDa IgE-binding component of F. proliferatum reacted with MoAb FUM20 against fungal vacuolar serine protease allergens. The cDNA of F. proliferatum vacuolar serine protease was subsequently cloned and expressed in E. coli. Moreover, the purified recombinant protein obtained reacted with human IgE antibodies (Fig. 2) and demonstrated dose-dependent inhibition of IgE- and FUM20-binding to the 36.5-kDa component of F. proliferatum (Fig. 3). Our results obtained indicate that the 36.5-kDa major allergen (Fus p 9.0101) is a vacuolar serine protease of F. proliferatum.
We have previously identified that the alkaline and/or vacuolar serine proteases are major allergens in 8 prevalent airborne Penicillium and Aspergillus species, including P. citrinum (Pen c 13), P. chrysogenum (Pen ch 13, Pen ch 18), P. oxalicum (Pen o 18), P. brevicompactum (Pen b 13), A. fumigates (Asp f 13, Asp f 18), A. flavus (Asp fl 13), A. oryzae (Asp o 13), and A. nigeri (Asp n 18).5,6,7,8,9,16 In addition, the vacuolar serine protease is also identified as a major allergen of C. cladosporioides (Cla c 4.0101)10 and R. mucilaginosa (Rho m 2).21 Thus, our results obtained indicate that serine proteases belong to the classes of highly conserved important pan-fungal allergens. Furthermore, results from Gupta et al.22 also demonstrated serine protease as a major allergen of Curvularia lunata (Cur l 1). IgE reactivity to Cur l 1 was detectable in sera from 80% of C. lunata hypersensitive patients.22
In this study, rPen ch 18 demonstrated inhibition of IgE binding to Fus p 9.0101 (Fig. 4), which suggests the presence of IgE cross-reactivity between the vacuolar serine protease allergens from Fusarium and Penicillium fungi. IgE cross-reactivity among alkaline/vacuolar serine protease major allergens from Penicillium, Aspergillus, Rhodotorula, and Cladosporium species has previously been reported.5,9,10,21 In the present study, Fusarium vacuolar serine protease may also display IgE cross-reactivity to other fungal serine protease major allergens through its cross-reactivity to the corresponding Penicillium Pen ch 18 allergen. The result that inhibition of IgE binding to rFus p 9.0101 was not detected when the same serum was pre-absorbed with 10 µg of the rFus p 9.0101 K88A and the rPen ch 18 K89A mutants confirms that K88 is a critical amino acid in IgE binding to Fus p 9.0101 and plays a significant role in IgE cross-reaction between Fus p 9.0101 and Pen ch 18 vacuolar serine protease fungal allergens.
Results from our previous study showed that the alkaline serine protease from P. chrysogenum (Pen ch 13) degrades the tight junction protein occludin and stimulates release of proinflammatory mediators from human bronchial epithelial cells.23 In addition to induction of IgE and inflammatory airway responses, the alkaline serine protease allergen from A. fumigatus (Asp f 13) has also been shown to have synergistic effects on Asp f 2-induced immune response in mice.24 Furthermore, Pen ch 13 major fungal allergen has been found to decrease CD44 expression in human bronchial epithelial cells,25 which may contribute to atopic diseases by influencing the resolution of lung inflammation and by prolonging the repair response of damaged bronchial epithelial cells. Results from Tripathi et al.26 demonstrated the serine protease activity of Cur l 1 from C. lunata augments Th2 response in mice. Recently, Namvar et al.27 found that the Aspergillus fumigatus proteases Asp f 5, and Asp f 13 are essential for airway inflammation and remodelling in a murine inhalation model.
In conclusion, in this study, the vacuolar serine protease was identified as a major allergen of F. proliferatum and confirmed that serine proteases are important IgE cross-reactive pan-fungal allergens. In addition to providing important bases for the clinical diagnosis of fungal allergy, studies of these serine protease major allergens may elucidate diverse allergic disease mechanisms and facilitate development of better therapeutic strategies.
ACKNOWLEDGMENTS
This work was supported by grants from the Ministry of Science and Technology, R.O.C. (Grant NSC101-2320-B-075-005-MY2) and the Taipei Veterans General Hospital (Grant V103C-034), Taipei, Taiwan, R.O.C. We thank Miss Yu-Sen Chen for her technical assistance.
Footnotes
There are no financial or other issues that might lead to conflict of interest.
References
- 1.Sharpe RA, Bearman N, Thornton CR, Husk K, Osborne NJ. Indoor fungal diversity and asthma: a meta-analysis and systematic review of risk factors. J Allergy Clin Immunol. 2015;135:110–122. doi: 10.1016/j.jaci.2014.07.002. [DOI] [PubMed] [Google Scholar]
- 2.Twaroch TE, Curin M, Valenta R, Swoboda I. Mold allergens in respiratory allergy: from structure to therapy. Allergy Asthma Immunol Res. 2015;7:205–220. doi: 10.4168/aair.2015.7.3.205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Esch RE. Manufacturing and standardizing fungal allergen products. J Allergy Clin Immunol. 2004;113:210–215. doi: 10.1016/j.jaci.2003.11.024. [DOI] [PubMed] [Google Scholar]
- 4.Jeong KY, Lee JH, Kim EJ, Lee JS, Cho SH, Hong SJ, et al. Current status of standardization of inhalant allergen extracts in Korea. Allergy Asthma Immunol Res. 2014;6:196–200. doi: 10.4168/aair.2014.6.3.196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shen HD, Tam MF, Chou H, Han SH. The importance of serine proteinases as aeroallergens associated with asthma. Int Arch Allergy Immunol. 1999;119:259–264. doi: 10.1159/000024202. [DOI] [PubMed] [Google Scholar]
- 6.Chou H, Lai HY, Tam MF, Chou MY, Wang SR, Han SH, et al. cDNA cloning, biological and immunological characterization of the alkaline serine protease major allergen from Penicillium chrysogenum. Int Arch Allergy Immunol. 2002;127:15–26. doi: 10.1159/000048165. [DOI] [PubMed] [Google Scholar]
- 7.Shen HD, Chou H, Tam MF, Chang CY, Lai HY, Wang SR. Molecular and immunological characterization of Pen ch 18, the vacuolar serine protease major allergen of Penicillium chrysogenum. Allergy. 2003;58:993–1002. doi: 10.1034/j.1398-9995.2003.00107.x. [DOI] [PubMed] [Google Scholar]
- 8.Shen HD, Lin WL, Tam MF, Chou H, Wang CW, Tsai JJ, et al. Identification of vacuolar serine proteinase as a major allergen of Aspergillus fumigatus by immunoblotting and N-terminal amino acid sequence analysis. Clin Exp Allergy. 2001;31:295–302. doi: 10.1046/j.1365-2222.2001.01026.x. [DOI] [PubMed] [Google Scholar]
- 9.Shen HD, Tam MF, Tang RB, Chou H. Aspergillus and Penicillium allergens: focus on proteases. Curr Allergy Asthma Rep. 2007;7:351–356. doi: 10.1007/s11882-007-0053-8. [DOI] [PubMed] [Google Scholar]
- 10.Chou H, Tam MF, Lee LH, Chiang CH, Tai HY, Panzani RC, et al. Vacuolar serine protease is a major allergen of Cladosporium cladosporioides. Int Arch Allergy Immunol. 2008;146:277–286. doi: 10.1159/000121462. [DOI] [PubMed] [Google Scholar]
- 11.Chou H, Wu KG, Yeh CC, Tai HY, Tam MF, Chen YS, et al. The transaldolase, a novel allergen of Fusarium proliferatum, demonstrates IgE cross-reactivity with its human analogue. PLoS One. 2014;9:e103488. doi: 10.1371/journal.pone.0103488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lin WL, Chou H, Tam MF, Huang MH, Han SH, Shen HD. Production and characterization of monoclonal antibodies to serine proteinase allergens in Penicillium and Aspergillus species. Clin Exp Allergy. 2000;30:1653–1662. doi: 10.1046/j.1365-2222.2000.00922.x. [DOI] [PubMed] [Google Scholar]
- 13.Sams JT, Smith RE. Cross-antigenicity of common mold antigens. Ann Allergy. 1968;26:55–60. [PubMed] [Google Scholar]
- 14.Prince HE, Morrow MB. Skin reaction patterns to dematiaceous mold allergens. Ann Allergy. 1971;29:535–538. [PubMed] [Google Scholar]
- 15.Cheng TT, Tam MF, Chou H, Tai HY, Shen HD. Lys89, Lys90, and Phe91 are critical core amino acid residues of the Pen ch 18 major fungal allergen recognized by human IgE antibodies. Biochem Biophys Res Commun. 2008;375:671–674. doi: 10.1016/j.bbrc.2008.08.097. [DOI] [PubMed] [Google Scholar]
- 16.Shen HD, Wang CW, Lin WL, Lai HY, Tam MF, Chou H, et al. cDNA cloning and immunologic characterization of Pen o 18, the vacuolar serine protease major allergen of Penicillium oxalicum. J Lab Clin Med. 2001;137:115–124. doi: 10.1067/mlc.2001.112096. [DOI] [PubMed] [Google Scholar]
- 17.Moehle CM, Tizard R, Lemmon SK, Smart J, Jones EW. Protease B of the lysosomelike vacuole of the yeast Saccharomyces cerevisiae is homologous to the subtilisin family of serine proteases. Mol Cell Biol. 1987;7:4390–4399. doi: 10.1128/mcb.7.12.4390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nebes VL, Jones EW. Activation of the proteinase B precursor of the yeast Saccharomyces cerevisiae by autocatalysis and by an internal sequence. J Biol Chem. 1991;266:22851–22857. [PubMed] [Google Scholar]
- 19.Frederick GD, Rombouts P, Buxton FP. Cloning and characterisation of pepC, a gene encoding a serine protease from Aspergillus niger. Gene. 1993;125:57–64. doi: 10.1016/0378-1119(93)90745-o. [DOI] [PubMed] [Google Scholar]
- 20.Shen HD, Chua KY, Lin WL, Hsieh KH, Thomas WR. Characterization of the house dust mite allergen Der p 7 by monoclonal antibodies. Clin Exp Allergy. 1995;25:416–422. doi: 10.1111/j.1365-2222.1995.tb01072.x. [DOI] [PubMed] [Google Scholar]
- 21.Chou H, Tam MF, Lee SS, Tai HY, Chang CY, Chou CT, et al. A vacuolar serine protease (Rho m 2) is a major allergen of Rhodotorula mucilaginosa and belongs to a class of highly conserved pan-fungal allergens. Int Arch Allergy Immunol. 2005;138:134–141. doi: 10.1159/000088435. [DOI] [PubMed] [Google Scholar]
- 22.Gupta R, Sharma V, Sridhara S, Singh BP, Arora N. Identification of serine protease as a major allergen of Curvularia lunata. Allergy. 2004;59:421–427. doi: 10.1046/j.1398-9995.2003.00378.x. [DOI] [PubMed] [Google Scholar]
- 23.Tai HY, Tam MF, Chou H, Peng HJ, Su SN, Perng DW, et al. Pen ch 13 allergen induces secretion of mediators and degradation of occludin protein of human lung epithelial cells. Allergy. 2006;61:382–388. doi: 10.1111/j.1398-9995.2005.00958.x. [DOI] [PubMed] [Google Scholar]
- 24.Kurup VP, Xia JQ, Shen HD, Rickaby DA, Henderson JD, Jr, Fink JN, et al. Alkaline serine proteinase from Aspergillus fumigatus has synergistic effects on Asp-f-2-induced immune response in mice. Int Arch Allergy Immunol. 2002;129:129–137. doi: 10.1159/000065882. [DOI] [PubMed] [Google Scholar]
- 25.Tai HY, Tam MF, Chou H, Perng DW, Shen HD. Pen ch 13 major fungal allergen decreases CD44 expression in human bronchial epithelial cells. Int Arch Allergy Immunol. 2010;153:367–371. doi: 10.1159/000316347. [DOI] [PubMed] [Google Scholar]
- 26.Tripathi P, Kukreja N, Singh BP, Arora N. Serine protease activity of Cur l 1 from Curvularia lunata augments Th2 response in mice. J Clin Immunol. 2009;29:292–302. doi: 10.1007/s10875-008-9261-9. [DOI] [PubMed] [Google Scholar]
- 27.Namvar S, Warn P, Farnell E, Bromley M, Fraczek M, Bowyer P, et al. Aspergillus fumigatus proteases, Asp f 5 and Asp f 13, are essential for airway inflammation and remodelling in a murine inhalation model. Clin Exp Allergy. 2015;45:982–993. doi: 10.1111/cea.12426. [DOI] [PubMed] [Google Scholar]