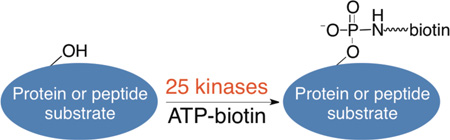
Abstract
Kinase-catalyzed protein phosphorylation is involved in a wide variety of cellular events. Development of methods to monitor phosphoproteins in normal and diseased states is critical to fully characterize cell signalling. Towards phosphoprotein analysis tools, our lab reported kinase-catalyzed labeling where γ-phosphate modified ATP analogs are utilized by kinases to label peptides or protein substrates with a functional tag. In particular, the ATP-biotin analog was developed for kinase-catalyzed biotinylation. However, kinase-catalyzed labeling has been tested rigorously with only a few kinases, preventing use of ATP-biotin as a general tool. Here, biotinylation experiments, gel or HPLC-based quantification, and kinetic measurements indicated that twenty-five kinases throughout the kinome tree accepted ATP-biotin as a cosubstrate. With this rigorous characterization of ATP-biotin compatibility, kinase-catalyzed labeling is now immediately useful for studying phosphoproteins and characterizing the role of phosphorylation in various biological events.
Keywords: Kinase Enzymes, ATP analogs, Biotinylation, Kinome Tree
Graphical Abstract
1. Introduction
Phosphorylation is a protein post-translational modification that is a key regulator of cell signaling networks.1 The activity of the protein changes often after phosphorylation, which is the basis of signaling in the cell. Kinases are the enzymes responsible for protein phosphorylation and aberrant kinase activity is linked to a variety of disease states, including cancer and immunosuppression.2, 3 In fact, a variety of kinase inhibitor drugs have been approved to treat diseases, predominantly cancer.4 With a fundamental role in cell biology and disease progression, the characterization of cellular phosphorylation in normal and disease states represents a critical aspect of biomedical research.
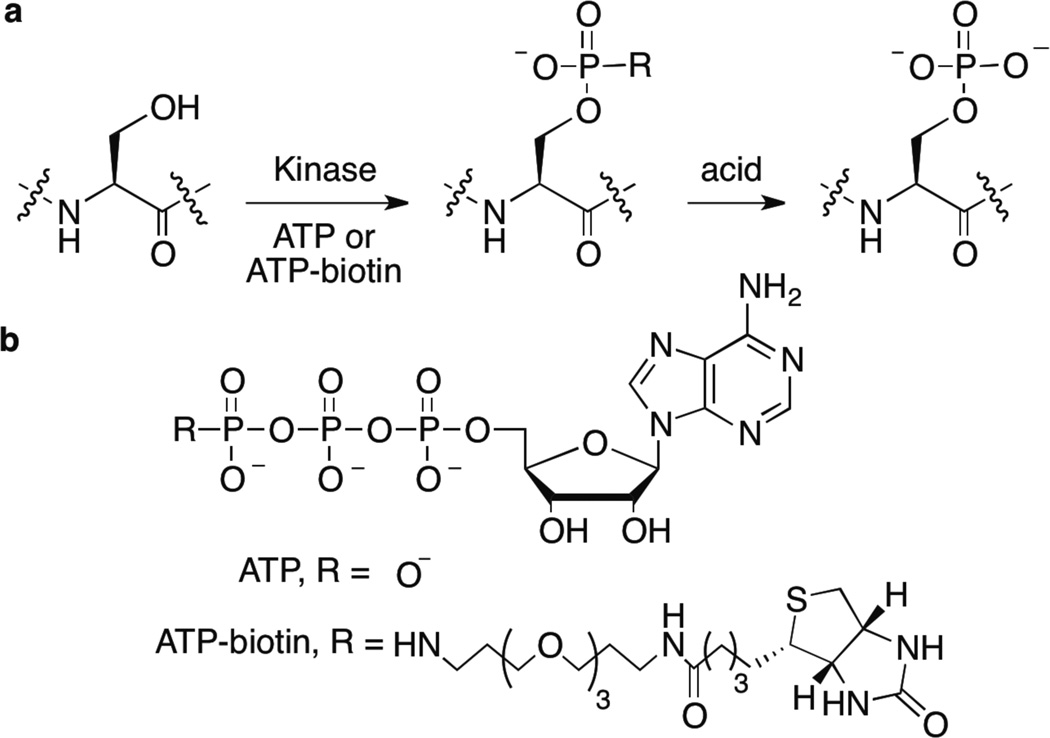
In phosphorylation, adenosine 5’-triphosphate (ATP) acts as the universal cosubstrate, with the kinase assisting in the transfer of phosphoryl group from the γ position of ATP to the peptide or protein substrate (Figure 1).2 Historically, phosphoproteins were monitored using [γ-32P]ATP labeling and subsequent analysis of the radioactive products by gel or HPLC methods.5 Recently, kinase-catalyzed labeling using γ-phosphate modified ATP analogs has emerged as a tool for studying phosphorylation (Figure 1a).6 A diversity of functional tags has been attached to γ-phosphate of ATP and transferred to proteins/peptide substrates by kinases, including biotin,6–10 dansyl,11 ferrocene,12, 13 BODIPY,14 azide,15, 16 alkyne,16 thiol,17, 18 acetyl, and sulfonyl groups.19 Among these analogs, ATP-biotin (Figure 1b) is an attractive probe for characterizing phosphorylation by affixing a biotin handle onto phosphoproteins, which can be used for subsequent visualization.20, 21 In addition, kinase-catalyzed biotinylation coupled with avidin affinity purification offers a powerful phosphoprotein enrichment strategy.22
Figure 1. Kinase-catalyzed labeling.
a) Kinase-catalyzed phosphorylation (ATP) or biotinylation (ATP-biotin) of hydroxyl containing amino acids (only serine is shown) on protein or peptide substrates. Acidic conditions (50% trifluoroacetic acid) cleave the phosphoramidate bond to create a phosphorylated product and release biotin amine (Scheme S1). b) The structures of ATP and ATP-biotin.
Several γ-phosphate modified ATP analogs, including ATP-biotin, have been qualitatively and quantitatively tested with select kinases and the kinase activity in cellular lysates.6, 11, 15, 19 However, a systematic study with the greater kinase family is lacking. The generality of ATP analog-mediated labeling with additional kinase family members must be established to support general use of kinase-catalyzed labeling for phosphoprotein analysis. We report here testing of ATP-biotin with twenty-five select kinases throughout the kinome family tree to rigorously characterize kinase-catalyzed labeling. Phosphorylbiotinylation was assessed qualitatively and quantitatively, and the combined data indicated that all twenty-five tested kinases accept ATP-biotin as a cosubstrate. The data establish that γ-phosphate modified ATP analogs are compatible with diverse kinases and suggest that kinase-catalyzed labeling is a useful tool for phosphoprotein analysis and cell biological research.
2. Material and Methods
2.1. Preparation of ATP-biotin
The synthesis of ATP-biotin was published elsewhere.24
2.2. Kinase-Catalyzed Phosphorylation and Biotinylation
Phosphorylated or phosphorylbiotinylated peptide or proteins were created by incubating ATP or ATP-biotin (2 mM) with kinase enzyme (Figure 2b, concentrations shown in Table S1) and corresponding substrate (Figure 2b, 0.25 mM) in the manufacturer provided buffer (1×). As a control, reactions were performed without addition of kinase. The final volumes of the reactions were 25 µL. The reaction mixtures were incubated at 30 °C for 2 hours without shaking. After reaction, the mixtures were separated using 16% SDS-PAGE, transferred on to Immobilion-P PVDF membrane (Invitrogen), and visualized with sypro ruby staining (Life Technologies) or SA-Cy5 conjugate (Fisher) according to the manufacturers protocol. In the case of peptide substrates, crude reactions were separated using a 16% Tristricine gel, transferred to Immobilion-PSQ PVDF membrane (Invitrogen), and visualized with Coomassie staining (Fisher) or SA-Cy5 conjugate according to the manufacturers protocol. In the case of SA-Cy5 visualization with proteins, membranes were incubated with the SA-Cy5 conjugate for no more than 1 hour in the presence of 5–10% lowfat dry milk to avoid nonspecific protein detection (Figure S3). SA-Cy5 stained gels were scanned using a Typhoon scanner (Amersham Biosciences) using an excitation wavelength of 633 nm and an emission at 670 nm. Sypro Ruby stained gels were scanned using a Typhoon scanner (Amersham Biosciences) using an excitation wavelength of 532 nm and an emission at 610 nm. Coomassie gels were scanned using HP Scanjet G4010. Three independent biotinylation reactions were performed for each kinase, which are shown in Figures S1 and S2.
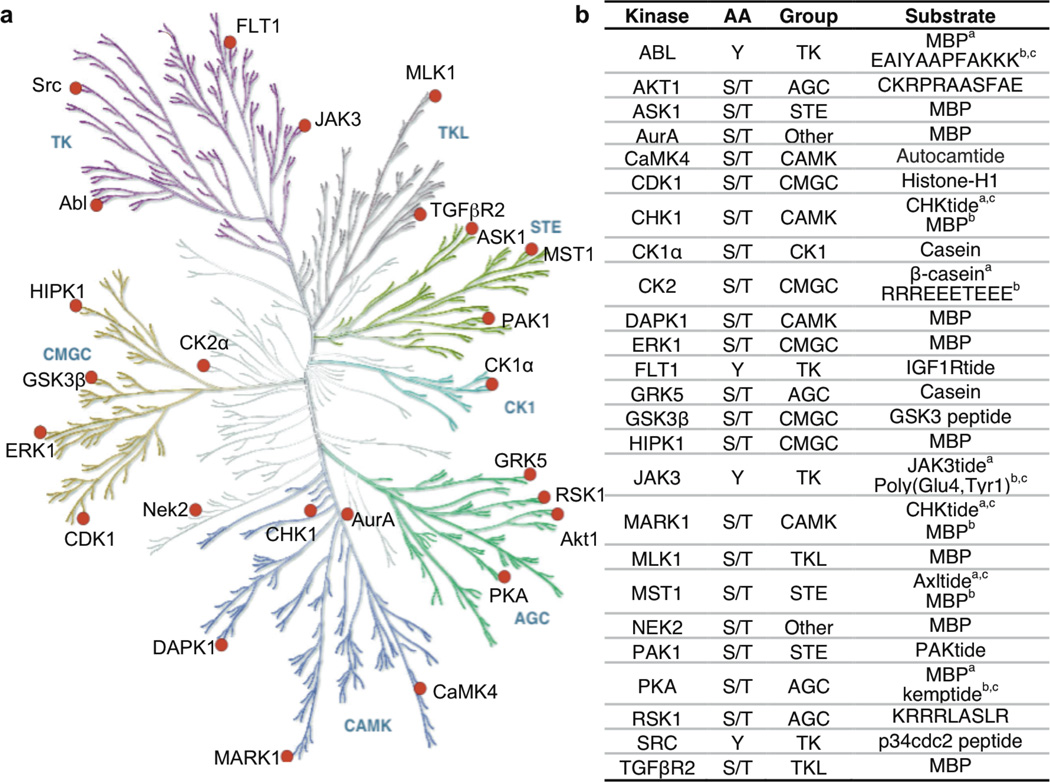
Figure 2. Select Kinases Used in This Study.
a) The human kinome tree23 with selected kinases tested in this work shown as red circles. Reprinted with permission from AAAS. Illustration reproduced courtesy of Cell Signaling Technology, Inc. (www.cellsignal.com) using Kinome Render. TK: Tyrosine kinase; TKL: Tyrosine kinase-like; STE: homologs of yeast Sterile 7, Sterile 11, Sterile 20 kinases; CK1: Casein kinase 1; AGC: Containing PKA, PKG, PKC families; CaMK: Calcium/calmodulin-dependent protein kinase; CMGC: Containing CDK, MAPK, GSK3, CLK families; Other: other kinases. b) The 25 kinases tested, along with amino acid specificity, group identity, and peptide, polypeptide, or protein substrate(s) used in kinase reactions. In some cases, different substrates were used in biotinylation, quantification, and kinetics experiments. abiotinylation, bHPLC or ProQ quantification, ckinetics. MBP- myelin basic protein; Autocamtide-KKALRRQETVDAL-amide; CHKtide- KKKVSRSGLYRSPSMPENLNRPR; IGF1Rtide- KKKSPGEYVNIEFG; JAK3tide- GGEEEEYFELVKKKK; GSK3 peptide-YRRAAVPPSPSLSRHSSPHQ(pS)EDEEE; Axltide- KKSRGDYMTMQIG; PAKtide- RRRLSFAEPG; kemptide- LRRASLG; p34cdc2 peptide-KVEKIGEGTYGVVYK-amide.
2.3. Quantitative Gel Analysis
After phosphorylated or phosphorylbiotinylated proteins were generated as described above, trifluoroacetic acid (TFA, 25 µL to make a 50% final concentration) was added to both phosphorylated and biotinylated samples and incubated for 15 minutes at 45°C with shaking (400 rpm). Acid cleavage of the phosphoramidate bond of the biotinylated proteins (Figure 1a) was necessary to create the same phosphoprotein product as the ATP reactions and allow quantitative comparison. After TFA incubation, the samples were evaporated to dryness using a speed vac (Thermo scientific, Savant SPD131DDA) and subsequently resuspended and neutralized in final concentration of 0.15 M Tris-base (25 µL). Phosphoproteins were separated by SDS-PAGE and visualized using Pro-Q Diamond Stain (Life Technologies) and sypro ruby stain according to the manufacturers protocol. Phosphoprotein gel bands from Pro-Q staining were detected on a Typhoon imager (excitation wavelength of 532 nm and emission at 580 nm). Band intensities were quantified using ImageQuant 5.1 or ImageJ (ASK1 only) by drawing the same sized rectangular shape around comparable bands. A conversion percentage was calculated for each kinase by dividing the band intensity of the ATP-biotin reaction by the band intensity of the ATP reaction, and multiplying by 100. The percentages and standard errors displayed in Figure 4 and Table 1 were obtained from three independent trials (Figure S4).
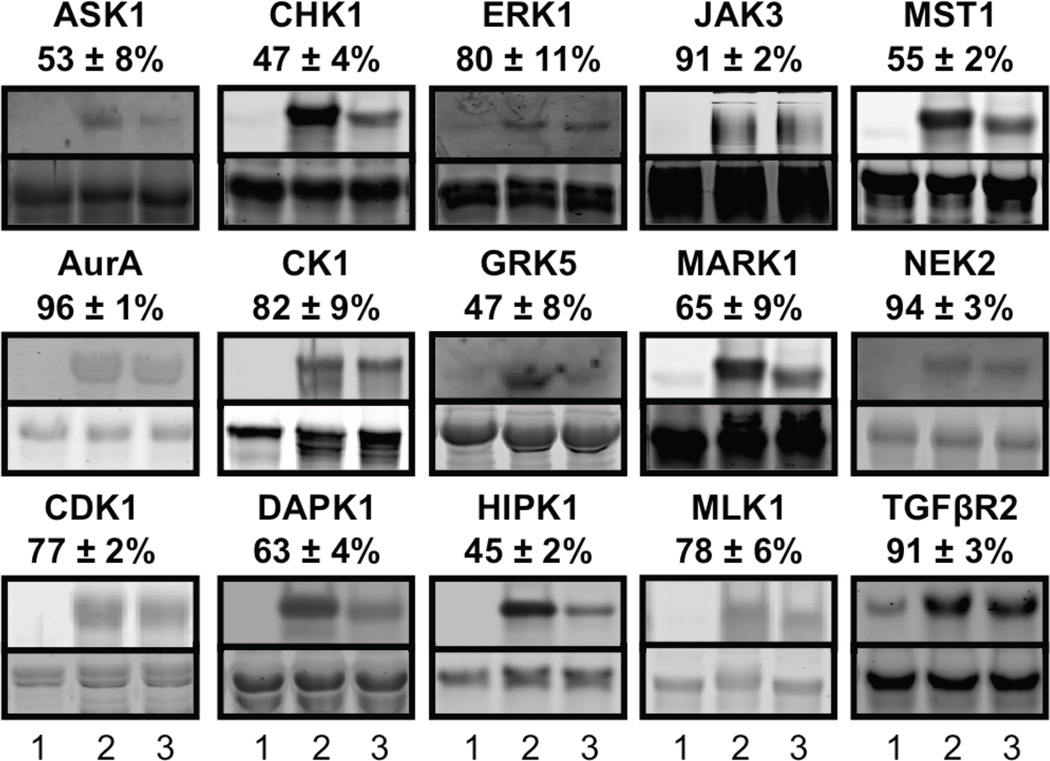
Figure 4. Quantitative Analysis of Protein Labeling.
Protein substrates (lane 1) were subjected to kinase-catalyzed phosphorylation with ATP (lane 2) or phosphorylbiotinylation with ATP-biotin (lane 3) before incubated with 50% TFA for 15 minutes, followed by SDS-PAGE separation and visualization with Pro-Q Diamond Stain (top gel) and sypro ruby stain (bottom gel). The kinase used in each experiment along with the calculated percentage conversion with ATP-biotin compared to ATP is shown above each gel. Full gel images are shown in Figure S4.
Table 1.
Quantitative conversion percentages and kinetic constants for kinases with ATP and ATP-biotina
| Kinase | Conversion | KMapp (µM)b | kcatapp (s−1)b | kcat/KMapp (mM s)−1 | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| ATP | ATP-biotin | Ratioc | ATP | ATP-biotin | Ratiod | ATP | ATP-biotin | Ratiod | ||
| ABLe | 85 ± 6% | NA | NA | NA | NA | NA | NA | NA | NA | NA |
| AKT1 | 51 ± 4% | 17 ± 2 | 24 ± 3 | 1.4 | ≥0.79 ± 0.04 f | 0.16 ± 0.01 | ≥4.9 | ≥47 | 6.7 | ≥7 |
| ASK1 | 53 ± 8% | 12 ± 2 | 10 ± 2 | 1.2 | ≥0.38 ± 0.02 f | 0.019 ± 0.001 | ≥20 | ≥31 | 1.9 | ≥16 |
| Aurora | 96 ± 1% | ≤24 ± 1g | 42 ± 2 | ≥1.7 | 1.0 ± 0.02 | 0.64 ± 0.02 | 1.6 | ≥42 | 15 | ≥2.7 |
| CaMK4 | 63 ± 3% | 13 ± 1 | 14 ± 2 | 1.1 | 0.15 ± 0.01 | 0.047 ± 0.003 | 3.1 | 11 | 3.4 | 3.4 |
| CDK1 | 77 ± 2% | 19 ± 3 | 28 ± 3 | 1.5 | 2.5 ± 0.2 | 1.5 ± 0.1 | 1.7 | 133 | 52 | 2.6 |
| CHK1 | 47 ± 4% | 15 ± 2 | 20 ± 3 | 1.3 | 0.64 ± 0.03 | 0.38 ± 0.02 | 1.7 | 42 | 19 | 2.2 |
| CK1 | 82 ± 9% | 4.5 ± 0.6 | 11 ± 1 | 2.3 | 0.062 ± 0.002 | 0.068 ± 0.002 | 0.9 | 14 | 6.5 | 2.1 |
| CK2 | 51 ± 4% | 7.4 ± 1.5 | 9.2 ± 3.3 | 1.2 | 0.81 ± .06 | 0.29 ± 0.04 | 2.8 | 110 | 32 | 3.5 |
| DAPK1 | 63 ± 4% | 3.9 ± 0.9 | 11 ± 1 | 2.9 | 0.098 ± 0.005 | 0.095 ± 0.003 | 1.0 | 25 | 8.3 | 3.0 |
| ERK1 | 80 ± 11% | 64 ± 5 | 48 ± 4 | 1.3 | 0.35 ± 0.01 | 0.091 ± 0.003 | 3.8 | 5.5 | 1.9 | 2.9 |
| FLT1 | 87 ± 20% | 32 ± 6 | 36 ± 4 | 1.1 | 0.057 ± 0.004 | 0.023 ± 0.001 | 2.4 | 1.8 | 0.64 | 2.8 |
| GRK5 | 47 ± 8% | 9.7 ± 1.3 | 6.6 ± 1.8 | 1.5 | 0.0072 ± 0.0003 | 0.0028 ± 0.0002 | 2.6 | 0.74 | 0.43 | 1.7 |
| GSK3βh | 44 ± 6% | 4.6 ± 0.6 | ND | ND | 0.21 ± 0.01 | ND | ND | 47 | ND | ND |
| HIPK1 | 45 ± 2% | 4.8 ± 0.4 | 15 ± 3 | 3.1 | 0.71 ± 0.01 | 0.59 ± 0.03 | 1.2 | 148 | 39 | 3.8 |
| JAK3 | 91 ± 2% | 12 ± 3 | 15 ± 2 | 1.2 | 0.54 ± 0.04 | 0.48 ± 0.02 | 1.1 | 44 | 32 | 1.4 |
| MARK1 | 65 ± 9% | 9.4 ± 1.0 | 13 ± 1 | 1.4 | 1.2 ± 0.1 | 0.72 ± 0.02 | 1.7 | 127 | 55 | 2.3 |
| MLK1 | 78 ± 6% | 24 ± 3 | 28 ± 3 | 1.2 | 0.61 ± 0.03 | 0.16 ± 0.01 | 3.8 | 26 | 5.7 | 4.5 |
| MST1 | 55 ± 2% | 17 ± 3 | 25 ± 4 | 1.5 | 0.20 ± 0.01 | 0.12 ± 0.01 | 1.7 | 12 | 4.8 | 2.5 |
| NEK2 | 94 ± 2% | 25 ± 4 | 21 ± 3 | 0.8 | 0.32 ± 0.02 | 0.23 ± 0.01 | 1.4 | 13 | 11 | 1.2 |
| PAK1 | 63 ± 2% | 23 ± 3 | 21 ± 2 | 0.9 | 0.023 ± 0.001 | 0.018 ± 0.001 | 1.3 | 1.0 | 0.88 | 1.1 |
| PKA | 61 ± 6% | 11 ± 4 | 37 ± 14 | 3.4 | 7.5 ± 0.6 | 3.8 ± 0.6 | 2.0 | 686 | 102 | 6.7 |
| RSK1 | 76 ± 7% | 25 ± 2 | 34 ± 4 | 1.4 | 1.8 ± 0.1 | 1.0 ± 0.1 | 1.8 | 74 | 30 | 2.5 |
| SRC | 96 ± 7% | 15 ± 3 | 16 ± 2 | 1.1 | 0.46 ± 0.02 | 0.30 ± 0.01 | 1.5 | 30 | 19 | 1.6 |
| TGFβR2 | 91 ± 3% | 1.2 ± 0.4 | 4.1 ± 0.9 | 3.4 | 0.015 ± 0.001 | 0.013 ± 0.001 | 1.2 | 13 | 3.2 | 3.9 |
Conversion percentages were determined from the quantitative ATP-biotin reactions in Figures 4 and 5. The ADP-Glo™ assay was used for the kinetics measurements with all kinases, except CK2 and PKA where the NADH-dependent enzyme coupled assay was used. Rate plots and curve fits for all kinases are shown in Figure S7.
KMapp and kcatapp were calculated due to limiting substrate concentrations.
Ratios were calculated by dividing the value with ATP-biotin by the ATP value.
Ratios were calculated by dividing the value with ATP by the ATP-biotin value.
kinetics measurements are not available (NA) with recombinant Abl kinase because the manufacturer discontinued its production.
With AKT and ASK kinases, under conditions suitable for ATP-biotin kinetics determinations, the rate versus [ATP] plots at higher concentrations did not intersect the y axis near zero (Figure S7B and C), indicating that the concentrations were too high to observe the true initial rates. Therefore, the kcat values represent a lower limit.
In the case of Aurora kinases with ATP, the curvatures of the rate versus [ATP] plots were not linear (Figure S7C), allowing for an estimation of an upper limit for the KM value.
Rate data with GSK3® and ATP-biotin were most consistent with product inhibition, which prevented determination of kinetics values (ND = not determined, see Figure S7M).
2.4. HPLC Analysis
After phosphorylated or phosphorylbiotinylated peptides were generated as described above, the reactions were heated at 95 °C for 1 minute to denature kinase activity. Buffer A (50 µL; 99.9% water with 0.1% trifluoroacetic acid) was added to the denatured reaction mixture and incubated for 10–15 minutes to pre-equilibrate. Reverse phase chromatographic separation of peptides was performed using a C18 column (YMC America; 250×4.6 mm, 4µm, 8 nm) and Waters 1525 binary HPLC pump. The elution gradient started at 95% Buffer A in Buffer B (99.9% acetonitrile with 0.1% trifluoroacetic acid), and decreased to 70% Buffer A over 15 min. The flow rate was 1 mL/min and the peptide was detected at 214 nm. Under the acidic conditions of the HPLC elution, the phosphoramidate bond in the biotinylated peptides was cleaved to give a phosphopeptide product in the HPLC. For both ATP and ATP-biotin reactions, the percentage of observed phosphopeptide was calculated by dividing the area under the phosphopeptide peak by the sum of the areas under the phosphopeptide and unmodified peptide peaks. The percentages and standard errors displayed in Figure 5 were obtained from three independent trials (Figure S5). Full HPLC traces are shown in Figure S5.
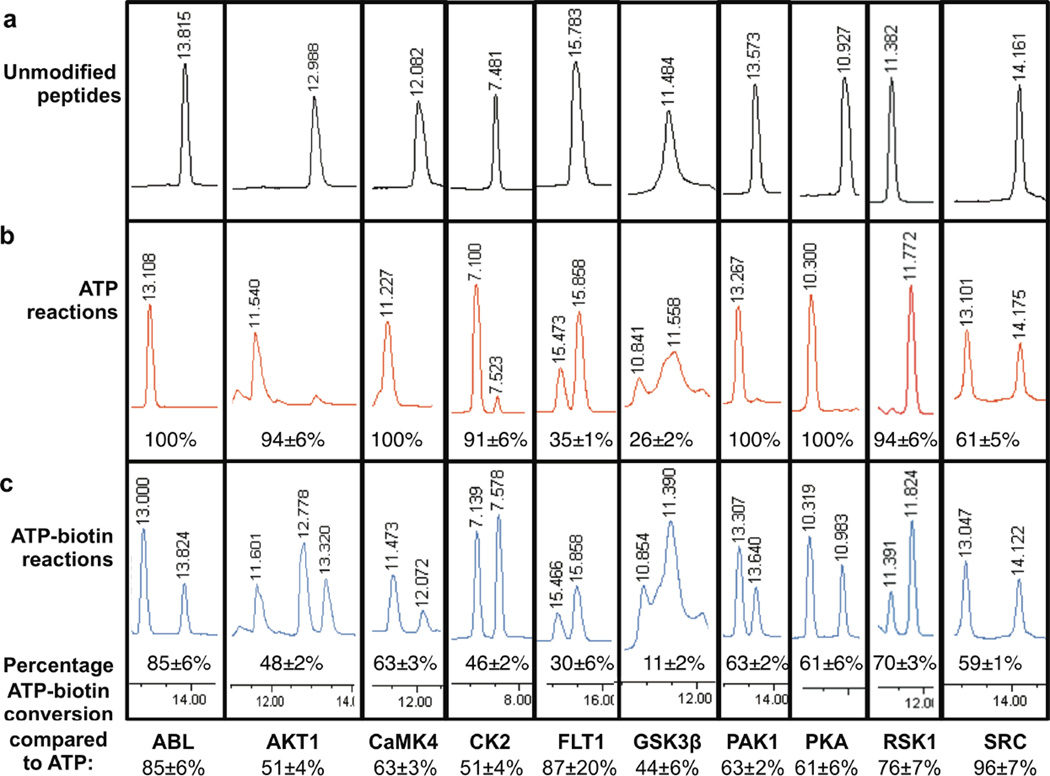
Figure 5. Quantitative Analysis of Peptide Labeling.
HPLC analysis of unmodified peptides (a), ATP phosphorylated peptides (b), and ATP-biotin phosphorylated peptides after acidic cleavage (c). For each ATP or ATP-biotin kinase reaction (b and c), the percentage of phosphopeptide compared to unmodified peptide is shown. The calculated percentage conversions of ATP-biotin relative to ATP ractions are shown below the HPLC traces, along with standard error (Table S2). ABL: peptide at ~13.8 min, phosphopeptide at ~13.1 min; AKT1: peptide at ~13.0 min, phosphopeptide at ~11.6 min, biotin-amine cleavage product at ~12.8min; CaMK4: peptide at ~12.1 min, phosphopeptide at ~11.3 min; CK2: peptide at ~7.5 min, phosphopeptide at ~7.1 min; FLT1: peptide at ~15.8 min, phosphopeptide at ~15.5 min; GSK3β: peptide at ~11.5 min, phosphopeptide at ~10.8 min; PAK1: peptide at ~13.6 min, phosphopeptide at ~13.3 min; PKA: peptide at ~10.9 min, phosphopeptide at ~10.3 min; RSK1 peptide at ~11.4 min, phosphopeptide at ~11.8 min; SRC peptide at ~14.2 min, phosphopeptide at ~13.1 min. The complete HPLC traces of three independent trials are shown in Figure S5.
2.5. Kinetics analysis
Kinetic analysis was performed using either the ADP-Glo™ kinase assay (Promega)25 or an NADH-dependent enzyme coupled assay.26 The NADH-dependent enzyme coupled assay was performed as previously described,11 except with 0.5 mM NADH, 24 units/mL of pyruvate kinase, 36 units/mL of lactic acid dehydrogenase, ATP or ATP-biotin final concentrations of 1, 3, 10, 30, and 100 µM, and absorbance at 360 nm taken every 30 second for 60 min. The concentrations of kinase and substrate used are shown in Table S1. For the ADP-Glo™ assay, white, flat-bottom 96-well half-area micro plates (Corning) were used. For kinase reactions, ATP or ATP-biotin were incubated with the corresponding peptide or protein substrate (Figure 2b, concentration shown in Table S1) in the manufacture’s provided buffer. The reaction was initiated by adding kinase (manufacturer’s recommended amount; see Table S1). The final volumes of the reactions were 20 µL. The reaction mixtures were incubated at 30°C for 5, 10, 20, 30, and 40 minutes for each concentration. After the incubation period, an equal amount of ADP-Glo™ reagent (20 µL) was added to the mixture and incubated for 40 minutes at room temperature. Then, an equal amount of kinase detection reagent (40 µL) was added to the reaction mixture and allowed to react for 60 minutes at room temperature. Finally, luminescence signal was monitored with a micro plate reader (GENios Plus, Tecan). The luminescence signal was background corrected by subtracting the signal of a reaction without substrate, which accounted for any non-enzymatic ATP hydrolysis. The concentration of ADP produced was determined using the background-corrected luminescence signal and a standard curve. The concentration of ADP or NADH produced/consumed in each reaction was plotted over the time and the initial rate of the reaction was obtained from the slope of the linear portion of the plot (Figure S7, right plot). The experimental results (rate vs. substrate concentration) were fit to the Michaelis-Menton equation (ν=Vmax[S]/(KM + [S]), where ν = rate of the reaction and [S] = substrate concentration) by nonlinear regression using Kaleidagraph software (Synergy Software) to obtain KMapp and Vmaxapp values (Figure S7, left plot). Vmax was divided by the concentration of enzyme (Table S1) to obtain the kcatapp value. Because the concentrations of peptide or protein substrate used in each reaction were less than 10-fold above the substrate KM, values in Table 1 are shown as KMapp and kcatapp. Data reported in Table 1 are the mean and standard error of three independent trials. Rate plots and curve fits for each kinase with ATP and ATP-biotin are shown in Figure S7.
2.6. Docking Analysis
Crystal structures of kinase enzymes were downloaded from RCSB Protein Data Bank (AKT1 pdb ID: 4EKK; GSK3b pdb ID: 1PYX; NEK2 pdb ID: 2W5B; SRC pdb ID: 2SRC). Pymol 1.5.0.5 (Schrodinger, LLC) was used to delete the co-crystalized peptide, ligand, and water. AutoDock Tools 1.5.6 was used to add all hydrogen atoms, Gasteiger charges and merging nonpolar hydrogen, followed by pdbqt output file generation.27 The charge of Mg (GSK3b and NEK2) or Mn (AKT1) was changed from zero to +2 manually. A grid box with a spacing of 0.375 Å, corresponding size and coordinates for the center of the grid box were used (Table S3). AutoGrid 4.2 was used to generate the grid maps file required for docking calculation. ATP and ATP-biotin were drawn in ChemBioDraw Ultra and MM2 energy minimization was done by Chem 3D Pro. Gasteiger charge computation, nonpolar hydrogens merge, and choice of torsions were done by AutoDock Tools 1.5.6 and pdbqt files were generated. AutoDock 4.2 was used to run docking calculations using the genetic algorithm and a DPF file was generated.27 The DPF files were set as a rigid macromolecule, and the genetic algorithm search parameters were set to 100 GA runs with a population size of 150, a maximum number of 2.5 × 105 energy evaluations, a maximum number of 2.7 × 104 generations, a mutation rate of 0.2, and a crossover rate of 0.8. Default docking parameters were used and the output DLG file was generated. The docking results and generated images were analyzed by AutoDock Tools 1.5.6.
3. Results and Discussion
To characterize the generality of kinase-catalyzed labeling, a diverse set of kinases was selected from the human kinome. Over 500 human protein kinases are known.23, 28 Kinases are generally classified by their substrate specificity, referred to as either a serine/threonine or tyrosine kinase according to their preferred amino acid substrate. The family is further organized into seven groups based on sequence similarity within the catalytic domain.23 Based on these classifications, a phylogenetic tree of kinases was created (Figure 2a), where sequence deviations between kinases are represented by the distance within the branches.23 Considering the phylogenetic analysis, twenty-five kinases distributed throughout the kinome tree were selected to assess the compatibility of the various family members (see red circles in Figure 2a). Based on the availability of commercial kinases and substrates, at least one kinase from each group was selected (Figure 2b). Given that tyrosine kinases represent 17% of the human kinome (TK family in Figure 2a),23 the majority of the kinases tested were serine/threonine kinases; four out of twenty-five enzymes were tyrosine kinases (16%). We also ensured that protein and peptides substrates were included in the study to possibly reveal substrate biases (Figure 2b).
3.1. Qualitative Analysis of Kinase-Catalyzed Biotinylation
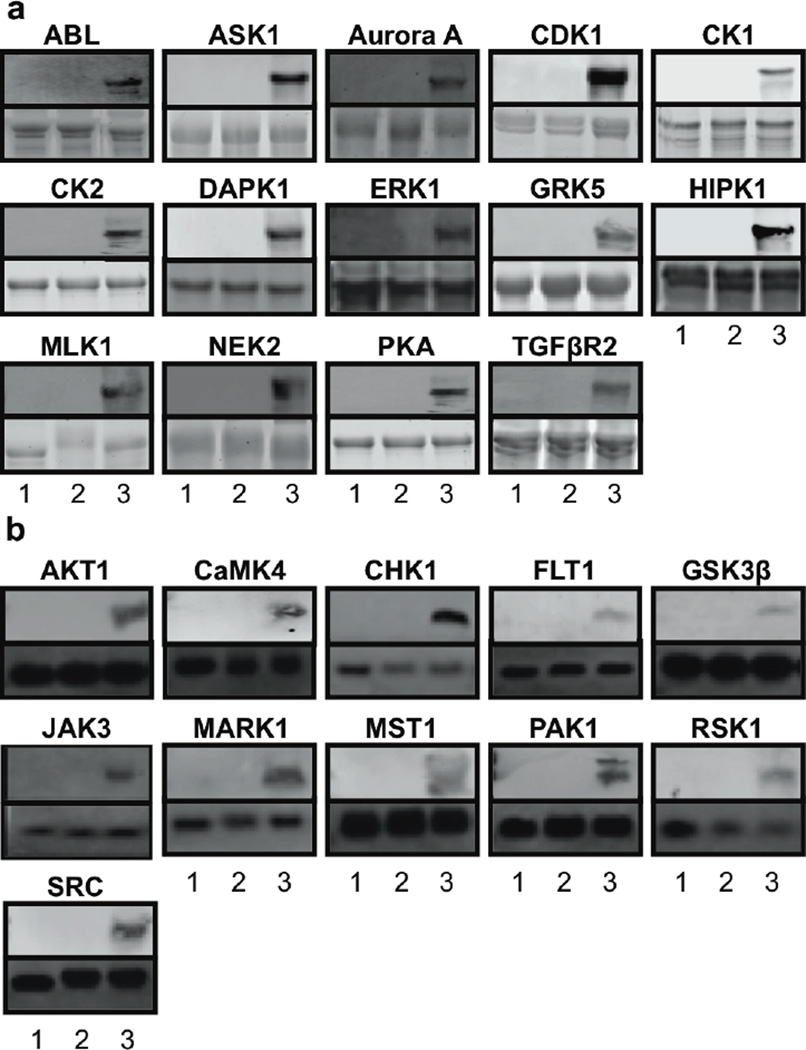
To initially probe the compatibility of ATP-biotin with the twenty-five kinases, substrate biotinylation was monitored using gel analysis. Biotinylated phosphopeptide or phosphoprotein products were generated by incubating ATP-biotin with kinase and corresponding substrate (Figure 2b). As a control, phosphorylated products were generated using ATP. To unambiguously show that biotinylation is kinase-dependent, ATP-biotin was also incubated with each substrate in the absence of kinase. The reaction products were separated by gel electrophoresis and visualized using a streptavidin-fluorophore (SA-Cy5) conjugate (Figure 3, top gels), due to the high affinity of biotin for streptavidin.29 Total peptide or protein was also visualized with coomassie or sypro ruby stain, which indicated equal loading of sample (Figure 3, bottom gels).
Figure 3. Kinase-catalyzed Biotinylation.
Protein (a) or peptide (b) substrates were incubated with ATP-biotin alone (lane 1), ATP and kinase (lane 2), or ATP-biotin and kinase (lane 3), prior to SDS-PAGE separation and visualization using SA-Cy5 (top gels) to observe biotinylation or sypro ruby (protein) or coomassie (peptide) stain (bottom gels) to assure equal sample loading. The kinase tested is indicated above each gel image and the substrate used is indicated in Figure 2b. Gel images are representative of three independent trials (Figure S1 and S2).
Biotinylated protein or peptide products were observed with all twenty-five kinases (Figure 3, lane 3, top gels for each kinase). No biotin labeling was observed in the absence of kinases (Figure 3, lane 1, top gels), indicating that biotinylation is kinase-catalyzed. In addition, no biotinylated products were detected using ATP as the cosubstrate (Figure 3, lane 2, top gel), which confirms dependence on the ATP-biotin probe. Protein (Figure 3a) and peptide (Figure 3b) substrates were biotinylated, indicating that kinase-catalyzed labeling occurs regardless of substrate size. Additional control reactions confirmed that high purity ATP-biotin10 and stringent blocking conditions during biotin visualization were required to assure kinase-dependent labeling (Figure S3).30 The data indicate that twenty-five diverse kinases were compatible with ATP-biotin as a cosubstrate in kinase-dependent biotinylation.
3.2. Quantitifaction of Kinase-Catalyzed Biotinylation
While the biotinylation studies indicated that ATP-biotin is utilized by all twenty-five kinases, we were interested in determining the efficiency of the labeling reactions. To quantify the extent of labeling, biotinylation with ATP-biotin was compared to phosphorylation with ATP. Peptide substrates were analyzed using HPLC, while gel analysis was used with protein substrates. For both methods, the biotin tag was removed from the biotinylated products using acidic conditions, which cleaved the phosphoramidate bond (Figure 1a); cleavage of the biotin tag creates a phosphorylated product identical to the product of ATP phosphorylation, allowing direct quantitative comparison.
Fifteen of the kinases were quantitatively tested for ATP-biotin compatibility using full length proteins or polypeptides along with a gel-based analysis, where levels of phosphorylation were assessed using Pro-Q diamond phosphoprotein stain, as previously reported.24 Initially, phosphorylated proteins or biotinylated phosphoproteins were generated by incubating ATP (Figure 4, lanes 2) or ATP-biotin (Figure 4, lanes 3) with kinase and substrate (Figure 2b). Pro-Q staining may differentially detect phosphorylated versus phosphorylbiotinylated proteins. For quantitative comparison, the biotin tag was removed by incubating with acid to yield identical phosphoprotein products in both reactions. After acid treatment, phosphoproteins were separated by SDS-PAGE and visualized with Pro-Q diamond phosphoprotein stain (Figure 4, top gels) or sypro ruby total protein stain (Figure 4, bottom gels). As a control, substrates alone were analyzed (Figure 4, lanes 1) to determine the levels of phosphorylation prior to reactions. Pro-Q staining indicated that reactions with ATP-biotin produced phosphoprotein products (Figure 4, lanes 3, top gel), which is consistent with earlier biotinylation studies (Figure 3).
To obtain a percentage conversion for kinase-catalyzed biotinylation with each kinase, the band intensity of the phosphoprotein product with ATP-biotin (Figure 4, lane 3) was compared to the product with ATP (Figure 4, lane 2). ATP phosphorylation was assumed to proceed to 100% conversion, with biotinylation efficiency calculated relative to the ATP reaction (Table 1 and S2). All band intensities were also background corrected to account for any low level of initial phosphorylation of the substrate (Figure 4, lanes 1). Reactions with four kinases (AurA, JAK3, NEK2, and TGFβR2) showed over 90% conversion with ATP-biotin. An additional six kinases demonstrated conversions between 63% and 82% (CDK1, CK1, DAPK1, ERK1, MARK1, and MLK1). Only five kinases displayed conversion efficiencies between 45% and 55% (ASK1, CHK1, GRK5, HIPK1, and MST1). While the kinases maintained a range of conversion percentages (44% to 96%), the quantitative analysis indicated that biotinylation occurs with efficiencies consistent with the earlier biotinylation studies (Figure 3).
Reaction quantification with ten of the kinases utilized peptide substrates (Figure 2b). In this case, after phosphorylation with either ATP or ATP-biotin, phosphorylated or phosphorylbiotinylated peptides were analyzed by HPLC (Figure 5). Because the HPLC buffer was acidic (~pH=2), the phosphoramidate bonds of the biotinylated phosphopeptides were cleaved to yield phosphopeptides identical to the product of ATP phosphorylation (Figure 1a) along with the biotin-amine byproduct (Scheme S1), which was observed at ~12.8min (see AKT1 in Figure 5c or Figure S5). Phosphoramidate bond cleavage was facilitated by heat denaturation of the kinase (95°C, 1 min) and equilibration with the acidic HPLC buffer prior to injection. HPLC analysis indicated that peptide substrates were phosphorylated with ATP (Figure 5b) with conversions ranging from 26% to 100%. While seven of the kinases demonstrated high 91 to 100% conversions (Abl, Akt1, CaMK4, CK2, PAK1, PKA, and RSK1), the remaining three kinases showed more modest conversions of 35% (FLT1), 26% (GSK3β), and 61% (SRC) with ATP. As expected, ATP-biotin reactions displayed reduced conversion percentages compared to ATP (Figure 5c compared to 5b). Overall conversion percentages with ATP-biotin were calculated by comparing to the percent conversions with ATP (bottom of Figure 5, Table 1, and Table S2). Similar to the protein substrate experiments, the percent conversion with ATP-biotin compared to ATP with peptide substrates ranged from 44% to 96%. Three kinases showed >85% conversion (ABL, FLT1, and SRC), four maintained conversions of 61–76% (CaMK4, PAK1, PKA, and RSK1), and three demonstrated roughly 50% conversions relative to ATP (AKT1, CK2, GSK3β). The data generated here with HPLC are consistent with prior analysis using an MS-based method, where PKA, CK2, and ABL showed 79%, 56%, and 80%, respectively6. In total, gel and HPLC-based quantitative data show that biotinylation occurs with reasonable conversion, which is consistent with the earlier biotinylation studies (Figure 3).6
3.3. Kinetic Analysis of Kinase-Catalyzed Biotinylation
Kinetics analyses were also performed to determine the efficiency of biotinylation with ATP-biotin. With most kinases, the ADP-Glo™ assay was used, where the ADP byproduct of phosphoryl transfer with either ATP or ATP-biotin was monitored using a series of enzymatic steps, resulting in a luminescence signal.25 To assure that ATP-biotin is compatible with the assay, a series of control experiments either with or without an initial kinase reaction were performed (Figure S6). ATP-biotin performed similarly to ATP and did not interfere with the enzyme-catalyzed steps of the reaction. Two kinases (CK2, and PKA) were tested using an enzyme coupled assay, as published previously.11 Using these two assays, kinetics measurements were taken with the kinases to determine apparent KM, kcat and kcat/KM values.
ATP-biotin displayed similar KMapp values compared to ATP for all kinases with only modest 0.6-fold to 2.9-fold changes in value (Table 1). A higher KMapp for ATP-biotin was observed with twenty-one kinases, although most with differences less than 1.5-fold compared to ATP (Table 1). Only five enzymes demonstrated greater than 2-fold elevated KMapp compared to ATP (CK1, DAPK, HIPK, PKA and TGFβR2). Two kinases displayed a modestly lower KMapp compared to ATP (NEK2, and PAK, 0.8- to 0.9-fold change). The analysis suggests that the γ-phosphate modification of ATP-biotin does not significantly affect cosubstrate KM values.
The kcatapp values from the kinetics analysis were also compared to assess the efficiency of phosphoramidyl transfer with ATP-biotin compared to phosphoryl transfer with ATP. Like with the KMapp values, many kinases showed similar kcatapp values with ATP-biotin compared to ATP (Table 1). Fourteen kinases displayed less than a 2-fold reduced kcatapp values compared to ATP, with CK1, DAPK1 and JAK3 showing roughly equivalent rates. Another eight kinases maintained kcatapp values between 2.0 and 4.9-fold reduced compared to ATP (AKT1, CaMK4, CK2, ERK1, FLT1, GRK5, MLK1, and PKA). Only one kinase showed significantly compromised kcatapp values; ASK1 displayed ≥20-fold reduced kcatapp values compared to ATP. While most kinases showed reduced catalytic efficiency with ATP-biotin compared to ATP, the reductions were generally modest. The data suggest that the unnatural phosphoramidyl transfer with ATP-biotin is slower than natural phosphoryl transfer with ATP. However, the majority of kinases showed only a modest <2-fold reduced catalytic rate.
The overall catalytic efficiencies (kcat/KMapp) for biotinylation and phosphorylation reactions were compared. As expected based on the analysis of KMapp and kcatapp values, the majority of kinases maintained kcat /KMapp values only modestly reduced compared to ATP; five kinases displayed less than 2-fold change efficiency, while another fifteen maintained between 2 and 4.5-fold changes. Consistent with the kcatapp analysis, ASK1 displayed the most unfavorable efficiencies with ≥16-fold change. In total, the kinetic analyses showed that the majority of kinases demonstrate reasonable kinetics with ATP-biotin, with the exception of ASK1.
The γ-phosphate modified ATP analog adenosine 5’-(γ-thio)triphosphate (ATP-γS) is widely used for protein labeling17, 18, 31–33 and also displays 8- to 30-fold reduced kcat/KM compared to ATP,11 which was primarily due to a lower kcat.34, 35 Another γ-phosphate modified ATP analog, ATP-dansyl, showed a 7- to 18-fold reduction in kcat/KM compared to ATP, which was also due to a significantly lowered kcat value.11 Similarly, the data here with twenty-five kinases and ATP-biotin showed that the reduced catalytic efficiencies were predominantly due to an effect on kcat. The combined data suggest that phosphoramidyl or phosphothiolyl transfer with γ-phosphate modified ATP analogs is less efficient than phosphoryl transfer with ATP. However, the fact that the kinases tested here demonstrated kcat/KM values with ATP-biotin that are similar to ATP-γS establishes kinase-catalyzed biotinylation as a tool for phosphoprotein labeling with diverse kinases.
A comparison of kinetic measurements to quantitative conversion data (Table 1 and S2) indicates that there is no direct correlation between kinetic and equilibrium efficiencies. For example, NEK1 and PAK1 demonstrated essentially identical kinetic efficiencies with ATP-biotin and ATP (1.1 and 1.2-fold reduction in kcat/KMapp), but quite different biotinylation under equilibrium conditions (94% and 63%, Table 1). On the other end of the spectrum, ASK1, which demonstrated the poorest catalytic efficiency with ATP-biotin relative to ATP (>16-fold reduction in kcat/KMapp), showed reasonable biotinylation under equilibrium condition (53%, Table 1). Taken together, there is a range of tolerance to ATP-biotin by the kinases under kinetic and equilibrium conditions. While the kinetic measurements allowed a more thorough assessment of kinase compatibility, it is noted that future phosphoprotein labeling studies utilizing kinase-catalyzed biotinylation will be performed under equilibrium labeling conditions. Therefore, the quantitative conversion values are likely to be more relevant for phosphoprotein analysis applications. Indeed, all kinases tested were compatible with ATP-biotin as a cosubstrate, with reasonable conversions and biotin labeling (Figure 3).
3.4. Docking Analysis
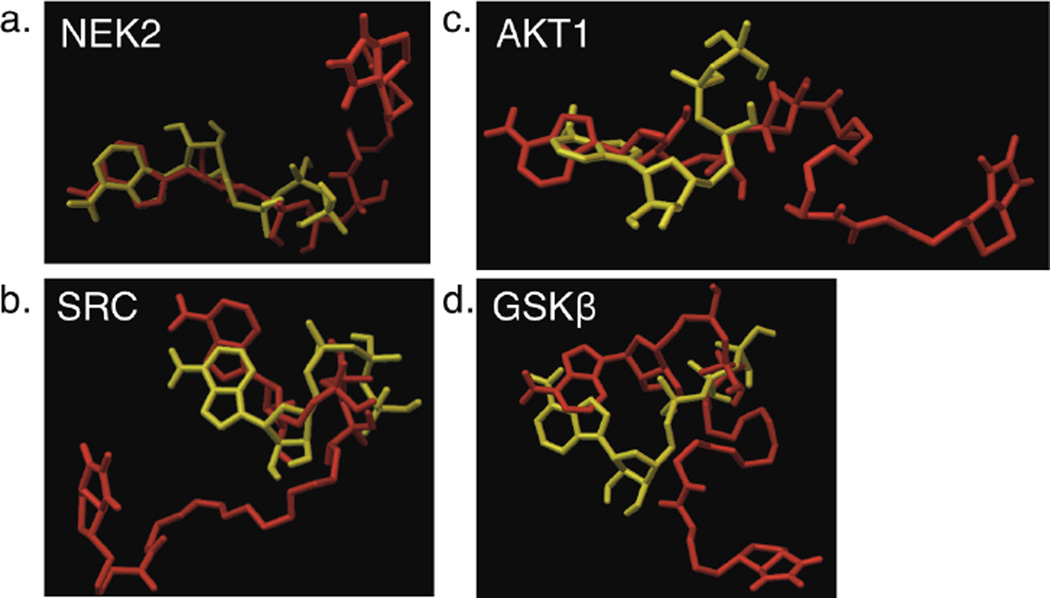
To rationalize the efficiency of the various kinases with ATP-biotin, a docking analysis was performed. We selected two kinases displaying excellent kinetics (1.2–1.6-fold reduced kcat/KMapp values compared to ATP) and equilibrium conversions (94–96%), NEK2 and SRC. We also selected two kinases demonstrating reduced kinetics (≥7-fold reduced kcat/KMapp values compared to ATP) and equilibrium efficiency (44–51%), AKT1 and GSK3β. Using the Autodock program,27 all four kinases produced ATP-biotin binding poses with reasonable energies, consistent with the fact that all four accepted ATP-biotin experimentally. However, differences in the binding were observed compared to ATP. The high efficiency kinases NEK2 and SRC bound ATP-biotin and ATP similarly, with significant overlap of the adenosines and triphosphates of the two cosubstrates in the active sites (Figure 6a and b). Indeed, similar distances between the triphosphate group and active site amino acids were observed between ATP and ATP-biotin (Figure S8a–d). On the other hand, overlap between the adenosines and triphosphates of ATP and ATP-biotin was reduced when bound with the lower efficiency kinases AKT1 and GSK3β, with twisting of the adenosine ring (Figure 6c and d) and lengthening of some bonds compared to ATP (Figure S8e–h). In total, the docking is consistent with the tolerance of all kinases to ATP-biotin. The variable binding of ATP-biotin compared to ATP in the kinase active sites roughly correlates with the different efficiencies of the selected kinases. With over 500 kinases known and many crystal structures available, the docking analysis can be a valuable tool to predict the efficiency of additional kinases to ATP-biotin labeling reactions.
Figure 6. Docking studies with ATP-biotin and select kinases.
Images of ATP (yellow) and ATP-biotin (red) bound to two highly active, NEK2 (a, PDB = 2W5B) and Src (b, PDB = 2SRC), and two less active, AKT1 (c, PDB = 4EKK) and GSK3β (d, PDB = 1PYX), enzymes in kinase-catalyzed biotinylation reactions.
In summary, the data indicate that a diverse range of kinases throughout the kinome accept ATP-biotin as a cosubstrate and allow biotinylation of protein or peptide substrates. Phosphoprotein labeling and analysis has historically involved radioactive labeling using [γ-32P]ATP,36 with subsequent analysis by gel or HPLC methods.5 However, [γ-32P]ATP provides only a radioactive visualization tag without the ability to purify labeled phosphoproteins. As an alternative, 5’-(γ-thio)triphosphate (ATPγS) was introduced where the thiophosphoryl group of the labeled protein product is incubated in a second step with an electrophilic group to promote purification and visualization31, 33. ATP-biotin maintains the purification advantage of ATPγS but avoids a second step reaction after labeling. The purification and visualization properties of biotinylated phosphoproteins make kinase-catalyzed labeling an ideal method for direct phosphoprotein analysis.
Taking a wider perspective, a variety of ATP analogs have been developed as kinase research tools, many of which utilize a phosphoramidyl group transfer in kinase-catalyzed labeling reactions.12–16 The data here suggest that this wider family of ATP analogs is compatible with diverse kinases. Therefore, beyond ATP-biotin, the data suggest that γ-modified ATP analogs are appropriate for a variety of kinase-catalyzed labeling reactions. These studies provide the foundation for application of kinase-catalyzed labeling and γ-modified ATP analogs to phosphoproteomics and cell signaling research.
Supplementary Material
Acknowledgments
We thank the National Institutes of Health (GM079529), National Science Foundation (1306493), and Wayne State University for funding, Promega for gifts of ADP-Glo™ reagent and select kinases, Tamara Hendrickson for technical advice, and P. Dedigama and C. Nanah for comments on the manuscript.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Additional experimental details, independent trials for the biotinylation and quantification experiments, curve fits for kinetic studies, control experiments with the ADP-Glo™ assay and biotin visualization, and docking studies are provided as supplementary information.
References and notes
- 1.Hunter T. Cell. 2000;100:113–127. doi: 10.1016/s0092-8674(00)81688-8. [DOI] [PubMed] [Google Scholar]
- 2.Adams JA. Chem. Rev. 2001;101:2271–2290. doi: 10.1021/cr000230w. [DOI] [PubMed] [Google Scholar]
- 3.Lahiry P, Torkamani A, Schork NJ, Hegele RA. Nat Rev Genet. 2010;11:60–74. doi: 10.1038/nrg2707. [DOI] [PubMed] [Google Scholar]
- 4.Cohen P, Alessi DR. ACS chemical biology. 2013;8:96–104. doi: 10.1021/cb300610s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hastie CJ, McLauchlan HJ, Cohen P. Nat. Protocols. 2006;1:968–971. doi: 10.1038/nprot.2006.149. [DOI] [PubMed] [Google Scholar]
- 6.Green KD, Pflum MH. J. Am. Chem. Soc. 2007;129:10–11. doi: 10.1021/ja066828o. [DOI] [PubMed] [Google Scholar]
- 7.Wang Z, Lee J, Cossins AR, Brust M. Anal. Chem. 2005;77:5770–5774. doi: 10.1021/ac050679v. [DOI] [PubMed] [Google Scholar]
- 8.Wang Z, Levy R, Fernig DG, Brust M. J. Am. Chem. Soc. 2006;128:2214–2215. doi: 10.1021/ja058135y. [DOI] [PubMed] [Google Scholar]
- 9.Gao X, Schutz-Geschwender A, Hardwidge PR. Biotechnology letters. 2009;31:113–117. doi: 10.1007/s10529-008-9824-0. [DOI] [PubMed] [Google Scholar]
- 10.Senevirathne C, Green KD, Pflum MKH. Kinase-Catalyzed Biotinylation of Peptides, Proteins, and Lysates, Current Protocols in Chemical Biology. John Wiley & Sons, Inc; 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Green KD, Pflum MH. ChemBioChem. 2009;10:234–237. doi: 10.1002/cbic.200800393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Song H, Kerman K, Kraatz HB. Chem Commun (Camb) 2008:502–504. doi: 10.1039/b714383d. [DOI] [PubMed] [Google Scholar]
- 13.Martić S, Gabriel M, Turowec JP, Litchfield DW, Kraatz H-B. J. Am. Chem. Soc. 2012;134:17036–17045. doi: 10.1021/ja302586q. [DOI] [PubMed] [Google Scholar]
- 14.Wilke KE, Francis S, Carlson EE. J Am Chem Soc. 2012;134:9150–9153. doi: 10.1021/ja3041702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Suwal S, Pflum MH. Angew Chem Int Ed Engl. 2010;49:1627–1630. doi: 10.1002/anie.200905244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee SE, Elphick LM, Anderson AA, Bonnac L, Child ES, Mann DJ, Gouverneur V. Bioorg Med Chem Lett. 2009;19:3804–3807. doi: 10.1016/j.bmcl.2009.04.028. [DOI] [PubMed] [Google Scholar]
- 17.Allen JJ, Lazerwith SE, Shokat KM. J. Am. Chem. Soc. 2005;127:5288–5289. doi: 10.1021/ja050727t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Allen JJ, Li M, Brinkworth CS, Paulson JL, Wang D, Hubner A, Chou W-H, Davis RJ, Burlingame AL, Messing RO, Katayama CD, Hedrick SM, Shokat KM. Nat. Methods. 2007;4:511–516. doi: 10.1038/nmeth1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Suwal S, Senevirathne C, Garre S, Pflum MK. Bioconjug Chem. 2012;23:2386–2391. doi: 10.1021/bc300404s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mann M, Ong S-E, Gronborg M, Steen H, Jensen ON, Pandey A. Trends in Biotechnology. 2002;20:261–268. doi: 10.1016/s0167-7799(02)01944-3. [DOI] [PubMed] [Google Scholar]
- 21.Fouda AE, Pflum MK. Angew Chem Int Ed Engl. 2015;54:9618–9621. doi: 10.1002/anie.201503041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dunn JD, Reid GE, Bruening ML. Mass Spectrometry Reviews. 2010;29:29–54. doi: 10.1002/mas.20219. [DOI] [PubMed] [Google Scholar]
- 23.Manning G, Whyte DB, Martinez R, Hunter T, Sudarsanam S. Science. 2002;298:1912–1934. doi: 10.1126/science.1075762. [DOI] [PubMed] [Google Scholar]
- 24.Senevirathne C, Pflum MK. ChemBioChem. 2013;14:381–387. doi: 10.1002/cbic.201200626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zegzouti H, Zdanovskaia M, Hsiao K, Goueli SA. Assay and drug development technologies. 2009;7:560–572. doi: 10.1089/adt.2009.0222. [DOI] [PubMed] [Google Scholar]
- 26.Cook PF, Neville ME, Vrana KE, Hartl FT, Roskoski R. Biochemistry. 1982;21:5794–5799. doi: 10.1021/bi00266a011. [DOI] [PubMed] [Google Scholar]
- 27.Morris GM, Huey R, Lindstrom W, Sanner MF, Belew RK, Goodsell DS, Olson AJ. J Comput Chem. 2009;30:2785–2791. doi: 10.1002/jcc.21256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hanks SK, Hunter T. FASEB journal : official publication of the Federation of American Societies for Experimental Biology. 1995;9:576–596. [PubMed] [Google Scholar]
- 29.Green NM. Biochem. J. 1963;89:585–591. doi: 10.1042/bj0890585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Arora DP, Boon EM. Biochem Biophys Res Commun. 2013;432:287–290. doi: 10.1016/j.bbrc.2013.01.115. [DOI] [PubMed] [Google Scholar]
- 31.Kwon SW, Kim SC, Jaunbergs J, Falck JR, Zhao Y. Mol Cell Proteomics. 2003;2:242–247. doi: 10.1074/mcp.M300039-MCP200. [DOI] [PubMed] [Google Scholar]
- 32.Blethrow JD, Glavy JS, Morgan DO, Shokat KM. Proc Natl Acad Sci U S A. 2008;105:1442–1447. doi: 10.1073/pnas.0708966105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sun IY, Johnson EM, Allfrey VG. J Biol. Chem. 1980;255:742–747. [PubMed] [Google Scholar]
- 34.Grace MR, Walsh CT, Cole PA. Biochemistry. 1997;36:1874–1881. doi: 10.1021/bi962138t. [DOI] [PubMed] [Google Scholar]
- 35.Sondhi D, Xu W, Songyang Z, Eck MJ, Cole PA. Biochemistry. 1998;37:165–172. doi: 10.1021/bi9722960. [DOI] [PubMed] [Google Scholar]
- 36.Krebs EG, Kent AB, Fischer EH. The Journal of biological chemistry. 1958;231:73–83. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.