Abstract
Background
Polymerase chain reaction (PCR) is useful for rapid microbial detection in body fluids with low microbial load. It is easier to use universal or broad range primers for the amplification of conserved stretches of DNA common to all bacteria like 16S rRNA gene, followed by restriction fragment length polymorphism (RFLP) of PCR products.
Methods
Forty samples of cerebrospinal fluid were collected. After DNA extraction, universal or broad range PCR was performed using two universal primers U1-5'-CCAGCAGCCGCGGTAATACG-3', corresponding to nucleotides 518 to 537 of the Escherichia coli 16S rRNA gene, and U2 − 5'-ATCGG(C/T)TACCTTGTTACGACTTC-3', corresponding to nucleotides 1513 to 1491 of the same gene. The PCR product was subjected to digestion by endonucleases- HaeIII, Mn11, BstB1 and Alu1. Restriction pattern obtained was compared with that of standard organisms to identify the pathogen. The results were compared with conventional methods.
Result
Universal PCR could detect pathogens in 20% samples within 13-18 hours as compared to 16% by conventional methods. The analytical sensitivity was 10 Gram negative and 250 Gram positive organisms per 200 μl sample. Overall sensitivity was 83.3% and specificity was 91.2%.
Conclusion
Universal PCR followed by RFLP of PCR product is a good alternative to conventional diagnosis of bacterial pathogens.
Key Words: Body fluids, Polymerase chain reaction, Restriction fragment length polymorphism
Introduction
Conventional methods of bacterial pathogen detection are time consuming and an optimal etiological concentration of 105 cfu/ml is offen required to achieve a reliable positive result. An extensive study over a period of 27 years revealed that culture might miss the diagnosis of bacterial meningitis in at least 13% of cases [1,2]. Regardless of the type of organism in the CSF; the percentage of positive microscopic results is only 25% with < 103 cfu/ml and 60% in the range of 103 to 105 cfu/ml [3]. Nucleic acid amplification technology using polymerase chain reaction (PCR) has opened new possibilities for rapid microbial detection and characterization. Many primer sets have been developed to detect species- specific sequences in simple PCRs [4, 5, 6]. However the use of species-specific primers is impractical for routine analysis of clinical samples that may contain several different pathogens. Therefore it is easier to use universal or broad range primers for the amplification of conserved stretches of DNA followed by sequence analysis of the PCR products [7], or by restriction fragment length polymorphism (RFLP) [8]. Since the 16S rRNA genes are common and conserved in all bacteria, it is possible to design PCR primers capable of amplifying this DNA [8]. The universal or broad range PCR followed by RFLP is a new and effective method of detection of bacterial pathogens in normally sterile body fluids. Most of the “culture-resistant”, fastidious, inactivated or slow-growing bacteria can be identified within 18 hours with the use of one restriction enzyme and the rest with the use of another three enzymes.
Material and Methods
Forty CSF samples were collected during the study period from patients suspected of having meningitis. All samples were subjected to the conventional methods of pathogen identification. An aliquot (200 µl) was processed for DNA extraction with QIAamp DNA Mini Kit (QIAGEN Inc., Chatsworth, Calif) as per the protocol provided by the manufacturer. The eluted DNA was stored at -20°C.
One pair of primers, designated U 1 and U 2, with sequences conserved among all bacteria under the study was selected. The sequence of primer U1 was 5'-CCAGCAGCCGCGGTAATACG-3', corresponding to nucleotides 518 to 537 of the E. coli 16S rRNA gene, and that of U2 was 5'-ATCGG(C/T)TACCTTGTT ACGACTTC-3', corresponding to nucleotides 1513 to 1491 of the same gene. PCR performed with these two primers is referred to as the universal PCR or broad range PCR in this study.
A reaction mixture containing approximately 5μl of template DNA, 5ul of 10X PCR buffer (10 mM Tris-HCl, pH 8.3; 50 mM KC1; 2.5 mM MgCl2; 0.001% gelatin), a 0.2 uM concentration of each PCR primer, a 0.2 mM concentration of each deoxynucleoside triphosphate and 2.5 U of Taq DNA polymerase in a total volume of 50 ul was prepared. After a 10 minute denaturation at 94°C, the reaction mixture was run through 35 cycles of denaturation for one minute at 94°C, annealing for one minute at 55°C and extension for two minutes at 72°C, followed by a final extension for 10 minutes at 72°C. Ten microlitres of PCR product was electrophoresed on a 1% agarose gel with ethidium bromide and bands were visualized by ultraviolet (UV) transillumination, to determine the size of the product. Both positive and negative controls were run with each PCR run.
Ten microlitres of the purified PCR product was digested with 10 U of HaeIII in presence of 2µl of appropriate buffer in a total volume of 20µl. Digestion was carried out at 37°C for three hours in a shaker water bath. The reaction was stopped by heating the reaction mixture at 65°C for 20 minutes. Restriction pattern was studied with electrophoresis using a 5% non-denaturing polyacrylamide gel (using a 30% acrylamide, 0.8% bis acrylamide stock) in TBE buffer. A 50ml mixture was prepared per gel and polymerization was initiated with addition of ammonium persulphate and TEMED. The gel was run at constant speed of 5-8volts/cm of the gel for 90 minutes. Gels were than stained in 0.5X TBE containing 0.7 % (W/V) ethidium bromide for 30 minutes prior to visualization in an UV transilluminator.
Result
Total of 40 CSF samples were collected. Out of these only six were positive by conventional culture while eight were positive by PCR. All parameters for meningitis were correlated with the culture and PCR results (Table 1).
Table 1.
Laboratory parameters of patients with bacterial meningitis
| Sex | Age | Bacterium identified by culture/PCR | Sugar (mg/dl) (10-30) | Protein (mg/dl) (12-60) | WBC (cells/μl) (<5) | CSF culture | CSF PCR |
|---|---|---|---|---|---|---|---|
| M | 5 | Escherichia coli | 10 | 210 | 1,000 | + | + |
| M | 13 | Pseudomonas aeruginosa | 15 | 295 | 2,400 | + | + |
| F | 32 | Staph aureus | 34 | 90 | 220 | + | + |
| M | 38 | Mycobacterium tuberculosis | 76 | 24 | 260 | − | + |
| F | 26 | Staphylococcus epidermidis | 60 | 42 | 4 | + | − |
| M | 70 | Streptococcus pneumoniae | 10 | 760 | 2,600 | − | + |
| M | 25 | Neisseria meningitidis | 15 | 180 | 270 | − | + |
| F | 06 | Haemophilius influenzae | 27 | 150 | 560 | + | + |
| M | 45 | Streptococcus pneumoniae | 24 | 185 | 1,220 | + | + |
When the PCR products were digested with Haelll, all PCR products, except for those of Staphylococcus aureus and Staphylococcus epidermidis, were digested into several fragments. Streptococcus pyogenes produced four bands at 460 bp, 330 bp, 135 bp, and 95 bp. Streptococcus pneumoniae and Enterococcus faecium produced identical patterns of bands at 460bp and 140 bp which could be further differentiated using Alu 1 digestion. Escherichia coli and Klebsiella pneumoniae produced similar six bands at 220bp, 210bp, 190bp, 160bp, I30bp, and 65bp (Fig.1). To differentiate them digestion by BstBl was used whereby Escherichia coli product was not digested by BstBl, while Klebsiella pneumoniae was digested into two fragments (876 and 120 bp) (Fig. 3). Pseudomonas aeruginosa produced a different pattern with bands at 390bp, 220bp, 175bp, 135bp, and 40bp (Fig.1). Mycobacterium tuberculosis also produced a different pattern with bands at 390bp, 195bp, 150bp, 105bp, and 65bp (Fig. 1). Haemophilus influenzae gave a unique pattern, with bands at 380bp, 270bp, 175bp and 135bp while Neisseria menigitidis gave at 710bp and 285bp (Fig 1).
Fig. 1.
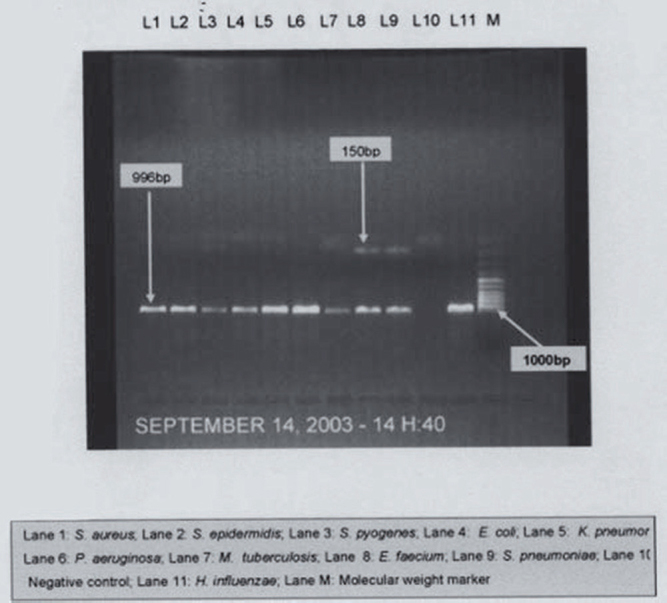
PCR of control strains with universal primers
Fig. 3.
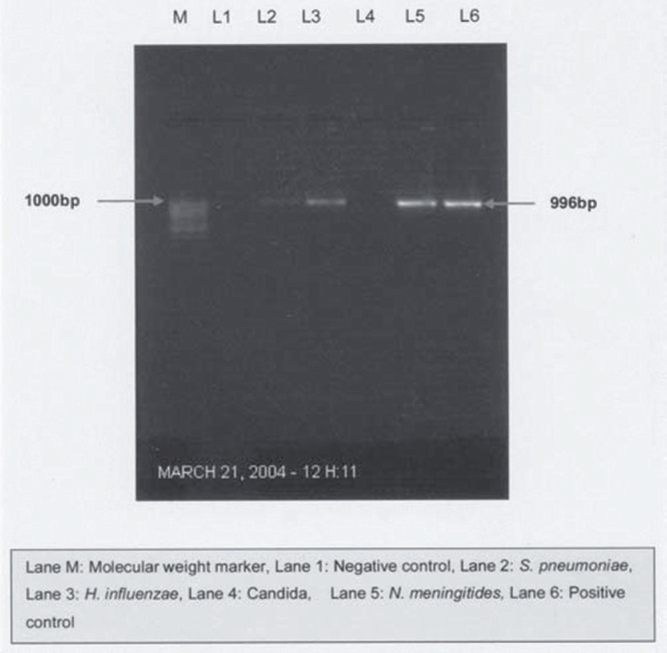
PCR with universal primers
The PCR products from Staphylococcus aureus and Staphylococcus epidermidis gave rise to different patterns when they were digested with MnlI. Staphylococcus aureus gave bands at 600bp, 485bp, 340bp, 250bp, 140bp, 110bp, 95bp, 25bp and Staphylococcus epidermidis produced bands at 485bp, 250bp, 110bp, 95bp, and 25bp (Fig. 2).
Fig. 2.
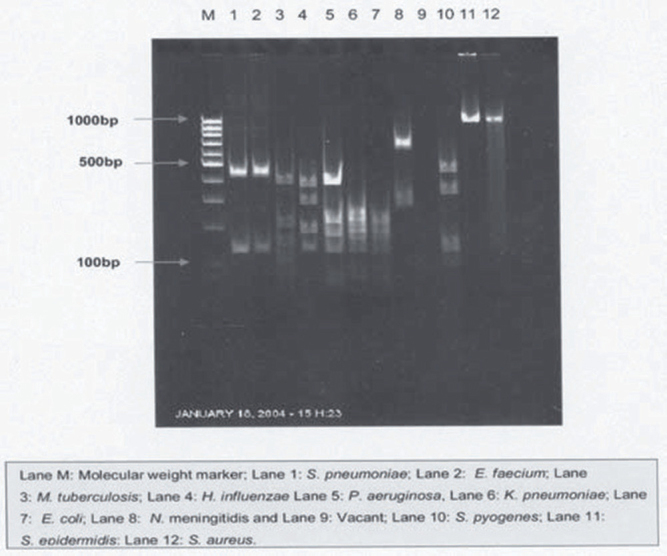
HaeIII digestion patterns of universal PCR products
Out of 40 CSF samples, the study yielded nine positive results for bacterial presence by culture/PCR. Out of these six were positive by conventional culture and eight were positive by PCR, the details of which are given in Table 1.
Discussion
Body fluids such as CSF, present a diagnostic challenge due to small sample volume and paucity of slow growing or uncultivable organisms that results in lower sensitivity. This study was carried out with the objectives to establish an alternative method of identifying bacterial pathogens in body fluids by means of broad range PCR followed by RFLP, and to compare it with conventional method of culture and identification.
Unlike culture, most molecular assays are designed specifically for one organism only. Broad-range assays, based on 16s rRNA genes, are designed to overcome this limitation. By using PCR primers that are targeted at conserved regions of 16S rRNA genes, it is possible to design broad-range PCRs capable of detecting DNA of almost any bacterial species. The identification of the bacterium is done by nucleotide sequencing of the PCR product [9] or by restriction endonuclease digestion [8].
Broad range PCR products from different bacteria had different restriction patterns, while PCR products from different isolates of the same bacteria had similar restriction pattern. These results formed the basis of identification of bacteria in this study.
Lu et al [8], have designed universal primers Ul and U2 for amplification of the bacteria found in CSF, which are then identified by restriction enzyme digestion on the amplified PCR product. Single set of universal primers for PCR followed by sequencing for identification of bacteria has been used by other workers also [10, 11, 12, 13, 14]. In this study, we detected bacteria with only one set of PCR primers and used restriction enzyme analysis, instead of species-specific probes or sequencing, to identify bacteria.
Out of 40 CSF samples, six (15%) were positive by culture and eight (20%) were positive by PCR. One specimen that was negative by PCR, had grown Staphylococcus epidermidis in culture, but the microscopy and biochemical tests were not in favour of bacterial infection (Table 1). Lu et al [8], had a culture positivity of 8.6% (13/150) and PCR positivity of 10%.
Biochemical and cytological studies were carried out on the CSF specimens to corroborate the findings of culture and PCR. Out of the nine positive samples, there were two isolates of Streptococcus pneumoniae and one isolate each of Escherichia coli. Staphylococcus aureus, Staphylococcus epidermidis, Pseudomonas aeruginosa, Neisseria meningitides, Mycobacterium tuberculosis and Haemophilus influenzae. The CSF glucose level varied from 10-76 mg/dl, protein levels from 24 to 760 mg/dl and WBC levels from 4-2,600 cells/μl. The biochemical and cytological parameters of the specimen that grew Staphylococcus epidermidis were within normal limits. Hence this isolate was considered to be a culture contamination. Three CSF specimens were negative by culture and positive by PCR.
History revealed that two of these patients were treated with antibiotics previously and one was tubercular in origin, which correlates well with other studies that show CSF sterilization following antibiotic use [11]. Other studies have also shown the efficacy of broad range PCR in cases where cultures are rendered sterile by antibiotic administration [10, 12, 13].
The universal PCR had a lower detection limit of 10 Gram-negative bacteria (e.g., Escherichia coli) and 250 Gram-positive bacteria (e.g., Staphylococcus aureus) as shown by Lu et al. [8]. Carroll et al [15], reported a sensitivity of the nested PCR between 10 fg and 1 pg of bacterial DNA, depending on the species tested, which was equivalent to between 24 and 4 live bacteria spiked in 100 µl water [15]. When mechanical disruption was used as the method of cell lysis, the sensitivity of the PCR was estimated to be 10-100 cfu per reaction for both Escherichia coli and Staphylococcus aureus [9]. Kiausegger et al [16], reported a sensitivity of 10 cfu of E coli per PCR for the Gram-negative-specific PCR and 10 cfu for Gram-positive-specific PCR using Staphylococcus aureus. Schuurman et al [17], had a lower detection limit of 100 and 200 for Gram positive and Gram negative organisms respectively. Our protocol had sensitivity below 10 cfu/ml, which is considered as the infection threshold in 85% cases of meningitis [3].
Our sample size was small to arrive at a significant statistical correlation. However comparing the study with culture as the gold standard; five out of six bacterial culture CSF specimens were positive by the universal PCR in our study. The sensitivity of PCR in our study was 83.3% and specificity 91.2%. The positive predictive value is 62.5%, and the negative is 96.8%. The lower positive predictive value is partly due to inefficiency of culture. Lu et al [8], reported a sensitivity of 92.3%. Richardson et al [18], have described a PCR assay for the diagnosis of meningococcal meningitis, which had a sensitivity of 97% as compared to a CSF culture sensitivity of 55%. Schuurman et al [17], found broad range PCR was more useful in meningitis with sensitivity of 86%, specificity of 97%, positive predictive value of 80% and negative predictive value of 98% as compared to conventional culture. Xu et al [19], recommended use of highly resolving polyacrylamide matrix for separation of amplicons and digestion fragments.
Thus, universal PCR followed by RFLP is an alternative, rapid, simple and effective method for detection of bacterial pathogens in CSF.
Conflicts of Interest
None identified
References
- 1.Durand ML, Calderwood SB, Weber DJ. Acute bacterial meningitis in adults, a review of 493 episodes. N Engl J Med. 1993;328:21–28. doi: 10.1056/NEJM199301073280104. [DOI] [PubMed] [Google Scholar]
- 2.Tunkel AR, Scheld WM. Acute bacterial meningitis. Lancet. 1995;346:1675–1680. doi: 10.1016/s0140-6736(95)92844-8. [DOI] [PubMed] [Google Scholar]
- 3.La scolea L, Jr, Dryja D. Quantification of bacteria in CSF and blood of children with meningitis and its diagnostic significance. J Clin Microbiol. 1984;19:187–190. doi: 10.1128/jcm.19.2.187-190.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Anzai T, Eguchi M, Sekizaki T. Development of a PCR test for rapid diagnosis of contagious equine metritis. J Vet Med Sci. 1999;61:1287–1292. doi: 10.1292/jvms.61.1287. [DOI] [PubMed] [Google Scholar]
- 5.Sellon DC, Walker K, Suyemoto M, Altier C. NA amplification for rapid detection of Rhodococcus equi in equine blood and tracheal wash fluids. Am J Vet Res. 1997;58:1232–1237. [PubMed] [Google Scholar]
- 6.Tang YW, Procop GW, Persing DH. Molecular diagnostics of infectious diseases. Clin Chem. 1997;43:2021–2038. [PubMed] [Google Scholar]
- 7.Mitterer G, Huber M, Leidinger E. Microarray-based identification of bacteria in clinical samples by solid phase PCR amplification of 23S ribosomal DNA sequences. J Clin Microbiol. 2004;42:1048–1057. doi: 10.1128/JCM.42.3.1048-1057.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lu J-J, Perng CL, Lee SY, Wan CC. Use of PCR with universal primers and RFLP for detection and identification of common bacterial pathogens in CSF. J Clin Microbiol. 2000;38:2076–2080. doi: 10.1128/jcm.38.6.2076-2080.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Harris KA, Hartley JC. Development of broad-range 16S rDNA PCR for use in the routine diagnostic clinical microbiology service. J Med Microbiol. 2003;52:685–691. doi: 10.1099/jmm.0.05213-0. [DOI] [PubMed] [Google Scholar]
- 10.Rantakokko-Jalava K, Nikkari S, Jalava J. Direct amplification of rRNA genes in diagnosis of bacterial infections. J Clin Microbiol. 2000;38:32–39. doi: 10.1128/jcm.38.1.32-39.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bashir HE, Laundy M, Booy R. Diagnosis and treatment of bacterial meningitis. Arch of Dis in Child. 2003;88:615–620. doi: 10.1136/adc.88.7.615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kanegaye JT, Soliemanazaden P, Bradley JS. Lumbar puncture in paediatric bacterial meningitis: defining the time interval for recovery of CSF pathogens after parenteral antibiotic treatment. Paediatrics. 2001;108:1169–1174. [PubMed] [Google Scholar]
- 13.Ley BE, Linton CJ, Bennett DMC, Jalal H, Foot ABM, Millar MR. Detection of bacteraemia in patients with fever and neutropenia using 16S rRNA gene amplification by PCR. Euro J Clin Microbiol & Infect Dis. 1998;17:247–253. doi: 10.1007/BF01699981. [DOI] [PubMed] [Google Scholar]
- 14.Javela JM, Ekblad ML. Bacterial 16S rDNA PCR in the detection of intra-amniotic infection. Br J Obstet Gynaecol. 1996;103:664–669. doi: 10.1111/j.1471-0528.1996.tb09835.x. [DOI] [PubMed] [Google Scholar]
- 15.Carroll NM, Jaeger EEM, Choudhury S. Detection of and discrimination between Gram-positive and Gram-negative bacteria in intraocular samples by using nested-PCR. J Clin Microbiol. 2000;38:1753–1757. doi: 10.1128/jcm.38.5.1753-1757.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Klausegger A, Hell M, Berger A, Zinober K. Gram typespecific broad-range PCR amplification for rapid detection of 62 pathogenic bacteria. J Clin Microbiol. 1999;37:464–466. doi: 10.1128/jcm.37.2.464-466.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schuurman T, de Boer RF, Kooistra-Smid AMD. Prospective study of use of PCR amplification and sequencing of 16S ribosomal DNA from CSF for diagnosis of bacterial meningitis in a clinical setting. J Clin Microbiol. 2004;42:734–740. doi: 10.1128/JCM.42.2.734-740.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Richardson DC, Louie L, Louie M, Simor AE. Evaluation of a rapid PCR assay for diagnosis of meningococcal meningitis. J Clin Microbiol. 2003;41:3581–3583. doi: 10.1128/JCM.41.8.3851-3853.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xu J, Millar BC, Moore JE. Employment of Broad range 16S rRNA PCR to detect aetiological agents of infection from clinical specimens in patients with acute meningitis- rapid separation of 16S rRNA amplicons without the need for cloning. Journal of Applied Microbiology. 2003;94:197–206. doi: 10.1046/j.1365-2672.2003.01839.x. [DOI] [PubMed] [Google Scholar]


